Abstract
Krüppel-like zinc finger transcription factors compose the largest transcription factor family in the mammalian genome. However, the functions for the majority of these transcription factors as well as their in vivo downstream targets are not clear. We have functionally characterized a novel KRAB domain zinc finger transcription factor ZNF431 using both in vitro and in vivo assays. ZNF431 is a nuclear transcriptional repressor whose repressive activity depends on its association with HDAC1 and -2. Using the limb mesenchymal cell line MPLB, we identified Patched1 as a direct transcriptional target of ZNF431. Promoter analyses revealed three ZNF431 binding sites that bind to ZNF431 both in vitro and in vivo as revealed by gel-shift and chromatin immunoprecipitation, respectively. Mutations of these three sites abolished ZNF431 repression in transient transfection assays. Moreover, overexpressing ZNF431 in MPLB cells or in Xenopus and mouse embryos strongly repressed Patched1 expression. On the other hand, shRNA knockdown of ZNF431 in MPLB cells elevated Patched1 expression. Finally, hedgehog signaling readout was reduced in ZNF431 overexpression but elevated in ZNF431 knockdown MPLB cells. Our results indicate that ZNF431 directly represses Patched1 expression and likely functions to repress the hedgehog response in cells.
Keywords: Protein-DNA Interaction, Signal Transduction, shRNA, Transcription Regulation, Transgenic, KRAB Zinc Finger, Ptch1, Hedgehog, Transcription Repression
Introduction
The hedgehog (Hh)2 signaling pathway plays key roles both in embryonic development and during carcinogenesis (1). Three hedgehog ligands exist in mammals, Desert hedgehog (Dhh), Indian hedgehog (Ihh), and Sonic hedgehog (Shh) (2). Shh is the best studied ligand in the hedgehog pathway, and it functions as a morphogen involved in the development of many organs such as the limb, external genitalia, neural tube, bone, craniofacial, tooth, and hair follicle (3–8). Shh binds to Patched1 (Ptch1), a 12-pass transmembrane protein, to activate signal transduction. In the absence of Shh, Patched1 inhibits Smoothened (Smo) and its downstream signaling cascades. Upon Shh binding, Patched1 releases its repression on Smo, which leads to activation of Gli transcription factors. Activated Glis translocate to the nucleus where they regulate downstream target gene expression (9). One of these targets is Ptch1, which upon induction generates a negative feedback loop to avoid sustained activation of this pathway. Both genetic and biochemical evidence suggest that Gli1 interacts directly with the Gli binding sites within the Ptch1 proximal promoter through its zinc finger domain (10). However, how basal Ptch1 expression level is controlled in the absence of Shh signaling remains unclear.
Among all zinc finger transcription factors in the mammalian genome, C2H2 zinc finger is the most common type of zinc finger that binds to DNA. C2H2 zinc finger motifs contain the consensus sequence YCX2–4CX3YX5YX2HX3,4H, where X represents any amino acid, and Y represents a hydrophobic residue (11). The two cysteines and histidines coordinate a zinc ion and fold the domain into a finger-like structure to interact with DNA (12). Many zinc finger transcription factors contain multiple zinc fingers, and each zinc finger recognizes three nucleotides (13). However, not all zinc fingers bind to DNA at the same time, as post-translational modifications and interactions with other proteins could interfere with their DNA binding capabilities.
Of all the C2H2 zinc finger transcription factors, roughly one-third contain the Krüppel-associated box (KRAB) domain, which is only present in tetrapods (14). KRAB-containing C2H2 zinc finger proteins are involved in the regulation of cell differentiation, proliferation, apoptosis, and neoplastic transformation (15–17). The KRAB domain spans 50–75 amino acids and is further subdivided into KRAB A and B boxes. The KRAB A box is more conserved and plays a key role in transcription repression by binding to corepressors, whereas the B box plays a supportive role (18). Corepressors for the KRAB zinc finger protein include Kap1, Tif1b, or Krip1 (19–21). The current model suggests that KRAB zinc finger proteins bind to their cognitive DNA sequences, recruit co-repressors, and form a facultative heterochromatin environment with HDACs, HP1, and Setdb1 on target promoters to silence gene expression (12).
In this paper we demonstrate that a previously uncharacterized KRAB zinc finger protein ZNF431 directly represses Ptch1 expression by binding to three response elements in the Ptch1 1b promoter. This repression occurs both in MPLB cells and in Xenopus and mouse embryos. In addition, ZNF431 is also necessary for controlling basal Ptch1 expression as short hairpin RNA (shRNA)-mediated knockdown of ZNF431 led to elevated Ptch1 expression in MPLB cells. Finally we show that Hh signaling response was decreased in ZNF431-overexpressing cells but elevated in ZNF431 knockdown cells. Together, these results implicate ZNF431 in controlling both Ptch1 basal expression and cellular response to Hh signaling.
EXPERIMENTAL PROCEDURES
Plasmids
The ZNF431 full-length cDNA in pCMV-Sport6 (IMAGE clone #3710576, Invitrogen) was digested with HindIII and cloned in-frame into pEGFP-c1 (Clontech, Mountain View, CA), pGAL1, and pCMV-HA (Clontech) vectors to generate N-terminal ZNF431 fusion proteins to EGFP, GAL4DBD, and HA tag, respectively. The ZNF431ΔN was generated from ZNF431 cDNA with EarI (nucleotides 162–1913) and cloned in-frame into pMX-VP16 to generate pMX-VP16-ZNF431ΔN. ZNF431 coding sequence was amplified using oligos HAZNF431F and ZNF431RNAiR (supplemental Table S1) and cloned into the pGEM-T-Easy vector (Promega, Madison, WI) which was then released by SalI and SacII double digestion and cloned into the pIRES2-EGFP vector to generate the ZNF431 RNAi rescue construct. A 1.7-kb mouse Ptch1 promoter fragment was amplified with oligos Ptch1PF and Ptch1PR (supplemental Table S1) and subsequently cloned into the pGEM-T-Easy vector. The Ptch1 promoter fragment was released by NcoI and SalI and ligated into the SmaI site in the pGL3-Promoter vector (Promega) upstream of a firefly luciferase gene. A series of 5′ promoter deletions were generated by further restricting the 1.7-kb Ptch1 genomic DNA with SmaI and NcoI, KpnI and NcoI, and NotI and NcoI, which were then subcloned into the pGL3-Promoter vector. ZNF431 response elements were mutated on the Ptch1 promoter using site-directed mutagenesis with primer pairs Ptch1PMut1F and Ptch1PMut1R, Ptch1PMut2F and Ptch1PMut2R, and Ptch1PMut3F and Ptch1PMut3R (supplemental Table S1). SV40 β-galactosidase vector and Gli-Luc vector were previously described and kindly provided by Dr. Towler and Dr. Long (Washington University).
Cell Lines and Transfections
HEK293T and MPLB cell lines were maintained in DMEM with 10% Cosmic Calf Serum (Hyclone, Logan, UT). Transient transfections were performed using Lipofectamine 2000 (Invitrogen). In all transfection experiments SV40 LacZ plasmid was cotransfected to normalize transfection efficiency, and the total amount of DNA in each transfection was equalized by irrelevant vector DNA. Briefly, 2 × 105 cells per well were cultured in a 6-well plate for overnight. Two micrograms of DNA and 10 μl of Lipofectamine 2000 were diluted into 100 μl of Opti-MEM medium. DNA and Lipofectamine were then mixed and applied to the medium directly.
In Vitro Transcription and Xenopus Injection
Capped mRNA encoding ZNF431 was transcribed in vitro (mMessage mMachine kit, Ambion, Austin, TX). RNA was injected into one blastomere of a two-cell stage embryo along with β-galactosidase mRNA. Embryos were collected at stage 15 and fixed in 100% ethanol, and X-gal staining was performed to visualize the injected side. For X-gal staining, embryos were fixed in MEMFA buffer (0.1 m MOPS, pH 7.4, 2 mm EGTA, 1 mm MgSO4, 3.7% formaldehyde) for 1 h and stained in X-gal staining buffer (7.2 mm Na2HPO4, 2.8 mm NaH2PO4, 144 mm NaCl, 1 mm MgCl2, 3.05 mm K3(FeCN6) and 3.05 mm K4(FeCN6)) and stored in 100% ethanol at −20 °C and processed by whole mount in situ hybridization as previously described (22).
Co-immunoprecipitation
MPLB cells with or without HA-ZNF431 transfection were cultured for 24 h and lysed in lysis buffer (1% Nonidet P-40, 140 mm NaCl, 5 mm MgCl2, 20 mm Tris pH 7.5, 0.5 mm PMSF, and protease inhibitor mixture) on ice for 15 min. Lysates were centrifuged and precleared with protein G-Sepharose beads (Fisher). Lysates were precipitated with a mouse monoclonal anti-HA antibody (HA-11, Covance, Alice, TX), a rabbit polyclonal anti-ZNF431 antibody (ab87232, Abcam, Cambridge, MA), and anti-HDAC1, HDAC2 antibodies (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). Precipitates were separated on SDS-PAGE and blotted with indicated antibodies.
Electrophoretic Mobility Shift Assay (EMSA)
EMSA was performed with in vitro translated ZNF431 proteins following a standard protocol (23). The sequences of EMSA probes (EMSA1 forward and reverse (F and R), EMSA2 F and R, and EMSA3 F and R) and competitors (EMSA1C F and R, EMSA2C F and R, and EMSA3 F and R) are listed in supplemental Table S1. Supershift assays were performed by adding the HA11 antibody (Covance) into the binding reaction.
Chromatin Immunoprecipitation (ChIP) Assay
A ChIP assay was used to detect direct interactions between ZNF431 and response elements in the Ptch1 promoter and was carried out as previously described (24). Briefly, MPLB cells were transfected with either pMX-HA-ZNF431 or empty vector, and chromatins were collected 48 h after transfection. Proteins and DNA were cross-linked with 1% formaldehyde for 15 min at room temperature. Cross-linked protein-DNA complexes were sonicated to an average size of 600 bp with Fisher dismembrator sonicator (Model 150) at 50% power for six 10-s pulses separated by 30-s intervals. Protein-DNA complexes were reverse-cross-linked at 65 °C for 5 h in IP elution buffer (50 mm NaHCO3, 1% SDS, and 0.2 mm NaCl), and DNA was recovered by phenol-chloroform extraction and ethanol precipitation. DNA was then amplified using oligo ChiP1F and ChiP1R to amplify BS1 at −266 and ChiP2F and ChiP2R to amplify BS2 and 3 at −131 and −39.
RNA Interference and Rescue
shRNA duplexes (mission lentiviral transduction particles) targeting ZNF431 were purchased either from Sigma (Clone ID TRCN0000173219, TRCN0000174737, TRCN0000174850, TRCN0000176030) or from Washington University (the Stewart laboratory, supplemental Table S1). Cells were infected at a multiplicity of infection of 10 using the standard protocol. RNA samples were collected using the RNA STAT-60 kit, and cDNAs were generated using Superscript II (Invitrogen). Rescue experiment was performed by transfecting pIRES2-EGFP-ZNF431 together with shRNA2 in a 1:1 ratio into MPLB cells as previously described (25). ZNF431 and Ptch1 expressions were examined by real time RT-PCR with primers ZNF431F and ZNF431R and Ptch1F and Ptch1R (supplemental Table S1).
Whole Mount RNA in Situ Hybridization
Embryos were fixed in 4% paraformaldehyde, and whole mount in situ hybridization was performed using digoxigenin-UTP-labeled probes as described (26). Xenopus ptch1 (Xptc1) probe was kindly provided by Dr. Katja Koebernick (Göttingen Center for Molecular Biosciences, Germany), and the mouse Ptch1 probe was kindly provided by Dr. Fanxin Long (Washington University).
Construction of the β-Actin::HAZNF431 Transgene
To generate the transgenic construct, HA-tagged ZNF431 full-length cDNA was excised from pMX-HA-ZN431 and cloned into the BglII site of pCAGGS-CreERTM. Subsequently, fragment containing HA-ZNF431 along with the β-globin intron was released by StuI and BamHI double digest and inserted into the EcoRI site of the pMES-Stop vector. Finally vector sequences were removed by XmnI and XmaI double digest, and DNA was purified for pronuclear injection. The β-actin::HAZNF431 transgenic animals were maintained in a Washington University animal facility with an approved protocol and genotyped with primers CbapF and StopR (supplemental Table S1). Msx2-Cre transgenic mice were described previously (27). Upon crossing β-actin::HAZNF431 male mice to Msx2-Cre females, E10.5 embryos carrying both transgenes were identified by genotyping. Expression of HA-ZNF431 was then confirmed by RT-PCR using primers HAZNF431F and HAZNF431R and a Western blot using the HA11 antibody.
In Vitro Translation
HA-tagged ZNF431, HDAC1, and HDAC2 proteins were translated using the TNT Coupled Reticulocyte Lysate Transcription/Translation kit (Promega) following a standard protocol. Briefly, the pGEM-HA-ZNF431 plasmid was used for TNT reaction, which was carried out at 30 °C for 90 min in the presence of [35S]methionine and SP6 RNA polymerase. HDAC1 expression vector was purchased from Invitrogen (clone ID 4976514) and FLAG-tagged HDAC2 expression vector kindly provided by Dr. Jing Hu at the University of Pittsburgh. A reaction with pGEM-HA template was used as a control.
Statistical Analysis
All data were analyzed by SPSS using either Student's t test or one-way analysis of variance. Significance is indicated between different samples. Error bars shown are S.D.
RESULTS
ZNF431 Gene and Protein Structures
The mouse ZNF431 (per Sigma, Mouse Genome Informatics 2310001H12rik) gene contains 4 exons spanning over 20 kb on chromosome 5. Alternative splicing generates four mRNA isoforms that code for different proteins. We focused on characterizing one isoform (BC012405; supplemental Fig. S1A), which encodes an N-terminal Krüppel-like (KRAB) domain followed by 15 tandem C2H2 zinc fingers at the C terminus (supplemental Fig. S1B). Alignment of the ZNF431 N terminus to other KRAB zinc finger transcription factors revealed that ZNF431 contains a full-length KRAB A domain and a less conserved B-domain (supplemental Fig. S1C), suggesting that ZNF431 may function as a Krüppel-like zinc finger transcriptional repressor. By RT-PCR analysis, we found that ZNF431 is ubiquitously expressed in all adult mouse tissues tested: brain, heart, lung, thymus, spleen, lymph node, liver, kidney, muscle, testis, ovary, skin, and uterus. Its expression was also detected during organogenesis in tooth, submandibular glands, thymus, thyroid, vibrissa follicles, and many other organs in E14.5 mouse embryos by in situ hybridization (supplemental Fig. S2).
ZNF431 Is a Nuclear Transcriptional Repressor
To determine whether ZNF431 functions as a transcription factor, we first examined its subcellular localization. We created an EGFP-ZNF431 fusion construct and transiently transfected a limb mesenchymal cell line, MPLB (28), that expresses endogenous ZNF431 (see Fig. 5A). Twenty-four hours post-transfection, cells were fixed and visualized by fluorescence microscopy. Although cells transfected with EGFP (pEGFP-c1) alone showed fluorescence both in the nucleus and in the cytoplasm (Fig. 1A, a–c), only nuclear fluorescence was observed in cells transfected with EGFP-ZNF431 fusion protein, which colocalizes with DAPI staining (Fig. 1A, d–f). The nuclear localization of ZNF431 protein is consistent with its role as a transcription factor.
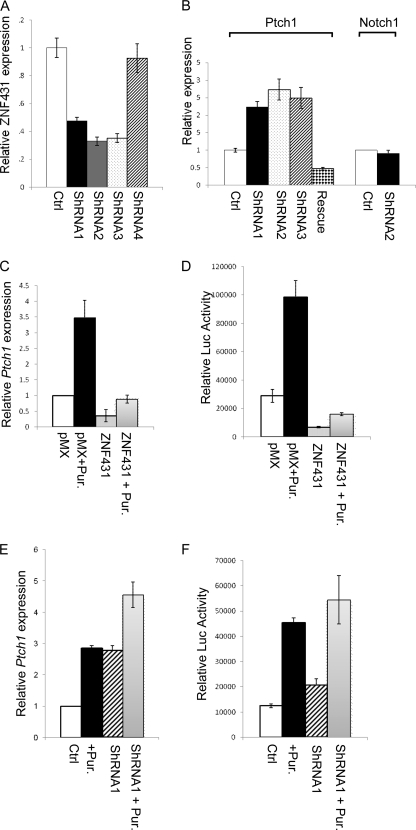
FIGURE 5.
Effects of ZNF431 on Ptch1 expression and Hh responsiveness. A, shRNA knockdown of ZNF431 is shown. Three shRNA constructs (shRNA1, -2, and -3) knocked ZNF431 expression down to less than 50%. B, knockdown of ZNF431 elevated Ptch1 expression in MPLB cells but had no effect on Notch1 expression. Elevated Ptch1 expression induced by shRNA2 was abolished when co-transfected with an RNAi rescue construct that is insensitive to shRNA2 knockdown. C–F, ZNF431 levels affect cellular responsiveness to Hh signaling. Overexpressing ZNF431 reduced both the basal expression levels of Ptch1 and Gli-Luc as well as their induction by Hh stimulation (+Pur.). C and D, on the contrary, knocking down ZNF431 elevated both Ptch1 and Gli-Luc basal expression, but the magnitude of their induction in response to Hh stimulation was reduced likely because the system is saturated (E and F).
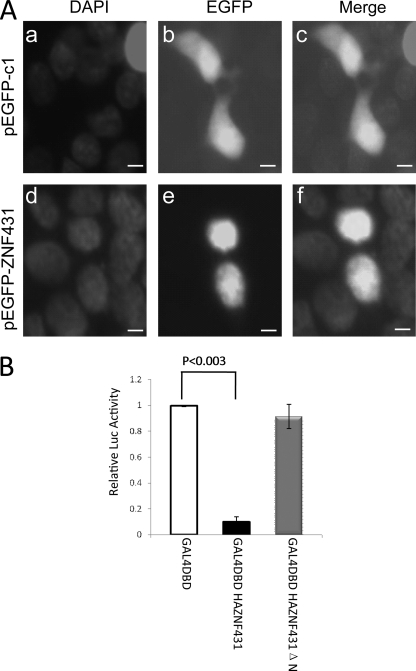
FIGURE 1.
Intracellular localization of EGFP-ZNF431 fusion protein in MPLB cells. A, EGFP alone was found both in the cytoplasm and nucleus (b and c), whereas EGFP-ZNF431 fusion protein was confined to the nucleus (e and f). Cells were counterstained with DAPI to show the nucleus (a and d). Scale bars are 10 μm. B, ZNF431 represses transcription of an artificial promoter. Transfection of GAL4DBDHAZNF431 significantly repressed transcription from a GAL4 responsive promoter. In contrast, transfection of either GAL4DBD or GAL4DBDHAZNF431ΔN did not repress transcription.
To address whether ZNF431 functions as a transcriptional activator or repressor, we fused GAL4 DNA binding domain (GAL4DBD) in-frame onto the N terminus of full-length ZNF431. This fusion construct was co-transfected with a GAL4-responsive luciferase reporter construct into either MPLB or HEK293T cells. Luciferase activity was measured 48 h after transfection, and transfection efficiency was normalized by SV40-LacZ. As clearly shown in Fig. 1B, co-transfection of GAL4DBDHAZNF431 strongly suppressed the basal transcription of the reporter construct compared with that co-transfected with GAL4DBD in MPLB cells. Similar results were obtained in HEK293T cells. This suppression of transcription by ZNF431 was dose-dependent, indicating that this effect was not caused by “squelching” of the basal transcription machinery (supplemental Fig. S3). To determine whether the KRAB A domain is responsible for this repressive activity, we deleted the N-terminal half of the KRAB A domain in ZNF431 and repeated the experiment. Consistent with the known function of the KRAB domain, removal of this domain abolished the repressive activity of ZNF431 on reporter gene expression (Fig. 1B). A Western blot using a GAL4 antibody showed higher expression of GAL4DBDHAZNF431ΔN protein than that of GAL4DBDHAZNF431 (supplemental Fig. S4), suggesting that the failure of GAL4HAZNF431ΔN to repress transcription is not due to a lower expression level. These results indicate that ZNF431 can function as a transcriptional repressor, and its repressive activity is dependent on the KRAB A domain.
ZNF431 Represses Transcription by Binding to Histone Deacetylases (HDACs)
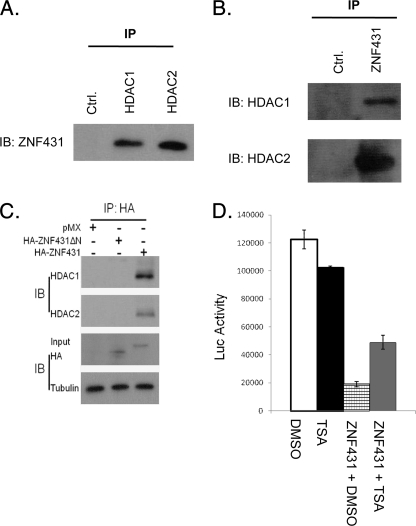
KRAB zinc finger proteins have been shown to repress transcription through interaction with the NuRD complexes (29). To test whether ZNF431 functions similarly, we examined whether ZNF431 represses DNA transcription by recruiting histone deacetylases. We collected MPLB cell lysates and immunoprecipitated ZNF431, HDAC1, or HDAC2 protein complexes using respective antibodies. Western blot demonstrated that HDAC1 and HDAC2 co-precipitated with ZNF431 and vice versa (Fig. 2, A and B), indicating that ZNF431 forms a protein complex with these two proteins in vivo. To determine whether ZNF431 can directly interact with HDAC1/2, we synthesized ZNF431, HDAC1, and HDAC2 in vitro and performed co-IP experiments. HDAC2 but not HDAC1 showed a direct interaction with ZNF431 (supplemental Fig. S5). To determine whether the KRAB A domain is responsible for this interaction, we repeated the co-IP experiment with transfected HA-tagged full-length or KRAB A-deleted ZNF431ΔN constructs. Both HDAC proteins co-precipitated with HA-ZNF431 but not with HA-ZNF431ΔN (Fig. 2C), indicating that the KRAB A domain is required for interaction with the HDACs. Previous studies showed that Trichostatin A (TSA), a specific inhibitor of HDACs, could alleviate the repressive activity of some KRAB zinc finger transcription factors (30). We, therefore, tested whether ZNF431 repressive activity depends on HDAC activity. The addition of TSA alone did not affect the basal Luc activity (Fig. 2D). Co-transfection with ZNF431 repressed reporter activity to one-sixth that of control (Fig. 2D). However, upon TSA treatment (50 ng/ml), luciferase reporter activity was elevated to 2.5-fold compared with DMSO control (Fig. 2D), indicating that TSA can partially relieve ZNF431 repression. These results demonstrate that ZNF431-mediated repression depends, at least in part, on HDAC activity.
FIGURE 2.
ZNF431 forms a repressive complex with HDACs. A, MPLB cell lysates were immunoprecipitated with IgG, HDAC1, or HDAC2 antibodies and blotted with the ZNF431 antibody. B, lysates were immunoprecipitated with a ZNF431 antibody and Western-blotted (IB) for HDAC1 and -2. C, MPLB cells were transfected with an empty control vector (pMX), full-length HA-tagged ZNF431 (HA-ZNF431), or N-terminal deleted ZNF431 (HA-ZNF431ΔN). Cell lysates were immunoprecipitated with the HA antibody and immunoblotted for HDAC1 and HDAC2. Tubulin was blotted as a loading control in all Western blots. D, TSA treatment (50 ng/ml) partially rescued ZNF431-mediated transcriptional repression (column 4 versus 3) but did not affect transcription in the absence of ZNF431 overexpression (column 1 versus 2).
Identification of Ptch1 as a ZNF431 Target
We next sought to understand the biological function of ZNF431. Uncovering ZNF431 target genes will help us understand its in vivo function. We chose MPLB cells for this study because endogenous ZNF431 is expressed in these cells, and therefore, the regulation on target genes could be physiologically relevant. However, the endogenous ZNF431 might have already efficiently suppressed target gene transcription in these cells, and thus, overexpressing more ZNF431 may have a minimal effect on target gene transcription. To overcome this, we converted ZNF431 from a transcriptional repressor into an activator by replacing the KRAB A domain with the VP16 activation domain. The resulting plasmid, pMX-VP16-ZNF431ΔN, or the backbone plasmid pMX-VP16 was transfected into MPLB cells. RNA samples were collected 48 h after transfection, reverse-transcribed, and hybridized to the Mouse Exonic Evidence Based Oligonucleotide (MEEBO) microarray chip (Illumina). Ptch1 was one of the genes showing the greatest change in expression by microarray. We, therefore, focused our work on Ptch1 regulation given its critical role in Hh signaling. Real-time PCR quantification showed an 11-fold induction on endogenous Ptch1 expression upon pMX-VP16-ZNF431ΔN transfection in MPLB cells (Fig. 3A). To determine whether ZNF431 normally functions to repress Ptch1 expression, we transfected HA-ZNF431 into MPLB cells, and indeed Ptch1 expression was reduced to 50% 12 h after ZNF431 transfection as assayed by real-time RT-PCR (Fig. 3A). To exclude the possibility that ZNF431 is a nonspecific global repressor of transcription, we examined Notch1 and Ptch2 expression and found no significant reduction after ZNF431 overexpression (Fig. 3B and supplemental Fig. S6), suggesting that Ptch1 is one of the specific targets for ZNF431.
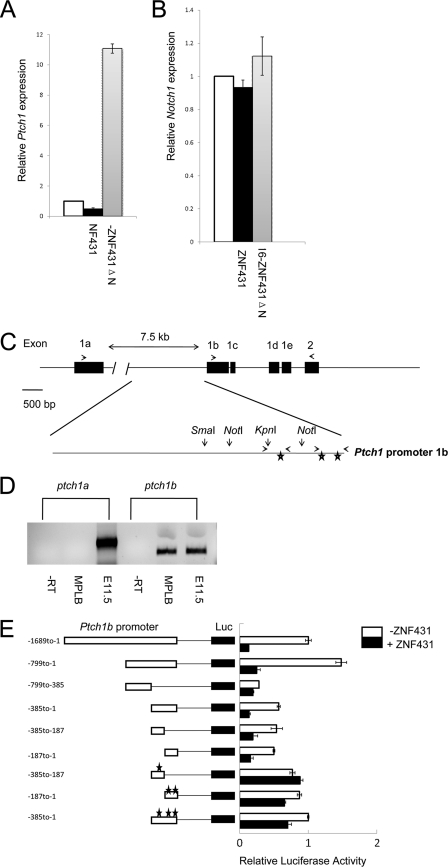
FIGURE 3.
ZNF431 directly regulates Ptch1 expression. A and B, HA-ZNF431 repressed, whereas VP16-ZNF431ΔN strongly activated, endogenous Ptch1 expression but had minimal effects on the Notch1 promoter as assayed by real-time PCR. C, a schematic diagram of the mouse Ptch1 promoter is shown. Exons are marked in black. Primers used to amplify the 1a and 1b isoforms were indicated by arrows above exons. The 1.7-kb Ptch1 promoter upstream of exon 1b is magnified. Restriction sites as well as primers used for ChIP assays are indicated. The three stars show the relative positions of the three ZNF431 binding sites. D, RT-PCR revealed that the 1b but not the 1a isoform is expressed in MPLB cells. Total RNA from E10.5 embryos were used as a positive control for PCR, whereas no RT (−RT) RNA sample was used as a negative control. E, deletion analysis of the Ptch1 promoter is shown. A schematic diagram of the Ptch1 promoter deletion constructs and their responses to ZNF431 transfection was shown. The transcription start site is designated as +1. Filled bars represent cells receiving ZNF431 co-transfection, whereas open bars represent control cells receiving empty vectors. Mutated ZNF431 response elements are indicated by stars.
ZNF431 Directly Regulates Ptch1 Expression
Because the kinetics of ZNF431 repression of Ptch1 expression is rapid (within 12 h after transfection), it is likely that ZNF431 can directly interact with the Ptch1 promoter to repress transcription. To test this hypothesis, we sought to identify ZNF431 response elements in the Ptch1 proximal promoter. Five alternative first exons have been identified in the mouse Ptch1 gene (31). Exon1a is located ∼8 kb upstream of exon 2, whereas exons 1b-e are clustered within a 1.5-kb fragment upstream of exon 2 (Fig. 3C). It has been suggested that Ptch1 transcription can start from two promoters; one 5′ of exon1a and the other 5′ of exon1b. To determine which promoter is active in MPLB cells, we collected RNA samples from MPLB cells and examined the expression of 1a versus 1b isoforms by RT-PCR. The results clearly showed that Ptch1b is the only isoform expressed in MPLB cells (Fig. 3D). To identify the promoter region(s) responsive to ZNF431 regulation, a 1.7-kb Ptch1 proximal promoter upstream of exon1b was cloned into the pGL3P vector containing an SV40 basal promoter upstream of the luciferase reporter. Co-transfection of this construct along with ZNF431 resulted in a 6-fold repression on reporter gene expression (Fig. 3E), whereas ZNF431 has no effect on a 4.7 kb Notch1 promoter in the same vector (supplemental Fig. S7). This result suggested that the 1.7-kb proximal promoter contained one or more ZNF431 response elements. To identify this element, a series of deletion constructs were made, and transfection experiments were repeated. Serial deletion analysis revealed a 385-bp fragment immediately upstream of exon1b to be important both for basal promoter activity as well as for ZNF431 repression (Fig. 3E). Further truncations of this fragment revealed two smaller fragments (−385 to −187 and −187 to −1) that were both repressed by ZNF431, suggesting that more than one ZNF431 response element exists in the Ptch1 promoter (Fig. 3E).
To identify putative ZNF431 binding sites within the Ptch1 promoter, we compared fragments −385 to −187 and −187 to −1 and found 3 islands of similar sequences (BS1–3, Fig. 4A). To investigate whether these sequences can bind ZNF431, EMSA were performed by incubating in vitro translated HA-ZNF431 with radiolabeled oligonucleotide probes. Slower migrating complexes were observed with all three probes tested, and the binding was specific, as the complexes were effectively competed away by 100× excess cold oligos but not with mutated oligos in which the core binding sites were mutated (Fig. 4A). To test whether the shifted complexes contained HA-ZNF431, we added the HA11 monoclonal antibody and showed that the addition of the HA antibody induced a supershifted band indicating the presence of HA-ZNF431 in these complexes (Fig. 4A). To examine the functional relevance of these ZNF431 binding sites, we mutated the core sequence from GCGCCC to GaGCtC in all three sites and asked whether mutation of these sites abolished transcription repression by ZNF431. Indeed, reporter constructs carrying these mutated sites no longer responded to ZNF431 transfection, indicating that these sites are required for ZNF431 to bind and repress Ptch1 expression (Fig. 3E).
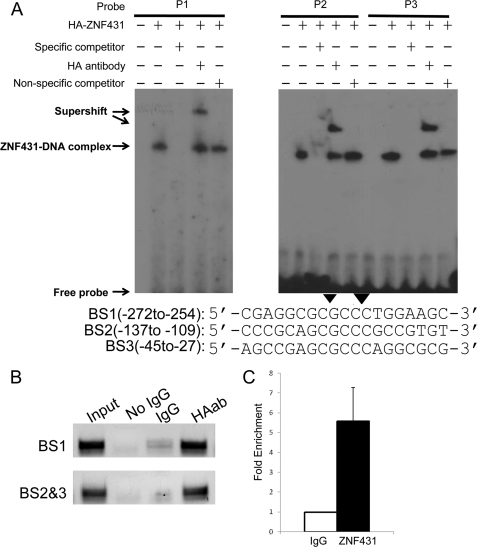
FIGURE 4.
ZNF431 binds directly to the Ptch1 promoter. A, EMSA was performed by incubating 32P-labeled probes with in vitro translated HA-ZNF431. The sequences and positions of the three probes are shown in A with the core ZNF431 binding region shaded. Components in EMSA binding reactions are indicated, and results are visualized by autoradiography. Competition assays were performed by adding 100× molar excess of either unlabeled probes (specific competitor) or unlabeled mutant probe (nonspecific competitor, arrowheads indicate mutated bases). Supershift assays were performed by adding the anti-HA monoclonal antibody in the binding reactions. B, a ChIP assay shows in vivo binding of ZNF431 to two fragments in the Ptch1 promoter containing the three ZNF431 binding sites (BS). Primers used for amplification were described under “Experimental Procedures” and are indicated in Fig. 3C. Enrichment for both fragments was observed in the HA antibody pulldown lane but not in the IgG only negative control lane. HAab, HA antibody. C, real-time PCR quantification of the ChIP result on BS2 and -3 is shown. The results are shown as -fold of relative enrichment. Quantification of BS1 was hampered by the high GC content in this fragment.
To confirm that ZNF431 binds to these sites in vivo, we performed chromatin immunoprecipitation assays. HA-ZNF431 expression vector was transfected into MPLB cells, and 48 h after transfection, cells were cross-linked and harvested. The precipitated chromatin was analyzed by PCR using real-time PCR for quantification. Two PCR fragments were amplified; one containing BS1 (−266 to −260) and the other containing BS2 and 3 (−131 to −125 and −39 to −33, respectively). PCR results showed that both fragments were enriched in anti-HA11 antibody pulldown samples compared with IgG controls (Fig. 4B). Real-time PCR quantification confirmed the above result and showed that the fragment containing BS2 and -3 was enriched by ∼5.5-fold in HA antibody-precipitated chromatin compared with controls (Fig. 4C). These results indicate that ZNF431 binds to the three binding sites in vivo to regulate Ptch1 expression.
Knockdown of ZNF431 Elevates Ptch1 Expression in MPLB Cells
We showed that overexpressing ZNF431 was sufficient to repress Ptch1 expression in MPLB cells. To address whether ZNF431 is required to regulate Ptch1 basal expression, we used shRNA to knock down ZNF431 expression in MPLB cells. To test the efficacy of shRNAs, we transfected MPLB cells with 5 ZNF431 shRNA constructs, and 3 were able to knock ZNF431 expression levels down to less than 50%, whereas the other 2 had no significant effects (Fig. 5A and supplemental Table S1). To control for off-target effects of shRNA, we examined Notch1 expression and showed that it was not affected by shRNA2, consistent with a gene-specific knockdown (Fig. 5B). Next, we examined Ptch1 expression upon ZNF431 knockdown by quantitative real-time PCR and found that Ptch1 mRNA levels were elevated by more than 2-fold 48 h after ZNF431 shRNA viral transduction (Fig. 5B). To demonstrate that this up-regulation results directly from reduced ZNF431 expression, we co-transfected a ZNF431 rescue construct that has the 3′-UTR removed and, thus, is insensitive to shRNA2 silencing along with shRNA2. ZNF431 expression was restored by co-transfecting the rescue construct (supplemental Fig. S8). As a result, elevated Ptch1 expression induced by shRNA2 was abolished, and Ptch1 expression was reduced to ∼50% of basal levels due to ZNF431 overexpression (Fig. 5B). These results demonstrate that ZNF431 is both necessary and sufficient to regulate Ptch1 basal expression in MPLB cells.
ZNF431 Represses Hh Signaling Response
Previous studies showed that Ptch1 expression is directly up-regulated by Hh signaling (32). We, thus, asked whether ZNF431 is involved in this regulation. First, we tested whether activation of Hh signaling alters ZNF431 expression in MPLB cells. Treatment of MPLB cells with purmorphamine activated the Hh pathway (33) but did not alter ZNF431 expression (supplemental Fig. S9). Next, we asked whether ZNF431 expression levels change cell response to Hh signaling by examining both Ptch1 expression and an Hh-responsive Gli-Luc reporter. MPLB cells were transfected with either HA-ZNF431 or empty vector and 24 h later exposed to purmorphamine to activate Hh signaling. Cells were harvested 48 h later, and Ptch1 expression was quantified by real-time RT-PCR. In MPLB cells, purmorphamine treatment induced Ptch1 expression to 3.5-fold (Fig. 5C). However, when ZNF431 was overexpressed, basal Ptch1 expression was reduced to only 37%, and further purmorphamine treatment only led to a 2.3-fold induction over basal, which is still lower than the basal Ptch1 expression level in MPLB cells (Fig. 5C). Similar effects were observed on the Hh-responsive Gli-Luc reporter upon ZNF431 transfection (Fig. 5D). Next, we examined Hh response in ZNF431 knockdown cells. Purmorphamine was added to the culture media 24 h after ZNF431 knockdown by shRNA2. In this experiment Ptch1 expression was elevated to 2.7-fold after ZNF431 knockdown; however, purmorphamine was only able to induce Ptch1 expression to 1.7-fold on top of the elevated basal level, which could saturate the system (Fig. 5E). Similar results were obtained with all three shRNA constructs (supplemental Fig. S10). The effect of purmorphamine on Gli-Luc reporter activity in ZNF431 knockdown cells was similar to that of Ptch1 (Fig. 5F). To probe into the mechanism underlying the dampening effect of ZNF431 on Hh response, we examined regulation of Hh pathway components by ZNF431. Expression of five genes, Smo, Kif7, and Gli1, -2, and -3, is repressed by ZNF431 overexpression and elevated to various degrees by ZNF431 knockdown (supplemental Figs. S11 and S12). Together, these results suggest that ZNF431 represses both Ptch1 basal expression and the cellular response to Hh signaling, the latter likely mediated through repression of Hh signal component expression.
Overexpression of ZNF431 Represses Ptch1 Expression in Vivo
Our data clearly demonstrated that ZNF431 directly binds to the Ptch1 promoter and represses its transcription in MPLB cells. However, it is not clear whether this regulation occurs in vivo. To determine the in vivo functional relevance of our findings, we utilized two in vivo overexpression systems: Xenopus embryo injection and transgenic mice. First, we injected HA-ZNF431 mRNA into one of the two cells of Xenopus laevis embryos at the two-cell stage. At neurula stage (Stage 15), embryos were harvested, and Xptc1 expression was determined by whole mount in situ hybridization. On the uninjected side, strong Xptc1 expression was detected in the developing somites (Fig. 6A, arrows). In contrast, Xptc1 expression was strongly suppressed on the injected side (Fig. 6A). Expression of Xptc1 was reduced in 95% of injected embryos (18/19 in three experiments).
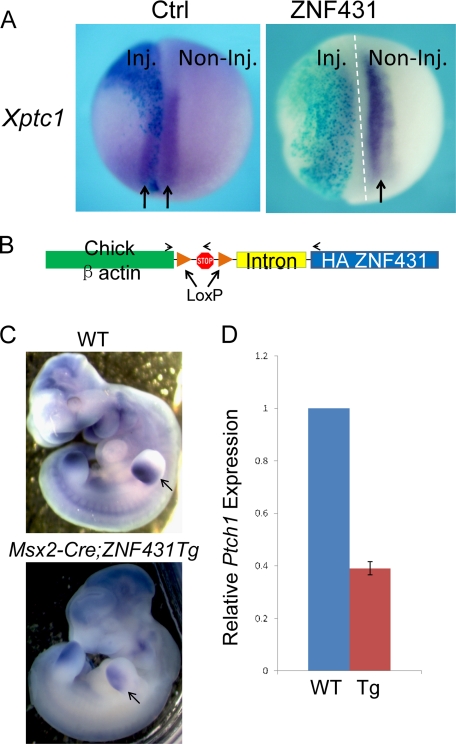
FIGURE 6.
Overexpression of ZNF431 repressed Ptch1 expression in Xenopus and transgenic mouse embryos. A, shown are whole mount in situ hybridization results of Xptc1 expression. The injected halves (Inj.) are indicated by X-gal staining. B, a schematic diagram of the ZNF431 transgene is shown. HA-tagged ZNF431 was driven by the chicken β-actin promoter preceded by a LoxP-flanked transcriptional stop cassette and a β-globin intron. The relative positions of genotyping primers are indicated by arrows. C, whole mount in situ hybridization of Ptch1 expression in E10.5 wild-type (upper) or HA-ZNF431 transgenic embryos (lower) is show. Ptch1 staining in the limb bud is indicated by arrows. D, real-time PCR quantification of Ptch1 expression in wild-type or HA-ZNF431 transgenic embryos is shown. The results were shown as relative Ptch1 expression. Tg, transgenic.
To determine whether overexpression of ZNF431 can also repress Ptch1 expression in mice, we created transgenic mouse lines containing the chicken β-actin promoter driving HA-ZNF431 preceded by a stop cassette (Fig. 6B). We obtained three transgenic founders that had normal lifespans and were able to pass on the transgenes. All three lines were mated to female Msx2-Cre transgenic mice to remove the stop cassette from the germ line, which resulted in ZNF431 overexpression in all cells in the body. Bigenic embryos (βactin::HA-ZNF431; Msx2-Cre) were collected at E10.5, and Ptch1 expression was examined by whole mount in situ hybridization and compared with littermate controls without the HA-ZNF431 transgene. Ptch1 expression was found significantly reduced in ZNF431-overexpressing transgenic embryos in all three transgenic lines (n > 3 for each line). In situ hybridization showed that Ptch1 expression was reduced globally, especially obvious in the developing limb bud and somites (Fig. 6C). Real-time PCR on mRNAs isolated from E10.5 bigenic embryos showed that Ptch1 expression was reduced to 40% compared with littermate control embryos (Fig. 6D). These results combined with those from the Xenopus overexpression study indicate that ZNF431 can repress Ptch1 expression during embryonic development.
DISSCUSSION
More than 350 KRAB C2H2 zinc finger protein family members have been identified both in mice and in humans, yet the biological function for most of these proteins remains unclear (34, 35). The C2H2 Zinc fingers have been shown to bind DNA in a sequence-specific manner, and the KRAB domain represses DNA transcription (15, 36, 37). However, only a few in vivo targets have been identified for these proteins. In this study we characterized the mouse ZNF431, a C2H2 zinc finger protein with N-terminal KRAB domains and 15 tandem C2H2 zinc fingers at the C-terminus. We showed that ZNF431 is a nuclear transcriptional repressor and the N-terminal KRAB-A domain is required for this repressive activity through its association with HDAC1 and HDAC2. In addition, we identified Ptch1 as a direct transcriptional target for ZNF431 in MPLB cells. Three ZNF431 binding sites were identified in the mouse Ptch1 promoter with a consensus sequence of GCGCCC. Chromatin immunoprecipitation demonstrated that ZNF431 binds to these sites in vivo. We validated our cell culture findings both in Xenopus embryos and in transgenic mice showing that overexpression of ZNF431 during embryo development suppressed endogenous Ptch1 expression. Finally, we showed that ZNF431 expression level can affect cellular response to Hh stimulation. Together, these results implicate an important role for ZNF431 in Hh signal transduction.
Like other KRAB C2H2 zinc finger proteins, ZNF431 represses gene transcription by binding to target promoters and likely first recruits HDAC2 and then HDAC1 to alter local chromatin structure. HDAC1 and -2 are components of two well studied repressive complexes, NuRD and Sin3A (38). At present, we do not know which repressive complex is responsible for interacting with ZNF431. Nonetheless, our data showed that the N-terminal ZNF431 KRAB A domain was absolutely required both for interaction with this complex as well as for transcription repression. Furthermore, we demonstrated that this transcription repression was dependent on HDAC enzyme activity as TSA treatment partially relieved this repressive effect both on an artificial GAL4-responsive promoter and on the endogenous Ptch1 promoter (supplemental Fig. S13). Other than the N-terminal KRAB domain, ZNF431 contains 15 tandem zinc fingers. Based on crystallographic studies on other tandem zinc finger proteins, each zinc finger can interact with a 3-bp sequence through a one-to-one interaction between individual amino acids within the recognition helix to DNA bases (39). Thus, tandem zinc finger proteins can theoretically increase their target specificity by adding more zinc fingers. However, not all zinc fingers in tandem bind DNA. Our EMSA results identify a 6-base pair consensus sequence of ZNF431, suggesting that only two zinc fingers are critical for DNA binding, whereas other zinc fingers may be functionally synergistic although structurally independent.
We identified Ptch1 as one of the direct transcription targets of ZNF431. We showed that ZNF431 repressed Ptch1 expression both in MPLB cells and in Xenopus and mouse embryos. In MPLB cells only promoter 1b is active, and promoter analysis on the Ptch1 1b promoter revealed three ZNF431 binding sites. All three sites can bind to ZNF431 in an EMSA assay, and mutating all three sites is required to abolish ZNF431-mediated repression. In addition, mutating all three sites also elevated the basal activity of the Ptch1 1b promoter, suggesting that endogenous ZNF431 also regulates the 1b promoter through these sites. These ZNF431 binding sites do not overlap with the consensus Gli binding site located between −923 and −914, suggesting that the two zinc finger proteins regulate Ptch1 expression independently (32). In the mouse embryo, global overexpression of ZNF431 leads to down-regulation of Ptch1 in all tissues where endogenous Ptch1 is expressed. The Ptch1 1b is the major isoform expressed in all tissues, and ZNF431 overexpression specifically down-regulated the 1b but not the 1a isoform (supplemental Fig. S14). However, given that ZNF431 also reduces Hh responsiveness in MPLB cells, we cannot exclude the possibility that reduced Hh signaling also contributes to the observed Ptch1 reduction in vivo. In Xenopus embryos, overexpression of the mouse ZNF431 could also repress Xptc1 expression. This implies that ZNF431 response element should also exist in the Xptc1 promoter. A search of the Xptc1 proximal promoter for ZNF431 binding sites revealed one consensus sequence (GCGCCC) at −315 relative to the transcription start site. Thus, it is possible that ZNF431 represses Xptc1 expression through binding to this site. These data suggest that ZNF431 function might be conserved in Xenopus; however, our effort to identify its homolog in Xenopus was hampered by an overwhelming number of putative homologous proteins in our blast search. In addition, the fact that presumably only 2 of the 15 zinc fingers may be responsible for binding to DNA makes it much more difficult to pinpoint a Xenopus homolog at this time. Nonetheless, the regulatory pathway between ZNF431 and Ptch1 appears to be conserved between Xenopus and mouse.
If ZNF431 is a negative regulator of Ptch1 expression, the two genes should exhibit complementary patterns of expression. In general, ZNF431 expression was detected where Ptch1 was expressed. In a few places such as the developing tooth buds and vibrissa follicles, we observed stronger ZNF431 expression in the epithelial compartment compared with the mesenchyme, whereas Ptch1 was mainly expressed in the mesenchyme (supplemental Fig. S2) (40). On the other hand, ZNF431 showed uniform expression in the developing neural tube, whereas Ptch1 exhibited a gradient expression. Thus, even though ZNF431 is a direct regulator of Ptch1 expression, it cannot be the sole transcriptional regulator governing Ptch1 basal expression.
As Ptch1 is a direct target of Hh signaling and functions in a negative feedback loop to inhibit Hh signaling, reducing Ptch1 expression by ZNF431 overexpression should in theory increase Hh signaling. However, that is not the case. We observed that Hh responsiveness was also decreased upon ZNF431 overexpression and elevated upon ZNF431 knockdown. Consistently, expression of several components of Hh signaling cascade was suppressed by ZNF431. Moreover, when ZNF431 is overexpressed, these genes showed no response to Hh stimulation (supplemental Fig. S15). These results suggest that ZNF431 dampens Hh responsiveness by reducing Hh signal transduction, which may not be related to its repressive activity on Ptch1 expression. Together, our data indicate that ZNF431 protein represses both Ptch1 expression as well as the cellular response to Hh signaling. These results suggest that the normal function of ZNF431 may be to control Hh responsiveness within physiological range for signal transduction.
Supplementary Material
Acknowledgments
We thank Dr. Michael Rauchman for HDAC1 and HDAC2 antibodies, Dr. Katjia Koebernick for XPtc1 probe, Dr. Jing Hu for FLAG-tagged HDAC2 plasmid, and Dr. Fanxin Long and members of the Ma laboratory for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants ES014482 and ES016597.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S15.
- Hh
- hedgehog
- Shh
- Sonic hedgehog
- Ptch1
- Patched1
- Smo
- Smoothened
- KRAB
- Krüppel-associated box
- X-gal
- 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside
- DBD
- DNA binding domain
- HDAC
- histone deacetylases
- Xptc1
- Xenopus ptch1
- IP
- immunoprecipitation
- Ctrl
- control
- EGFP
- enhanced green fluorescent protein.
REFERENCES
- 1. Ingham P. W., McMahon A. P. (2001) Genes Dev. 15, 3059–3087 [DOI] [PubMed] [Google Scholar]
- 2. Lum L., Beachy P. A. (2004) Science 304, 1755–1759 [DOI] [PubMed] [Google Scholar]
- 3. Niswander L., Jeffrey S., Martin G. R., Tickle C. (1994) Nature 371, 609–612 [DOI] [PubMed] [Google Scholar]
- 4. Ahn S., Joyner A. L. (2005) Nature 437, 894–897 [DOI] [PubMed] [Google Scholar]
- 5. Lin C., Yin Y., Veith G. M., Fisher A. V., Long F., Ma L. (2009) Development 136, 3959–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Helms J. A., Kim C. H., Hu D., Minkoff R., Thaller C., Eichele G. (1997) Dev. Biol. 187, 25–35 [DOI] [PubMed] [Google Scholar]
- 7. Hardcastle Z., Mo R., Hui C. C., Sharpe P. T. (1998) Development 125, 2803–2811 [DOI] [PubMed] [Google Scholar]
- 8. St-Jacques B., Dassule H. R., Karavanova I., Botchkarev V. A., Li J., Danielian P. S., McMahon J. A., Lewis P. M., Paus R., McMahon A. P. (1998) Curr. Biol. 8, 1058–1068 [DOI] [PubMed] [Google Scholar]
- 9. Kovacs J. J., Hara M. R., Davenport C. L., Kim J., Lefkowitz R. J. (2009) Dev. Cell 17, 443–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yoon J. W., Kita Y., Frank D. J., Majewski R. R., Konicek B. A., Nobrega M. A., Jacob H., Walterhouse D., Iannaccone P. (2002) J. Biol. Chem. 277, 5548–5555 [DOI] [PubMed] [Google Scholar]
- 11. Pabo C. O., Peisach E., Grant R. A. (2001) Annu. Rev. Biochem. 70, 313–340 [DOI] [PubMed] [Google Scholar]
- 12. Urrutia R. (2003) Genome Biol. 4, 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Miller J. C., Holmes M. C., Wang J., Guschin D. Y., Lee Y. L., Rupniewski I., Beausejour C. M., Waite A. J., Wang N. S., Kim K. A., Gregory P. D., Pabo C. O., Rebar E. J. (2007) Nat. Biotechnol. 25, 778–785 [DOI] [PubMed] [Google Scholar]
- 14. Bellefroid E. J., Poncelet D. A., Lecocq P. J., Revelant O., Martial J. A. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 3608–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jheon A. H., Ganss B., Cheifetz S., Sodek J. (2001) J. Biol. Chem. 276, 18282–18289 [DOI] [PubMed] [Google Scholar]
- 16. Hennemann H., Vassen L., Geisen C., Eilers M., Möröy T. (2003) J. Biol. Chem. 278, 28799–28811 [DOI] [PubMed] [Google Scholar]
- 17. Hering T. M., Kazmi N. H., Huynh T. D., Kollar J., Xu L., Hunyady A. B., Johnstone B. (2004) Exp. Cell Res. 299, 137–147 [DOI] [PubMed] [Google Scholar]
- 18. Vissing H., Meyer W. K., Aagaard L., Tommerup N., Thiesen H. J. (1995) FEBS Lett. 369, 153–157 [DOI] [PubMed] [Google Scholar]
- 19. Friedman J. R., Fredericks W. J., Jensen D. E., Speicher D. W., Huang X. P., Neilson E. G., Rauscher F. J., 3rd. (1996) Genes Dev. 10, 2067–2078 [DOI] [PubMed] [Google Scholar]
- 20. Kim S. S., Chen Y. M., O'Leary E., Witzgall R., Vidal M., Bonventre J. V. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 15299–15304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Moosmann P., Georgiev O., Le Douarin B., Bourquin J. P., Schaffner W. (1996) Nucleic Acids Res. 24, 4859–4867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Harland R. M. (1991) Methods Cell Biol. 36, 685–695 [DOI] [PubMed] [Google Scholar]
- 23. Fried M., Crothers D. M. (1981) Nucleic Acids Res. 9, 6505–6525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Solomon M. J., Larsen P. L., Varshavsky A. (1988) Cell 53, 937–947 [DOI] [PubMed] [Google Scholar]
- 25. Fasano C. A., Dimos J. T., Ivanova N. B., Lowry N., Lemischka I. R., Temple S. (2007) Cell Stem Cell 1, 87–99 [DOI] [PubMed] [Google Scholar]
- 26. Seo S., Herr A., Lim J. W., Richardson G. A., Richardson H., Kroll K. L. (2005) Genes Dev. 19, 1723–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pan Y., Lin M. H., Tian X., Cheng H. T., Gridley T., Shen J., Kopan R. (2004) Dev. Cell 7, 731–743 [DOI] [PubMed] [Google Scholar]
- 28. Treviño C., Anderson R., Landry M., König G., Tonthat B., Shi C., Muneoka K. (1993) Prog. Clin. Biol. Res. 383A, 295–304 [PubMed] [Google Scholar]
- 29. Schultz D. C., Friedman J. R., Rauscher F. J., 3rd (2001) Genes Dev. 15, 428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matsuda E., Agata Y., Sugai M., Katakai T., Gonda H., Shimizu A. (2001) J. Biol. Chem. 276, 14222–14229 [DOI] [PubMed] [Google Scholar]
- 31. Nagao K., Toyoda M., Takeuchi-Inoue K., Fujii K., Yamada M., Miyashita T. (2005) Genomics 85, 462–471 [DOI] [PubMed] [Google Scholar]
- 32. Agren M., Kogerman P., Kleman M. I., Wessling M., Toftgård R. (2004) Gene 330, 101–114 [DOI] [PubMed] [Google Scholar]
- 33. Sinha S., Chen J. K. (2006) Nat. Chem. Biol. 2, 29–30 [DOI] [PubMed] [Google Scholar]
- 34. Ding G., Lorenz P., Kreutzer M., Li Y., Thiesen H. J. (2009) Nucleic Acids Res. 37, D267–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Groner A. C., Meylan S., Ciuffi A., Zangger N., Ambrosini G., Dénervaud N., Bucher P., Trono D. (2010) PLoS Genet. 6, e1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bellefroid E. J., Lecocq P. J., Benhida A., Poncelet D. A., Belayew A., Martial J. A. (1989) DNA 8, 377–387 [DOI] [PubMed] [Google Scholar]
- 37. Gebelein B., Urrutia R. (2001) Mol. Cell. Biol. 21, 928–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ahringer J. (2000) Trends Genet. 16, 351–356 [DOI] [PubMed] [Google Scholar]
- 39. Pavletich N. P., Pabo C. O. (1991) Science 252, 809–817 [DOI] [PubMed] [Google Scholar]
- 40. Goodrich L. V., Johnson R. L., Milenkovic L., McMahon J. A., Scott M. P. (1996) Genes Dev. 10, 301–312 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.