Abstract
The Atg4 cysteine proteases are required for processing Atg8 for the latter to be conjugated to phosphatidylethanolamine on autophagosomal membranes, a key step in autophagosome biogenesis. Notably, whereas there are only one atg4 and one atg8 gene in the yeast, the mammals have four Atg4 homologues and six Atg8 homologues. The Atg8 homologues seem to play different roles in autophagosome biogenesis, and previous studies had indicated that they could be differentially processed by Atg4 homologues. The present study provided the first detailed kinetics analysis of all four Atg4 homologues against four representative Atg8 homologues. The data indicated that Atg4B possessed the broadest spectrum against all substrates, followed by Atg4A, whereas Atg4C and Atg4D had minimal activities as did the catalytic mutant of Atg4B (C74S). On the other hand, GATE-16 seemed to be the overall best substrate for Atg4 proteases. The kinetics parameters of Atg4B were also affected by its structure and that of the substrates, indicating a process of induced fit. The determination of the kinetics parameters of the various Atg4-Atg8 pairs provides a base for the understanding of the potential selective impact of the reaction on autophagosome biogenesis.
Keywords: Autophagy, Cysteine Protease, Enzyme Catalysis, Mammal, Signal Transduction
Introduction
Macroautophagy plays multiple roles in mammalian cells, although it is primarily a stress response to nutrient deficiency in the yeast. More than 30 genes have been defined that participate in autophagy or an autophagy-related process in the yeast, many of which have mammalian homologues (1, 2), whose heterogeneity could contribute to functional diversity.
The core autophagy machinery is built around two ubiquitin-like conjugation systems (3). In one system, the ubiquitin-like protein, Atg12, is conjugated to Atg5 through a covalent bond with the participation of Atg7 and Atg10. The Atg5-Atg12 complex interacts with Atg16 to form a multimer complex, which is localized to membranes of early autophagosomes. In another system, the ubiquitin-like protein, Atg8, is first cleaved by a cysteine protease, Atg4, to expose the conserved C-terminal glycine. Atg8 is then conjugated to phosphatidylethanolamine with the participation of Atg7 and Atg3 (3–6). Recent studies have also indicated the participation of an Atg12-Atg5-Atg16 complex in the lipidation of Atg8 (7).
The unconjugated form of Atg8 is in the cytosol or loosely associated with the membranes, whereas the phosphatidylethanolamine-conjugated form is tightly associated with the membranes (5, 8). This association of Atg8 with the autophagosomal membrane is considered to be important for membrane extension, membrane fusion, and the eventual enclosure of the membrane to form the vesicles (9–12). The deletion of Atg8 in the yeast results in much smaller autophagosomes (13).
Yeast cells express a single Atg8 gene, whereas in humans, there are six Atg8 homologues belonging to two subfamilies: the MAP1A/B (microtubule-associated proteins 1 A/B)/LC3 (light chain 3) subfamily (LC3A, LC3B, and LC3C) and the γ-aminobutyric acid receptor-associated protein (GABARAP) subfamily (GABARAP, GATE-16 (Golgi-associated ATPase enhancer-16)/GABARAPL2, and Atg8L/GABARAPL1) (11, 14–17). The conserved glycine, which is proximal to the scissile bond, is present in yeast Atg8 and all of the mammalian homologues.
Although LC3B has been most extensively studied in mammalian cells and has been widely used as a mammalian autophagosome marker, the differences among different mammalian Atg8 homologues have not been well characterized. However, a recent study (16) indicated that these homologues could contribute to autophagosome biogenesis in different ways. LC3s are involved in phagophore elongation, whereas GABARAP molecules are important for later stage autophagosome maturation (16).
Atg4 is a cysteine protease of the C54 family (18). There is one single member in yeast, and deletion of it arrests the autophagy process (8). There are four homologues, Atg4A, Atg4B, Atg4C, and Atg4D, in humans and mice (18), whose individual contribution to autophagy is not well characterized. However, genetic deletion of Atg4B (19, 20), but not Atg4C (21) resulted in notable defects in autophagy, suggesting that there are functional variations among the Atg4 homologues.
The specificity of various Atg4 homologues versus various Atg8 homologues in mammalian cells and their individual significance has just begun to be revealed. Studies have indicated that Atg4B are able to cleave LC3B, GATE-16, GABARAP, and Atg8L (11, 17, 22, 23). On the other hand, GABARAP (11) and GATE-16 (11, 24) seemed to be better substrates than LC3B (11) or Atg8L (17) for Atg4A. It is not clear whether native Atg4C and Atg4D could process any of the mammalian Atg8 homologues (17, 18, 25). Overall, systemic studies of all four Atg4 homologues together have not been conducted to quantitatively investigate and compare their catalytic properties against the mammalian Atg8 homologues, which was the aim of the present work.
Our results indicated that Atg4B had the broadest substrate spectrum with similar affinity and catalytic efficiency toward each of the Atg8 substrates tested. Although Atg4A was the second best enzyme, Atg4C and Atg4D had minimal activities, as did the Atg4B catalytic mutant (C74A). On the other hand, GATE-16 was the better substrate for all Atg4 homologues. Kinetics studies also indicated that conformation changes of Atg4B induced by its substrate was important for the catalysis. These studies thus indicated the broad diversity in the catalytic efficiency among different Atg4-Atg8 pairs, which could affect autophagy in mammalian cells in ways different from that seen in the yeast in order to meet the functional diversity.
EXPERIMENTAL PROCEDURES
Chemicals, Antibodies, and Peptides
Chemical reagents were obtained from Sigma unless otherwise stated. The mouse anti-FLAG and anti-GAPDH antibodies were obtained from Sigma. The anti-Atg4 antibodies were from Abgent (San Diego, CA). HRP-conjugated goat anti-mouse secondary antibodies were from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA). LC3B peptides were customarily ordered from Chi Scientific (Boston, MA). All peptides were protected at the N terminus by an acetyl group and chemically linked to a fluorochrome, 7-amino-4-trifluoromethyl coumarin (AFC),2 in their C termini.
Bacteria Strains, DNA, and Plasmids
The Escherichia coli strain BL21(DE3) (Invitrogen) was used as the host for prokaryotic production of proteins. The plasmids pET-28a(+) (Novagen, Madison, WI), pGEX-6P-1 (GE Healthcare), and pcDNA3.1(+) (Invitrogen) were used for DNA cloning.
The open reading frames of the following human Atg4 homologues and Atg8 homologues were amplified by high fidelity PCR using RNA extracted from HEK-293A cells: Atg4A (GenBankTM accession number NP_443168), Atg4B (NP_037457), Atg4C (NP_116241), Atg4D (NP_116274), MAP 1A/1B Light Chain 3B (LC3B, NP_073729), GABA(A) receptor-associated protein (GABARAP, NP_009209), GABA(A) receptor-associated protein-like 1 (Atg8L, NP_113600), and GABA(A) receptor-associated protein-like 2 (GATE-16, NP_009216). ATG4A, ATG4B, and ATG4C were cloned into pET-28a(+) in fusion with the N-terminal His6 tag. ATG4D was cloned into pGEX-6P-1 in fusion with the N-terminal GST tag. All Atg8 homologues (Atg8s) were first cloned into pET-28a(+) in fusion with the His6 tag at the N terminus. The DNA fragment encoding GST was then amplified from the pGEX-6P-1 plasmid and inserted in frame at the C termini of the Atg8s in the above constructs. The mutant Atg4BC74S was constructed using the ExSite in vitro site-directed mutagenesis system (Stratagene, La Jolla, CA). Atg4B with the deletion of the N-terminal first 24 amino acids (Atg4BΔ1–24) was constructed by amplifying the corresponding ATG4B sequence.
To express Atg4s in mammalian cells, ATG4 genes were cloned into the mammalian expression vector pcDNA3.1(+) with the insertion of the FLAG tag at their N termini. All DNA constructs were confirmed by sequencing.
Protein Expression and Purification
The plasmids were introduced into BL21(DE3). Protein expression was induced by isopropyl β-d-1-thiogalactopyranoside. Atg4D was purified by affinity chromatography using glutathione-Sepharose (GE Healthcare), whereas all other Atg4s and Atg8s were purified by affinity chromatography using Ni2+-NTA resin (U.S. Biochemical Corp.). All preparations were further purified by gel filtration using a Superdex 75 column (GE Healthcare). The purity and the apparent molecular weight of the purified proteins were verified by SDS-PAGE and Coomassie Brilliant Blue (CBB) staining.
Kinetics Analysis of Atg4 with Atg8s-GST
Purified Atg4s and Atg8s-GST at the concentrations indicated in the figure legends were incubated together in Buffer A (150 mm NaCl, 1 mm EDTA, 1 mm DTT, and 50 mm Tris-HCl, pH 7.2) in a volume of 20 μl at 37 °C for the designated times. Reactions were stopped by adding the Laemmli sample buffer and resolved by SDS-PAGE, followed by CBB staining. The amounts of substrates (Atg8s-GST) and the cleaved products (Atg8s and GST) were quantified by densitometry and the use of a GST standard curve.
The percentage of substrate that remained at each reaction time point was equal to ODAtg8s-GST/(ODAtg8s-GST + ODGST + ODAtg8s) × 100%. This value was then plotted again the reaction time. For subsequent kinetic analysis, the optimal time was determined, at which the reaction became linear under the given experiment conditions.
Subsequently, Atg8s-GST at different concentrations were incubated with a given Atg4 enzyme for the period of this optimal time. The initial velocity (mm/s, y axis) was defined as the change in the concentration of GST (one of the cleavage products of GST-Atg8s), which was plotted against the concentration of the substrate (mm, x axis). The curves were then fitted using the non-linear regression method (SigmaPlot 10.0, Systat Software, San Jose, CA), from which the Vmax and Km (Michaelis constant) for each enzyme-substrate reaction were derived. kcat (catalytic constant) was determined by dividing Vmax by the concentration of the enzyme. Catalytic efficiency is defined as kcat/Km (mol−1 liter s−1).
Kinetics Analysis of Atg4B with LC3B Peptides
LC3B-AFC peptides (0.1 mm) were incubated with Atg4s in Buffer B (10% sucrose, 0.1% CHAPS, 100 mm NaCl, 1 mm EDTA, 50 mm Tris-HCl, pH 7.2) in a total volume of 100 μl at 37 °C in opaque 96-well plates. Enzyme activity was determined by the release of the AFC using a fluorescence plate reader (GENios, TECAN, San Jose, CA) at λEx = 400 nm/λEm = 510 nm.
The relative fluorescence units (RFU) from the released AFC group were recorded up to 48 h. In the kinetics analysis, the RFU was converted to the concentration of AFC using an AFC standard. In these assays, 100 RFU was equivalent to 10 nm free AFC. The initial velocities (in the first 20 h of reaction) were calculated as the change of AFC concentration/min, which was plotted against the concentration of the peptide substrate. The curve was fitted using the non-linear regression method (SigmaPlot 10.0). The kinetic parameters were determined as described above for the full-length substrates.
Cleavage of Atg8s-GST by Cell Lysate
25 μg of cell lysate of HEK-293A with or without Atg4 transfection were mixed with 5 μg of Atg8s-GST. After 3 min of incubation, the reactions were stopped by adding the Laemmli sample buffer and resolved by SDS-PAGE and CBB staining. The density of the product GST was determined by densitometry, from which the percentage of cleavage of the substrates was calculated.
Cell Culture, Transfection, and Immunoblotting Assay
HEK-293A cells were grown in DMEM containing 10% fetal bovine serum and 1% penicillin/streptomycin. They were transfected with the indicated constructs using Lipofectamine 2000 (Invitrogen). Transfected cells were harvested 24 h later and lysed using Buffer C (150 mm NaCl, 1 mm EDTA, 0.1% Triton, 50 mm Tris-HCl, pH 8.0) with freshly prepared protease inhibitor (1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin, and 20 μg/ml PMSF).
For immunoblot assay, lysates were separated by SDS-PAGE and transferred to PVDF membranes. After the application of the appropriate antibodies, signals were developed using an Immobilon Western detection kit (Millipore, Billerica, MA) according to the manufacturer's instructions.
RESULTS
Atg4 Homologues Are Different in Their Capability of Processing Atg8 Substrates
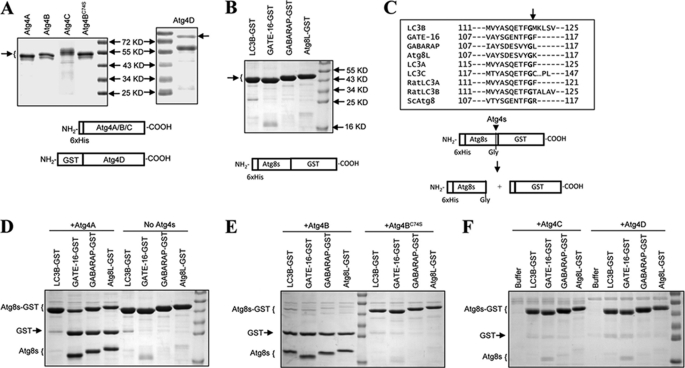
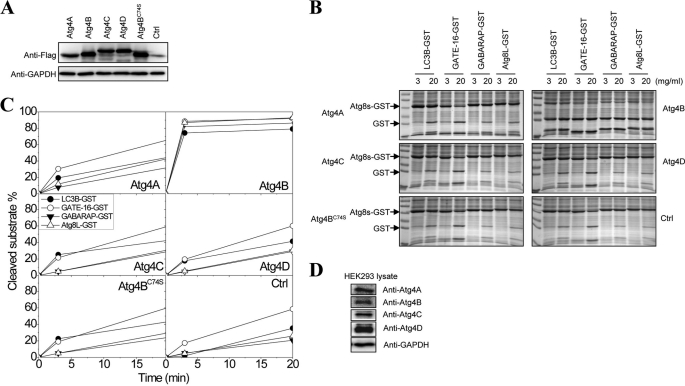
To determine and to compare the enzymatic characteristics of the Atg4 homologues, we prepared recombinant proteins using a bacterial expression system. Atg4A, Atg4B and its catalytic mutant, Atg4BC74S, and Atg4C could be readily expressed and purified when fused with the His6 tag at the N terminus. Atg4D was not soluble when fused with the His6 tag, but a workable amount of the protein was expressed in the soluble fraction when it was fused with GST at the N terminus. These proteins were then purified using an Ni2+-NTA column and glutathione beads, respectively (Fig. 1A). Similarly, the four major Atg8 homologues fused with the His tag were purified using an Ni2+-NTA column (Fig. 1B).
FIGURE 1.
Cleavage of Atg8 homologues by Atg4 homologues. A, His-tagged human Atg4A, Atg4B, Atg4BC74S, and Atg4C and GST-tagged human Atg4D were expressed in E. coli, and purified using Ni2+-NTA resins or glutathione-Sepharose, respectively. Five micrograms of each purified protein were verified by SDS-PAGE and CBB staining. The arrow indicates the major species purified. Atg4C seemed to exist in several species with different electrophoretic mobility. The 73 kDa band in Atg4D preparation was confirmed by immunoblot assay (data not shown). B, four human Atg8 homologues were fused with the His tag at the N terminus and GST at the C terminus, expressed in E. coli, and purified using Ni2+-NTA resins. Ten micrograms of each purified protein were verified by SDS-PAGE and CBB staining. The arrow indicates the major species purified. C, alignment of the amino acid sequences at the cleavage site of the four Atg8 homologues examined here and five others in the literature. Cleavage occurs C-terminal to the glycine (P1). The lower panel shows the scheme of the cleavage of the Atg8-GST by Atg4, which results in two small fragments, His-tagged Atg8s (∼17 kDa) and GST (∼26 kDa). D and E, the substrates Atg8s-GST (0.5 mg/ml) was incubated with vehicle control (D) or 0.0125 mg/ml of Atg4A (D), Atg4B (E), or Atg4BC74S (E), respectively, in a volume of 20 μl for 1 h at 37 °C. The reaction was stopped, and the mixture was resolved by SDS-PAGE and CBB staining. F, the substrates Atg8s-GST (0.5 mg/ml) were incubated with 0.025 mg/ml Atg4C or Atg4D, respectively, in a volume of 20 μl for 3 h at 37 °C. The reaction was stopped, and the mixture was resolved by SDS-PAGE and CBB staining.
The GST-Atg8s, when cleaved by Atg4 at the conserved P1 glycine, would result in products (GST and Atg8s) that could be easily separated from the full-length substrates by SDS-PAGE (Fig. 1C), which constitutes an easy and reproducible way to analyze the reaction. Using this assay, we found that Atg4A was able to cleave GATE-16, GABARAP, and Atg8L but not LC3B (Fig. 1D), whereas Atg4B could hydrolyze all four substrates (Fig. 1E). The activity depended on the catalytic cysteine so that Atg4BC74S lost most of the activities (Fig. 1E). On the other hand, Atg4C and Atg4D could only weakly cleave LC3B and GATE-16 but not GABARAP or Atg8L (Fig. 1F). These results indicated that the four Atg4 homologues had diverse abilities to process the various Atg8 homologues, consistent with some of the findings in previous work (11, 14–17, 26).
Quantitative Analyses Reveal Kinetics Differences in Different Pairs of Atg4 and Atg8
To better define the unique characteristics of each Atg4 homologue, we decided to determine their enzymatic kinetics toward the various Atg8 homologues.
Whereas the amount of substrates and the enzymes in a particular reaction could be well controlled, the amount of products generated from such reactions had to be determined by a particular analytic method. Although different approaches could be taken, we decided to measure the products by SDS-PAGE followed by CBB staining, which allowed a direct visualization of the products. Toward that point, we determined that the amount of GST generated from the Atg8s-GST substrates could be reproducibly and quantitatively measured. The molecular mass of GST is around 26 kDa, which allows its migration in a standard SDS-PAGE to a relative stable position in the middle section of the gel. The optical density of the CBB-stained GST band was a linear function of the amount of the protein (data not shown) and was minimally affected by the electrophoresis conditions, as compared with the smaller bands of Atg8s. With a standard curve constructed for each experiment, the amount of GST generated could be determined by a careful densitometry analysis.
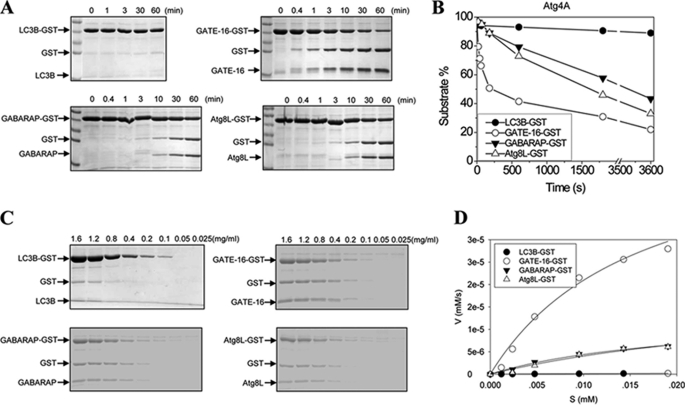
We first examined the kinetics of Atg4A (Fig. 2). Atg4A could rapidly cleave GATE-16-GST. About 50% of the GATE-16-GST proteins were cleaved in 3–4 min in the present experimental conditions (Fig. 2, A and B). On the other hand, cleavage of GABARAP and Atg8L were much slower, and it took about 30 and 40 min, respectively, to digest up to 50% of these substrates. Consistently, Atg4A did not seem to digest LC3B to any significant levels up to 300 min (Fig. 2A) (data not shown), suggesting that the reaction, if it ever occurred, was extremely slow, under these conditions.
FIGURE 2.
Determination of the catalytic kinetics of Atg4A. A, the substrates Atg8s-GST (0.5 mg/ml) were incubated with Atg4A (0.0125 mg/ml) in a volume of 20 μl for the indicated time. The reaction was stopped, and the mixture was resolved by SDS-PAGE and CBB staining. B, the gels were subjected to densitometry analysis, and the densities of the substrates, Atg8s-GST, and of the two products, GST and Atg8s, were determined. The percentage of substrate that remained at each reaction time point was equal to ODAtg8s-GST/(ODAtg8s-GST + ODGST + ODAtg8s) × 100%. The time at which 50% of substrates were cleaved was 200 s for GATE-16, 1,700 s for Atg8L, and 2,400 s for GABARAP, respectively. C, different concentrations of Atg8s-GST as indicated were incubated with Atg4A (0.0125 mg/ml) in a 20-μl volume for 5 h (for LC3B-GST), 1 min (for GATE-16-GST), or 5 min (for GABARAP-GST and Atg8L-GST), respectively. The reactions were stopped, and the mixture was resolved by SDS-PAGE and CBB staining. D, the initial velocity (V, y axis) was defined as the change of the concentration of GST/s (mm/s) (see “Experimental Procedures”), which was then plotted against the concentration of the substrate (S (mm), x axis). The curves were fitted using the non-linear regression method (SigmaPlot 10.0).
Based on this analysis and the consideration of experimental feasibility and reproducibility, we determined that the preferred reaction times for further kinetic analysis of Atg4A were 1, 5, 5, and 300 min for GATE-16, GABARAP, Atg8L, and LC3B, respectively. The amount of the substrates that would be cleaved during the designated period of time was estimated to be less than 20, 10, 10, and 5% for the four substrates, respectively.
Having set the reaction time, a fixed amount of Atg4A was then mixed with different amounts of Atg8 homologues. The amount of GST generated was determined (Fig. 2C), and the velocity was calculated at each substrate concentration as detailed under “Experimental Procedures.” This plot (Fig. 2D) allowed the calculation of Km and kcat/Km of Atg4A toward the four substrates (Table 1). Remarkably, the affinity (1/Km) of Atg4A toward the four Atg8 homologues was very similar, although that for GATE-16 was the highest. GATE-16 was also the best substrate for Atg4A, followed by GABARAP and Atg8L, with LC3B being the last in the list. The catalytic efficiency (kcat/Km) of Atg4A for GATE-16 was around 220-fold that for LC3B.
TABLE 1.
The kinetic parameters of Atg4 enzymes against full-length Atg8s-GST
The Vmax (mol liter−1 s−1) and Km (mol/liter) for each enzyme-substrate pair was derived from the curve fitting (Figs. 2–4, and 6) using the non-linear regression method (SigmaPlot 10.0). kcat (catalytic constant, s−1) was derived by dividing Vmax by the concentration of the enzyme in each reaction. ND, experiment not done.
| LC3B-GST | GATE-16-GST | GABARAP-GST | Atg8L-GST | |
|---|---|---|---|---|
| Km (10−5 mol/liter) | ||||
| Atg4A | 3.39 ± 0.51 | 1.57 ± 0.46 | 2.08 ± 0.75 | 3.67 ± 0.30 |
| Atg4B | 0.51 ± 0.05 | 0.61 ± 0.07 | 0.58 ± 0.05 | 0.44 ± 0.23 |
| Atg4C | 4.75 ± 0.24 | 1.24 ± 0.13 | ND | ND |
| Atg4D | 1.31 ± 0.27 | 0.72 ± 0.36 | ND | ND |
| Atg4BC74S | 0.68 ± 0.04 | 0.44 ± 0.08 | 0.84 ± 0.09 | 0.35 ± 0.06 |
| Atg4BΔ1−24 | 0.59 ± 0.06 | 0.94 ± 0.13 | ND | ND |
| kcat/Km (mol−1 liter s−1) | ||||
| Atg4A | 58.5 | 12,800 | 2,460 | 1,960 |
| Atg4B | 89,600 | 107,000 | 78,900 | 88.800 |
| Atg4C | 12.2 | 20.3 | ND | ND |
| Atg4D | 34.2 | 32.8 | ND | ND |
| Atg4BC74S | 43.6 | 65.9 | 10.9 | 16.5 |
| Atg4BΔ1−24 | 118,000 | 121,000 | ND | ND |
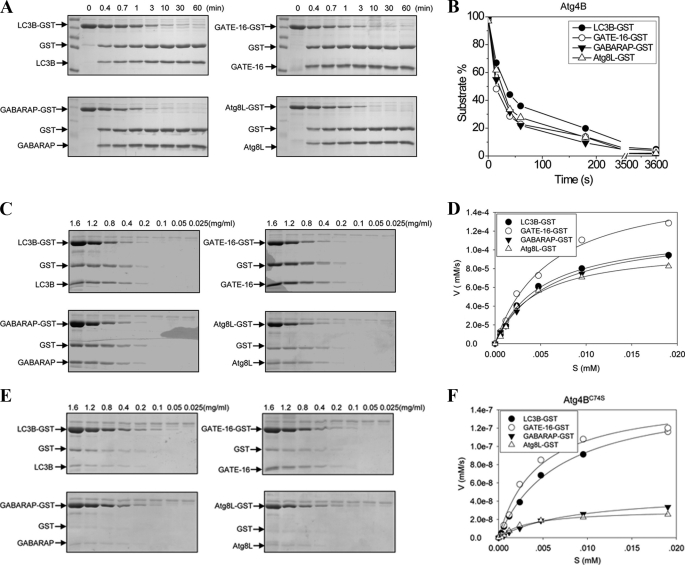
Using the same approach, we determined the Km and kcat/Km for Atg4B (Fig. 3) (Table 1), Atg4C, and Atg4D toward different substrates (Fig. 4) (Table 1). Atg4B was able to bind to the four Atg8 homologues with similar affinities, which were higher than those of Atg4A toward these substrates. The catalytic efficiency was also similar for all four substrates with that for GATE-16 being the best. Compared with Atg4A, Atg4B had higher kcat/Km values toward the three common substrates, GATE-16, GABARAP, and Atg8L, ranging from 7-fold higher for GATE-16 to 44-fold higher for Atg8L (Table 1).
FIGURE 3.
Determination of the catalytic kinetics of Atg4B and Atg4BC74S. A, the substrates Atg8s-GST (0.5 mg/ml) were incubated with Atg4B (0.0125 mg/ml) in a volume of 20 μl for the indicated time. The reaction was stopped, and the mixture was resolved by SDS-PAGE and CBB staining. B, the percentage of substrate that remained at each reaction time point was determined as in Fig. 2B. The time at which 50% of substrates were cleaved was 35 s for LC3B, 23 s for GATE-16, 25 s for GABARAP, and 26 s for Atg8L, respectively. C, different concentrations of Atg8s-GST as indicated were incubated with Atg4B (0.0125 mg/ml) in 20 μl for 20 s for all substrates. The reactions were stopped, and the mixture was resolved by SDS-PAGE and CBB staining. D, the initial velocity of the reaction at each substrate concentration was plotted, and the kinetics was determined as described in the legend to Fig. 2D. E, different concentrations of Atg8s-GST as indicated were incubated with Atg4BC74S (0.025 mg/ml) in a volume of 20 μl for 5 h for all substrates. The reactions were stopped, and the mixture was resolved by SDS-PAGE and CBB staining. F, the initial velocity of the reaction at each substrate concentration was plotted, and the kinetics was determined as described in the legend to Fig. 2D.
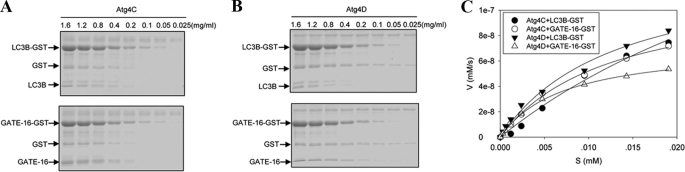
FIGURE 4.
Determination of the catalytic kinetics of Atg4C and Atg4D. A and B, different concentrations of LC3B-GST and GATE-16-GST as indicated were incubated with 0.025 mg/ml Atg4C (A) or Atg4D (B) in a volume of 20 μl for 5 h. The reactions were stopped, and the mixture was resolved by SDS-PAGE and CBB staining. C, the initial velocity of the reaction at each substrate concentration was plotted, and the kinetics was determined as described in the legend to Fig. 2D.
The catalytic mutant (C74S) of Atg4B had dramatically reduced ability to cleave all four substrates, with the kcat/Km values smaller than 0.1% of those of the wild type enzyme (Fig. 3, E and F, and Table 1). As anticipated, the affinities of this mutant to all four substrates were generally not changed; thus, the reduced kcat/Km largely reflected the decreased catalytic rate.
Atg4C and Atg4D possessed the lowest activity toward the four Atg8 homologues (Figs. 1F and 4 and Table 1). No activities for GABARAP and Atg8L could be detected in the assay conditions with prolonged reaction time up to 5 h and increased concentrations of enzyme or substrates (data not shown). Very weak activities toward LC3B and GATE-16 could be detected in prolonged reactions. Interestingly, the affinities of Atg4C and Atg4D toward GATE-16 were very similar to those of Atg4A and Atg4B, respectively (Table 1), yet the kcat/Km values of both reactions were in the same range as that of Atg4BC74S toward the same substrate. Although other factors had to be considered regarding these differences, the data would suggest that Atg4C and Atg4D were catalytically more or less similar to Atg4BC74S.
Selective Activities of Atg4 Homologues Can Be Confirmed in ex Vivo Conditions
To determine whether the various Atg4 homologues had the same catalytic profile in cells as that defined in the in vitro assay, we transfected these molecules tagged with FLAG into HEK-293A cells. Western blot using an anti-FLAG antibody indicated that the four Atg4 homologues and the Atg4B catalytic mutant (C74S) were all expressed at similar levels (Fig. 5A), making it possible to compare their enzymatic activity against the same set of substrates.
FIGURE 5.
Determination of the cleavage activities of Atg4 homologues in mammalian cells. A, each of the four FLAG-Atg4 homologues were transfected into HEK-293A cells for 24 h, and the lysates were prepared for the immunoblot analysis using an anti-FLAG antibody. The expression of the Atg4s was comparable among different constructs. B, HEK-293A cell lysates (1 μg/ml) with or without the overexpressed Atg4s were incubated with different Atg8s-GST (0.25 mg/ml) in a volume of 20 μl for 3 or 20 min. The reactions were resolved by SDS-PAGE and CBB staining. The optic densities of the substrates (Atg8s-GST) and the products (GST and Atg8s) were determined by densitometry. C, the percentage of cleaved substrates was calculated as the ratio of ODGST/(ODAtg8s-GST + ODGST + ODAtg8s). D, HEK-293A cell lysates were prepared for the immunoblot analysis using antibodies against different Atg4 homologues and GAPDH.
Lysates from these cells were prepared and admixed with the four GST-fused Atg8 homologues individually. The cleavage assay was characterized for the generation of the cleaved product, GST (Fig. 5B). Quantitative analysis indicated that transfection of Atg4B resulted in increased activities against all four substrates, whereas transfection of Atg4A resulted in an increased activity mainly against GATE-16 (Fig. 5C). Transfection of Atg4C, Atg4D, and Atg4BC74S led to no more activities than the endogenous background level. These observations were consistent with the results from the in vitro assay, indicating Atg4B as the most potent with the broadest substrate specificity, followed by Atg4A.
We noted that the endogenous Atg4 activity was readily detectable against GATE-16 and LC3B as well as, to a lesser extent, the other two substrates (Fig. 5C). This activity was probably contributed by the endogenous Atg4B and Atg4A (Fig. 5D). The observation that transfection of Atg4A resulted in a noticeable difference in the activity only against GATE-16 seemed to reinforce the notion that Atg4A was mainly specific to GATE-16, as indicted by the in vitro assay (Table 1). On the other hand, transfection of Atg4B caused increases in activities against all substrates, supporting the in vitro finding that this homologue had the broadest activity.
The Catalytic Efficiency of Atg4B Is Affected by Its Structure and Interaction with the Substrates
Crystallographic studies of Atg4B alone (27) or in association with LC3B (28) suggested several structural elements of Atg4B that could affect its reaction with LC3B. The binding of LC3B could trigger a conformation change in Atg4B to induce a better fit of the substrate in the catalytic site. In addition, the N-terminal part of Atg4B may hamper the exit of cleaved LC3B from the active site, and deletion of this part seemed to enhance the activity.
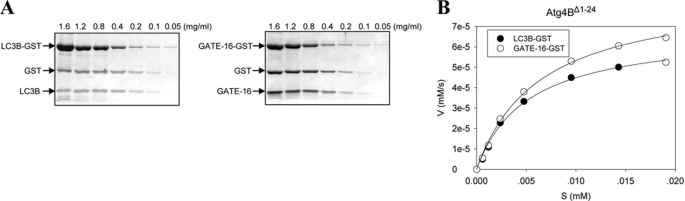
To examine these issues from the kinetics aspect, we prepared the recombinant Atg4B that lacked the N-terminal tail (Δ1–24) and determined the kinetics of this variant against the two most sensitive substrates, LC3B and GATE-16 (Fig. 6, A and B). When compared with the wild type Atg4B (Table 1), Atg4BΔ1–24 had a significantly increased kcat/Km in processing LC3B, an approximately 31.7% increase. But this enhancement was less noticeable with GATE-16 with only a 13.1% increase in kcat/Km (Fig. 6B). The affinity (1/Km) of Atg4BΔ1–24 was reduced slightly toward LC3B (0.59 × 10−5 mol/liter mutant versus 0.51 × 10−5 mol/liter wild type) but quite notably toward GATE-16 (0.94 × 10−5 mol/liter mutant versus 0.61 × 10−5 mol/liter wild type), which might explain the more significant increase in the catalytic efficiency of Atg4BΔ1–24 toward LC3B.
FIGURE 6.
The impact of the N-terminal sequences on the catalytic efficiency of Atg4B. A, different concentrations of LC3B-GST or GATE-16-GST were incubated with Atg4BΔ1–24 (5 μg/ml) in a volume of 20 μl for 30 s. The reactions were stopped and resolved by SDS-PAGE and CBB staining. B, the initial velocity of the reaction at each substrate concentration was plotted, and the kinetics was determined as described in the legend to Fig. 2D.
We then examined the impact of conformational change of Atg4B on LC3B cleavage in terms of the differences in the kinetics. The crystallographic study indicates that a regulatory loop (amino acids 259–262) in Atg4B covers the entrance of the catalytic site, preventing its interaction with the C-terminal tail of LC3B (28). Upon binding of LC3B, however, this regulatory loop is lifted, allowing the formation of a groove along which the LC3B tail could enter the catalytic site. Although Phe119 of LC3B is critical in causing this process, overall conformation changes require multiple interfaces of Atg4B-LC3B interaction. Moreover, it had been suggested that a rat LC3 peptide (amino acids 116–124) containing the conserved P1 glycine 120 could not bind to and be cleaved by human Atg4B (27).
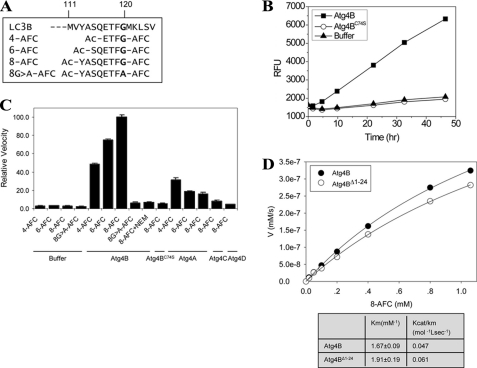
The above finding, however, could have been affected by the less sensitive detection method used in the assay and the presence of only 5 amino acids preceding the P1 glycine. We thus designed a series of peptide composed of 4, 6, or 8 amino acids, including the P1 glycine 120 (Fig. 7A). These peptides were coupled with AFC, which allowed a much more sensitive analysis by monitoring the fluorescence signals of the released AFC.
FIGURE 7.
The catalytic efficiency of Atg4 for LC3B peptides. A, based on the sequence of LC3B, three peptides ranging from 4 to 8 amino acids long were designed. In addition, a G120A mutant of the octapeptide was synthesized. All peptides were chemically linked with AFC at their C termini. B, the peptide, 4-AFC (0.1 mm), was incubated with Buffer B, 10 μg of Atg4B, or 20 μg of Atg4BC74S in a total volume of 200 μl for different times as indicated. The RFU from the released AFC group were recorded, which was correlated to the activities of Atg4B and was not detected in the presence of buffer only or Atg4BC74S. C, the LC3B peptides (0.1 mm) were incubated with Buffer B, Atg4A (20 μg), Atg4B (10 μg), Atg4BC74S (50 μg), Atg4C (50 μg), or Atg4D (50 μg) in a final volume of 100 μl for 40 h at 37 °C. For inhibition of the enzyme, 10 μm N-ethylmaleimide was premixed with Atg4B for 30 min before the peptide was included in the reaction. The relative velocity (RV) of each reaction was defined as the RFU change/h (RFUt) relative to the reaction of Atg4B + 8-AFC (i.e. RV = RFUtAtg4s + AFC substrates/RFUtAtg4B + 8-AFC). D, different concentrations of LC3B peptide (8-AFC, 0–1.2 mm) were incubated with 5 μg of Atg4B or 3 μg of Atg4BΔ1–24 in a total volume of 100 μl for 20 h. The released AFC was measured, and the RFU was converted to the concentration of AFC using an AFC standard. In this assay, 100 RFU was equivalent to 10 nm free AFC. Their initial velocities (in the first 20 h of reaction) were calculated as the change in AFC concentration, which was plotted against the concentration of the substrate peptide. Vmax and Km values were derived as described above.
Indeed, with this sensitive assay, cleavage of the peptides was detected (Fig. 7, B and C). The octapeptide (8-AFC) yielded a higher signal level than the hexapeptide (6-AFC) and the tetrapeptide (4-AFC). Mutation of the P1 glycine to alanine abolished the cleavage. Inclusion of N-ethylmaleimide, a general cysteine protease inhibitor, or the mutation of the catalytic cysteine (C74S) almost completely inhibited the cleavage, particularly for 8-AFC. In addition, the octapeptide was more specific than the tetrapeptide. It was only minimally cleaved by Atg4A and was not cleaved by Atg4C or Atg4D (Fig. 7C).
It was noted, however, that the reaction had a very slow kinetics (Fig. 7B). The amount of AFC generated was calculated using pure AFC as the standard. The results showed that both the affinity and the catalytic efficiency for the octapeptide were dramatically diminished, compared with the full-length LC3B, in the case of wild type Atg4B (kcat/Km = 0.047 mol−1 liter s−1) and Atg4BΔ1–24 (kcat/Km = 0.061 mol−1 liter s−1) (Fig. 7D). Thus, despite the fact that the octapeptide contained Phe119, which is important for lifting the regulatory loop, and included amino acids 113–120, which would fully occupy the groove leading to the active site on Atg4B, it could not fully induce Atg4B to the completely activated status as the full-length LC3B did. The catalytic efficiency of Atg4B for the full-length LC3B was 1.9 × 106-fold that for the octapeptide. Together, these data provide the kinetics evidence for the importance of the inducible activation of Atg4B by its full-length substrates.
DISCUSSION
Several approaches had been taken to assess the enzymatic activity of Atg4. Cleavage of Atg8 byAtg4 in yeast was evident from the altered size of Atg8 in immunoblot (8). In the mammalian system, Hemelaar et al. (22), by adding a reactive chemical group to the substrates, determined that Atg4B could react with several Atg8 homologues. Most of the recent assays used fusion constructs that tag the Atg8 homologues, mainly LC3B, at the C terminus. The tag is released after the cleavage and is detected as an indication of Atg4 activity.
The GST tag had been used in many studies (11, 17, 23, 24). The cleavage is determined by SDS-PAGE, which separates the cleaved GST from the cleaved Atg8s and the uncleaved Atg8s-GST. We have found this approach to be highly specific with low background and to be very reproducible. We selected this method for our kinetic analysis also because all previous major work regarding the recognition of Atg8s by Atg4s was performed with this method, which provided the base for comparisons.
However, it is possible that kinetic analysis using different detection methods will cause disparity in the results. Thus, it is important to consistently use the same method when comparing the kinetics parameters among different Atg4 homologues or among different Atg8 substrates.
Does the kinetics measured using GST-fused Atg8s truly represent the real cleavage of Atg8s? Although a firm conclusion could not be drawn without the actual comparison, co-crystallographic study of Atg4B and LC3B does not suggest the importance of the C-terminal end of LC3B in the binding to Atg4B or in the catalysis by Atg4B (28). Comparisons of the cleavage of LC3B with fusion partners of different sizes at the C terminus do not indicate any obvious changes in cleavage efficiency (27). It is thus likely that the fusion with GST or other tags at the C terminus of Atg8s would not affect the kinetics analysis. We thus do not attempt to differentiate Atg8s and Atg8s-GST in our analysis.
Kinetic Variations among Different Atg4 Homologues
Using the GST-tagged Atg8 homologues, our kinetic analysis indicated that Atg4B was most potent among Atg4 homologues, followed by Atg4A. Thus, Atg4B was able to cleave all four Atg8s-GST (GATE-16 > LC3B ≥ Atg8L ≥ GABARAP) with the catalytic efficiency (kcat/Km) ranging from 1.07 × 105 mol−1 liter s−1 to 7.9 × 104 mol−1 liter s−1.
Atg4A could not cleave LC3B-GST, and it also has a lower affinity and lower catalytic efficiency than Atg4B toward the other three substrates. Still, it had a quite high catalytic efficiency (kcat/Km from 1.28 × 104 to 1.9 × 103 mol−1 liter s−1) toward these substrates (GATE-16 > GABARAP ≥ Atg8L) (Table 1). Although Atg4C and Atg4D could process LC3B and GATE-16, their activities seemed to be minimal, at a level comparable with that of Atg4BC74S (Table 1).
Previous studies (11, 17, 22–24), although collectively leading to comparable conclusions, could cause misinterpretation without the measurement of the kinetics. For example, it was reported that Atg4A could not process Atg8L (17). Kinetic studies showed that Atg4A could process Atg8L, but the catalytic efficiency was only about one-sixth of that for GATE-16. Another finding was that recombinant Atg4C was shown to be unable to process mouse Atg8L (17) but able to hydrolyze a synthetic peptide ([(7-methoxycoumarin-4-yl)acetyl]-Thr-Phe-Gly-Met-[N-3-(2,4-dinitrophenyl)-l-α,β-diaminopropionyl)]-NH2) derived mainly from the yeast Atg8 (18), resulting in doubts about the true capability of Atg4C. Our kinetic studies suggested that Atg4C and Atg4D were not capable of cleaving Atg8L or GABARAP under comparable experimental conditions, although they did possess very low activities toward LC3B and GATE-16 (Table 1).
Most importantly, kinetic analysis could reveal novel information about the relationships of Atg4 enzymes with the various substrates. Atg4B is not only able to cleave all four substrates, but it also demonstrates a similar affinity and catalytic efficiency toward these substrates (Table 1), suggesting that it has the structural flexibility to bind and catalyze all of these substrates.
On the other hand, Atg4A does not bind to these substrates as well as Atg4B, which may explain why the catalytic efficiency is lower too. However, the most surprising finding from the kinetics point of view is that Atg4A could not hydrolyze LC3B. Notably, whereas the Km of Atg4A for LC3B (3.39 × 10−5 mol/liter) was comparable with that for Atg8L (3.67 × 10−5 mol/liter), the catalytic efficiency toward LC3B (58.5 mol−1 liter s−1) was only about 3% of that toward Atg8L (1,960 mol−1 liter s−1). When compared with the cleavage of GATE-16, Atg4A processed LC3B with a catalytic efficiency of only 0.5% of that toward GATE-16, but the affinity of binding was reduced by about 50% (Table 1). It is thus possible that structural characteristics unrelated to binding capability contribute to the differences in catalytic efficiency. This notion is further supported by the comparison with the kinetic parameters of Atg4BC74A, which had a very similar kcat/Km value toward LC3B as Atg4A.
The crystal structure of human Atg4B has been resolved (27, 29). The active site is composed of elements from Cys74, Asp278, and His280. This is the geometry observed in the canonical catalytic triad of cysteine protease. Mutagenesis studies confirmed that they are indeed required for Atg4B enzymatic activity (27).
The crystal structure of Atg4A has not been resolved. However, Atg4A shares a significant similarity in sequences and in the catalytic triad (Cys77/Asp279/His281) with Atg4B (18). Other structure elements may affect its recognition of LC3B as an effective substrate. Structure studies indicate that all the residues of Atg4B that interact with LC3B are also conserved in Atg4A, except Leu232. Atg4A instead possesses Ile233 at the corresponding position. When Ile233 was changed to Leu, the mutated Atg4A acquired a notable ability to cleave LC3B (28). However, although the affinity (1/Km) of Atg4A toward LC3B was only about one-seventh of that of Atg4B, the magnitude of reduction of catalytic efficiency was more significant than one might expect, compared with other substrates (Table 1), again indicating that other factors could contribute to the catalytic efficiency.
Interestingly, Atg4C and Atg4D bound to LC3B with similar affinity as Atg4A but had an even lower catalytic efficiency toward this substrate. They bound to GATE-16 better than Atg4A. In fact, Atg4D binds to GATE-16 in the same affinity range as Atg4B, but both Atg4C andAtg4D could only minimally catalyze LC3B or GATE-16. Thus, Atg4C and Atg4D were very similar to Atg4BC74S, which bound to all four Atg8s-GST as well as Atg4B did but possessed a catalytic efficiency of as little as 0.01% of that of wild type Atg4B (Table 1).
The crystal structures of Atg4C and Atg4D are not available yet, but a significant sequence similarity is shared with Atg4B (18). The catalytic triad is conserved (Cys110/Asp345/His347 for Atg4C and Cys134/Asp356/His358 for Atg4D). It is of note that Atg4C and Atg4D have longer sequences than Atg4A and Atg4B, and the catalytic triad may be spatially positioned differently, rendering it difficult for the substrates to access it. Interestingly, deletion of the N-terminal 63 amino acids of Atg4D by a caspase-mediated cleavage stimulated its catalytic activity toward Atg8L (25), confirming that structural changes could improve substrate access to the active site.
Substrate-induced Conformation Change in Atg4 for Maximal Catalytic Efficiency
The Atg4B-LC3B crystallography study indicates that binding of LC3B with Atg4B through the initial interactions between the ubiquitin domain of LC3 and a concave surface at the left side of the conserved region of Atg4 induces conformation changes in Atg4B, which involves the relocation of the regulatory loop (amino acids 259–262) and allows the catalytic site accessible to the LC3B C-terminal tail (28). In addition, the N-terminal region of 24 amino acids of Atg4B is positioned at the back of the active site, which could affect the exit of the cleaved substrate (28). Thus, deletion of this N-terminal domain was found to lead to an increased activity of Atg4B (28). Our kinetic analysis confirmed these findings. Atg4BΔ1–24 had a 31.7% increase in catalytic efficiency toward LC3B but only a 13.1% increase toward GATE-16 (Table 1). A slightly improved catalytic efficiency of this mutant was also observed in another study in the presence of Na2-citrate (30). However, this deletion actually reduced the binding affinity, particularly for GATE-16, which might counteract any improvement in catalytic efficiency. It is also possible that the improved catalytic efficiency of Atg4BΔ1–24 may be more significant for membrane-bound substrates, which may demand more stringent conditions for substrate exit (28).
The more notable difference was found in the use of peptide substrates. Although Atg4B can specifically cleave an octapeptide of LC3B (amino acids 113–120), the catalytic efficiency (kcat/Km = 0.047 mol−1 liter s−1) was even lower than that of its catalytic mutant (C74S) toward the full-length LC3B (kcat/Km = 43.6 mol−1 liter s−1). Such a cleavage was detected essentially because of a prolonged reaction time, an increased enzyme concentration, and the use of a sensitive fluorescent tag (AFC). The low catalytic efficiency did not seem to be related to the actual composition of the amino acids at the P2–P4 positions (30) or the length (Fig. 7C) of the peptide. On the contrary, the lack of the ubiquitin domain of LC3B could lead to a very inefficient binding of the LC3B C-terminal tail to the catalytic site of Atg4B due to the lack of induced conformation change. It is conceivable that the regulatory loop (amino acids 259–262) would still mask the access to the active site. Nonetheless, deletion of this regulatory loop did not improve catalytic efficiency (27) but led to the loss of the activity completely (30).
The apparent importance of the inducible activation of Atg4B by LC3B may also suggest that in cases where substrate hydrolysis was ineffective, such as cleavage of LC3B by Atg4A, there might be a lack of such conformation changes, in addition to other structural concerns (see above).
Kinetic Variations among Different Atg8 Substrates
Interestingly, although LC3B was most frequently examined in the mammalian autophagy process, GATE-16 seemed to be better processed by Atg4 enzymes. The order of substrate preference for Atg4A is GATE-16 > GABARAP ≥ Atg8L ≫ LC3B, and the order for Atg4B is GATE-16 > LC3B ≥ Atg8L ≥ GABARAP in terms of catalytic efficiency. A similar order of substrate preference for Atg4B had been reported in another study (30), which had also examined LC3C but not Atg8L. LC3C was not as good of a substrate as LC3B for Atg4B. The catalytic efficiency for LC3C fell between that for LC3B and that for GABARAP. Combining the data from the two studies, the relative order of the catalytic efficiency of Atg4B for the various substrates seems to be the following: GATE-16 > LC3B > Atg8L/LC3C > GABARAP. Although the third member of the LC3 subfamily, LC3A, had not been experimentally examined, it has the highest sequence homology to LC3B among all other Atg8 homologues, implying that it might be processed by Atg4B with a similar efficiency.
The structural basis of the substrate preference by different Atg4 homologues is unclear. Crystal and solution structures of LC3B, GABARAP, and GATE-16 have been reported (31–35). The tertiary structures of these Atg8 homologues are very similar. The structures include an N-terminal subdomain composed of two α-helices and a C-terminal subdomain that adopts a ubiquitin fold. The N-terminal subdomain of cleaved LC3B (LC3-I) makes contact with the surface of the C-terminal subdomain, and as such, they adopt a single compact conformation in solution. The electronic potential surfaces of the α-helices differ among the Atg8 homologues, potentially contributing to the selection of specific binding partners. However, the binding affinity between different Atg4-Atg8 pairs did not seem to be correlated with the catalytic efficiency (Table 1), although overall, these Atg8 molecules prefer Atg4B to other Atg4 homologues.
The glycine residue at the scissile site is conserved in yeast Atg8 and all of its mammalian homologues and is crucial for the hydrolysis by Atg4 family protease (5, 8) (Fig. 1C). In addition, a conserved aromatic residue at the P2 position (Phe or Tyr) is also required for the hydrolysis, as shown in the case of LC3B (27) and confirmed in a positional scanning study (30). The latter study also indicated that the optimal amino acid at the P3 position had a longer side chain (Thr, Leu, or Val) and that the optimal amino acid at the P4 position was Gly, followed by Ser and Asn.
It is interesting to note that among the six mammalian Atg8 homologues, only GATE-16 has the exact same P4–P1 sequence (NTFG) as the yeast Atg8 (ScAtg8), implying that it might be evolutionarily more ancient from catalytic point of view and providing a possible explanation for why it is a more preferred substrate kinetically.
Potential Physiological Significance of Unique Atg4-Atg8 Reactions
At the expression level, Atg4B and Atg4C possess broader tissue distribution than Atg4A and Atg4D (18, 21). At the functional level, Atg4B and Atg4C, but not Atg4A and Atg4D, could complement yeast Atg4 in autophagy (18). However, this does not mean that Atg4A and Atg4D cannot be functional in mammalian cells. Notably, although the deletion of the single Atg4 gene in yeast arrests autophagy (8), genetic deletion of Atg4B (19, 20) and Atg4C (21) in mice indicated a much more significant role of Atg4B in autophagy, although the defect seems to be only partial. These results are consistent with the biochemical properties of Atg4B and Atg4C, with the former being more potent in substrate selection and catalytic efficiency. On the other hand, the results also suggest that the function of individual Atg4 enzymes could be compensated to some degrees so that the defect of Atg4B deletion was not lethal, unlike the case of Atg7 or Atg5 deletion (36, 37).
All four Atg8 homologues examined in this study have been shown to be not only cleavable by one or more Atg4 homologues but are also found to be conjugated to phosphatidylethanolamine and to be associated with membranes (4–6, 11, 15, 17, 23). Their individual roles in autophagosome biogenesis were more directly examined recently (16). It was found that LC3B, GATE-16, and GABARAP had different roles in the early and later stage of autophagosome biogenesis. This suggests that Atg8 homologues could play different roles in autophagy. Genetic deletion of LC3B (38), however, did not lead to arrest of autophagy, suggesting that either other LC3 subfamily members (LC3A or LC3C) or the GABARAP subfamily molecules can compensate for the loss of LC3B.
The expression pattern of LC3, GABARAP, and GATE-16 in various rat tissues has been determined and found to be broad, although LC3 is most enriched in the brain, GABARAP is most enriched in the spleen, and GATE-16 is most enriched in the heart and brain (15). Thus, it is possible that variations in the expression of Atg4 homologues and Atg8 homologous, coupled with the kinetics differences between different Atg4-Atg8 pairs, may affect autophagosome biogenesis in a temporal and spatial way.
This work was supported, in whole or in part, by National Institutes of Health Grants R01CA 83817 and R01 CA111456 (to X.-M. Y.).
- AFC
- 7-amino-4-trifluoromethyl coumarin
- Atg4s
- mammalian Atg4 homologues
- Atg8s
- mammalian Atg8 homologues
- CBB
- Coomassie Brilliant Blue
- GABARAP
- γ-aminobutyric acid receptor-associated protein
- RFU
- relative fluorescence unit(s).
REFERENCES
- 1. Klionsky D. J., Cregg J. M., Dunn W. A., Jr., Emr S. D., Sakai Y., Sandoval I. V., Sibirny A., Subramani S., Thumm M., Veenhuis M., Ohsumi Y. (2003) Dev. Cell 5, 539–545 [DOI] [PubMed] [Google Scholar]
- 2. Mizushima N. (2007) Genes Dev. 21, 2861–2873 [DOI] [PubMed] [Google Scholar]
- 3. Ohsumi Y., Mizushima N. (2004) Semin. Cell Dev. Biol. 15, 231–236 [DOI] [PubMed] [Google Scholar]
- 4. Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M., Noda T., Ohsumi Y. (2000) Nature 408, 488–492 [DOI] [PubMed] [Google Scholar]
- 5. Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Oh-oka K., Nakatogawa H., Ohsumi Y. (2008) J. Biol. Chem. 283, 21847–21852 [DOI] [PubMed] [Google Scholar]
- 7. Fujita N., Itoh T., Omori H., Fukuda M., Noda T., Yoshimori T. (2008) Mol. Biol. Cell 19, 2092–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kirisako T., Ichimura Y., Okada H., Kabeya Y., Mizushima N., Yoshimori T., Ohsumi M., Takao T., Noda T., Ohsumi Y. (2000) J. Cell Biol. 151, 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Suzuki K., Kubota Y., Sekito T., Ohsumi Y. (2007) Genes Cells 12, 209–218 [DOI] [PubMed] [Google Scholar]
- 10. Xie Z., Nair U., Klionsky D. J. (2008) Mol. Biol. Cell 19, 3290–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kabeya Y., Mizushima N., Yamamoto A., Oshitani-Okamoto S., Ohsumi Y., Yoshimori T. (2004) J. Cell Sci. 117, 2805–2812 [DOI] [PubMed] [Google Scholar]
- 12. Nakatogawa H., Ichimura Y., Ohsumi Y. (2007) Cell 130, 165–178 [DOI] [PubMed] [Google Scholar]
- 13. Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T., Ohsumi Y. (1999) J. Cell Biol. 147, 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tanida I., Komatsu M., Ueno T., Kominami E. (2003) Biochem. Biophys. Res. Commun. 300, 637–644 [DOI] [PubMed] [Google Scholar]
- 15. Tanida I., Ueno T., Kominami E. (2004) Int. J. Biochem. Cell Biol. 36, 2503–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weidberg H., Shvets E., Shpilka T., Shimron F., Shinder V., Elazar Z. (2010) EMBO J. 29, 1792–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tanida I., Sou Y. S., Minematsu-Ikeguchi N., Ueno T., Kominami E. (2006) FEBS J. 273, 2553–2562 [DOI] [PubMed] [Google Scholar]
- 18. Mariño G., Uría J. A., Puente X. S., Quesada V., Bordallo J., López-Otín C. (2003) J. Biol. Chem. 278, 3671–3678 [DOI] [PubMed] [Google Scholar]
- 19. Mariño G., Fernández A. F., Cabrera S., Lundberg Y. W., Cabanillas R., Rodríguez F., Salvador-Montoliu N., Vega J. A., Germanà A., Fueyo A., Freije J. M., López-Otín C. (2010) J. Clin. Invest. 120, 2331–2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Read R., Savelieva K., Baker K., Hansen G., Vogel P. (2011) Vet. Pathol., in press [DOI] [PubMed] [Google Scholar]
- 21. Mariño G., Salvador-Montoliu N., Fueyo A., Knecht E., Mizushima N., López-Otín C. (2007) J. Biol. Chem. 282, 18573–18583 [DOI] [PubMed] [Google Scholar]
- 22. Hemelaar J., Lelyveld V. S., Kessler B. M., Ploegh H. L. (2003) J. Biol. Chem. 278, 51841–51850 [DOI] [PubMed] [Google Scholar]
- 23. Tanida I., Sou Y. S., Ezaki J., Minematsu-Ikeguchi N., Ueno T., Kominami E. (2004) J. Biol. Chem. 279, 36268–36276 [DOI] [PubMed] [Google Scholar]
- 24. Scherz-Shouval R., Sagiv Y., Shorer H., Elazar Z. (2003) J. Biol. Chem. 278, 14053–14058 [DOI] [PubMed] [Google Scholar]
- 25. Betin V. M., Lane J. D. (2009) J. Cell Sci. 122, 2554–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Scherz-Shouval R., Shvets E., Fass E., Shorer H., Gil L., Elazar Z. (2007) EMBO J. 26, 1749–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sugawara K., Suzuki N. N., Fujioka Y., Mizushima N., Ohsumi Y., Inagaki F. (2005) J. Biol. Chem. 280, 40058–40065 [DOI] [PubMed] [Google Scholar]
- 28. Satoo K., Noda N. N., Kumeta H., Fujioka Y., Mizushima N., Ohsumi Y., Inagaki F. (2009) EMBO J. 28, 1341–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kumanomidou T., Mizushima T., Komatsu M., Suzuki A., Tanida I., Sou Y. S., Ueno T., Kominami E., Tanaka K., Yamane T. (2006) J. Mol. Biol. 355, 612–618 [DOI] [PubMed] [Google Scholar]
- 30. Shu C. W., Drag M., Bekes M., Zhai D., Salvesen G. S., Reed J. C. (2010) Autophagy 6, 936–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kouno T., Mizuguchi M., Tanida I., Ueno T., Kanematsu T., Mori Y., Shinoda H., Hirata M., Kominami E., Kawano K. (2005) J. Biol. Chem. 280, 24610–24617 [DOI] [PubMed] [Google Scholar]
- 32. Sugawara K., Suzuki N. N., Fujioka Y., Mizushima N., Ohsumi Y., Inagaki F. (2004) Genes Cells 9, 611–618 [DOI] [PubMed] [Google Scholar]
- 33. Paz Y., Elazar Z., Fass D. (2000) J. Biol. Chem. 275, 25445–25450 [DOI] [PubMed] [Google Scholar]
- 34. Knight D., Harris R., McAlister M. S., Phelan J. P., Geddes S., Moss S. J., Driscoll P. C., Keep N. H. (2002) J. Biol. Chem. 277, 5556–5561 [DOI] [PubMed] [Google Scholar]
- 35. Stangler T., Mayr L. M., Willbold D. (2002) J. Biol. Chem. 277, 13363–13366 [DOI] [PubMed] [Google Scholar]
- 36. Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. (2004) Nature 432, 1032–1036 [DOI] [PubMed] [Google Scholar]
- 37. Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., Kominami E., Tanaka K., Chiba T. (2005) J. Cell Biol. 169, 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cann G. M., Guignabert C., Ying L., Deshpande N., Bekker J. M., Wang L., Zhou B., Rabinovitch M. (2008) Dev. Dyn. 237, 187–195 [DOI] [PubMed] [Google Scholar]