Abstract
The crystal structure of the microbial transglutaminase (MTGase) zymogen from Streptomyces mobaraense has been determined at 1.9-Å resolution using the molecular replacement method based on the crystal structure of the mature MTGase. The overall structure of this zymogen is similar to that of the mature form, consisting of a single disk-like domain with a deep active cleft at the edge of the molecule. A major portion of the prosequence (45 additional amino acid residues at the N terminus of the mature transglutaminase) folds into an L-shaped structure, consisting of an extended N-terminal segment linked with a one-turn short helix and a long α-helix. Two key residues in the short helix of the prosequence, Tyr-12 and Tyr-16, are located on top of the catalytic triad (Cys-110, Asp-301, and His-320) to block access of the substrate acyl donors and acceptors. Biochemical characterization of the mature MTGase, using N-α-benzyloxycarbonyl-l-glutaminylglycine as a substrate, revealed apparent Km and kcat/Km values of 52.66 mm and 40.42 mm−1 min−1, respectively. Inhibition studies using the partial prosequence SYAETYR and homologous sequence SQAETYR showed a noncompetitive inhibition mechanism with IC50 values of 0.75 and 0.65 mm, respectively, but no cross-linking product formation. Nevertheless, the prosequence homologous oligopeptide SQAETQR, with Tyr-12 and Tyr-16 each replaced with Gln, exhibited inhibitory activity with the formation of the SQAETQR-monodansylcadaverine fluorophore cross-linking product (SQAETQR-C-DNS). MALDI-TOF tandem MS analysis of SQAETQR-C-DNS revealed molecular masses corresponding to those of NSQAETQC-C-DNS and C-DNS-NQRC sequences, suggesting the incorporation of C-DNS onto the C-terminal Gln residue of the prosequence homologous oligopeptide. These results support the putative functional roles of both Tyr residues in substrate binding and inhibition.
Keywords: Enzyme Inactivation, Enzyme Inhibitors, Enzyme Kinetics, Enzyme Structure, Enzymes
Introduction
Transglutaminase (TGase3; EC 2.3.2.13) catalyzes the formation of a cross-link between the γ-carboxamide group of a peptide-bound glutaminyl residue (an acyl donor) and a variety of primary amines (acyl acceptors), including the amino group of lysine, through an acyl transfer reaction. This enzyme nonspecifically polymerizes proteins, either intra- or intermolecularly, making them highly resistant to proteolytic degradation. TGases are widely distributed in plants, microorganisms, and vertebrates and are involved in many physiological functions, including blood clotting, wound healing, epidermal keratinization, and stiffening of the erythrocyte membrane in vertebrates (1). TGase also plays a role in stabilization of photosynthetic complexes in the chloroplast, modification of cytoskeletal proteins, abiotic and biotic stresses, aging, and programmed cell death in plants (2). Currently, TGase is used extensively in the food industry as a binding agent to improve the texture of protein-rich foods in a variety of processes, including the production of processed meat and fish products, such as surimi and ham (3). In other sectors, the potential applications of TGase include the building of collagen- or gelatin-based scaffolds for the construction of bioartificial organs, site-specific protein conjugation with DNA in biotechnology and materials science, and textile and leather processing (4).
The biochemical functions of microbial TGase (MTGase) from Streptomyces mobaraense (BCRC 12165, ATCC 29032; Streptoverticillium mobaraense, Streptomyces ladakanum, and Streptoverticillium ladakanum) have been studied extensively (5–7). This Ca2+-independent enzyme is secreted as a zymogen with an additional prosequence that consists of 45 amino acid residues (DNGAGEETKSYAETYRLTADDVANINALNESAPAASSAGPSFRAP) at the N terminus. This zymogen becomes active after proteolytic cleavage of the prosequence (8). The crystal structure of the mature form of MTGase, as determined by Kashiwagi et al. (9), reveals that the protein is folded into a disk-like shape that has a deep active cleft at the edge. This structure is distinct from those of other TGases, such as human factor XIII and fish TGase, which consist of four sequential domains: a β-sandwich, a core domain, and two barrels. The catalytic Cys-His-Asp triad is in the core domain. Apart from their structural similarities, those TGases that are homologous to human factor XIII share its Ca2+-dependent catalytic activity (10–13). Interestingly, the catalytic triad of MTGase and the structure of the active site superimpose well with those of the factor XIII-like TGases. However, the residues of the catalytic triad take the sequence Cys-Asp-His. Studies have also shown that MTGase has a higher reaction rate, a broader substrate specificity for the acyl donor, and a lower level of activity for deamidation than the factor XIII-like TGases (14).
To provide insights into the structural information of the prosequence, we determined the x-ray structure of the zymogen form of MTGase from S. mobaraense at 1.9 Å and compared it with the crystal structure of the mature MTGase. In addition, several synthetic oligopeptides (either prosequence or prosequence homologous oligopeptides) were synthesized and subjected to biochemical characterization of the possible mode of action on substrate recognition/binding and inhibition. The information obtained will be useful in biomedical engineering, materials science, and the textile- and leather-processing industries.
EXPERIMENTAL PROCEDURES
Enzyme Expression and Purification
The gene coding for full-length MTGase was cloned into the pET21d vector (Invitrogen) at the NcoI and XhoI cutting sites. The vector was transformed into BL21(DE3)* competent cells (Novagen) for protein overexpression. Protein production was induced by the addition of isopropyl β-d-thiogalactopyranoside to a final concentration of 0.1 mm, and the culture was incubated at 22 °C for an additional 24 h before harvest. The cells were collected by centrifugation and then resuspended in 50 ml of buffer A (100 mm Tris-HCl (pH 8.0), 0.3 m NaCl, 10 mm β-mercaptoethanol, and 0.01% NaN3) The mixture was sonicated, and the cell debris was removed by centrifugation at 30,000 × g for 50 min at 4 °C. The supernatant was applied to a 60-ml nickel-nitrilotriacetic acid affinity column previously equilibrated with buffer A containing 10 mm imidazole. The protein-loaded column was first washed with 0.5 column volumes of buffer A containing 10 mm imidazole and then with 4, 2, and 1 column volumes of buffer A containing 50, 150, and 400 mm imidazole, respectively. The purified proteins were eluted in buffer A containing 150 mm imidazole. Fractions containing MTGase activity were pooled and dialyzed against 100 mm Tris-HCl (pH 5.5), 50 mm NaCl, 2 mm β-mercaptoethanol, 0.01% NaN3, and 2 mm EDTA; concentrated to 17 mg/ml; and stored at −80 °C.
Crystallization and Data Collection
Crystallization of MTGase was performed at 293 K by the hanging-drop vapor diffusion method against a reservoir solution containing 30% PEG 8000, 50 mm NaCl, 1 mm EDTA, 1 mm β-mercaptoethanol, 0.01% NaN3, 100 mm cacodylic acid (pH 5.0), and 2% glycerol. A rod-shaped crystal appeared within 7 days and grew to ∼0.2 mm in size. Diffraction data were collected to 1.9-Å resolution using a Rigaku RUH3R generator equipped with a copper rotating anode generator operating at 5.0 killowatts (50 kV, 100 mA). The data were processed to 1.9 Å using HKL98 and SCALEPACK (1).
Structural Determination and Refinement
The apo form of the TGase structure was solved by molecular replacement using the structure of the active form (Protein Data Bank code 1IU4) as the search model in the phasing procedure using MOLREP of the CCP4 Program Suite (15). A subsequent annealing omit map simulated using the CNS program (2) showed the missing electron density of the N terminus clearly. The missing amino acids from the N terminus (residues 9–33) of apo-TGase were fitted into the electron density map using the graphics program O (16). Rigid-body refinement and 10 cycles of maximum likelihood refinement using REFMAC 5.0 were carried out, and the calculated composite 2Fo − Fc and Fo − Fc maps were examined for misfitted residues (17). Solvent molecules having the appropriate stereochemistry with their hydrogen-bonding partners were added using the program wARP (17). Final model building and refinement yielded an Rwork of 0.210 and an Rfree of 0.244 for the reflection data from 20.0 to 1.9 Å. The model was verified using MolProbity (18) and PROCHECK (19). The data collection and refinement statistics of the final refined structure are listed in Table 1. The full-length 379-amino acid MTGase zymogen has been numbered from 1 to 379. In contrast to the numbering scheme of the mature MTGase, residues 47–377 correspond to residues 1–331 of the mature MTGase.
TABLE 1.
Data collection and refinement statistics
Numbers in parentheses refer to the statistics of the highest resolution structure. r.m.s.d., root mean square deviation.
| Data collection | |
| Space group | P212121 |
| Unit cell dimensions (Å) | a = 64.3, b = 67.1, c = 84.0 |
| Resolution range (Å) | 20.0–1.9 (1.99–1.90) |
| Total reflections | 240,943 |
| Unique reflections | 29,324 |
| Data redundancy | 8.2 (8.0) |
| Completeness (%) | 100 (99.9) |
| I/σ(I) | 39.2 (8.1) |
| Rmergea | 4.3 (23.8) |
| Rrim | 4.1 (23.6) |
| Rpim (%)b | 1.5 (8.4) |
| Refinement statistics | |
| R factor (%)c | 21.0 |
| Rfree factor (%)d | 24.4 |
| No. of residues | 354 (from 385) |
| No. of waters | 150 |
| Ramachandran core region (%) | 98.6 (2.4% in allowed region) |
| r.m.s.d. bond length (Å) | 0.01 |
| r.m.s.d. bond angle | 1.1° |
| Average B factors (Å2) for protein | 29.2 |
| Average B factors (Å2) for waters | 31.5 |
a Rmerge = ΣhklΣi|Ii(hkl) − 〈Ihkl〉|/ΣhklΣiIi(hkl), where Ii(hkl) is the ith observation of reflection hkl, and 〈Ihkl〉 is the weighted average intensity for all observations i of reflection hkl.
b Rrim = Σhkl(N/(N − 1)1/2)Σi|Ii(hkl) − Ihkl|/ΣhklΣiIi(hkl), and Rpim = Σhkl(1/(N − 1)1/2)Σi|Ii(hkl) − Ihkl|/ΣhklΣiIi(hkl).
c R = Σ|FP − FP(calc)|/ΣFP.
d R factor for a subset of 10% of the reflection data that were excluded from the structural refinement.
Enzymatic Activity Assay and Inhibition Studies of MTGase
The enzymatic activity was assayed according to the method described by Folk and Cole (20), which is based on the chromogenic hydroxamate procedure using N-α-benzyloxycarbonyl-l-glutaminylglycine (Cbz-Gln-Gly) as a substrate. For determination of Km and Vmax values, substrate concentrations of 0.9–30.0 mm and a suitable time point that shows the linear relationship of the concentration of the ferric complex of l-glutamic acid γ-monohydroxamate as a function of time were used. The data collected were next applied to the Lineweaver-Burk equation. All reactions were carried out in triplicate.
Inhibition assays were performed by preincubating MTGase (0.63 μm) with various amounts of synthetic oligopeptide for 1 h at 37 °C before adding a mixture of 250 mm Tris acetate buffer (pH 8.0), 0.1 m hydroxylamine, and 15 mm Cbz-Gln-Gly. Four oligopeptides, Pep-1 to Pep-4, were synthesized and subjected to inhibition assay. The reaction was terminated by the addition of 15% (w/v) TCA containing 5% (w/v) FeCl3 after 10 min of incubation. The absorbance at 540 nm was measured and used to evaluate the degree of inhibition of TGase activity. IC50 values for the inhibitors were determined by TGase residual activity versus inhibitor concentration.
HPLC Assay for the MTGase-catalyzed Reaction Product
The reaction product of MTGase-catalyzed incorporation of the amine-containing fluorophore monodansylcadaverine (C-DNS) into various acyl donors was measured using an Agilent 1100 series HPLC system coupled with a fluorescence detector. A reaction mixture containing 3 mm Cbz-Gln-Gly (or various acyl donors), 0.5 mm C-DNS, 5 mm CaCl2, and 400 mm Tris-HCl (pH 7.5) in a total volume of 50 μl at 37 °C was added with 100 milliunits of MTGase to start the reaction. At intervals of 3, 6, 10, 20, 40, 60, and 120 min, 5 μl of solution was removed, and the reaction was stopped by the addition of 45 μl of 0.1% TFA. 40-μl solutions in each time course were assayed by HPLC. Separation of the cross-linking product Cbz-Gln-Gly-C-DNS or acyl donor-C-DNS was carried out using an Intersil ODS-3 column with 0.1% aqueous TFA (solvent A) and acetonitrile (solvent B) in a linear gradient from 0 to 50% over 20 min at a flow rate of 1 ml/min. Fluorescence was monitored with an excitation wavelength of 260 nm and an emission wavelength of 538 nm.
MALDI-TOF MS Identification of the MTGase-catalyzed Reaction Product
The Cbz-Gln-Gly-C-DNS or acyl donor-C-DNS product was collected from the HPLC system and subjected to MALDI-TOF MS determination of the exact molecular mass. The tandem MS (MS/MS) analyses were performed on a Bruker autoflexTM III MALDI-TOF mass spectrometer with an MTP 384 target plate polished steel T F and a pulsed 337-nm nitrogen laser. The Cbz-Gln-Gly-C-DNS or acyl donor-C-DNS solution was mixed directly with α-cyano-4-hydroxycinnamic acid matrix solution at a ratio of 1:1 and subjected to MS/MS spectrum determination.
RESULTS AND DISCUSSION
Overall Structure of the MTGase Zymogen
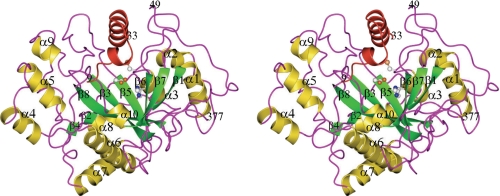
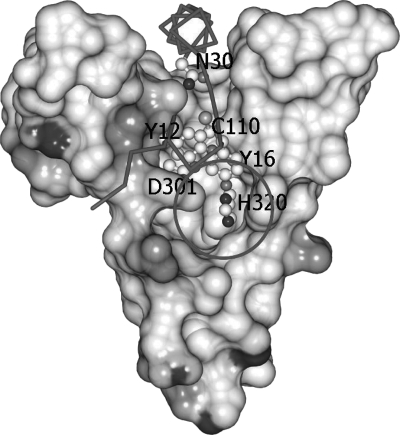
The full structure of the TGase zymogen from S. mobaraense is shown in Fig. 1. The first eight N-terminal amino acids (MDNGAGEE-), residues 34–48, the last two C-terminal amino acids (-LE), and the artificial His6 tag on the C terminus are disordered in the structure. The side chains of 18 surface amino acids, mainly lysine, arginine, and glutamate, are disordered and therefore were modeled as alanine. The molecule folds into a disk-like shape with overall dimensions of 65 × 59 × 41 Å. The deep cleft at the edge of the disk originally found in the mature MTGase structure is filled by the 25 N-terminal amino acids (residues 9–33). The active-site cleft that is located at the top of the structure is formed by two protrusions that are composed of two loops separated by ∼13 Å. The Cys-110, Asp-301, and His-320 catalytic triad is located at the bottom of the cleft. The N-terminal proregion of the zymogen forms an L-shaped structure in which a long α-helix (residues 20–33) on the C terminus intercalates with the side wall of the open end of the active cleft. The other loose structure, consisting of residues 9–20 and a short one-turn α-helix, splines on the other open end of the active cleft. The zymogen, with this 45-amino acid prosequence at the N terminus, has no enzymatic activity (8). Therefore, it is evident that the ordered structural part of the prosequence covers the active-site cleft and blocks the substrates from accessing the active site.
FIGURE 1.
Stereo view of the overall structure of the MTGase zymogen. The Cys-Asp-His catalytic triad has been drawn in ball-and-stick format (as it has been in Figs. 2–4 also). The visible portion of the prosequence (residues 9–33) of the zymogen has been colored red. Figs. 1–5 were generated with the program CCP4MG.
Structural Comparison with the Mature MTGase
The crystal structures of the MTGase zymogen and mature MTGase were compared to investigate the conformational changes that take place on activation and/or substrate binding. Except for alternate conformations of some residues that have a flexible side chain, the overall structures of these two proteins are essentially the same. A comparison of the topology of the mature form with that of the zymogen shows that the best average root mean square deviation over 328 Cα atoms is 0.56 Å. Further close comparison of the active-site cleft between these two structures shows that one of the protrusion loops (Asn-285–Asn299) moved slightly aside from its original position in the mature MTGase. The maximum shift that occurred at the tip of the loop is 4.7 Å (Gly-294). The average B-factor of this loop (Arg-288-Gly–296) is 34.6 Å2 (over all atoms). This compares with 50 Å2 for the mature MTGase. This small overall positional shift of the protrusion loop did not perturb the overall conformation of the active site of the zymogen compared with that of the mature MTGase (9). The MTGase is secreted from the cytoplasmic membrane as a zymogen and is activated by proteolytic processing. The current structure reveals that the active-site cleft and the overall protein structure were not perturbed greatly after the activation by proteolytic cleavage.
Prosequence Binding in the Active-site Cleft
The presumptive active-site cleft that is found at the edge of the disk-like structure of the mature MTGase remained unchanged in the current structure. The catalytic residues at the bottom of the cleft (Cys-110, Asp-301 and His-320) superimpose well on the catalytic Cys-His-Asp triad of the factor XIII-like TGases, even though the overall structures of these proteins are completely different, and sequence similarity is low (9).
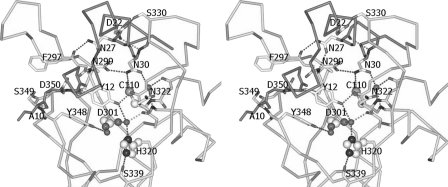
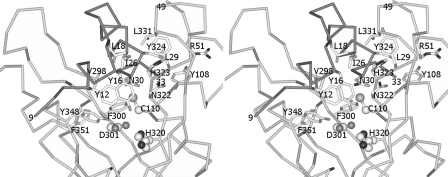
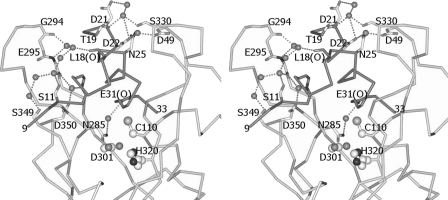
The prosequence folds into an L-shape and covers the active-site cleft in a complementary manner, with a buried surface area of 1927 Å2. Two key residues on the short α-helix of the prosequence, Tyr-12 and Tyr-16, stack close to but do not form direct interactions with the catalytic triad. Instead, the –OH group and the amide nitrogen atom of Tyr-12 form hydrogen bonds with the main chain oxygen atom of Asn-322 and the Oδ1 atom of Asp-350, respectively (Fig. 2). In addition, the Cδ2 atom of Tyr-12 and the Cϵ2 atom of Tyr-16 are in van der Waals contacts with the Cϵ1 atom of Tyr-348 (distance of 3.5 Å) and the Cβ atom of Asn-322 (distance of 3.5 Å), respectively (Fig. 3). Two water molecules are immobilized near the Cys-His-Asp catalytic triad. The –OH group of Tyr-12, the Oδ1 atom of Asp-301, the main chain oxygen atom of Gly-321, and the Nδ1 atom of His-320 are tetrahedrally coordinated within hydrogen-bonding distance to a water molecule. The Oδ2 atom of the catalytic Asp-301 residue also forms water-mediated hydrogen bonds with Tyr-348. A water molecule that is close to the catalytic Cys-110 residue (distance 3.5 Å, with a slightly distorted tetrahedral geometry) interacts with the Oδ1 atom of Asn-30, the amide nitrogen atoms of Cys-110 and Val-111, and the main chain oxygen atom of Tyr-108 (Fig. 4).
FIGURE 2.
Stereo view of the direct and water-mediated hydrogen-bonding interactions between the prosequence of MTGase zymogen and the residues in the active-site cleft. Note the hydrogen bonds that are also formed between the main chain oxygen of Ala-10 and the main chain NH of Ser-349 and between the main chain NH of Tyr-12 and the Oδ1 of Asp-350, respectively. The side chains of Arg-72 and Tyr-70 are shifted compared with the structure of the mature MTGase because of the shift in the position of His-320 to form a hydrogen bond with the Oγ atom of Ser-339. This has not been shown, as it was not possible to generate a clear view.
FIGURE 3.
Stereo view of the hydrophobic interactions between the prosequence of the MTGase zymogen and the residues in the active cleft. The criterion that we adopted for the presence of a van der Waals interaction is that the distance between the atoms had to be <4.0 Å. The van der Waals contacts in this structure are weak, as the interaction distance is generally ∼3.5 Å.
FIGURE 4.
Stereo view of the water-mediated hydrogen bonds on the right- and left-hand sides of the prosequence of the MTGase zymogen in the active cleft. The criterion that we used for the presence of a hydrogen bond is that the distance between the donor and acceptor had to be ≤3.3 Å. The side chains of those residues that formed hydrogen bonds by using the main chain amide nitrogen or oxygen atom, such as Leu-18 and Glu-31, have not been shown.
Based on our crystal structure (Fig. 5), it is tempting to suggest that the binding mode of the prosequence can be used as a general model for substrate binding to MTGase. The wide active-site cleft, which is wide enough to accommodate the α-helix, explains why the MTGase had less need for a specific acyl donor substrate and has a higher reaction rate than the factor XIII-like TGases (14). Residues 34–48 are disordered in this structure and are probably not important for the binding of the prosequence in the active cleft. Interestingly, the overall interactions of the prosequence within the active cleft are weak and asymmetric on either side of the prosequence, with fewer hydrogen bonds and weak van der Waals contacts and water-mediated hydrogen bonds on the apparently flexible protrusion loop (Asn-285–Asn-299). It is reasonable to suggest that the role of these generally weak and asymmetric interactions is to facilitate the docking of the prosequence in the active cleft and to provide less hindrance to the activation of the MTGase zymogen by proteolytic cleavage of the prosequence and to its subsequent departure from the active cleft.
FIGURE 5.
Electrostatic molecular surface of the active cleft and the Cα main chain trace of the prosequence of the MTGase zymogen. Only the active cleft region that encompasses the prosequence is drawn here. Tyr-12 and Tyr-16 are supposed to block the acyl donor and acceptor during the cross-linking reaction. Asn-30 was used to mark the position of the long helix of the prosequence in the active cleft. The circle marks the solvent-exposed region of the catalytic triad (Cys-Asp-His).
The current structure also supports the catalytic mechanism that has been proposed by Kashiwagi et al. (9). In their model, Cys-110 and Asp-301 play essential roles in the nucleophilic attack for deamination of the acyl donor substrate and for the cross-linking reaction of the acyl acceptor (such as the side chain of lysine residues). A mutation experiment with Asp-301 (D301A) that drastically reduces the catalytic activity of the enzyme supports the involvement of Asp-301 (10). In the structure presented here, Tyr-12 of the MTGase zymogen is in close proximity to these two residues and can block the reaction. On the other hand, His-320 of the proposed Cys-Asp-His catalytic triad is sufficiently exposed to the solvent and can react with the substrate (Fig. 5). His-320 is 4.5 Å (3.1 Å in the mature MTGase) from the catalytic Asp-301 residue and uses water-mediated hydrogen bonds to reach Asp-301. Combined with the fact that an MTGase mutant (H320A) still retains ∼50% activity relative to the wild type (10), this suggests that the MTGase mutant may play a role in stabilizing the oxyanion intermediate of Asp-301 and facilitating the preferable conformation of the acyl acceptor in the active site for the cross-linking reaction. Further stabilization of the oxyanion intermediates may be provided by water-mediated hydrogen bonds, as there is evidence of water molecules near Cys-110 and Asp-301 in this structure, and there are no other hydrogen bond donors nearby. Based on our structure, it is plausible that Tyr-16, which stacks close to Tyr-12, would hinder the access of the acyl acceptor substrate to the active site for the cross-linking reaction.
Biochemical Characterization and Inhibition Studies of MTGase
The biochemical properties of the mature MTGase were characterized further using the purified enzyme to investigate kinetic parameters, inhibition, and product identity. The apparent Km, Vmax, and kcat/Km of the mature MTGase, using Cbz-Gln-Gly as a substrate, were determined to be 52.66 mm, 49.67 μmol/min/mg, and 40.42 mm−1 min−1, respectively. In addition, the mature MTGase showed a specific activity of 10.59 units/mg and a kcat of 2128.57 min−1. Comparatively, the TGase from guinea pig liver exhibits specific activity, Vmax, and Km values of 25 units/mg, 37 μmol/min/mg, and 66 mm, respectively (20).
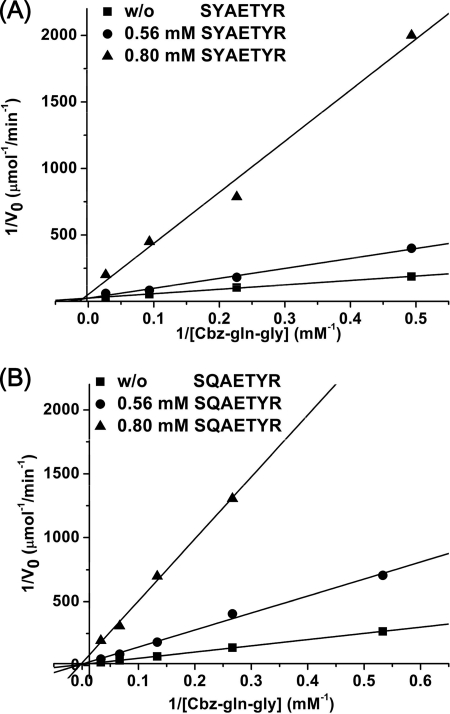
To shed light on the inhibitory activity of the MTGase-catalyzed transglutamination reaction, a series of oligopeptides (either prosequence or prosequence homologous oligopeptides) were designed and synthesized. Pep-1 (SYAETYR) is the prosequence, whereas Pep-2 (SQAETYR) has Tyr-12 substituted with Gln. Pep-3 (SQAETQR), with Tyr-12 and Tyr-16 each substituted with Gln, was also synthesized to characterize possible substrate binding or recognition. Pep-4 (GLGQGETYR), a previously identified substrate of TGase from guinea pig liver, fish, Pagrus major, and liver, was also synthesized and used as positive control, in addition to Cbz-Gln-Gly (5). As expected, Pep-1 and Pep-2 showed inhibitory activity with IC50 values of 0.75 and 0.65 mm, respectively. Interestingly, a noncompetitive mechanism of inhibition was determined for both Pep-1 and Pep-2 (Fig. 6). The results indicate that these prosequence homologous oligopeptides may occupy the position above the enzyme's active cleft, similar to that of the prosequence in MTGase prior to proteolytic maturation, thus inhibiting the enzymatic activity.
FIGURE 6.
Double-reciprocal Lineweaver-Burk plots of S. mobaraense MTGase inhibited by synthetic oligopeptides. A, Pep-5 (SYAETYR); B, Pep-6 (SQAETYR). The S. mobaraense TGase activities were measured in the absence or presence of various concentration of peptide. ■, absence of inhibitor; ●, 0.56 mm peptide; ▴, 0.80 mm peptide.
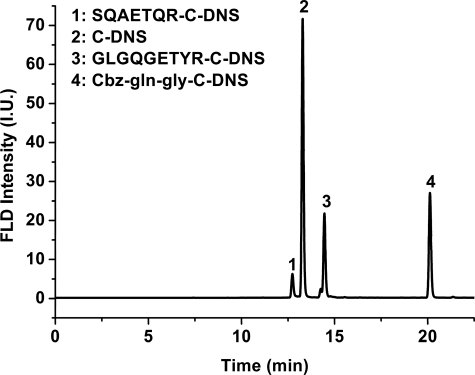
We next characterized the cross-linking position of the product using C-DNS, an amine-containing fluorophore, as a reporter and an HPLC system coupled with a fluorescence detector (21). The above-mentioned oligopeptides, Pep-1 to Pep-4, in addition to Cbz-Gln-Gly, were used to investigate the possible incorporation of C-DNS into the MTGase-catalyzed transglutamination. As shown in Fig. 7, new fluorescent product peaks could be detected from the HPLC profile when MTGase-catalyzed transglutamination of Pep-3, Pep-4, and Cbz-Gln-Gly with C-DNS occurred. Conversely, no new product peak could be detected for Pep-1 and Pep-2, consistent with previous UV-visible determination in inhibition studies. Furthermore, the loss of activity and the absence of product for Pep-1 and Pep-2 might indicate a binding site similar to that of the prosequence prior to proteolytic cleavage, thus blocking the substrates from accessing the active site.
FIGURE 7.
HPLC profile of S. mobaraense MTGase-catalyzed C-DNS incorporation reactions. Peak 1, retention time of 12.13 min, SQAETQR-C-DNS; peak 2, 13.28 min, C-DNS; peak 3, 14.45 min, GLGQGETYR-C-DNS; peak 4, 20.12 min, Cbz-Gln-Gly-C-DNS. FLD, field; I.U., intensity units.
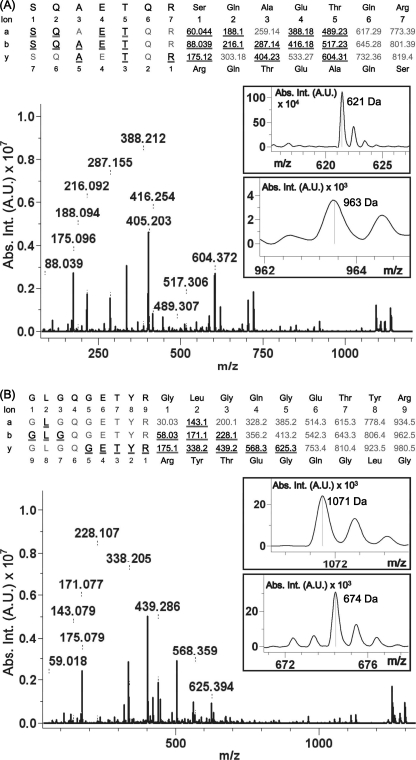
To clearly characterize the Pep-3 and Pep-4 reactions, the reaction products were subjected to molecular mass and modification site determinations using MALDI-TOF MS. As expected, singly charged peptides at m/z 1137.59 and 1298.71, which correspond to Pep-3-C-DNS and Pep-4-C-DNS, respectively, could be detected from the MALDI-TOF mass spectra (data not shown). Further MS/MS analysis of Pep-3-C-DNS confirmed the complete amino acid sequence SQAETQR and the C-DNS-modified Gln residue near the position of the C terminus based on the combination of molecular mass differentiation in both b-fragment ion and y-fragment series and partly supported by a-ion series (Fig. 8). Specifically, the position of C-DNS modification was determined by the mass fragments of 963 and 621 Da, which correspond to the masses of peptide sequence NSQAETQC-C-DNS in the b-ion series and peptide sequence C-DNS-NQRC in the y-ion series, respectively. Furthermore, the MS/MS data also support the intact Gln residue at a position close to the N terminus of Pep-2 and Pep-3. Similarly, the mass fragments of 1071 and 674 Da, which correspond to the masses of peptide sequence C-DNS-NQGETYRC in the y-ion series and peptide sequence NGLGQC-C-DNS in the b-ion series, respectively, also support the Pep-4-C-DNS complete amino acid sequence and the C-DNS-modified Gln residue. Therefore, these oligopeptides might interact with the critical functional residues and thus possess the ability to interfere in the catalytic activity of the mature MTGase.
FIGURE 8.
MALDI-TOF tandem mass spectra of MTGase-cross-linked products. A, SQAETQR-C-DNS; B, GLGQGETYR-C-DNS. The sequence is shown using single-letter amino acids confirmed by the combination of y-ion, b-ion, and a-ion series in the tandem mass spectrum. Underlined ions indicate fragment ions observed by the mass spectrometer. The insets show the observed molecular masses of the C-DNS-modified fragments. Abs. Int., absorbance intensity; A.U., absorbance units.
In summary, we have determined the crystal structure of the S. mobaraense MTGase zymogen at 1.9 Å and found an overall structure similar to that of the mature MTGase, except for the prosequence region. Interestingly, the prosequence folds into an L-shaped structure with its two key residues, Tyr-12 and Tyr-16, located on top of the putative Cys-His-Asp catalytic triad to putatively block access of the substrate acyl donors and acceptors. Synthetic oligopeptides (prosequence or prosequence with Tyr-12 substituted with Gln) showed a noncompetitive mode of inhibition of the mature MTGase-catalyzed reactions. Furthermore, MALDI-TOF MS/MS identification of the SQAETQR-C-DNS adduct, but not SQAETYR, from mature MTGase-catalyzed C-DNS incorporation supports the putative functional roles of both Tyr residues in substrate recognition/binding and inhibition. Therefore, these results support the putative functions of the prosequence in covering the active-site cleft and subsequently blocking the substrates from accessing the active site.
This work was supported in part by intramural funding from the Institute of Biochemistry, National Chung Hsing University, and by the Ministry of Education Aiming for Top University Plan, the National Chiao Tung University, and National Science Council of the Republic of China Contract NSC 99-2113-M-009-004-MY2.
The atomic coordinates and structure factors (code 3IU0) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- TGase
- transglutaminase
- MTGase
- microbial TGase
- Cbz-Gln-Gly
- N-α-benzyloxycarbonyl-l-glutaminylglycine
- C-DNS
- monodansylcadaverine
- MS/MS
- tandem MS.
REFERENCES
- 1. Aeschlimann D., Paulsson M. (1994) Thromb. Haemost. 71, 402–415 [PubMed] [Google Scholar]
- 2. Serafini-Fracassini D., Del Duca S. (2008) Ann. Bot. 102, 145–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yokoyama K., Nio N., Kikuchi Y. (2004) Appl. Microbiol. Biotechnol. 64, 447–454 [DOI] [PubMed] [Google Scholar]
- 4. Zhu Y., Tramper J. (2008) Trends Biotechnol. 26, 559–565 [DOI] [PubMed] [Google Scholar]
- 5. Ohtsuka T., Ota M., Nio N., Motoki M. (2000) Biosci. Biotechnol. Biochem. 64, 2608–2613 [DOI] [PubMed] [Google Scholar]
- 6. Ohtsuka T., Umezawa Y., Nio N., Kubota K. (2001) J. Food Sci. 66, 25–29 [Google Scholar]
- 7. Hemung B. O., Li-Chan E. C., Yongsawatdigul J. (2008) J. Agric. Food Chem. 56, 7510–7516 [DOI] [PubMed] [Google Scholar]
- 8. Pasternack R., Dorsch S., Otterbach J. T., Robenek I. R., Wolf S., Fuchsbauer H. L. (1998) Eur. J. Biochem. 257, 570–576 [DOI] [PubMed] [Google Scholar]
- 9. Kashiwagi T., Yokoyama K., Ishikawa K., Ono K., Ejima D., Matsui H., Suzuki E. (2002) J. Biol. Chem. 277, 44252–44260 [DOI] [PubMed] [Google Scholar]
- 10. Chung S. I., Folk J. E. (1972) Proc. Natl. Acad. Sci. U.S.A. 69, 303–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lorand L., Barnes N., Bruner-Lorand J. A., Hawkins M., Michalska M. (1987) Biochemistry 26, 308–313 [DOI] [PubMed] [Google Scholar]
- 12. Melino G., De Laurenzi V., Catani M. V., Terrinoni A., Ciani B., Candi E., Marekov L., Steinert P. M. (1998) Results Probl. Cell Differ. 24, 175–212 [DOI] [PubMed] [Google Scholar]
- 13. Ahvazi B., Kim H. C., Kee S. H., Nemes Z., Steinert P. M. (2002) EMBO J. 21, 2055–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shimba N., Yokoyama K., Suzuki E. (2002) J. Agric. Food Chem. 50, 1330–1334 [DOI] [PubMed] [Google Scholar]
- 15. Project C. C. (1994) Acta Crystallogr. Sect. D 50, 760–76315299374 [Google Scholar]
- 16. Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. Sect. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 17. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 18. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 20. Folk J. E., Cole P. W. (1966) J. Biol. Chem. 241, 5518–5525 [PubMed] [Google Scholar]
- 21. Pasternack R., Laurent H. P., Rüth T., Kaiser A., Schön N., Fuchsbauer H. L. (1997) Anal. Biochem. 249, 54–60 [DOI] [PubMed] [Google Scholar]