Abstract
Autophagy enables cells to degrade and recycle cytoplasmic materials both as a housekeeping mechanism and in response to extracellular stress such as nutrient deprivation. Recent studies indicate that autophagy also functions as a protective mechanism in response to several cancer therapy agents, making it a prospective therapeutic target. Few pharmacological inhibitors suitable for testing the therapeutic potential of autophagy inhibition in vivo are known. An automated microscopy assay was used to screen >3,500 drugs and pharmacological agents and identified one drug, verteporfin, as an inhibitor of autophagosome accumulation. Verteporfin is a benzoporphyrin derivative used in photodynamic therapy, but it inhibits autophagy without light activation. Verteporfin did not inhibit LC3/Atg8 processing or membrane recruitment in response to autophagic stimuli, but it inhibited drug- and starvation-induced autophagic degradation and the sequestration of cytoplasmic materials into autophagosomes. Transient exposure to verteporfin in starvation conditions reduced cell viability whereas cells in nutrient-rich medium were unaffected by drug treatment. Analysis of structural analogs indicated that the activity of verteporfin requires the presence of a substituted cyclohexadiene at ring A of the porphyrin core but that it can tolerate a number of large substituents at rings C and D. The existence of an autophagy inhibitor among FDA-approved drugs should facilitate the investigation of the therapeutic potential of autophagy inhibition in vivo.
Keywords: Anticancer Drug, Autophagy, Cancer Therapy, Drug Action, Tumor Metabolism, Benzoporphyrin Derivative, Chemical Screen, Sequestration
Introduction
Autophagy is a catabolic process responsible for the bulk degradation of cytoplasmic components (1). It is characterized by formation of sequestering membranes called phagophores that expand, engulf cytoplasmic material, and then fuse to form double-membraned autophagosomes. Fusion with lysosomes results in degradation of the autophagosomes and their contents by lysosomal acidic hydrolases and release of the products into the cytoplasm for macromolecular synthesis. Constitutive autophagy functions as a housekeeping mechanism by controlling the turnover of long lived proteins and organelles (2).
Autophagy also functions as an adaptive response to maintain cellular homeostasis; it is stimulated in response to cellular stresses such as nutrient depletion, oxidative stress, protein aggregation, and several cancer drugs (3, 4). By removing aberrant organelles and limiting the production of reactive oxygen species, autophagy is believed to protect cells from genotoxic stress that may lead to cancer pathogenesis (5). This is supported by the observation that Beclin 1 and UVRAG, which are involved in autophagosome assembly, also have tumor suppressor activity (6, 7). Furthermore, autophagy-deficient cells are susceptible to enhanced gene amplification and chromosomal instability, both tumorigenic characteristics (8). In established tumors, autophagy is enhanced compared with corresponding noncancerous tissue (9), particularly in the tumor center (10) which is generally poorly supplied with nutrients and oxygen. Although the dependence on autophagy for tolerating nutrient deprivation is variable among different cancer cell types (3, 11), it has been demonstrated that suppression of autophagy can sensitize cells to apoptosis in a starved environment (9).
Multiple cancer therapy agents, such as temozolamide (12), tamoxifen (13), imatinib (14), and also ionizing radiation, induce autophagy in human cancer cell lines (15, 16), and excessive autophagy has been implicated as a death mechanism named type II programmed cell death (17). These observations led to the proposal that stimulation of autophagy by cancer therapeutics contributes to cancer cell death (16, 18). An alternative explanation for the increased autophagic activity observed in response to cancer therapy is that it constitutes a protective response against drug-induced cellular stress or vascular effects by clearing damaged organelles or replenishing nutrients (16). This hypothesis was investigated in studies using RNA interference (RNAi) technology to decrease autophagy while exposing cells to autophagy-stimulating cancer agents. RNAi-mediated knockdown of the autophagy gene Atg5 resulted in decreased cell survival in tamoxifen-treated MCF-7 cells (19) and in glioma cells treated with a DNA-damaging agent (20) or the tyrosine kinase inhibitor imatinib (14). Inhibition of autophagy also sensitizes tamoxifen-resistant T47D cells to treatment (19), further supporting a prosurvival role. Therefore, inhibitors of autophagy are needed to elucidate the interplay among autophagy, tumor survival, and cancer therapy.
Many studies on inhibition of autophagy have been carried out using RNAi technology, but there is a lack of chemical inhibitors with comparable specificity (1, 21, 22). Most of the chemicals currently employed to inhibit autophagy act at a late stage of the process. For example, lysosomotropic agents (chloroquine), V-ATPase inhibitors (bafilomycin A1), and lysosomal protease inhibitors (pepstatin A) all interfere with lysosomal function. They prevent the degradation of autophagosomes by lysosomal enzymes, leading to cytoplasmic accumulation of abnormal autophagosomes, which can be toxic to cells (22). To our knowledge, the only previously described early stage inhibitors of autophagy are compounds that inhibit phosphatidylinosital 3- (PI3) kinases, such as 3-methyladenine (3-MA),2 wortmannin, and LY294002. As PI3-kinase inhibitors, these compounds affect a number of cellular processes and are toxic after prolonged exposure (1, 21). To identify pharmacologically suitable candidates, we developed an automated microscopy screen for inhibitors of autophagosome accumulation from libraries of off-patent drugs and chemicals with known pharmacological activity. Of ∼3,500 chemicals screened, the only active compound identified was verteporfin, a benzoporphyrin derivative used for photodynamic therapy of age-related macular degeneration (23). Inhibition of autophagy by verteporfin occurred in the dark and was therefore not a photodynamic effect. This study provides an initial characterization of the effects of verteporfin on autophagy. The identification of a drug already approved for use in humans as an autophagy inhibitor could have an important impact on the preclinical and clinical assessments of the potential of autophagy inhibition in cancer therapy.
EXPERIMENTAL PROCEDURES
Cell Culture and Starvation
MCF-7 cells and MCF-7 cells stably expressing EGFP-LC3 were maintained in RPMI 1640 supplemented with 100 units/ml penicillin/streptomycin, 1 mm Hepes, and 10% (v/v) FBS. EGFP-LC3 expressing cells were supplemented with 400 μg/ml G418 (Sigma, G8168). For starvation experiments, cells were washed twice with DPBS (Invitrogen, 14040) before incubation in the denoted starvation medium. Serum starvation was carried out by incubating cells in the previously described growth medium lacking FBS. Glucose starvation was carried out by incubating cells in DPBS supplemented with amino acids from a 10× mixture (24) and 10% FBS. Amino acid starvation was carried out in DPBS containing 11 mm glucose, and DPBS was used for serum, glucose, and amino acid starvation. Earle's balanced salt solution (EBSS) was purchased from Sigma.
Screening Assay for Inhibitors of Autophagosome Formation
MCF-7 cells stably expressing EGFP-LC3 (25) were seeded in PerkinElmer View 96-well plates at 20,000/well. Eighteen hours after seeding, chemicals from the Prestwick, Sigma LOPAC, Microsource Spectrum and BIOMOL natural products collections were added to plates at ∼10 μm using a Biorobotics Biogrid II robot equipped with a 0.4-mm diameter 96-pin tool. Chloroquine (30 μm) was added immediately after to all but negative control wells. Plates were incubated for 4 h at 37 °C. Punctate EGFP-LC3 was determined quantitatively using a Cellomics Arrayscan VTI automated fluorescence imager and the Compartmental Analysis Bioapplication as described in detail by Balgi et al. (25). Briefly, cells were photographed using a 20× objective in the Hoechst and GFP channels (XF-100 filter), and the compartmental analysis algorithm was used to identify the nuclei, apply a cytoplasmic mask, and to quantitate GFP spots above a fixed threshold. Cells were gated such that only those with an average GFP fluorescence intensity of 10 and above were analyzed. Spots were distinguished from the background using a spot kernel radius parameter of 6 and a fixed threshold set at 500-pixel intensity units. The punctate fluorescence indicates the total intensity of all pixels inside the spot within the cytoplasmic mask. Analysis of 1,000 cells/well resulted in a Z factor of 0.54. Images were examined to exclude toxic compounds.
Confocal Microscopy
MCF-7 cells stably expressing EGFP-LC3 were cultured on glass coverslips in 6-well plates. Eighteen hours after plating, drugs were added for 4 h. The cells were then fixed with 3% paraformaldehyde in PBS and stained with 500 ng/ml Hoechst 33342. The coverslips were mounted in DABCO and viewed using the 60× objective of an Olympus Fluoview FV1000 laser scanning microscope equipped with Olympus-selected Hamamatsu photomultiplier tubes. Images were analyzed using Olympus FV10-ASW1.7 software.
Cell Viability and Proliferation Assay
Cells were seeded at 8,000/well in 96-well plates and grown overnight. Cells were washed twice with DPBS and treated as described. The drugs were then removed, and cells were grown in complete medium for 48 h. Cell viability and proliferation were measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay (26).
SDS-PAGE and Western Blotting
Cells were lysed, protein was quantified, and EGFP-LC3 processing was assayed by Western blotting (25). Five micrograms of protein from each lysate was separated on a 12% polyacrylamide gel and electroblotted onto a polyvinylidene fluoride membrane, blocked in 5% (w/v) nonfat milk, and incubated in the appropriate antiserum. LC3 lipidation was assessed by resolving identical amounts of cell lysates (70–100 μg of protein) on a 16% polyacrylamide gel. Electroblotting as above was followed by cross-linking using 0.2% (v/v) glutaraldehyde in PBS + 0.02% (v/v) Tween 20 for 30 min.
Subcellular Fractionation
MCF-7 cells were treated as described for 4 h at 37 °C. After treatment, cells were washed once with cold PBS and collected in subcellular fractionation buffer (250 mm sucrose, 20 mm Hepes (pH 7.4), 10 mm KCl, 1.5 mm MgCl2, 1 mm EDTA, 1 mm EGTA). The lysate was passed through a 26-gauge 1/2 needle 10 times and incubated on ice for 20 min. The nuclear pellet was collected by centrifugation at 500 × g for 10 min. The supernatant was ultracentrifuged at 100,000 × g to obtain a cytosolic supernatant and a pellet containing total membrane (27). The pellet was resuspended in subcellular fractionation buffer containing 1% Triton X-100 and 10% glycerol. Identical amounts of the supernatant and the membrane fractions (∼30–50 μg of protein), respectively, were resolved on a 16% polyacrylamide gel and immunoblotted as described above.
Long Lived Protein Degradation Assay
MCF-7 EGFP-LC3 cells were seeded in a 6-well plate at 1,000,000 cells/well and grown overnight. Cells were incubated for 24 h at 37 °C with 0.2 μCi/ml l-[14C]valine (PerkinElmer Life Sciences, NEC291) in RPMI 1640 medium supplemented as described above. After radiolabeling, cells were washed three times with PBS and incubated in complete RPMI supplemented with 10 mm cold valine for 2 h. After this cold chase period where short lived proteins were degraded, the cells were treated for 6 h as described in either complete RPMI or EBSS, both supplemented with 10 mm cold valine. Protein degradation was measured as described previously (28).
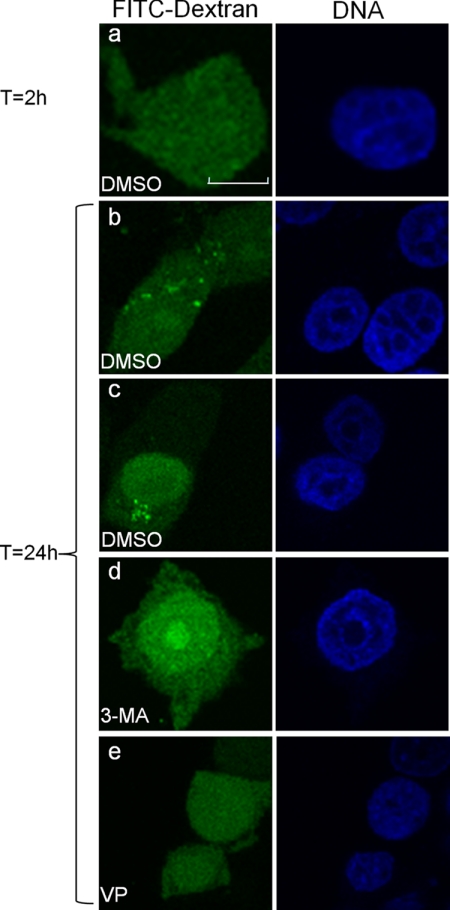
FITC-Dextran Sequestration Assay
FITC-dextran was scrape-loaded into MCF-7 cells as described (29), except cells were suspended in medium containing 0.1% DMSO, 10 mm 3-methyladenine, or 10 μm verteporfin after scrape loading. The suspended cells were plated on coverslips for 2 h or 24 h before fixation with 3% paraformaldehyde in PBS and DNA staining with Hoechst 33342. Slides were then analyzed using confocal microscopy.
Benzoporphyrin Derivatives
The ring A and B benzoporphyrin derivatives were synthesized using standard techniques (30–32). All derivatives were characterized using 1H NMR and elemental analysis. The verteporfin regioisomers were separated by reversed-phase HPLC. Using a 250 × 10-mm Phenomenex Synergi 4-μm MAX-RP 80A C-12 column and a mobile phase of 95% MeOH/0.01 m ammonium formate, verteporfin isomer A eluted at 11.3 min, and verteporfin isomer B eluted at 11.9 min.
Electron Microscopy
MCF-7 EGFP-LC3 cells were seeded onto BD 24-well cell culture inserts with PET track-etched membrane at 30,000 cells/well. Thirty-six hours after seeding, cells were treated as described. Medium in Transwells was replaced with fixative (1.5% paraformaldehyde, 1.5% glutaraldehyde, 0.1 m sodium cacodylate (pH 7.3)) at room temperature. After 30 min, membranes were cut from the Transwells and immersed for an additional 2.5–3 h in fixative. The fixative was replaced with buffer (0.1 m sodium cacodylate (pH 7.3)) and left overnight at room temperature. The next morning, the membranes were washed twice (10 min each wash) with buffer and then postfixed for 1 h on ice in 1% OsO4 in 0.1 m sodium cacodylate (pH 7.3). Membranes were washed three times for 10 min each at room temperature with dH2O, stained en bloc with 1% aqueous uranyl acetate for 1 h, washed three times with dH2O, and then dehydrated through an ascending concentration series of ETOH. The membranes were treated three times for 10 min in 100% ETOH, twice with 100% propylene oxide (15 min each), and then infiltrated overnight in 1:1 propylene oxide:EMbed-812 (Electron Microscope Sciences, Hatfield, PA). After two changes of EMbed-812 (2 h each), the membranes were embedded and the resin polymerized for 24 h at 60 °C. Thin sections were stained with uranyl acetate and lead citrate and photographed on a Philips 300 electron microscope operated at 60 kV.
RESULTS
Automated Microscopy Screen for Chemical Inhibitors of Autophagosome Formation
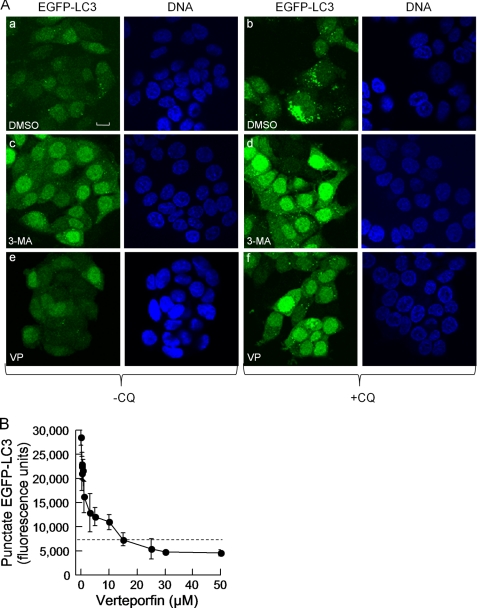
To monitor the inhibition of autophagosome formation, we used MCF-7 breast cancer cells stably expressing LC3 tagged to EGFP at its N terminus (MCF-7 EGFP-LC3). LC3 is a cytosolic protein that is recruited to autophagosomes via cleavage of its C terminus to expose a glycine residue that is then conjugated to phosphatidylethanolamine, enabling its insertion into autophagosomal membranes (27). In cells grown in complete medium, EGFP-LC3 was distributed diffusely throughout the cytoplasm with a few EGFP-LC3 spots, demonstrating a low basal level of autophagy (Fig. 1A). Stimulation of autophagy by exposure to medium lacking serum or to the mTORC1 inhibitor rapamycin increased the punctate EGFP-LC3 fluorescence intensity, but this increase was not sufficiently high to identify inhibitors in a screening assay. We overcame this problem by using chloroquine (CQ), a lysosomotropic agent that neutralizes the acidic pH of lysosomes (33), thereby preventing autophagic protein degradation and causing autophagosome accumulation (34–36). Incubation of cells with 30 μm CQ caused a large increase in the number of autophagosomes (Fig. 1A). 3-MA, a PI3-kinase inhibitor, is known to prevent autophagosome formation efficaciously but with low potency and selectivity (21). As expected, 10 mm 3-MA considerably reduced the ability of CQ to induce autophagosome accumulation (Fig. 1A).
FIGURE 1.
Cell-based assay for the identification of inhibitors of autophagosome accumulation and identification of verteporfin as an inhibitor of autophagy. A, MCF-7 EGFP-LC3 cells were exposed to 0.1% DMSO (a and b), 10 mm 3-MA (c and d), or 10 μm verteporfin (VP; e and f) without (a, c, and e) or with 30 μm chloroquine (b, d, and f) for 4 h in complete cell culture medium. The images were acquired by confocal microscopy. Scale bar, 10 μm. B, cells were exposed to 30 μm chloroquine and different concentrations of verteporfin for 4 h in complete medium. The cells were fixed and stained, and punctate EGFP-LC3 fluorescence was quantitated using a Cellomics Arrayscan VTI automated imager. The level of punctate EGFP-LC3 fluorescence observed in control DMSO-treated cells is indicated by the dotted line (mean ± S.D. (error bars), n = 3).
We wished to search for chemicals that, like 3-MA, inhibit autophagosome formation. Cellular autophagosomal content was detected quantitatively by automated fluorescence microscopy in 96-well plates (25). CQ caused a 5–10-fold increase in punctate EGFP-LC3 fluorescence over that of untreated control cells. Cells were exposed for 4 h to CQ and chemicals from a collection of 3,584 drugs and pharmacological agents at a concentration of 10 μm. Compounds were designated as active if they decreased CQ-induced punctate fluorescence by ≥50% and induced <20% cell death during a 4-h incubation period. Verteporfin was the only active compound identified in this screen. It showed concentration-dependent inhibition of CQ-induced punctate EGFP-LC3 with an IC50 of 1 μm and reducing punctate fluorescence below that of DMSO controls at ≥15 μm (Fig. 1B).
Verteporfin is a benzoporphyrin derivative used clinically for photodynamic therapy of age-related macular degeneration. Red light irradiation causes the generation of oxygen radicals that nonselectively kill cells exposed to verteporfin. Verteporfin shows little or no cellular toxicity in the absence of light activation (37, 38). Importantly, the ability of verteporfin to inhibit CQ-induced autophagosome accumulation occurred in the absence of light and was consequently unrelated to its photodynamic properties. All experiments described in this study were conducted without direct light.
Characterization of Verteporfin as an Inhibitor of Autophagy
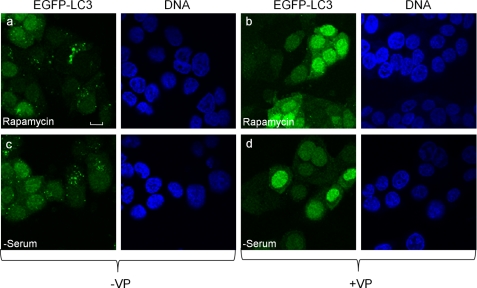
Having identified verteporfin as an inhibitor of autophagosome accumulation in the presence of CQ, we next asked whether this drug can inhibit autophagosome formation in response to well characterized autophagic stimuli such as rapamycin or serum deprivation. Cellular exposure to 30 nm rapamycin or to serum-free medium caused a substantial increase in punctate EGFP-LC3 fluorescence (Fig. 2 and supplemental Fig. 1). Simultaneous treatment with verteporfin considerably reduced both rapamycin-induced and starvation-induced punctate EGFP-LC3 fluorescence and increased diffuse cytoplasmic fluorescence (Fig. 2 and supplemental Fig. 1).
FIGURE 2.
Verteporfin inhibits autophagosome accumulation stimulated by rapamycin or serum starvation. MCF-7 EGFP-LC3 cells were exposed for 4 h to 30 nm rapamcycin without (a) or with 10 μm verteporfin (VP) (b) in complete cell culture medium or to medium lacking serum without (c) or with 10 μm verteporfin (d). Images were acquired by confocal microscopy. Scale bar, 10 μm.
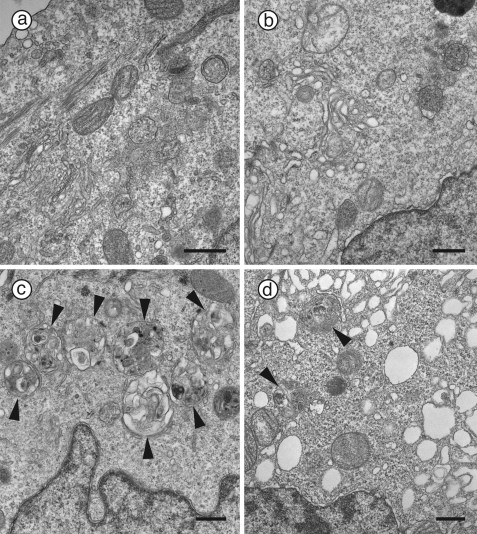
Electron microscopy was used to examine this effect at the ultrastructural level. Sections of control cells treated with DMSO rarely if ever contained any autophagosomes (Fig. 3a) whereas exposure to 75 μm CQ caused a significant perinuclear accumulation of autophagic vacuoles containing lamellar structures and undigested cytoplasmic material (Fig. 3c, arrowheads), as expected. Noticeably, incubation with 10 μm verteporfin alone caused the distinct appearance of small empty rounded single-membraned vesicles; however, it did not alter the integrity of most distinguishable organelles, such as mitochondria and multivesicular bodies (Fig. 3b). Cells exposed to both 75 μm CQ and 10 μm verteporfin contained significantly fewer autophagosomes (Fig. 3d, arrowheads) than those exposed to CQ alone. Interestingly, these cells also contained numerous empty single-membraned vesicles that were larger in size than in cells treated with verteporfin alone (Fig. 3d). There was no clear accumulation of structures resembling elongation membranes in any of the samples; however, these are difficult to identify (39).
FIGURE 3.
Ultrastructural examination of inhibition of CQ-induced autophagosome accumulation by verteporfin. MCF-7 EGFP-LC3 cells were exposed for 4 h to 0.1% DMSO (a and b) or 75 μm CQ (c and d) without (a and c) or with 10 μm verteporfin (b and d). Images were acquired by transmission electron microscopy. Arrowheads point to autophagic vesicles. Scale bar, 0.5 μm.
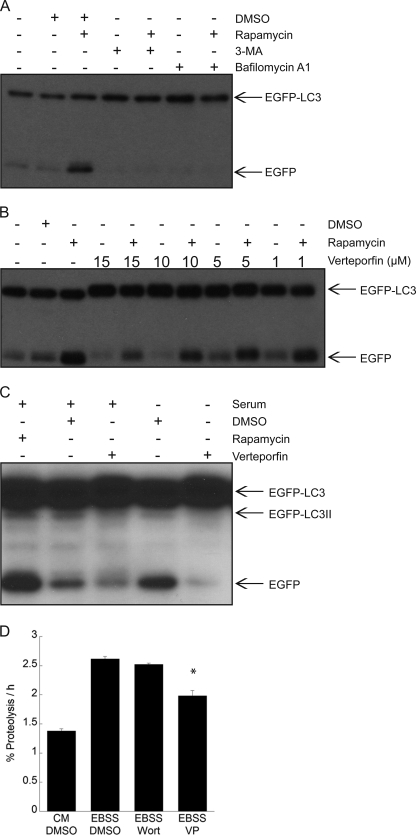
EGFP-LC3 recruited to the membranes of autophagosomes is degraded upon fusion with lysosomes, but EGFP is less sensitive to lysosomal proteases than the LC3 moiety, leading to transient EGFP accumulation. Therefore, the relative levels of EGFP-LC3 and free EGFP reflect autophagic flux (25). MCF-7 cells treated with 30 nm rapamycin for 4 h showed a significant increase in free EGFP levels compared with controls, consistent with its stimulation of autophagy (Fig. 4A). As expected, co-treatment with 3-MA prevented LC3 degradation and thus the appearance of the free EGFP band. Bafilomycin A1, a V-ATPase inhibitor that prevents lysosomal acidification and lysosomal protein degradation (35), also prevented the appearance of the free EGFP band in cells treated with rapamycin (Fig. 4A). Having established these conditions with well characterized compounds, we then tested the effects of different combinations of verteporfin and rapamycin on EGFP-LC3 processing. Exposure of cells to verteporfin alone led to a decrease in the intensity of the free EGFP band in a concentration-dependent manner, indicating that verteporfin inhibits basal autophagy (Fig. 4B). Verteporfin also caused a concentration-dependent decrease in the intensity of the free EGFP band when co-incubated with rapamycin (Fig. 4B). The effects of verteporfin on autophagic flux were also tested in serum starvation conditions. Cells exposed to serum-free medium for 4 h showed a significant increase in the free EGFP band compared with controls, demonstrating that starvation induces autophagic degradation of EGFP-LC3 (Fig. 4C). When cells were exposed to 10 μm verteporfin in serum-free conditions, the intensity of the free EGFP band decreased, showing that verteporfin inhibits starvation-induced autophagic degradation.
FIGURE 4.
Inhibition of EGFP-LC3 degradation and long lived protein degradation by verteporfin. A–C, MCF-7 EGFP-LC3 cells were treated for 4 h with 30 nm rapamycin without or with 10 mm 3-MA or 100 nm bafilomycin A1 in complete medium (A); different concentrations of verteporfin without or with 30 nm rapamycin in complete medium (B); 10 μm verteporfin in complete medium or in serum-free medium (C). A–C, cells were exposed to 0.1% DMSO as a vehicle control, and EGFP-LC3 processing and degradation were monitored by Western blotting with anti-GFP antibody. D, the amount of [14C]valine-labeled long lived protein degradation was measured in MCF-7 EGFP-LC3 cells treated with 0.1% DMSO, 10 μm verteporfin (VP), or 100 nm wortmannin (Wort) in EBSS, and cells were exposed to complete cell culture medium for 6 h (mean ± S.D. (error bars), n = 3). *, p < 0.05 versus corresponding DMSO treatment.
The effect of verteporfin on autophagic flux was verified and quantified by monitoring the degradation of long lived proteins. Cells were incubated with [14C]valine for 24 h followed by an additional 2-h cold chase to allow degradation of short lived proteins before treatment. Over 6 h, the basal long lived protein degradation in complete medium was 1.4%/h whereas cells exposed to EBSS, which lacks serum and amino acids, showed more proteolysis at 2.6%/h, consistent with previous studies (e.g. 35). Exposure to verteporfin significantly reduced starvation-induced proteolysis (Fig. 4D). In this cell line exposure to the PI3-kinase inhibitor, wortmannin only showed minimal inhibition of long lived protein degradation. These results further demonstrate that verteporfin inhibits degradation by starvation-induced autophagy.
Verteporfin Inhibits the Sequestration of FITC-Dextran
Having demonstrated that verteporfin inhibits autophagosome accumulation and autophagic protein degradation, we next investigated whether it affected the sequestration of cytoplasmic material. An early step in autophagy involves the expansion of phagophores into bowl-shaped structures that surround cytoplasmic material and capture it when the edges of the phagophores fuse to form double-membraned autophagosomes (40, 41). This process may be monitored experimentally by examining the transfer of fluorescently labeled dextran from the cytoplasm into autophagic vesicles (29). FITC-dextran was introduced into the cytoplasm of MCF-7 cells by scrape-loading at a temperature of 4 °C to prevent uptake by fluid phase endocytosis. Two hours after loading, FITC-dextran was localized diffusely throughout the cytoplasm (Fig. 5a), but it redistributed to punctate structures within 24 h, reflecting autophagosomal sequestration (Fig. 5, b and c). When cells were treated with 3-MA, FITC-dextran remained diffuse in the cytoplasm 24 h after loading (Fig. 5d), consistent with its demonstrated ability to inhibit autophagic sequestration (42). In cells treated with verteporfin, FITC-dextran remained completely diffuse in the cytosol (Fig. 5e), showing that it too inhibits sequestration of cytoplasmic material into autophagosomes.
FIGURE 5.
Inhibition of the sequestration of cytosolic FITC-dextran into vesicles by verteporfin. FITC-dextran was scrape-loaded into MCF-7 cells, and its distribution was analyzed by confocal microscopy. DMSO-treated MCF-7 cells were fixed 2 h (a) or 24 h (b and c) after FITC-dextran loading. MCF-7 cells loaded with FITC-dextran were incubated in 10 mm 3-MA (d) or 10 μm verteporfin (VP; e) for 24 h in complete medium. Cells were fixed, DNA was stained, and images were acquired by confocal microscopy. Scale bar, 10 μm.
Verteporfin Does Not Inhibit LC3 Processing or Lipidated LC3 (LC3II) Membrane Association
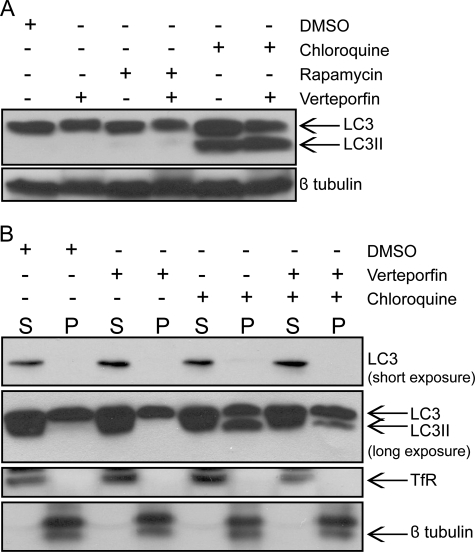
Having established that verteporfin inhibits autophagic vacuole formation, sequestration, and degradation, we tested whether verteporfin inhibits the processing, lipidation, and membrane association of LC3 in MCF-7 cells. LC3II associates with the isolation membrane of nascent autophagosomes and is believed to participate in phagophore expansion (43). LC3II was not detected in either vehicle- or rapamycin-treated MCF-7 cells, consistent with previous observations in murine embryonic fibroblast and HeLa cell lines (25, 44). Whereas rapamycin caused an increase in autophagosome accumulation (Fig. 2 and supplemental Fig. 1) and induction of autophagic degradation (Fig. 4), a 4-h treatment did not cause a detectable accumulation of LC3II, perhaps due to the transient nature of LC3II as a kinetic intermediate. However, treatment with CQ inhibited autophagic degradation, causing significant LC3II accumulation (Fig. 6A). Interestingly, cells treated with both CQ and verteporfin showed a similar increase in LC3II despite inhibition of autophagosome accumulation, indicating that LC3 becomes lipidated. These results suggest that verteporfin acts downstream of LC3 processing.
FIGURE 6.
Verteporfin does not inhibit LC3 processing or membrane association. A, MCF-7 cells were exposed to 0.1% DMSO, 30 nm rapamycin, or 75 μm chloroquine without or with 10 μm verteporfin for 4 h in complete medium. Endogenous LC3 processing was examined by Western blotting with anti-LC3 antibody, and protein loading was monitored with anti-β-tubulin antibody. B, MCF-7 cells were exposed to 0.1% DMSO or 75 μm chloroquine without and with 10 μm verteporfin for 4 h in complete medium. Lysates were subjected to ultracentrifugation, and the resulting supernatant (S) and pellet (P) fractions were immunoblotted with anti-LC3 antibody. Subcellular fractionation and protein loading were monitored by immunoblotting for the membrane marker, transferrin receptor (TfR), and the cytosolic marker, β-tubulin.
Because verteporfin prevents autophagosome accumulation without inhibiting LC3 lipidation, we sought to establish whether verteporfin affects the intracellular distribution of LC3II. LC3II has been shown to associate specifically with autophagosome membranes whereas unprocessed LC3 is cytosolic (27). Lysates of treated cells were subjected to ultracentrifugation to separate membrane and cytosolic fractions. In both DMSO and verteporfin-treated cells, LC3 was predominantly in the supernatant cytosolic fraction, and no LC3II was detected in either cytosolic or pellet membrane fractions (Fig. 6B). In CQ-treated cells, there was a large increase in LC3II, all of which was found in the membrane fraction as expected (Fig. 6B). Interestingly, in cells co-treated with verteporfin and CQ, LC3II was also only detected in the membrane fraction (Fig. 6B), demonstrating that verteporfin does not prevent LC3II membrane association. In a majority of experiments, the amount of LC3II detected in the membrane pellet was similar between cells treated with CQ and those co-treated with verteporfin and CQ; however, in some experiments, there was a noticeable LC3II decrease in the membrane fraction of co-treated cells. This variability was most likely caused by washing the membrane pellet and may indicate that LC3II is not bound as tightly in the co-treated cells, resulting in a loss of material during the wash.
Verteporfin Sensitizes Cells to Starvation
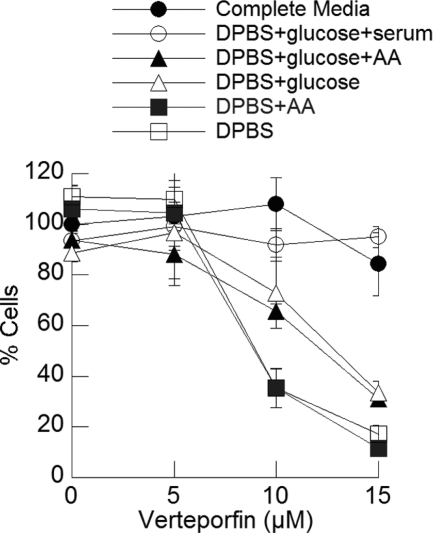
Having identified verteporfin as an inhibitor of autophagy, we investigated its effect on cell survival and proliferation. MCF-7 cells were exposed to increasing concentrations of verteporfin in complete medium for 8 h. Verteporfin was then washed away, and the cells were incubated in complete medium for 48 h to monitor their ability to recover and proliferate. Verteporfin had no effect on viability, suggesting that transient inhibition of autophagy under nutrient-rich conditions does not affect MCF-7 proliferation and viability (Fig. 7).
FIGURE 7.
Effect of verteporfin on cell survival in different starvation conditions. MCF-7 cells were incubated for 8 h with different concentrations of verteporfin in complete medium or in the different starvation conditions shown. Chemicals and medium were washed away, and the cells were incubated for 40 h in complete medium without chemicals before measuring cell viability using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay (mean ± S.D. (error bars), n = 4).
Similar experiments were then carried out using different starvation conditions. Exposure of cells to medium lacking glucose, amino acids or serum (DPBS) for 8 h resulted in only a small decrease in cell numbers after 48 h, showing that transient exposure to starvation did not affect cell survival. However, when cells were exposed to DPBS and verteporfin, cell survival at 48 h decreased substantially and in a concentration-dependent manner (Fig. 7). Supplementing DPBS with amino acids did not rescue cells. However, supplementation of DPBS with glucose considerably increased cell survival, and further addition of serum resulted in complete cell survival. Therefore, inhibiting autophagy with verteporfin in nutrient-rich conditions does not affect MCF-7 cell proliferation and viability, but verteporfin sensitizes cells to glucose and serum deprivation.
Structural Requirements for Inhibition of Autophagy by Benzoporphyrin Derivatives
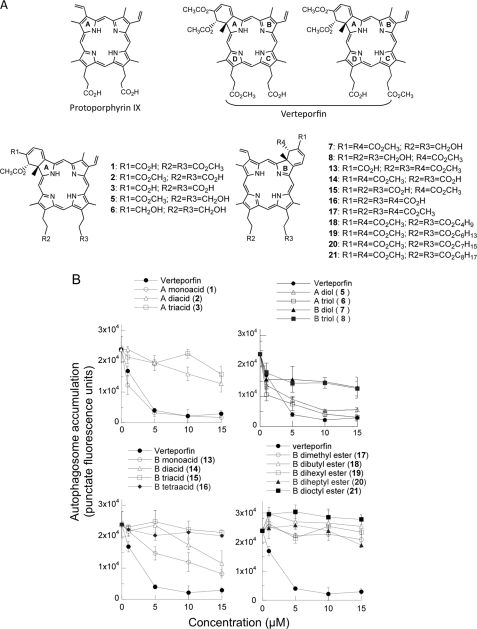
Verteporfin is composed of an equal mixture of two regioisomers (Fig. 8A), each of which consists of a pair of enantiomers. The regioisomers were separated by HPLC and tested in the automated microscopy assay. They were equally active (supplemental Fig. 2), indicating that the propionic acid and propionic acid methyl ester on rings C and D could be interchanged without affecting activity. Verteporfin may be described as a derivative of protoporphyrin IX (apo-heme), bearing modifications to rings A, C, and D (Fig. 8A). Protoporphyrin IX itself showed no inhibition of autophagy (data not shown), demonstrating the dependence on one or more of these modifications for activity. To examine this question, a number of analogs modified at these positions (Fig. 8A) were tested at different concentrations for inhibition of autophagy (Fig. 8B).
FIGURE 8.
Effect of selected verteporfin analogs on chloroquine-induced autophagosome accumulation. A, structures of protoporphyrin IX, verteporfin, and benzopophyrin derivatives. B, MCF-7 EGFP-LC3 cells were incubated for 4 h in complete medium with different concentrations of each verteporfin analog in the presence of 75 μm chloroquine. Punctate EGFP-LC3 was quantitated using the automated microscopy assay (mean ± S.D. (error bars), n = 3).
In protoporphyrin IX, two propionic acid groups are attached to rings C and D whereas verteporfin has a propionic acid methyl ester at one of these positions. Verteporfin analog 1 with two propionic acid methyl esters at rings C and D was fully active, but analog 2, with two propionic acid groups, was essentially inactive, showing that the presence of one carboxylic acid is tolerated, but not two. Verteporfin analogs 5 and 6 with propyl alcohol functionalities attached to rings C and D remained active. To determine whether additional C and D ring substitutions would affect activity, a number of different groups were also incorporated at these positions. Inhibition of autophagy by verteporfin derivatives was not affected by the presence of various large substituents on rings C and D, including some bearing distal OH groups (data not shown). However, the presence of two carboxylic acid groups close to the C and D rings prevented activity.
Verteporfin also differs from protoporphyrin IX by the presence, fused to ring A, of a cyclohexadiene bearing two methanoic acid methyl esters. Analog 1 with one methanoic acid methyl ester and one methanoic acid group retained full activity, suggesting that a variety of substitutions are likely tolerated at this position.
Thirteen analogs of verteporfin were generated with a disubstituted cyclohexadiene fused to ring B instead of ring A. Interestingly, 11 derivatives showed no activity, and the remaining two were only weakly active: analog 13 (one methanoic acid on ring B) was much less potent than its corresponding ring A analog 1 whereas analog 14 (two methyl esters on ring B and propionic acids attached to C and D) was comparable in activity with its corresponding ring A derivative 2. These data show that fusing the substituted cyclohexadiene to ring B instead of ring A almost completely abolishes activity.
DISCUSSION
This study describes a screening assay for inhibitors of autophagosome formation and the identification and characterization of verteporfin as an early stage inhibitor of autophagy. The chemical collections screened comprised mainly approved drugs and pharmacological agents and yielded verteporfin as the only active compound. The same collection yielded >30 chemical stimulators of autophagosome formation3 (25), suggesting that, unlike stimulators of autophagy, few inhibitors of autophagosome formation exist among known drugs.
We showed that verteporfin inhibits the basal level of autophagy in fed cells, as demonstrated by reduced numbers of EGFP-LC3 puncta, reduced EGFP-LC3 degradation by Western blot analysis, and reduced long lived protein degradation. Verteporfin also inhibited autophagy stimulated by serum starvation, a physiological stimulus, and by rapamycin, the chemical inhibitor of mTORC1. Moreover, verteporfin inhibited autophagy induced by all other chemicals tested to date, including rottlerin, amiodarone, perhexiline, and niclosamide (data not shown). The observation that verteporfin inhibits autophagy triggered by a wide variety of stimuli suggests that it targets the autophagic process itself rather than an upstream control mechanism.
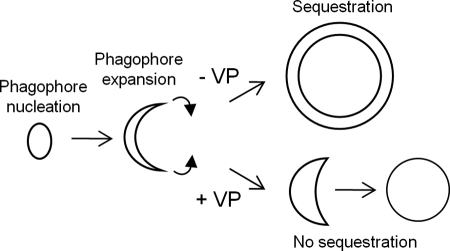
Autophagy may be arbitrarily divided into four steps: phagophore nucleation, phagophore elongation, sequestration of cytoplasmic constituents, and lysosomal degradation. Electron microscopy analysis did not reveal the presence of partially formed phagophores or phagophores with unfused tips. Rather, the cytoplasm of cells treated with verteporfin alone contained small empty vesicles with a single membrane. Verteporfin also inhibited the sequestration of fluorescent dextran loaded into the cytoplasm. Based on this evidence, we speculate that verteporfin binds to the membrane of expanding phagophores or to a factor involved in phagophore expansion and prevents expanding phagophores from adopting their characteristic cup shape, so they are unable to capture cytoplasmic cargo and only form empty single-membrane vesicles, as depicted in the model presented in Fig. 9. The number and size of these vesicles were increased in cells treated with both verteporfin and CQ. Although CQ is a known lysosomal inhibitor, it is believed to cause autophagosome accumulation not only by inhibiting lysosomal degradation but also by stimulating the expression of certain autophagy-associated genes including LC3 (45). In our proposed model, verteporfin would also affect the formation of CQ-induced phagophores, resulting in an increase in the size and number of these empty vesicular structures compared with verteporfin treatment alone.
FIGURE 9.
Hypothetical model for inhibition of autophagosome formation by verteporfin. Verteporfin prevents phagophore expansion and maturation, which causes the double membrane structure to lose its shape, thus forming an empty single-membrane vesicle.
In yeast, the processing and lipidation of LC3/Atg8 are required for phagophore elongation (46, 47). Localization of lipidated LC3 to the phagophore appears necessary for expansion into bowl-shaped structures that engulf cytoplasmic materials and generate the characteristic double membrane of the autophagosome (27). Verteporfin did not inhibit the generation of the LC3II band, nor did it inhibit membrane association of LC3II. However, the subcellular fractionation data suggest that LC3II may be less tightly bound to membranes in the presence of verteporfin than in its absence. Whether lipidated LC3 is associated with the membranes of the abnormal cytoplasmic vesicles seen by electron microscopy remains to be determined. What is clear at present is that verteporfin inhibits autophagosome formation via a mechanism not previously described that is downstream of the widely used PI3-kinase inhibitors, which prevent LC3 lipidation, and upstream of lysosomal inhibitors, which cause the accumulation of autophagosomes containing undigested materials, making verteporfin a useful pharmacological tool to study autophagy in cells and animals.
Verteporfin was used to examine the effect of inhibiting autophagy on cell survival under starvation. In the presence of glucose, amino acids, and serum, transient exposure of cells to verteporfin for 8 h had essentially no effect on proliferation and viability. In such nutrient-rich conditions, autophagy occurs at a basal level, and down-regulation of autophagy is not expected to be deleterious, as demonstrated in previous studies using siRNA knockdown to target components of autophagy machinery including Atg5, Atg7, and Beclin-1 (19, 35). However, in starvation conditions that stimulate autophagy, verteporfin significantly, and dose-dependently, reduced cell proliferation and viability after transient exposure. These results are consistent with a considerable body of evidence that autophagy is a mechanism of cellular protection under nutrient-limiting conditions (10, 35). Cell death was not rescued by the addition of amino acids, was partially rescued by the addition of glucose, and was completely rescued by the addition of glucose and serum, suggesting that cell death depends on the interaction between the energy state of the cell, the nutrients available, and the degree of autophagy inhibition.
Verteporfin is a benzoporphyrin derivative used clinically for photodynamic therapy of age-related macular degeneration. It is administered systemically, and irradiation of the retina with red laser light causes the generation of oxygen radicals that damage the vascular endothelium and block blood vessels (48). By contrast, verteporfin inhibits autophagy in the absence of light and therefore by a different mechanism. Our examination of verteporfin-related compounds in the EGFP-LC3 screening assay provided a clearer picture of the structural elements important for activity and the possible mode of binding of verteporfin to its putative target. The data showed that the “tails” attached to rings C and D are unlikely to interact with the putative binding site and might simply jut out into the solvent. Acid moieties positioned close to the planar porphyrin ring hindered activity, but moving them to more distal positions did not. The presence of propanol groups did not abolish activity. These data suggest that verteporfin inserts into a region that does not tolerate the presence of charged groups. One of the methyl ester groups attached to the cyclohexadiene moiety fused to the A ring is attached to an sp3 hybridized carbon and is oriented substantially out of plane from the otherwise flat verteporfin molecule. This group can be replaced with other substituents without affecting activity, indicating that it does not make important contacts with the binding site, and it could project outside the binding pocket. Finally, the observation that none of the ring B benzoporphyrin derivatives shows significant activity indicates that the overall shape of the molecule is important for binding to its target. Even though analogous ring A and B derivatives have the same elemental composition, the “bulk volume” and orientation of that volume are substantially different. Whether verteporfin inhibits autophagy by binding to a protein or a membrane target cannot be predicted from these data.
Evidence suggesting that autophagy promotes tumor growth in a nutrient-deprived environment and in response to several cancer therapy agents makes inhibition of autophagy an attractive strategy for cancer treatment. The facts that verteporfin is not toxic to cells under nutrient-rich conditions and in the absence of light irradiation, that it selectively kills cells under metabolic stress, that it is well tolerated provided that patients are not exposed to intense light (48), and that it is already approved for clinical use, make it a prime candidate for testing of its therapeutic potential.
Supplementary Material
Acknowledgments
We thank the Canadian Chemical Biology Network for access to screening chemicals; David Dolphin for benzoporphyrin derivates; Lindsay Heller and the Centre for Drug Research and Development for help with confocal microscopy; and Aruna Balgi, Bruno Fonseca, and Karen Lam for helpful discussion.
This work was supported by the Canadian Breast Cancer Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
A. Balgi and M. Roberge, unpublished data.
- 3-MA
- 3-methyladenine
- CQ
- chloroquine
- DMSO
- dimethyl sulfoxide
- DPBS
- Dulbecco's phosphate-buffered saline
- EBSS
- Earle's balanced salt solution
- LC3II
- lipidated LC3.
REFERENCES
- 1. Klionsky D. J., Cuervo A. M., Seglen P. O. (2007) Autophagy 3, 181–206 [DOI] [PubMed] [Google Scholar]
- 2. Levine B., Klionsky D. J. (2004) Dev. Cell 6, 463–477 [DOI] [PubMed] [Google Scholar]
- 3. Ogier-Denis E., Codogno P. (2003) Biochim. Biophys. Acta 1603, 113–128 [DOI] [PubMed] [Google Scholar]
- 4. Meijer A. J., Codogno P. (2004) Int. J. Biochem. Cell Biol. 36, 2445–2462 [DOI] [PubMed] [Google Scholar]
- 5. Jin S. (2006) Autophagy 2, 80–84 [DOI] [PubMed] [Google Scholar]
- 6. Furuya N., Yu J., Byfield M., Pattingre S., Levine B. (2005) Autophagy 1, 46–52 [DOI] [PubMed] [Google Scholar]
- 7. Liang X. H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. (1999) Nature 402, 672–676 [DOI] [PubMed] [Google Scholar]
- 8. Mathew R., Karp C. M., Beaudoin B., Vuong N., Chen G., Chen H. Y., Bray K., Reddy A., Bhanot G., Gelinas C., Dipaola R. S., Karantza-Wadsworth V., White E. (2009) Cell 137, 1062–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sato K., Tsuchihara K., Fujii S., Sugiyama M., Goya T., Atomi Y., Ueno T., Ochiai A., Esumi H. (2007) Cancer Res. 67, 9677–9684 [DOI] [PubMed] [Google Scholar]
- 10. Degenhardt K., Mathew R., Beaudoin B., Bray K., Anderson D., Chen G., Mukherjee C., Shi Y., Gélinas C., Fan Y., Nelson D. A., Jin S., White E. (2006) Cancer Cell 10, 51–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Izuishi K., Kato K., Ogura T., Kinoshita T., Esumi H. (2000) Cancer Res. 60, 6201–6207 [PubMed] [Google Scholar]
- 12. Kanzawa T., Germano I. M., Komata T., Ito H., Kondo Y., Kondo S. (2004) Cell Death Differ. 11, 448–457 [DOI] [PubMed] [Google Scholar]
- 13. Bilir A., Altinoz M. A., Erkan M., Ozmen V., Aydiner A. (2001) Pathobiology 69, 120–126 [DOI] [PubMed] [Google Scholar]
- 14. Ertmer A., Huber V., Gilch S., Yoshimori T., Erfle V., Duyster J., Elsässer H. P., Schätzl H. M. (2007) Leukemia 21, 936–942 [DOI] [PubMed] [Google Scholar]
- 15. Paglin S., Hollister T., Delohery T., Hackett N., McMahill M., Sphicas E., Domingo D., Yahalom J. (2001) Cancer Res. 61, 439–444 [PubMed] [Google Scholar]
- 16. Kondo Y., Kanzawa T., Sawaya R., Kondo S. (2005) Nat. Rev. Cancer 5, 726–734 [DOI] [PubMed] [Google Scholar]
- 17. Levine B. (2005) Cell 120, 159–162 [DOI] [PubMed] [Google Scholar]
- 18. Bursch W., Ellinger A., Kienzl H., Török L., Pandey S., Sikorska M., Walker R., Hermann R. S. (1996) Carcinogenesis 17, 1595–1607 [DOI] [PubMed] [Google Scholar]
- 19. Qadir M. A., Kwok B., Dragowska W. H., To K. H., Le D., Bally M. B., Gorski S. M. (2008) Breast Cancer Res. Treat. 112, 389–403 [DOI] [PubMed] [Google Scholar]
- 20. Katayama M., Kawaguchi T., Berger M. S., Pieper R. O. (2007) Cell Death Differ. 14, 548–558 [DOI] [PubMed] [Google Scholar]
- 21. Caro L. H., Plomp P. J., Wolvetang E. J., Kerkhof C., Meijer A. J. (1988) Eur. J. Biochem. 175, 325–329 [DOI] [PubMed] [Google Scholar]
- 22. Schneider P., Korolenko T. A., Busch U. (1997) Microsc. Res. Tech. 36, 253–275 [DOI] [PubMed] [Google Scholar]
- 23. Chen B., Pogue B. W., Hoopes P. J., Hasan T. (2005) Int. J. Radiat. Oncol. Biol. Phys. 61, 1216–1226 [DOI] [PubMed] [Google Scholar]
- 24. Tee A. R., Fingar D. C., Manning B. D., Kwiatkowski D. J., Cantley L. C., Blenis J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13571–13576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Balgi A. D., Fonseca B. D., Donohue E., Tsang T. C., Lajoie P., Proud C. G., Nabi I. R., Roberge M. (2009) PLoS One 4, e7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Curman D., Cinel B., Williams D. E., Rundle N., Block W. D., Goodarzi A. A., Hutchins J. R., Clarke P. R., Zhou B. B., Lees-Miller S. P., Andersen R. J., Roberge M. (2001) J. Biol. Chem. 276, 17914–17919 [DOI] [PubMed] [Google Scholar]
- 27. Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bauvy C., Meijer A. J., Codogno P. (2009) Methods Enzymol. 452, 47–61 [DOI] [PubMed] [Google Scholar]
- 29. Hariri M., Millane G., Guimond M. P., Guay G., Dennis J. W., Nabi I. R. (2000) Mol. Biol. Cell 11, 255–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Richter A. M., Kelly B., Chow J., Liu D. J., Towers G. H., Dolphin D., Levy J. G. (1987) J. Natl. Cancer Inst. 79, 1327–1332 [PubMed] [Google Scholar]
- 31. Richter A. M., Cerruti-Sola S., Sternberg E. D., Dolphin D., Levy J. G. (1990) J. Photochem. Photobiol. B 5, 231–244 [DOI] [PubMed] [Google Scholar]
- 32. Tovey A. (1994) Third-Generation Photosensitizers Synthesis, Characterization, and Liposome Interaction of Promising New Benzoporphyrin Derivatives. Ph.D. thesis, University of British Columbia, Vancouver [Google Scholar]
- 33. de Duve C., de Barsy T., Poole B., Trouet A., Tulkens P., Van Hoof F. (1974) Biochem. Pharmacol 23, 2495–2531 [DOI] [PubMed] [Google Scholar]
- 34. Wibo M., Poole B. (1974) J. Cell Biol. 63, 430–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Boya P., González-Polo R. A., Casares N., Perfettini J. L., Dessen P., Larochette N., Métivier D., Meley D., Souquere S., Yoshimori T., Pierron G., Codogno P., Kroemer G. (2005) Mol. Cell Biol. 25, 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kawai A., Uchiyama H., Takano S., Nakamura N., Ohkuma S. (2007) Autophagy 3, 154–157 [DOI] [PubMed] [Google Scholar]
- 37. Belzacq A. S., Jacotot E., Vieira H. L., Mistro D., Granville D. J., Xie Z., Reed J. C., Kroemer G., Brenner C. (2001) Cancer Res. 61, 1260–1264 [PubMed] [Google Scholar]
- 38. Granville D. J., Hunt D. W. (2000) Curr. Opin. Drug. Discov. Dev. 3, 232–243 [PubMed] [Google Scholar]
- 39. Eskelinen E. L. (2008) Autophagy 4, 257–260 [DOI] [PubMed] [Google Scholar]
- 40. Mari M., Reggiori F. (2007) Nat. Cell Biol. 9, 1125–1127 [DOI] [PubMed] [Google Scholar]
- 41. Eskelinen E. L. (2005) Autophagy 1, 1–10 [DOI] [PubMed] [Google Scholar]
- 42. Seglen P. O., Gordon P. B. (1982) Proc. Natl. Acad. Sci. U.S.A. 79, 1889–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Reggiori F., Klionsky D. J. (2005) Curr. Opin. Cell Biol. 17, 415–422 [DOI] [PubMed] [Google Scholar]
- 44. Thoreen C. C., Kang S. A., Chang J. W., Liu Q., Zhang J., Gao Y., Reichling L. J., Sim T., Sabatini D. M., Gray N. S. (2009) J. Biol. Chem. 284, 8023–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kimura N., Kumamoto T., Kawamura Y., Himeno T., Nakamura K. I., Ueyama H., Arakawa R. (2007) Pathobiology 74, 169–176 [DOI] [PubMed] [Google Scholar]
- 46. Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. (2001) EMBO J. 20, 5971–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kirisako T., Ichimura Y., Okada H., Kabeya Y., Mizushima N., Yoshimori T., Ohsumi M., Takao T., Noda T., Ohsumi Y. (2000) J. Cell Biol. 151, 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Michels S., Schmidt-Erfurth U. (2001) Semin. Ophthalmol. 16, 201–206 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.