Abstract
Costameres are cellular sites of mechanotransduction in heart and skeletal muscle where dystrophin and its membrane-spanning partner dystroglycan distribute intracellular contractile forces into the surrounding extracellular matrix. Resolution of a functional costamere interactome is still limited but likely to be critical for understanding forms of muscular dystrophy and cardiomyopathy. Dystrophin binds a set of membrane-associated proteins (the dystrophin-glycoprotein complex) as well as γ-actin and microtubules and also is required to align sarcolemmal microtubules with costameres. Ankyrin-B binds to dystrophin, dynactin-4, and microtubules and is required for sarcolemmal association of these proteins as well as dystroglycan. We report here that ankyrin-B interactions with β2 spectrin and dynactin-4 are required for localization of dystrophin, dystroglycan, and microtubules at costameres as well as protection of muscle from exercise-induced injury. Knockdown of dynactin-4 in adult mouse skeletal muscle phenocopied depletion of ankyrin-B and resulted in loss of sarcolemmal dystrophin, dystroglycan, and microtubules. Moreover, mutations of ankyrin-B and of dynactin-4 that selectively impaired binary interactions between these proteins resulted in loss of their costamere-localizing activity and increased muscle fiber fragility as a result of loss of costamere-associated dystrophin and dystroglycan. In addition, costamere-association of dynactin-4 did not require dystrophin but did depend on β2 spectrin and ankyrin-B, whereas costamere association of ankyrin-B required β2 spectrin. Together, these results are consistent with a functional hierarchy beginning with β2 spectrin recruitment of ankyrin-B to costameres. Ankyrin-B then interacts with dynactin-4 and dystrophin, whereas dynactin-4 collaborates with dystrophin in coordinating costamere-aligned microtubules.
Keywords: Cytoskeleton, Microtubules, Muscular Dystrophy, Protein Targeting, Skeletal Muscle, Ankyrin, Dynactin, Dystrophin, Dystrophin-Glycoprotein Complex, Spectrin
Introduction
A central challenge for reductionist biology is to explain complex phenomena through molecular interactions. The potential payoff includes understanding and modifying disease processes. Unfortunately, protein interactomes derived from in vitro data frequently include an unmanageable number of binary interactions with uncertain functional significance. Moreover, a gene knock-out approach can be complicated by multiple gene functions and unanticipated adaptations. We present here a strategy to determine critical nodes in protein interaction networks in vivo in skeletal muscle by acutely exchanging wild type proteins for mutated forms selectively impaired in specific binary interactions. We focus on costameres, which are protein assemblies associated with the plasma membranes of striated muscle that protect skeletal and heart muscle from contraction injury (1). Costameres are of clinical interest because mutations in costamere-associated proteins such as dystrophin and components of the dystrophin-glycoprotein complex result in progressive muscular dystrophies and cardiomyopathies (2, 3).
Pioneering studies by Craig and colleagues (4, 5) first defined costameres in skeletal muscle as sites at the junction between myofibrils and the sarcolemma that contained γ-actin, spectrin, intermediate filament proteins, and vinculin. Costameres also contain dystrophin, the protein mutated in Duchenne muscular dystrophy, which associates with dystroglycan and other glycoproteins that are missing in the absence of dystrophin (1, 6, 7). Dystrophin also associates with γ-actin (8) as well as microtubules (9) and is required for alignment of sarcolemmal microtubules with costameres (9, 10). Ankyrin-G associates with dystrophin and dystroglycan, whereas ankyrin-B also associates with dystrophin. Both ankyrins cooperate in coordinating costamere association of dystrophin and dystroglycan (11). Ankyrin-B also interacts directly with microtubules (12, 13), binds to dynactin-4 of the dynactin complex, and is required for costamere association of dynactin-4 and microtubules (11). Even though these are only a partial list of protein components and interactions at costameres (1), it should be clear that the number of potential protein interactions is enormous, although their functional significance in many cases remains to be determined.
We have used an approach of acute siRNA knockdown of endogenous proteins and substitution with binding site-specific mutated versions of these proteins to address the role of individual protein-protein interactions involving ankyrin-B in adult mouse skeletal muscle. We demonstrate that interactions of ankyrin-B with dynactin-4 of the dynactin complex, and with β2 spectrin are required for proper organization and function of dystrophin and dystroglycan in skeletal muscle and for costamere-associated microtubules.
EXPERIMENTAL PROCEDURES
Molecular Biology
The siRNA-targeting sequences were designed by the criteria of Elbashir et al. (14) using the Whitehead Institute siRNA design program. Two optimal target sites in the spectrin binding domains of ankyrin-B were selected to maximize the ability to knock down multiple splice forms. The sequences 5′-AGCTTCAAGTGATGTCATG-3′ and 5′-GAGTGGCCAACATCATATA-3′ were targeted within the ankyrin-B spectrin binding domain. Using the same strategy, for dynactin-4 knockdown, the two sequences 5′-GTCAGCTGAAGCCAAATTA-3′ and 5′-GACCCTGATAATATCAACA-3′ were targeted, and for b2-spectrin, the two sequences 5′-CCGTGAGAGAATCATTTAT-3′ and 5′-CGGCGGCTCTTTGATGCAAAT-3′ were targeted. 59-Nucleotide oligonucleotides bearing the 19-nucleotide siRNA sequence along with sequences coding for a stem loop structure (shRNA) were cloned into the pFIV-Venus plasmid vector. The shRNA are transcribed from an H1 polymerase III promoter. The pFIV-Venus plasmid was constructed from the pFIV-H1-puro plasmid (System Biosciences) by replacing the puromycin gene with the cDNA encoding Venus (15). HA-ankyrin-B was prepared based on an ankyrin-B-GFP template in a pEGFP-N1 vector (16). GFP was cut out between the PmeI and the NotI sites and replaced with a 2XHA-tag encoding sequence (YPYDVPDYA) with a PmeI site on its 5′ end and a stop codon followed by a NotI site on the 3′ end that was synthesized by PCR. For the preparation of HA-ankyrin-B-DD1320AA, the DD1320AA double mutation was introduced into the HA-ankyrin-B construct by site-directed mutagenesis using QuikChange II XL Site-directed mutagenesis kit (Stratagene). For the preparation of HA-dynactin-4, GFP was first cut out of pEGFP-N1 vector between HindIII and NotI, and in its place, we introduced a triple HA sequence that was synthesized by PCR and ligated into the vector. To a recombinant ORF sequence of dynactin-4, we added by PCR a 5′ XhoI site, and a HindIII site on its 3′ end. The dynactin-4 construct was then cloned into the modified 3XHA-containing vector. All constructs described here were verified by DNA sequencing. Point mutation constructs were also resubcloned into fresh vector before use.
Yeast Two-hybrid Assays
The spectrin-binding-domain/death domain/C-terminal domain of 220-kDa ankyrin-B, either in a wild type form or containing the DD1320 double point mutation, was PCR-amplified and ligated into pAS2–1 (Clontech) to create the GAL4-ankyrin-B. The bait plasmid containing either wild type ankyrin-B or ankyrin-B-DD1320AA was transformed into the yeast strain AH109 (ADE2, HIS3, lacZ selection) using lithium acetate. Autoactivation did not occur for any bait. After confirming full-length expression of the bait protein, dynactin-4 and b2-spectrin in pACT2 (Clontech) was transformed into the yeast strain harboring the bait plasmid. Double transformants were selected on a medium lacking adenine, leucine, and tryptophan. Positive clones were tested for positive growth on a medium lacking histidine. TD1–1 and pLAM5 (Clontech) were negative interaction controls, and pVA3 and TD1–1 were positive interaction controls. For screening of dynactin-4 truncations and mutations (see supplemental Fig. 1 for details), the different dynactin-4 constructs were cloned into pACT2 and co-transformed with either ankyrin-B or β2 spectrin as above.
Antibodies
Affinity-purified antibodies against ankyrin-B and ankyrin-G were described previously (11, 16). Ankyrin-B antibody was preabsorbed on a purified ankyrin-G column, and ankyrin-G antibody was preabsorbed on a purified ankyrin-B column to eliminate cross-reactivity. Mouse anti-α-tubulin (Sigma), rabbit anti-Arp1 (Sigma), affinity-purified rabbit antibody against β-dystroglycan cytoplasmic domain and β2 spectrin all generated in our laboratory (17, 18), affinity-purified rabbit anti-dystrophin (Lab Vision, Fremont, CA; used in all dystrophin immunofluorescence images), mouse anti-His (Penta-His, Qiagen; Valencia, CA). Secondary antibodies were donkey- and goat-labeled with Alexa Fluor 488 and 568 (Invitrogen).
In Vivo Transfections and Fiber Preparation
Tibialis anterior (TA)2 muscles of 3–4-week-old C57BL/6 mice were injected with 15 units of hyaluronidase (30 ml in saline) (Worthington, Lakewood, NJ) 2 h prior to transfection (19) under isofluorane anesthesia. TA muscles were injected under isofluorane anesthesia with a mixture of 10 μg of either the two ankyrin-B siRNAs, dynactin-4 siRNAs, or the β2 spectrin siRNAs (5 mg each) in 30 μl of saline. Controls were injected with 10 μg of pFIV-Venus vector. For rescue experiments, depending on the experiment (see “Results”), 10 mg of either HA-ankyrin-B, HA-ankyrin-B-DD1320AA, HA-dynactin-4, or HA-dynactin-4-N331A in saline were injected. Immediately after injection muscles were placed between paddle electrodes and a three-pulse train (75 V, 20 ms, and 1 Hz) was delivered followed by a second train in the opposite direction (square wave BTX ECM 830 pulse generator; Harvard Apparatus, Holliston, MA). Conductive gel was applied on electrodes (Signa Gel, Parker Laboratories; Fairfield, NJ). Transfection conditions, DNA amounts, and electroporation settings were empirically tested and calibrated to deliver high transfection efficiency (determined by extent of Venus expression) using the least amounts of DNA and lowest voltages. To prevent electroporation-induced muscle damage, we applied the electrodes externally on the skin surrounding the TA muscle and not directly on the exposed muscle and determined that under our condition and after testing a range of 50–200 volts, low voltage 75-V pulses were sufficient for efficient transfection. TA muscles were harvested for fiber isolation, processing, and imaging 4 days after ankyrin-B and dynactin-4 siRNA and rescue transfections and 5 days after β2 spectrin siRNA transfections. Fibers were imaged for α-tubulin, dynactin-4, and Arp1 as follows. TA muscles were harvested and incubated in 4% paraformaldehyde, 0.5% saponin in PBS at room temperature for 2 h, washed in PBS, and teased with fine forceps. Fibers were incubated overnight at 4° C in primary antibody in 3% BSA, 0.05% saponin in PBS. Fibers were washed twice in PBS and incubated 2 h at 4° C in secondary antibody in 3% BSA, 0.05% saponin in PBS. All other fiber preparations for immunofluorescence were done as follows. TA muscles were removed and placed in ice-cold 2% PFA in PBS, fixed for 20 min on ice, and washed in PBS. Fibers were isolated, permeabilized in 0.1% Triton X-100 in 3% BSA in PBS for 20 min at 4° C and washed twice in cold PBS. Fibers were then incubated overnight in primary antibody in 3% BSA, 0.2% Tween in PBS. Fibers were washed twice in PBS and incubated with secondary antibody in 3% BSA, 0.2% Tween in PBS for 2 h at 4° C.
Exercise and EBD Experiments
Mice were treadmill exercised (LE 8706 Letica; Barcelona, Spain) twice a day for 3 days (30 min, 10° uphill angle, 30 cmü. 12 h before final exercise, Evans blue dye (EBD) (Sigma) was injected i.p. (1 mg per 0.1 ml per 10 g of body weight). After the last running session, TA muscles were removed, snap frozen in OCT, and cross-sectioned (20-mm sections). Cross-sectioning was performed throughout the length of the muscle to evaluate the extent of EBD uptake, where sections were imaged in 200-mm intervals (every tenth section).
Microscopy and Imaging
Microscopy was performed with a Zeiss LSM510 Meta confocal microscope. Single fiber imagining was dome with a ×100 numerical aperture 1.45 objective. EBD uptake in muscle sections was visualized using a 568-nm channel.
Protein Purification and Binding Assays
Full-length histidine-tagged ankyrin-B was expressed using a BacPak baculovirus protein expression system (Clontech) and purified as described (18). Histidine-tagged ankyrin-B-Zu5-DD and an ankyrin-B Zu5-DD1320DD/AA mutant were cloned into the pMAL-c4G vector (New England Biolabs) with an amino-terminal maltose binding protein tag, and a C-terminal histidine tag. pMAL-ankyrin-B-Zu5-DD and GST-dynactin-4-His7 were expressed in BL21 bacteria. The dual-tagged fusion proteins were first isolated on nickel-Sepharose beads (High Performance Ni-Sepharose, GE Healthcare), washed with 50 mm sodium phosphate, pH 8.0, 0.3 m sodium bromide, 1 mm sodium azide, 0.2 mm βME, and 20 mm imidazole, and eluted (wash solution with 0.3 m imidazole). Eluted dynactin-4 was adjusted to 0.1 mm zinc sulfate and immediately incubated with glutathione-Sepharose beads for immobilization and washed and equilibrated in binding assay buffer (see below). Ankyrin-B-Zu5-DD WT and with the 1320 DD/AA mutation were first isolated on nickel-Sepharose beads followed by an amylose affinity column (New England Biolabs) washed with wash buffer (20 mm Tris, pH 7.4, 0.2 mm NaCl, 1 mm DTT, 0.5 mm EGTA) then eluted with the addition of 10 mm maltose. Protein was dialyzed against binding buffer (see below), and in vitro protein interactions studies were performed as described (9). Briefly, purified GST-Dyn4 or GST alone were immobilized on glutathione-Sepharose beads during an overnight incubation at 4° C in binding assay buffer. Beads were washed three times with binding assay buffer and evaluated for protein loading by SDS-PAGE. Protein-bearing beads, normalized to contain equivalent levels of polypeptides, were incubated with the candidate binding partner overnight at 4° C in binding buffer (20 mm HEPES, pH 7.3, 60 mm NaCl, 0.5 mm EGTA, 1 mm NaN3, 0.1% Tween 20, 0.1 mm zinc sulfate) in a final volume of 50–100 μl. Following incubation, the glutathione-Sepharose beads were pelleted (5000 × g for 15 min) through a 20% glycerol barrier. Tubes were frozen on dry ice, and tips containing the pelleted beads were resolved by SDS-PAGE. Polypeptides were visualized either directly by Coomassie Blue staining or by immunoblotting with an anti-His Ig.
Computer Modeling of Ankyrin-B-ZU5-UPA-DD
Structure of the UNC5b netrin receptor cytoplasmic domain containing the ZU5-UPA-DD domains (20) was obtained from the Protein Data Bank and visualized using RasMol software (version 2.75) for windows. Modeling the location of ankyrin-B-DD1320 on the UNC5b structure was based on the alignment of ankyrin-B and UNC5b (20).
RESULTS
We optimized electroporation-promoted percutaneous transfection of adult mouse muscle under mild conditions to simultaneously express bi-cistronic plasmids encoding Venus as a fluorescent marker for transfected fibers and ankyrin-B-siRNAs, as well as a plasmid encoding HA-tagged human ankyrin-B resistant to siRNA (see “Experimental Procedures”) (Fig. 1D) (11). We selected TA muscle because it is amenable to evaluation of exercise-induced injury (see below and see Fig. 4). Knockdown of ankyrin-B to undetectable levels by immunofluorescence results within 4 days in loss of sarcolemmal dystrophin, dystroglycan, dynactin-4, and a subpopulation of microtubules aligned with costameres (Fig. 1D) (11). All of these proteins are restored to their wild type patterns by co-transfection with cDNA encoding HA-tagged wild type ankyrin-B resistant to mouse siRNA (Fig. 1D). It was important in these experiments to adjust plasmid DNA concentrations to achieve equivalent levels of expression endogenous and transfected ankyrin-B. The ability of co-expressed wild type ankyrin-B to prevent siRNA-induced loss of sarcolemmal dystrophin and other proteins is a valuable control for potential off-target effects of siRNAs and for possible tissue damage due to the transfection procedure.
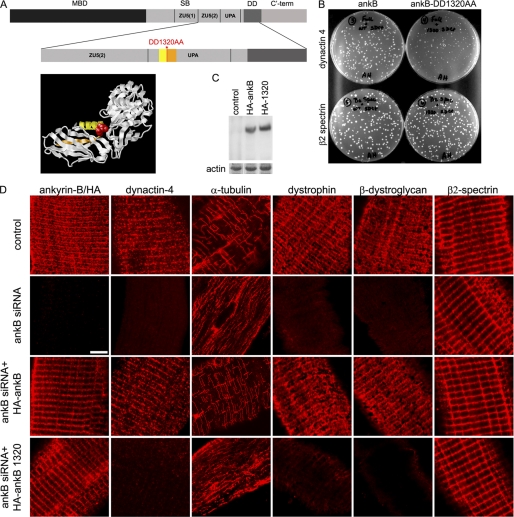
FIGURE 1.
Ankyrin-B requires dynactin-4 binding activity to promote association of microtubules, dystrophin, and dystroglycan with costameres. A, ankyrin-B domains (MBD, membrane binding domain; SB, spectrin binding domain; DD, death domain; ankB, ankyrin-B; C′-term, C′-terminal). The ankyrin-B-DD1320AA mutation (red) resides in an exposed loop in the UPA domain modeled from the UNC5b cytoplasmic domain structure (20). B, DD1320 is required for ankyrin-B-dynactin-4 but not β2 spectrin interaction in a yeast two-hybrid assay (as described under “Experimental Procedures”). C, representative immunoblot with HA antibody of homogenates of TA muscles depleted of endogenous ankyrin-B and transfected with either HA-ankyrin-B or HA-ankyrin-B-DD1320AA (as described under “Experimental Procedures”). D, single TA muscle fibers transfected in vivo with ankyrin-B siRNA and rescued with either wild type ankyrin-B or with ankyrin-B-DD1320AA (as described under “Experimental Procedures”). Rows (top to bottom), untransfected control, ankyrin-B siRNA (in both ankyrin-B staining is with ankyrin-B antibody), ankyrin-B siRNA co-transfected with HA-tagged ankyrin-B, and ankyrin-B siRNA co-transfected with HA-tagged ankyrin-B-DD1320AA (in both ankyrin-B staining is with an HA antibody). Scale bar, 5 microns.
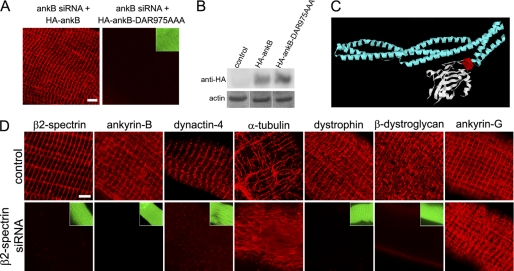
FIGURE 4.
Costamere association of ankyrin-B requires β2 spectrin. A, DAR975AAA mutation impairing spectrin binding prevents sarcolemmal localization of HA-ankyrin-B. Fibers were isolated and stained with an HA antibody. B, comparable expression levels (anti-HA immunoblot) of HA-ankyrin-B and HA-ankyrin-B-DAR975AAA in transfected TA muscles. D, β2 spectrin siRNA-transfected fibers exhibit loss of ankyrin-B (ankB), dystrophin, β-dystroglycan, dynactin-4, and costamere-associated microtubules while retaining ankyrin-G. Green insets show Venus expression in the knockdown fibers and cell boundaries. Scale bars, 5 microns.
Ankyrin-B Function Requires Dynactin-4 Binding Activity
Ankyrin-B binds directly to dystrophin as well as dynactin-4 of the dynactin complex (11). Moreover, a dystrophin mutation associated with muscular dystrophy impairs association of dystrophin-Dp71 with ankyrin-B and prevents association of dystrophin-Dp71 with costameres (11). Dystrophin itself also binds microtubules and is required for their grid-like co-alignment with costameres (9, 10). As further potential complications, ankyrin-B also binds to microtubules as well as to β2 spectrin (12, 13), and β2 spectrin associates with Arp1 of the dynactin-complex (21). We asked whether, among these multiple potential interactions, ankyrin-B requires dynactin-4 binding activity to promote sarcolemmal association of dystrophin, dystroglycan, and microtubules. Our strategy to address this question was to first identify a mutation that selectively eliminated dynactin-4 binding activity of ankyrin-B and then substitute this mutant ankyrin-B for wild type ankyrin-B in adult muscle.
We identified a DD1320AA mutation in the UPA domain of ankyrin-B by alanine scanning mutagenesis of adjacent charged residues (17) that had no effect on binding to β2 spectrin (Fig. 1B) but impaired association with dynactin-4 in yeast two-hybrid assays (Fig. 1B). The DD1320 site is separated from the first ZU5 domain, which contains β-spectrin-binding activity (22) and instead is located on a surface-exposed loop in the UPA domain, by analogy with the structure of the homologous UNC5b cytoplasmic domain (Fig. 1A) (20). We did not directly evaluate effects of the DD1320AA mutation on microtubule-binding activity of ankyrin-B. However, microtubules associate with ankyrin-R through its ankyrin repeats (13), and microtubule interactions with ankyrin-B are not likely to have been affected by mutation of the UPA domain. Similarly, the DD1320 mutation also is unlikely to affect association with dystrophin as Dp71 dystrophin associates with the membrane-binding domain and not the UPA domain of ankyrin-B (supplemental Fig. 1).
HA-tagged DD1320AA ankyrin-B was expressed as a full-length polypeptide in immunoblots of transfected muscle homogenates (Fig. 1C). Moreover, as anticipated from the full activity of the DD1320AA mutant in binding to β2 spectrin in yeast two-hybrid assays, HA-tagged DD1320AA ankyrin-B localized in a costamere pattern identical to endogenous ankyrin-B and to HA-tagged wild-type ankyrin-B (Fig. 1D). Strikingly, HA-ankyrin-B-DD1320AA did not rescue the sarcolemmal localization of dynactin-4, dystrophin, or β-dystroglycan (Fig. 1D). In addition, ankyrin-B-DD1320AA did not restore costamere-associated microtubules, which were instead present in an intracellular longitudinal pattern (Fig. 1D). Thus, expression of ankyrin-B DD1320AA, which localizes to costameres and binds to β2 spectrin but is impaired in its binding to dynactin-4, is equivalent to complete lack of ankyrin-B.
Dynactin-4 Associates with Costameres in Absence of Dystrophin
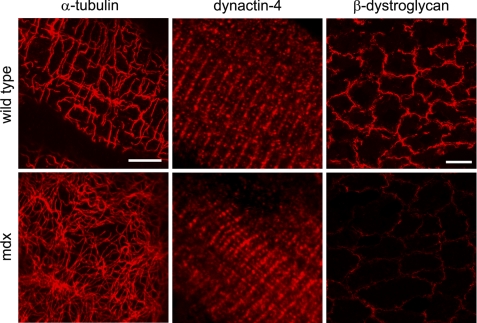
Ankyrin-B associates with costameres in mdx muscle lacking dystrophin (11) and therefore does not depend on dystrophin for its organization in skeletal muscle. However, it is conceivable that ankyrin-B and dynactin-4 act through dystrophin to organize sarcolemmal-associated microtubules into a costamere-associated grid (Fig. 2) (9, 10). We therefore evaluated the effect of the absence of dystrophin in the mdx mouse on localization of dynactin-4 in skeletal muscle (Fig. 2). Dynactin-4 localizes normally in costameres of mdx mutant mice lacking dystrophin and dystroglycan (Fig. 2). Dystrophin thus imposes a grid-like organization on sarcolemmal microtubules likely recruited to the membrane by dynactin-4 and ankyrin-B and may act independently or in collaboration with these proteins.
FIGURE 2.
Dystrophin is required for costamere patterning of microtubules but not for sarcolemmal association of microtubules and dynactin-4. Immunofluorescence labeling of muscle fibers (left and middle) and muscle cross-sections (right) of 5-week-old wild type, and mdx mice that lack dystrophin for localization of microtubules and dynactin-4. Left, grazing optical section at the level of the sarcolemma shows that in mdx mouse fibers, membrane-associated microtubules stained for α-tubulin are not organized in a costamere pattern, in contrast to in wild type fibers. Middle, dynactin-4 is localized along costameres in wild type as well as in mdx fibers. Right, cross-sections of TA muscles from wild type and mdx mice show loss of β-dystroglycan in the mdx muscle, establishing that neither dystrophin nor β-dystroglycan are required for costamere localization of dynactin-4 and for sarcolemmal association of microtubules. Scale bar for single fiber images (left and middle), 5 μm. Scale bar for muscle cross-sections (right), 20 μm.
Dynactin-4 Localization and Function Requires Ankyrin-B Binding Activity
These results implicate an ankyrin-B-dynactin-4 interaction in costamere-localization of microtubules but also are consistent with alternative interpretations such as loss of an additional ankyrin-B activity due to the DD1320 mutation. We therefore examined whether loss of dynactin-4 and/or substitution of mutant dynactin-4 lacking ankyrin-B-binding activity phenocopied loss of ankyrin-B in skeletal muscle. We observed that siRNA-mediated knockdown of dynactin-4 resulted in retention of ankyrin-B at costameres but loss of sarcolemmal dystrophin and dystroglycan (Fig. 3c). In contrast to loss of ankyrin-B, which affected only costamere microtubules, dynactin-4-depleted fibers exhibited loss of both costamere-associated microtubules as well as fragmentation of longitudinal microtubules. siRNA-mediated knockdown of dynactin-4 combined with expression of HA-tagged human dynactin-4 resistant to siRNA restored normal wild type-like localization of the HA-tagged dynactin-4, microtubules, and the dystrophin-glycoprotein complex proteins (Fig. 3C). Dynactin-4 thus stabilizes both ankyrin-B-dependent and -independent microtubules.
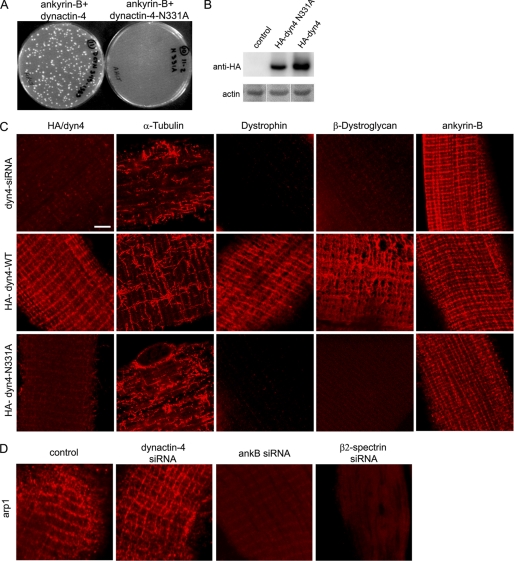
FIGURE 3.
Dynactin-4 requires ankyrin-B-binding activity for costamere localization of microtubules, dystrophin, and dystroglycan. A, N331A mutation of dynactin-4 impairs interaction with ankyrin-B (ankB) in yeast two-hybrid assays. B, shown are immunoblots (anti-HA) of HA-dynactin-4 and dynactin-4-N331A in transfected mouse TA muscles (as described under “Experimental Procedures”). C, single TA muscle fibers were transfected in vivo with a dynactin-4 siRNA and rescued with either wild type dynactin-4 or with dynactin-4-N331A (as described under “Experimental Procedures”). Rows from top to bottom, dynactin-4-siRNA-transfected muscle fibers (dynactin-4 detection with anti dynactin-4 antibody): middle row, dynactin-4 siRNA co-transfected with HA-wild type dynactin-4; bottom row, dynactin-4 siRNA cotransfected with HA-dynactin-4-N331A (middle and bottom rows, dynactin-4 detection with an anti-HA antibody). D, immunostaining for Arp1 in control, dynactin-4-, ankyrin-B-, and β2 spectrin-depleted muscle fibers (see “Experimental Procedures”). Scale bar, 5 microns.
We next identified a N331A mutation of dynactin-4 that impaired its binding to ankyrin-B in yeast two-hybrid assays (supplemental Fig. 2) (Fig. 3A). Interestingly, HA-dynactin-4-N331A in dynactin-4-depleted fibers does not localize to the sarcolemma at costameres and does not rescue the normal localization of microtubules and dystrophin-glycoprotein complex proteins, even though it was expressed as a full-length protein (Fig. 3B). Based on the mislocalization of dynactin-4 observed in muscles expressing either ankyrin-B-DD1320AA or dynactin-4-N331A, we conclude that direct interaction of ankyrin-B and dynactin-4 is critical for localization of dynactin-4 to skeletal muscle costameres and that dynactin-4 is required for localization of microtubules in a costamere pattern. Moreover, these results also suggest that costamere-associated microtubules are required for the presence of dystrophin and dystroglycan at the sarcolemma.
Experiments up to this point were consistent with a straightforward pathway where ankyrin-B binds to dynactin-4, which is believed to localize at the slow-growing end of the dynactin Arp1 protofilament, which in turn engages microtubules through p150 (23). An additional potential connection between Arp1 and costameres could result from interaction with β spectrin, which also is associated with costameres (4) and binds actin/Arp1 (21, 24). To test the functional significance of these interactions, we therefore determined the localization of Arp1 in skeletal muscle and its dependence on dynactin-4 and ankyrin-B. As expected, Arp1 is localized at costameres in skeletal muscle, where it potentially could interact with dynactin-4 and microtubules (Fig. 3D). We next determined effects of depletion of ankyrin-B or dynactin-4 on localization of Arp1 at costameres. Surprisingly, Arp1 remains associated with costameres in dynactin-4-depleted fibers (Fig. 3D). Given that costamere microtubules are lost in dynactin-4-depleted fibers that retain Arp1 (Fig. 2C), the Arp1 remaining in the absence of dynactin-4 is not sufficient to retain microtubules. We also found that Arp1 was missing from costameres in fibers depleted of ankyrin-B (Fig. 2C), but retaining β2 spectrin (Fig. 1C). Ankyrin-B thus directly or indirectly stabilizes costamere-associated Arp1 through a mechanism independent of dynactin-4 and β2 spectrin. Together, these results do not support a simple linear ankyrin-B-dynactin-4-Arp1 pathway. It will be important in the future to critically evaluate functional interactions of dynactin-4 and ankyrin-B with other components of the dynactin complex as well as possible direct interactions of dynactin-4 with microtubules.
β2 Spectrin Localizes Ankyrin-B at Costameres
Next, we investigated how ankyrin-B couples dynactin-4 to costameres. Ankyrin-B directs β2 spectrin to an intracellular compartment in cardiomyocytes, suggesting a similar role of ankyrin-B at costameres (25). However, β2 spectrin remains in costameres in the absence of ankyrin-B (Fig. 1). We therefore, determined the effect of mutating the β2 spectrin-binding site of ankyrin-B on its localization in skeletal muscle. The DAR975AAA mutation in the first ZU5 domain, which eliminates β spectrin binding (25), also abolishes ankyrin-B association with costameres (Fig. 4A). Immunoblots confirm that both wild type and mutated forms of ankyrin-B are comparably expressed (Fig. 4B). This result implies that β spectrin is required for ankyrin-B localization to skeletal muscle costameres.
Skeletal muscle contains both β1 and β2 spectrins (26), raising the question of which β spectrin is active in recruiting ankyrin-B to costameres. We directly evaluated the role of β2 spectrin in skeletal muscle by siRNA knockdown as described above for ankyrin-B and dynactin-4 (Fig. 4). We found that depletion of β2 spectrin results within 5 days in loss of sarcolemmal ankyrin-B, dynactin-4, dystrophin, β-dystroglycan, as well as costamere-associated microtubules (Fig. 4C). Interestingly, ankyrin-G, which also binds to β spectrins (as well as to dystrophin and β-dystroglycan (11)), remained at unaltered levels in costameres upon β2 spectrin depletion (Fig. 4C). Because we observe normal expression and localization of β2 spectrin in muscle fibers depleted of either ankyrin-B or dynactin-4, we conclude that localization of β2 spectrin is independent of these two proteins.
Role of Ankyrin-B Interactions in Preventing Exercise-induced Muscle Injury
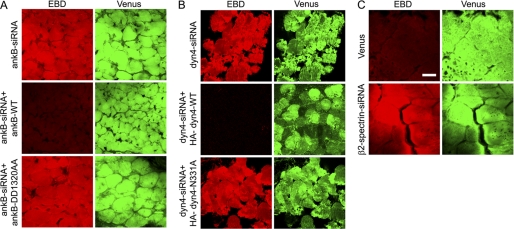
We next investigated the effect of perturbing interactions within the ankyrin-B-dynactin-4 interaction network on ability of skeletal muscle to withstand exercise injury. Loss of dystrophin results in sarcolemmal fragility, which can be assessed by permeability of damaged fibers to EBD (27). We tested the sarcolemmal integrity/fragility in TA muscles of mice that were transfected and then challenged after 4 days by treadmill exercise (see “Experimental Procedures”). Ankyrin-B-depleted muscles show extensive uptake of EBD that is prevented by co-transfection of the siRNA with human wild type ankyrin-B (Fig. 5A). Ankyrin-B-DD1320AA, with impaired dynactin-4 binding, fails to rescue ankyrin-B-depleted muscles, which show extensive EBD uptake (Fig. 5A). Knockdown of dynactin-4 in muscles of exercised mice also results in EBD uptake into muscle fibers, which can be rescued by the expression of recombinant wild type dynactin-4 but not by N331A dynactin-4, which is impaired in ankyrin-B binding (Fig. 5B). β2 spectrin knockdown also causes extensive EBD uptake following exercise (Fig. 5C).
FIGURE 5.
Ankyrin-B-dynactin-4 interaction is required to maintain sarcolemmal integrity following exercise. Mice were transfected (Figs. 1, 3, and 4), exercised on a treadmill, and injected with EBD (see “Experimental Procedures”), and muscle sections evaluated for EBD uptake to assess sarcolemmal fragility. Red, EBD; Green, Venus, which marks the fibers that expressed the Venus vector containing siRNA. A, ankyrin-B (ankB)-depleted muscles exhibit EBD uptake and can be rescued by co-transfection with wild type HA-ankyrin-B but exhibit sarcolemmal fragility and EBD uptake when cotransfected with HA-ankyrin-B-DD1320AA, which is impaired in dynactin-4 binding. B, dynactin-4 knockdown results in sarcolemmal fragility and EBD uptake, which can be rescued by co-transfection with HA-dynactin-4 but are not with HA-dynactin-4-N331A, which is impaired in ankyrin-B binding. C, β2 spectrin knockdown results in sarcolemmal fragility and EBD uptake. Scale bar, 20 microns.
DISCUSSION
We have applied a strategy of acute substitution in adult muscle of native proteins with mutant forms deficient in specific interactions to study proteins responsible for localization and function of dystrophin at costameres of skeletal muscle. We present evidence for physiological roles for ankyrin-B association with dynactin-4 and β2 spectrin that are required to protect muscle from exercise-induced injury through a dystrophin-based mechanism. We establish a functional hierarchy where β2 spectrin localizes ankyrin-B to costameres, and ankyrin-B in turn localizes dynactin-4 and dystrophin. Dynactin-4 stabilizes costamere-associated microtubules by a yet to be defined mechanism that likely also involves dystrophin.
Dynactin-4 (initially referred to as p62) was discovered based on its association with the slow-growing end of Arp1 filaments in the dynactin complex (28–31). Dynactin-4 is an ancient protein with a homologue in Neurospora (ropy-2) and predates evolution of ankyrins, which first appear in metazoans. Thus, it is likely that dynactin-4 has both ankyrin-dependent and -independent functions. In support of this idea, we observed that knockdown of ankyrin-B affected only the costamere-associated population of microtubules in skeletal muscle, whereas loss of dynactin-4 interrupted both costamere and longitudinal microtubules (Fig. 3). Dynactin-4 has been proposed to connect the dynactin complex to membrane surfaces (23). However, we observe that Arp1 remains associated with costameres in the absence of dynactin-4 (Fig. 3). Thus, dynactin-4 either has Arp1-independent function(s), and/or dynactin associates with membranes through multiple mechanisms.
The function of costamere-microtubules remains to be demonstrated but has been proposed to include routes of directed post-Golgi transport for dystroglycan and/or as mechanical supports for the sarcolemma (9–11). Microtubules in post-mitotic cells such as skeletal muscle, neurons, and epithelia typically are not associated with centrosomes and have attracted much less attention than centrosomal microtubules of cultured cells (32). Skeletal muscle has two populations of noncentrosomal microtubules: one associated with costameres and dependent on ankyrin-B interaction with dynactin-4 and the other extending longitudinally parallel to myofibrils and independent of ankyrin-B. It will be of interest to determine roles of ankyrin-B and dynactin-4 in organization and function of other noncentrosomal microtubules preferentially associated with membranes in neurons and epithelial cells.
Ankyrin-B requires β2 spectrin for its localization and function at costameres (Fig. 4), raising the question of how β2 spectrin sorts to costameres. Ankyrin-G persists at costameres in the absence of β2 spectrin (Fig. 4). Ankyrin-G also localizes independently of β spectrins at axon initial segments (33) and epithelial lateral membranes (17). Ankyrin-G thus potentially could provide the initial docking signal for β2 spectrin and hence ankyrin-B. However, ankyrin-B associates with costameres in ankyrin-G-depleted muscle fibers (11). Thus, the epistatic relationship between ankyrins and β spectrins at costameres is not described by a simple linear pathway. Further clues to costamere assembly mechanisms may be provided by direct imaging of dynamic behavior of fluorescently tagged ankyrins, β spectrins, dystrophin, and microtubules in vivo.
Supplementary Material
This work was supported in part by a grant from the Muscular Dystrophy Association.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- TA
- tibialis anterior
- EBD
- Evans blue dye.
REFERENCES
- 1. Ervasti J. M. (2003) J. Biol. Chem. 278, 13591–13594 [DOI] [PubMed] [Google Scholar]
- 2. Cohn R. D., Campbell K. P. (2000) Muscle Nerve 23, 1456–1471 [DOI] [PubMed] [Google Scholar]
- 3. Dalkilic I., Kunkel L. M. (2003) Curr. Opin. Genet. Dev. 13, 231–238 [DOI] [PubMed] [Google Scholar]
- 4. Craig S. W., Pardo J. V. (1983) Cell Motil. 3, 449–462 [DOI] [PubMed] [Google Scholar]
- 5. Pardo J. V., Siliciano J. D., Craig S. W. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 1008–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ervasti J. M., Ohlendieck K., Kahl S. D., Gaver M. G., Campbell K. P. (1990) Nature 345, 315–319 [DOI] [PubMed] [Google Scholar]
- 7. Ervasti J. M., Campbell K. P. (1991) Cell 66, 1121–1131 [DOI] [PubMed] [Google Scholar]
- 8. Rybakova I. N., Amann K. J., Ervasti J. M. (1996) J. Cell Biol. 135, 661–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Prins K. W., Humston J. L., Mehta A., Tate V., Ralston E., Ervasti J. M. (2009) J. Cell Biol. 186, 363–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Percival J. M., Gregorevic P., Odom G. L., Banks G. B., Chamberlain J. S., Froehner S. C. (2007) Traffic 8, 1424–1439 [DOI] [PubMed] [Google Scholar]
- 11. Ayalon G., Davis J. Q., Scotland P. B., Bennett V. (2008) Cell 135, 1189–1200 [DOI] [PubMed] [Google Scholar]
- 12. Davis J. Q., Bennett V. (1984) J. Biol. Chem. 259, 13550–13559 [PubMed] [Google Scholar]
- 13. Davis L. H., Otto E., Bennett V. (1991) J. Biol. Chem. 266, 11163–11169 [PubMed] [Google Scholar]
- 14. Elbashir S. M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. (2001) Nature 411, 494–498 [DOI] [PubMed] [Google Scholar]
- 15. Nagai T., Ibata K., Park E. S., Kubota M., Mikoshiba K., Miyawaki A. (2002) Nat. Biotechnol. 20, 87–90 [DOI] [PubMed] [Google Scholar]
- 16. Mohler P. J., Gramolini A. O., Bennett V. (2002) J. Biol. Chem. 277, 10599–10607 [DOI] [PubMed] [Google Scholar]
- 17. Kizhatil K., Yoon W., Mohler P. J., Davis L. H., Hoffman J. A., Bennett V. (2007) J. Biol. Chem. 282, 2029–2037 [DOI] [PubMed] [Google Scholar]
- 18. Kizhatil K., Davis J. Q., Davis L., Hoffman J., Hogan B. L., Bennett V. (2007) J. Biol. Chem. 282, 26552–26561 [DOI] [PubMed] [Google Scholar]
- 19. McMahon J. M., Signori E., Wells K. E., Fazio V. M., Wells D. J. (2001) Gene Ther. 8, 1264–1270 [DOI] [PubMed] [Google Scholar]
- 20. Wang R., Wei Z., Jin H., Wu H., Yu C., Wen W., Chan L. N., Wen Z., Zhang M. (2009) Mol. Cell 33, 692–703 [DOI] [PubMed] [Google Scholar]
- 21. Holleran E. A., Tokito M. K., Karki S., Holzbaur E. L. (1996) J. Cell Biol. 135, 1815–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ipsaro J. J., Mondragón A. (2010) Blood 115, 4093–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schroer T. A. (2004) Annu. Rev. Cell Dev. Biol. 20, 759–779 [DOI] [PubMed] [Google Scholar]
- 24. Lorenzo D. N., Li M. G., Mische S. E., Armbrust K. R., Ranum L. P., Hays T. S. (2010) J. Cell Biol. 189, 143–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mohler P. J., Yoon W., Bennett V. (2004) J. Biol. Chem. 279, 40185–40193 [DOI] [PubMed] [Google Scholar]
- 26. Zhou D., Ursitti J. A., Bloch R. J. (1998) Mol. Biol. Cell 9, 47–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Straub V., Rafael J. A., Chamberlain J. S., Campbell K. P. (1997) J. Cell Biol. 139, 375–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schafer D. A., Gill S. R., Cooper J. A., Heuser J. E., Schroer T. A. (1994) J. Cell Biol. 126, 403–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Eckley D. M., Gill S. R., Melkonian K. A., Bingham J. B., Goodson H. V., Heuser J. E., Schroer T. A. (1999) J. Cell Biol. 147, 307–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Garces J. A., Clark I. B., Meyer D. I., Vallee R. B. (1999) Curr. Biol. 9, 1497–1500 [DOI] [PubMed] [Google Scholar]
- 31. Karki S., Tokito M. K., Holzbaur E. L. (2000) J. Biol. Chem. 275, 4834–4839 [DOI] [PubMed] [Google Scholar]
- 32. Bartolini F., Gundersen G. G. (2006) J. Cell Sci. 119, 4155–4163 [DOI] [PubMed] [Google Scholar]
- 33. Jenkins S. M., Bennett V. (2001) J. Cell Biol. 155, 739–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.