Abstract
The mammalian ferlins are calcium-sensing, C2 domain-containing proteins involved in vesicle trafficking. Myoferlin and dysferlin regulate myoblast fusion and muscle membrane resealing, respectively. Correspondingly, myoferlin is most highly expressed in singly nucleated myoblasts, whereas dysferlin expression is increased in mature, multinucleated myotubes. Myoferlin also mediates endocytic recycling and participates in trafficking the insulin-like growth factor receptor. We have now characterized a novel member of the ferlin family, Fer1L5, because of its high homology to dysferlin and myoferlin. We found that Fer1L5 protein is expressed in small myotubes that contain only two to four nuclei. We also found that Fer1L5 protein binds directly to the endocytic recycling proteins EHD1 and EHD2 and that the second C2 domain in Fer1L5 mediates this interaction. Reduction of EHD1 and/or EHD2 inhibits myoblast fusion, and EHD2 is required for normal translocation of Fer1L5 to the plasma membrane. The characterization of Fer1L5 and its interaction with EHD1 and EHD2 underscores the complex requirement of ferlin proteins and mediators of endocytic recycling for membrane trafficking events during myotube formation.
Keywords: Development, Intracellular Trafficking, Muscular Dystrophy, Sarcolemma, Skeletal Muscle, EHD Proteins, Dysferlin, Myoblast, myogenesis
Introduction
The ferlin family is defined by its homology to FER1, a Caenorhabditis elegans gene required for the fusion of intracellular organelles to the plasma membrane and normal reproduction in worms (1, 2). The mammalian ferlin family has six members, and like C. elegans FER1, these proteins are defined by the presence of multiple C2 domains and a carboxyl-terminal membrane anchor. C2 domains are independently folding domains that can mediate protein-protein interactions as well as protein-membrane interactions. The C2 domains of synaptotagmins, the best characterized C2 domains, regulate the very rapid fusion of membrane-bound vesicles to the plasma membrane during fast exocytosis (3–5). The synaptotagmins, like many C2 domain-containing proteins, contain just two C2 domains. The mammalian ferlin family is unique in that members have six or seven C2 domains. In mammals, three ferlins have been characterized, and two have been linked to inherited disease in humans. Dysferlin is highly expressed in muscle, and loss of function mutations lead to a recessive form of limb girdle muscular dystrophy 2B and Miyoshi myopathy, a milder muscle disease (6, 7). Otoferlin is highly expressed in the inner ear, and loss of function mutations in otoferlin lead to nonsyndromic deafness (8).
In muscle, membrane fusion is important for repair of plasma membrane as well as for the fusion of myoblasts to form multinucleate myotubes. The absence of dysferlin renders the sarcolemma less able to reseal after laser-induced damage (9). Myoferlin is most related to dysferlin, showing homology within the C2 domains and in the intervening regions, and the loss of myoferlin leads to impaired myoblast fusion (10). Myoferlin-null mice have smaller muscle fibers and delayed muscle healing after damage (10). Three additional genes, fer1L4, fer1L5, and fer1L6, are present in the human and mouse genomes, but little is known about their expression at the protein level or their function.
Myogenesis occurs through the fusion of singly nucleated myoblasts into multinucleated myotubes, a process that is recapitulated during muscle regeneration. Myoblast fusion begins with the recognition of appropriate fusion partners followed by cell attachment and finally the merging of membrane bilayers of two apposing cells into a single bilayer. Prior to membrane coalescence in fusing myoblasts, an accumulation of vesicles at sites of fusion can be observed on electron micrographs of fusing muscle cells (11, 12). Vesicle accumulation on the cytoplasmic face of the plasma membrane has also been observed in C. elegans fer-1 mutant spermatozoa, as well as dysferlin-null and myoferlin-null myoblasts and skeletal muscle (1, 2, 9, 13, 14), suggesting a similar effect where the loss of ferlin function is associated with abnormal vesicle trafficking leading to an accumulation of intracellular vesicles.
Myoferlin directly interacts with EHD2, a carboxyl-terminal Eps15 homology domain-containing protein (14, 15). The EHD proteins regulate endocytosis of receptors and their recycling to the plasma membrane after internalization (16–23). EHD proteins are characterized by an amino-terminal ATPase domain as well as a carboxyl-terminal EH domain; the EH domain is an EF hand-like structure that interacts with proteins containing an asparagine-proline-phenylalanine (NPF) motif (24–27). Myoferlin harbors an NPF motif in its C2B domain, and this region was shown to mediate EHD2 binding (14). Reduction of EHD1 in human cells impairs transferrin recycling (28). Myoferlin-null myoblasts accumulate more labeled transferrin initially and are less efficient at recycling the transferrin receptor to the plasma membrane than control myoblasts (14).
We have now characterized Fer1L5, the only other mammalian ferlin to contain an NPF motif. We found that Fer1L5 is expressed in myoblasts undergoing fusion to myotubes and that Fer1L5 can bind both EHD1 and EHD2, two EHD family members that are also expressed in myoblasts. siRNA-mediated reduction of EHD1 and/or EHD2 expression leads to impaired myoblast fusion. Reduction of EHD2 protein levels inhibits normal transit of Fer1L5 through the secretory system to the plasma membrane. EHD proteins and ferlin proteins form discrete structures in myoblasts. From these data, we propose a model where multiple ferlin proteins interact with EHD proteins to mediate the cytoskeletal rearrangements necessary for proper membrane recycling and myoblast fusion.
EXPERIMENTAL PROCEDURES
Cell Culture
C2C12 cells were obtained from ATCC (catalogue number CRL-1772). The cells were grown in DMEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin in 7.5% CO2. The cells were differentiated in DMEM supplemented with 2% horse serum and 1% penicillin/streptomycin in 7.5% CO2. All of the tissue culture media and sera were from Invitrogen.
Immunoblotting and Immunostaining
For immunoblot time course analysis, C2C12 cells were plated at equal densities on 10-cm tissue culture plates and harvested at specified time points. The cultures were lysed in 1 ml of lysis buffer (150 mm NaCl, 50 mm Tris-HCl, pH 7.4, 1% Triton, 1% Halt protease inhibitor mixture (Pierce), and PMSF). The lysates were centrifuged at 14,000 × g for 15 min at 4 °C to remove cellular debris, and the protein concentration of the supernatant was determined using a Bio-Rad protein assay. Fifty μg of protein was separated on a 4–20% acrylamide gel stained with GelCode Blue stain reagent (Pierce) or transferred to PVDF Immobilon-P membrane (Millipore, Billerica, MA). The membrane was immunoblotted with rabbit polyclonal anti-myoferlin at 1:3000 (MYOF3) (10), mouse monoclonal anti-dysferlin at 1:3000 (NCL-Hamlet; Novocastra Ltd.), and rabbit polyclonal anti-Fer1L5 antibody (ab1005) at 1:3000. Peptides for anti-Fer1L5 antibody production were selected using MacVector. The peptide EQKDQPRKEMEKTRSWQPWK (amino acids 1031–1050) was synthesized, coupled to keyhole limpet hemocyanin, and injected into rabbits (Bethyl Laboratories, Montgomery, TA) to generate anti-Fer1L5 ab1005. A second antibody, anti-Fer1L5 ab412 was generated against the peptide sequence RGGKKPPFRTSEEGTCIMDA (amino acids 438–457). The specificity of the antibodies was tested by blocking the immunostaining from 2 μg of antibody with 40 μg of respective peptides. Other than this specificity test, ab412 was not used. Secondary antibodies, goat anti-rabbit and goat anti-mouse antibodies conjugated to horseradish peroxidase (Jackson ImmunoResearch, West Grove, PA), were used at a dilution of 1:5000. Blocking and antibody incubations were performed in StartingBlock T20 blocking buffer (catalogue number 37543; Pierce). ECL-Plus chemiluminescence (Amersham Biosciences) and Kodak Biomax MS film were used for detection.
For immunostaining analysis, C2C12 cells were plated at equal densities on NaOH-washed glass coverslips within 6-well plates. The cells were fixed in 4% paraformaldehyde for 10 min. Blocking and antibody incubations were performed in 1× PBS containing 5% fetal bovine serum. Rabbit polyclonal anti-Fer1L5 was used at 1:100, rabbit polyclonal MYOF3 was used at 1:100, mouse monoclonal NCL-Hamlet was used at 1:200, goat polyclonal anti-EHD2 (ab23935, Abcam, Cambridge, MA) was used at 1:500, rabbit polyclonal anti-EHD1 (17) was used at 1:100, and rabbit polyclonal anti-EHD4 was used at 1:100. Donkey anti-rabbit conjugated to Alexa 488, donkey anti-goat conjugated to Alexa 594, goat anti-rabbit conjugated to Alexa 594, goat anti-mouse conjugated to Alexa 488, phalloidin conjugated to Alexa 488, and phalloidin conjugated to Alexa 633 (Molecular Probes) were used at 1:2000. The coverslips were mounted using Vectashield with DAPI (Vector Laboratories, Burlingame, CA). The images were captured using a Leica TCS SP2 AOBS Laser Scanning Confocal (Leica Microsystems Inc., Bannockburn, IL) or a Zeiss Axiophot microscope and Axiovision software (Carl Zeiss, Maple Grove, MN). To quantify ferlin expression in a 4-day differentiated culture of C2C12 cells, nuclei within myoferlin-, Fer1L5-, and dysferlin-positive cells were counted and classified as singly nucleated, containing two to four nuclei or containing five or more nuclei using the Cell Counter Plugin for Image J. Statistical analysis was performed with Prism (Graphpad, La Jolla, CA) using a two-way analysis of variance (10).
Immunoprecipitation
C2C12 cells were grown to confluency in growth medium and differentiated for 4 days in differentiation media. The cultures were lysed in immunoprecipitation buffer (150 mm NaCl, 50 mm Tris-HCl, pH 7.4, 0.15% CHAPS, 1% Halt protease inhibitor mixture (Pierce), and PMSF),2 as described previously (14). 200 μg of cell lysate was precleared with protein G Plus/protein A-agarose suspension (Calbiochem) for 30 min at 4 °C and then incubated overnight with 75 μl of protein G Plus/protein A-agarose and 5 μg of anti-Fer1L5 antibody, 5 μg of anti-EHD1, 5 μg of anti-EHD2, or 5 μg of IgG. The beads were washed four times in immunoprecipitation buffer and boiled for 10 min in SDS loading buffer (50 mm Tris, pH 6.8, 100 mm dithiothreitol, 2% SDS, 0.1% bromphenol blue, 10% glycerol, 5% 2-mercaptoethanol), and the supernatant was loaded on a 7.5% or 12% polyacrylamide gel. The gels were transferred to a PVDF Immobilon-P membrane (Millipore) and immunoblotted with anti-Fer1L5 (1:1000), anti-EHD1 (1:1000), or anti-EHD2 (1:1000).
Construction of Expression Vectors
SMART (29) was used to identify C2 domains to construct expression vectors containing the following sequences: Fer1L5 C2A residues 1–98, Fer1L5 C2B residues 168–264, Fer1L5 C2C residues 325–422, Fer1L5 C2D residues 1078–1222, Fer1L5 C2E residues 1487–1586, Fer1L5 C2F residues 1659–1851, and myoferlin C2B residues 1–100 (nucleotides 852–1151 of GenBankTM accession number NM_001099634). Primers corresponding to each of these nucleotide regions were designed and used to amplify inserts from mouse Fer1L5 and myoferlin cDNA templates. The inserts were ligated into the EcoRI and NotI sites of pGEX4T-1 (Amersham Biosciences). Mutant versions of Fer1L5 C2B were generated by site-directed mutagenesis using Pfu Turbo polymerase and the following sets of primers: NPF to dysferlin C2B sequence SPL (forward, 5′-ATCAAGATGGGAAACTCTCCTCTCTTCAATGAGATATTC-3′; reverse, 5′-GAATATCTCATTGAAGAGAGGAGAGTTTCCCATCTTGAT-3′) and NPF to fer1L4 C2B sequence CPF (forward, 5′-ATCAAGATGGGAAACTGTCCTTTCTTCAATGA03′; reverse, 5′-TCATTGAAGAAAGGACAGTT-TCCCATCTTGAT-3′).
Primers were also designed to amplify the DYSF domain and the homologous region of anti-Fer1L5 1005 epitope from mouse dysferlin, Fer1L5, and myoferlin cDNA templates. The primers used were dysferlin (forward, 5′-GCGGAATTCCATCTGAGCTTCGTG-3′; reverse, 5′-TATGCGGCCGCTGTCTTCTGGCTTTG-3′), Fer1L5 (forward, 5′-GCGGAATTCAAGAGCCAGGTGCTG-3′; reverse, 5′-CATGCGGCCGCGATGTAGTAAATGAA-3′), and myoferlin (forward, 5′-GCGGAATTCCACACCGAGTTCACT-3′; reverse, 5′-TATGCGGCCGCAGTGGTTTTGCTTCG-3′). The inserts were ligated into the EcoRI and NotI sites of pGEX4T-1.
In Vitro Binding Experiments
Experiments were performed as described (14). Cultures of BL21 cells (Invitrogen) expressing glutathione S-transferase-Fer1L5 C2 domain and glutathione S-transferase-myoferlin C2B fusion proteins were induced for 3 h with isopropyl 1-thio-β-d-galactopyranoside at a final concentration of 350 mm. The cells were resuspended in 50 μl of 1× SDS loading buffer and boiled for 10 min. Fifteen μl of cell lysate was loaded on a 12% acrylamide gel. For each experiment, two gels were used; one was transferred to PVDF membrane used for the gel overlay, and the other was stained with GelCode Blue stain reagent (Pierce). The EHD1 cDNA was digested from the EHD1-DsRed construct (17) and ligated into pcDNA3.1(−) mycHisA (Invitrogen) using XhoI and HindIII. EHD2 was ligated into pCR T7/NT-TOPO (Invitrogen) and has been described previously (14). 35S-Labeled proteins were generated using in vitro transcription/translation of EHD1 and EHD2 using [35S]methionine with the TnT coupled reticulocyte lysate system (Promega, Madison, WI). PVDF membranes containing the GST fusion proteins of C2 domains were blocked for 2 h at 4 °C in blocking buffer (0.1% gelatin, 5% bovine serum albumin, 0.1% Tween in 1× PBS, pH 7.5) and then incubated overnight at 4 °C in overlay buffer (150 mm NaCl, 20 mm HEPES, 2 mm MgCl2, 5% bovine serum albumin, pH 7.5) containing the radioactive EHD1 and EHD2. The membrane was washed four times for 20 min at 4 °C in overlay buffer and exposed to film overnight at −80 °C.
Antibody Specificity
Cultures of BL21 cells expressing glutathione S-transferase-DysF domain + epitope regions from dysferlin, Fer1L5, and myoferlin were induced for 3 h with isopropyl 1-thio-β-d-galactopyranoside at a final concentration of 350 mm. The cells were resuspended in 50 μl of 1× SDS loading buffer and boiled for 10 min. Twenty-five μl of cell lysate from dysferlin and myoferlin cultures and 12.5 μl of cell lysate from Fer1L5 culture was separated on a 12% polyacrylamide gel. For each experiment, two gels were used; one was transferred to PVDF membrane used for the gel overlay, and the other was stained with GelCode Blue stain reagent (Pierce). The PVDF membrane was immunoblotted with rabbit polyclonal anti-Fer1L5 (antibody 1005) primary antibody 1:1000 and goat anti-rabbit conjugated to horseradish peroxidase secondary antibody 1:5000. Blocking and antibody incubations were performed in StartingBlock T20 blocking buffer (catalogue number 37543; Pierce). ECL-Plus chemiluminescence (Amersham Biosciences) and Kodak Biomax MS film were used for detection.
siRNA-mediated Reduction
A 650-bp region of Mus musculus EHD1 3′-UTR was amplified from C2C12 cDNA through RT-PCR using the primers 5′-CCACTTTCTAACCACCAGCTCCTTGTCTGC-3′ (forward) and 5′-GTCACTACAGAACACTAAGTAGCTTATGC-3′ (reverse). 500 bp of EHD1 3′-UTR and EHD2 3′-UTR were amplified from cDNA with T7 priming sites using the primers EHD1 (forward, 5′-GCGTAATACGACTCACTATAGGGAGAATTTTTCAAGGTCCATAAAG-3′; reverse, 5′GCGTAATACGACTCACTATAGGGAGAGACGTCATGGACTGTCTCGC-3′) and EHD2 (forward, 5′-GCGTAATACGACTCACTATAGGGAGACTGCCTGCCTACCCTGCCTG-3′; reverse, 5′-GCGTAATACGACTCACTATAGGGAGAGATGTCACACTCCTGAGAGT-3′). In addition, three regions of ∼500 bp each were amplified from Fer1L5 cDNA with T7 priming sites using the primers siRNA1 (forward, 5′-GCGTAATACGACTCACTATAGGGAGACCATCTGTGCCCTCG-3′; reverse, S5′-GCGTAATACGACTCACTATAGGGAGAAGTGCCTTCCTCTGA-3′), siRNA2 (forward, 5′-GCGTAATACGACTCACTATAGGGAGAACATCCTCAACAAGC-3′; reverse, 5′-GCGTAATACGACTCACTATAGGGAGAGGGCAACAAGTAAAT-3′), and siRNA3 (forward, 5′-GCGTAATACGACTCACTATAGGGAGACTATGCTTCATCCTCCTGAGCGAAA-3′; reverse, 5′-GCGTAATACGACTCACTATAGGGAGAACAGCTCTGCTCTTCTTGTCAGAAG-3′). The three Fer1L5 amplicons were pooled into one sample. Amplicons were in vitro transcribed using the TurboScript T7 Transcription Kit (Genlantis, San Diego, CA). EHD1, EHD2, and Fer1L5 double-stranded RNA fragments were digested into 22-bp siRNAs using the recombinant human Turbo dicer enzyme kit (Genlantis). Genlantis provided a nonspecific siRNA control, GFP siRNA. C2C12 cells were plated at a density of 50,000 cells/well in a 24-well plate and transfected with 500 ng of siRNA 16 h after plating. Transfected C2C12 cells were differentiated for 4 days. The cells were lysed, and the proteins were separated on a 12% polyacrylamide gel. The gel was transferred to PVDF and immunoblotted with rabbit polyclonal anti-EHD1 (1:1000), goat polyclonal anti-EHD2 (1:1000), rabbit polyclonal anti-Fer1L5 (antibody 1005) (1:1000), mouse monoclonal anti-desmin (1:1000), or mouse monoclonal anti-smooth muscle actin (1:1000). The cells were immunostained with mouse monoclonal anti-desmin (Sigma) and Sytox Green (Invitrogen), and nuclei within desmin-positive cells were counted and used to classify cells as mononucleated, binuclear, or containing three or more nuclei using the Cell Counter Plugin for Image J. Statistical analysis was performed with Prism using an unpaired t test as described (10).
RESULTS
Fer1L5 Is Related to Dysferlin and Myoferlin
The Fer1L5 gene was identified based on homology to dysferlin (fer1L1) and myoferlin (fer1L3) and verified by RT-PCR as well as IMAGE cDNA clones. The open reading frame of Fer1L5 sequence is 6117 bp and predicts a protein of 2038 amino acids with an estimated molecular mass of 236 kDa (Fig. 1A). The mammalian ferlin family members share characteristic conserved features, including a single transmembrane domain and six C2 domains. The Fer1L5 protein contains six predicted C2 domains: C2A at amino acids 1–98, C2B at 168–264, C2C at 325–422, C2D at 1078–1222, C2E at 1487–1586, and C2F at 1659–1851. Each C2 domain is directly homologous to those previously identified in dysferlin and myoferlin (7, 30). Ferlin C2 domains share stronger similarity to positionally equivalent domains in other ferlin family members than to C2 domains within the same molecule (6). Myoferlin, Fer1L5, and dysferlin are unique from the other ferlin family members in that they also contain a DysF domain present in the ferlins that is the hypothesized binding site for caveolin-3 (31). The DysF domain is also found in the human β-propeller protein DKFZP434B0335 and the yeast peroxisomal proteins Pex30p and Pex31p (31). A seventh C2 domain can be identified between C2D and C2E in dysferlin and myoferlin by the SMART algorithm and other protein prediction programs (29, 32); however, this domain is highly divergent from the standard C2 domain profile.
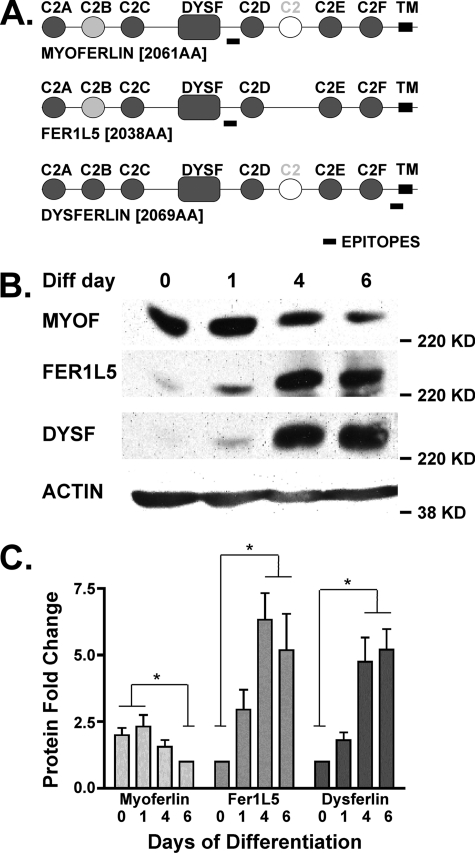
FIGURE 1.
A, schematic representation comparing myoferlin, Fer1L5, and dysferlin proteins. The ferlins are ∼230-kDa transmembrane proteins that contain at least six C2 domains. Myoferlin, Fer1L5, and dysferlin are the only members of the mammalian ferlin family that contain a DysF domain, a domain also found in yeast peroxisomal proteins. B, immunoblotting of C2C12 cell lysates at different stages of myoblast differentiation (Diff) demonstrates that different ferlin proteins are present at distinct stages. 0 represents the first day of serum withdrawal. Myoferlin is expressed early in differentiation, and dysferlin expression increases with differentiation. Fer1L5 is expressed within those extremes but closer to the pattern displayed by dysferlin. C, densitometric analysis of ferlin protein fold change throughout differentiation shown in the immunoblot in B. *, p < 0.05.
Fer1L5 Is Expressed in Differentiating Myoblasts
C2C12 cells are a transformed mouse myoblast-like cell line that can be induced to differentiate into myotubes upon serum starvation, mimicking the pattern of primary cultured myoblasts (33). We compared the expression patterns of myoferlin, dysferlin, and Fer1L5 throughout differentiation using antibodies specific for dysferlin and myoferlin (30) and a newly generated antibody to Fer1L5 protein. The anti-Fer1L5 antibody epitope is unique to Fer1L5, and correspondingly the homologous regions from myoferlin and dysferlin show no reactivity to this antibody (supplemental Fig. S1). Myoferlin was maximally expressed in early differentiating myoblasts (Fig. 1B, days 0 and 1, where day 0 represents the switch to differentiation media). Dysferlin was maximally expressed in mature differentiated myotubes (days 6+). We found that Fer1L5 protein was expressed in a pattern more similar to that of dysferlin than of myoferlin (Fig. 1C).
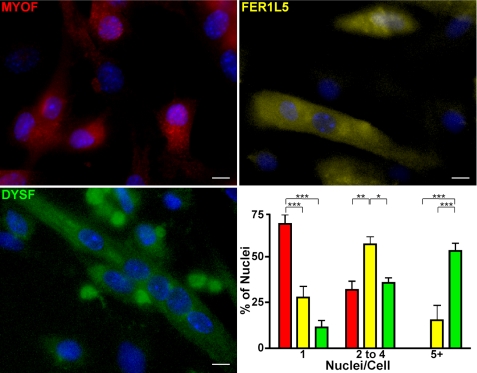
C2C12 cells grow and differentiate asynchronously, and individual cultures of cells can vary in terms of cell stages. For instance, a culture of mature, differentiated myotubes can also contain singly nucleated myoblasts as well as nascent microtubes, which we define as myotubes containing two to four nuclei. We performed immunofluorescence microscopy on C2C12 cells differentiated for 4 days using the same antibodies specific for ferlin proteins and correlated ferlin expression to cell stage by quantifying the number of nuclei in ferlin-stained cells (Fig. 2). Myoferlin-positive cells were mostly mononucleate. Fer1L5-positive cells showed a wider distribution with majority of cells containing two to four nuclei, consistent with the expression of Fer1L5 in microtubes, nascent myotubes having undergone a minimal number of fusion events. Fer1L5 was expressed less in cells containing either one nucleus or five or more nuclei. Dysferlin expression was most easily detected in the larger myotubes containing five or more nuclei. A minority of mononucleated and binucleated cells were dysferlin-positive, consistent with the mixed population of myoblasts and myotubes at this stage of culture and with the immunoblotting shown in Fig. 1A.
FIGURE 2.
Immunostaining of C2C12 cells differentiated in culture for 4 days for each ferlin protein. The majority of myoferlin-positive cells contain mostly one nucleus. The majority of Fer1L5-positive cells contain mostly two, three, or four nuclei. Dysferlin is mostly expressed in larger myotubes containing five nuclei or greater. Blocking and antibody incubation buffers contained 0.1% Triton X-100, which appears to disrupt the discrete ferlin foci. Scale bars, 10 μm. Red, MYOF; yellow, FER1L5; green, DYSF. ***, p < 0.001; **, p < 0.01; *, p < 0.05.
Fer1L5 Interaction with EHD2
An interaction between myoferlin and the Eps15 homology (EH) domain protein EHD2 was reported previously (14) and implicates ferlin proteins in the process of endocytic recycling. The EH domain of the EHD family of proteins recognizes and binds NPF motifs (34). When engaged, the NPF motif is predicted to project into a conserved hydrophobic pocket in the EH domain where a conserved tryptophan interacts with the asparagine (25, 26). This NPF motif is found in human and murine myoferlin in the C2B domain at amino acids 238–240. This NPF motif is also conserved in the Fer1L5 C2B domain at amino acids 206–208, and the proline residue in the NPF motif is conserved in mammalian ferlins, Drosophila misfire, and C. elegans fer-1. There are four mammalian EHD proteins defined by their carboxyl-terminal EH domain. EHD2, EHD1, and EHD4 are expressed in skeletal muscle; EHD4 is weakly expressed, and EHD3 has little or no expression (15, 35). Because of this expression pattern, we studied both EHD2 and EHD1 for their ability to interact with Fer1L5.
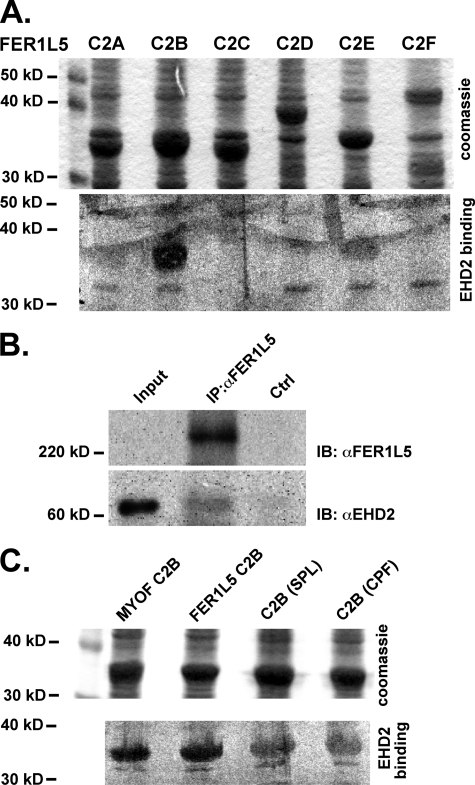
We expressed each individual C2 domain of murine Fer1L5 and the C2B domain of murine myoferlin as GST fusion proteins in Escherichia coli. We found that EHD2 binds directly to the C2B domain of Fer1L5 (Fig. 3A), similar to that shown for myoferlin (14). In addition, EHD2 demonstrated low level direct binding to C2E, but binding to C2A, C2C, C2D, or C2F (Fig. 3A) was undetectable. To confirm that this interaction occurs in cells, we performed an immunoprecipitation of endogenous proteins from C2C12 cells that were differentiated for 4 days, a stage where Fer1L5 expression is maximal. Using an anti-Fer1L5 antibody, we immunoprecipitated Fer1L5 protein from 4-day differentiated C2C12 cell extracts and immunoblotted with an anti-EHD2 antibody, confirming that EHD2 was immunoprecipitated along with Fer1L5 (Fig. 3B).
FIGURE 3.
A, a blot overlay shows the direct interaction between EHD2 and Fer1L5. All six Fer1L5 C2 domains were expressed in bacteria as fusion proteins with glutathione S-transferase, transferred to polyvinylidene difluoride membrane, and incubated with 35S-labeled EHD2. The upper panel shows the Coomassie-stained blots of GST-C2 fusion proteins. The lower panel shows the results after binding to radiolabeled EHD2. EHD2 strongly interacted with the C2B domain of Fer1L5. Less interaction was detected between EHD2 and the Fer1L5 C2E domain. Nonspecific bacterial proteins can be identified at 32kDa. B, an anti-Fer1L5 antibody was used to immunoprecipitate (IP) lysates from differentiated C2C12 cells, and these lysates were then immunoblotted (IB) with an anti-EHD2 and anti-Fer1L5 antibody. Immunoprecipitation with anti-Fer1L5 resulted in the precipitation with EHD2. The Input lane contains 25 μg of cellular lysate, which does not contain enough Fer1L5 for immunoreactivity. Immunoprecipitation used 200 μg of cellular lysate. C, mutation of the NPF motif within the C2B domain to SPL or CPF decreased, but did not abolish, binding of EHD2. Ctrl, control.
Because of the low level binding between EHD2 and the C2E domain of Fer1L5, a domain that does not contain an NPF motif, we queried the requirements of the NPF residues for this interaction. When the NPF motif of Fer1L5 was mutated to the SPL (the motif found in the C2B domain of dysferlin) or CPF (the motif found in fer1L4), the EHD2 binding to the C2B domain was reduced but not abolished (Fig. 3C). This low level interaction suggests that EHD2 may interact with dysferlin but in a reduced capacity. These data are corroborated by other in vitro binding studies using the DPF and GPF motifs that exhibit binding to EHD proteins (36).
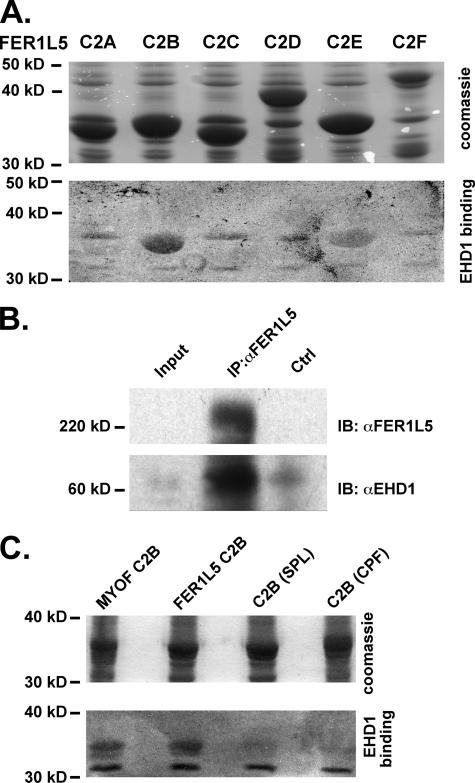
Fer1L5 Interacts with EHD1
Because EHD1 is also expressed in muscle, and EHD1 and EHD2 share 71% amino acid conservation, we queried whether EHD1 demonstrated a similar pattern of interaction (19). We expressed each individual C2 domain of murine Fer1L5 and the C2B domain of murine myoferlin. We performed a similar in vitro gel overlay assay using radiolabeled EHD1. Like EHD2, EHD1 also bound directly to the C2B of Fer1L5 (Fig. 4A). As with EHD2, we found lower level binding of EHD1 to the C2E domain of Fer1L5. Immunoprecipitation of Fer1L5 from lysates derived from 4-day differentiated C2C12 cell lysates showed co-immunoprecipitation of EHD1, indicating that Fer1L5 and EHD1 form a complex (Fig. 4B). Similarly, immunoprecipitation of EHD1 or EHD2 from differentiated C2C12 cell lysates demonstrated co-immunoprecipitation of Fer1L5 (data not shown). Mutation of the flanking residues of the NPF motif reduced but did not abolish EHD1 binding to Fer1L5 (Fig. 4C). Like EHD2, EHD1 bound avidly to the myoferlin C2B domain. These data underscore that multiple EHD proteins can interact with multiple ferlin family members.
FIGURE 4.
A, all six Fer1L5 C2 domains were fused to glutathione S-transferase, transferred to polyvinylidene difluoride membrane and, in this experiment, incubated with 35S-labeled EHD1. EHD1 bound to the C2B domain of Fer1L5 and had a smaller interaction with the C2E and C2F domains. Nonspecific bacterial proteins can be identified at 36 and 32 kDa. B, an anti-Fer1L5 antibody was used to immunoprecipitate (IP) C2C12 lysates, and immunoblotting (IB) with an anti-EHD1 antibody demonstrated that EHD1 associates with Fer1L5. The Input lane contains 25 μg of cellular lysate, which does not contain enough Fer1L5 for immunoreactivity. Immunoprecipitation used 200 μg of cellular lysate. C, mutation of the NPF motif within the C2B domain to SPL or CPF decreased but did not eliminate binding of EHD1. Ctrl, control.
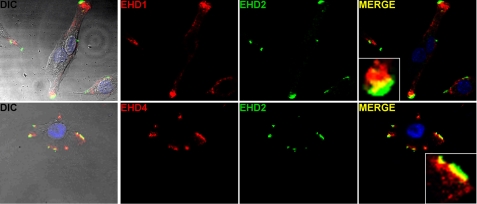
EHD1/EHD2/EHD4 Co-localize into Discrete Structures at the Myoblast Membrane
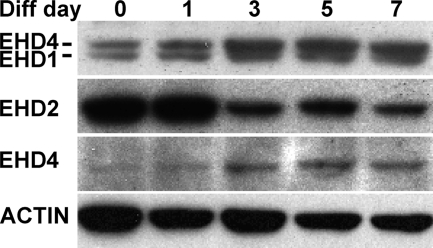
We examined EHD protein expression in myoblasts undergoing differentiation to myotubes using antibodies to EHD proteins. Immunoblotting with an anti-EHD antibody that cross-reacts with EHD4 showed that EHD1 protein expression levels did not appear to change throughout myoblast differentiation (Fig. 5). In contrast, EHD2 expression was highest in undifferentiated and nascent microtube cultures, with a reduced amount in the most differentiated cultures. EHD4, another member of the EHD family that is present in skeletal muscle, is enriched in cultures differentiated 4 days or more. We performed immunofluorescence microscopy using anti-EHD antibodies on undifferentiated C2C12 cells. EHD1 and EHD2 were found to align in discrete structures at the plasma membrane where EHD2 was localized most peripherally, and EHD1 was found just internal to EHD2 (Fig. 6, top row). In these images, it is likely that most of this pattern is derived from EHD1 because little EHD4 is expressed at this stage. Using an EHD4-specific antibody, we were able to detect that EHD4 and EHD2 showed a similar pattern where EHD4 was found just internal to EHD2 in discrete structures at the periphery of the cell (Fig. 6, bottom row).
FIGURE 5.
Immunoblotting of C2C12 cell lysates from different stages of myoblast differentiation with anti-EHD antibodies. EHD1 (top panel, bottom band) shows consistent expression throughout differentiation. The EHD1 antibody cross-reacts with EHD4 (top panel, top band); EHD4 is up-regulated at days 3–7 of differentiation. EHD2 is highly expressed early in differentiation, similar to the pattern of expression of myoferlin. A specific EHD4 antibody (third panel) confirms that EHD4 is up-regulated later in differentiation; the pattern of expression is similar to the expression patterns of Fer1L5 and dysferlin. Immunoblotting with anti-smooth muscle actin serves as the loading control.
FIGURE 6.
EHD1, EHD2, and EHD4 co-localize in undifferentiated C2C12 myoblasts. The upper row shows that EHD1 and EHD2 are found in discrete structures at the periphery. The merged images show that EHD2 was localized most peripherally, whereas EHD1 was aligned just internal to EHD2 with a thin layer of overlap (yellow in the merged images). The lower row shows a similar pattern where EHD4 localizes just internal to EHD2 in discrete structures. The DIC images include an overlay of the merged images.
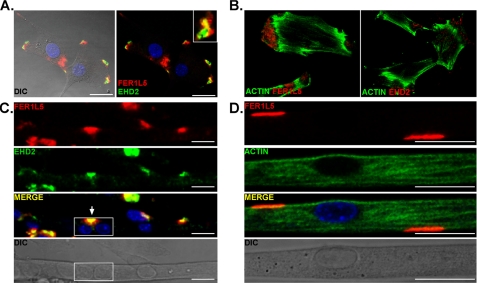
Ferl15 Co-localizes with EHD1 and 2 in Myoblasts and Myotubes
We found that Fer1L5 co-localized with EHD2 in the membrane-associated foci in undifferentiated myoblasts (Fig. 7A). The position of Fer1L5, just internal to EHD2, was the same pattern seen for EHD1 and EHD4 and strongly suggests that these proteins are co-localized. Because the anti-EHD1, anti-EHD4, and anti-Fer1L5 antibodies were all raised in rabbits, we could not assess directly the co-localization of EHD1, EHD4, and Fer1L5. The nature of these discrete structures in myoblasts and myotubes is suggestive of focal adhesions. However, Alexa 488-phalloidin staining suggested that these areas are actin-poor, a feature not consistent with focal adhesions (Fig. 7B). We examined the localization of Fer1L5 and EHD2 in differentiated, multinucleated myotubes to assess whether these discrete structures persisted. Interestingly, the EHD2/Fer1L5 foci persisted in the elongated myotubes (Fig. 7, C and D). Although the function of these foci is unknown, it is tempting to speculate that they are involved in mediating cell fusion events. These results support the in vitro binding data and demonstrate that the interaction between Fer1L5 and EHD1/EHD2/EHD4 occurs in cells.
FIGURE 7.
Fer1L5 and EHD2 co-localize in myoblasts and myotubes. A, myoblast co-localization of Fer1L5 and EHD2. EHD2 immunostaining is most peripheral, whereas FerlL5 staining appears just internal to and partially overlapping with EHD2. The Fer1L5 pattern was similar to that seen for EHD1 and EHD4 in relationship to EHD2. B, phalloidin staining was used to show the actin cytoskeleton of C2C12 myoblasts and demonstrates that Fer1L5/EHD structures are actin-poor. C, myotube expression of Fer1L5 and EHD2. An elongated myotube is shown, and the box highlights two adjacent nuclei within the same myotube. Just above this (arrow) is the similar co-localization where Fer1L5 and EHD2 show an aligned and partially overlapping pattern. Scale bars, 25 μm. D, Alexa 633 phalloidin and anti-Fer1L5 staining of C2C12 differentiated myotubes. The DIC images include an overlay of the merged images.
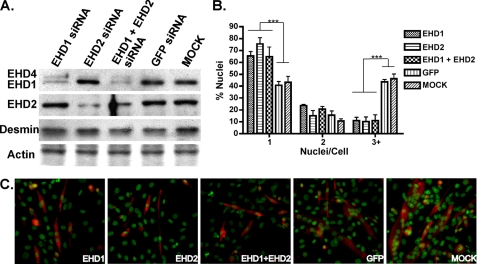
siRNA Reduction of EHD1 and EHD2 Expression Impairs Myoblast Fusion
We generated siRNAs specific to EHD1 or EHD2 and transfected these siRNAs into mononucleated C2C12 myoblasts to determine the role of EHD proteins in myoblast fusion. EHD1-specific siRNA reduced EHD1 protein expression, and EHD2-specific siRNA reduced EHD2 protein expression. Co-transfection of both EHD1- and EHD2-specific siRNA reduced both EHD1 and EHD2 protein expression (Fig. 8A). Interestingly, EHD4, which is recognized by the EHD1 antibody as the upper band in immunoblotted lysates, appeared to be up-regulated when EHD1 or EHD2 was reduced consistent with compensation. Desmin, a marker of myogenic differentiation, was normally expressed in siRNA-treated cells, consistent with normal differentiation. C2C12 cultures with reduced expression of EHD1, EHD2, or EHD1 plus EHD2 formed fewer multinucleated myotubes than C2C12 cultures transfected with a nonspecific siRNA or mock transfected (Fig. 8B). siRNA-treated and mock treated cells formed similar numbers of binucleated myotubes, indicating that the primary fusion of two myoblasts, a myo-initation step, can occur and consistent with at least some degree of normal differentiation. The inhibitory effect of EHD1 and EHD2 siRNA on myoblast fusion was not additive, indicating that reduction of either EHD1 or EHD2 expression is sufficient to mediate this effect. Thus, EHD1 and EHD2 appear to perform specific roles to contribute to myoblast fusion, and both are necessary to complete myogenesis. Similarly, a partial reduction of Fer1L5 with siRNA also demonstrated a significant effect on myoblast fusion (supplemental Fig. S2). Of note, siRNA reduction of Fer1L5 was inefficient.
FIGURE 8.
siRNA reduction of EHD1, EHD2, and EHD1 plus EHD2 inhibits myoblast fusion. A, immunoblot of C2C12 myoblast lysates 24 h after transfection with EHD1 siRNA, EHD2 siRNA, EHD1 plus EHD2 siRNA, a control GFP siRNA, or mock and probing with anti-EHD1 and anti-EHD2 antibodies. The actin band on a Coomassie-stained gel (bottom) demonstrates that lanes were equally loaded. EHD1 was significantly reduced after transfection with EHD1 siRNA and with EHD1 plus EHD2 siRNA but not with EHD2 siRNA or the two control transfections (top panel). EHD4 (top band) is up-regulated after transfection with EHD1 siRNA, EHD2 siRNA, and EHD1 plus EHD2 siRNA. Also, EHD2 was significantly reduced after transfection with EHD2 siRNA and with EHD1 plus EHD2 siRNA, but not with EHD1 siRNA or the two control transfections (second panel). Immunoblotting of desmin suggests that myogenic differentiation is similar between siRNA and mock treated myotubes. B, quantification of the number of nuclei found within desmin-stained C2C12 cells after 4 days of differentiation. Transfection with EHD1, EHD2, or EHD1 plus EHD2 siRNA significantly reduced the number of multinucleated myotubes with three or greater nuclei. An increase in differentiated mononuclear cells was found in cultures of C2C12 cells transfected with EHD siRNA. However, there was no apparent additive effect of EHD1 plus EHD2 siRNA. In addition, the number of binuclear cells is similar between siRNA and mock treated cells, suggesting that myoblast differentiation is not impaired. *, p < 0.001. C, C2C12 myoblasts were transfected with siRNA, differentiated with 4 days, and immunostained with anti-desmin antibody and Sytox Green. EHD siRNA inhibited multinucleated myotube fusion, whereas the control siRNA and mock transfection had no effect on myoblast fusion.
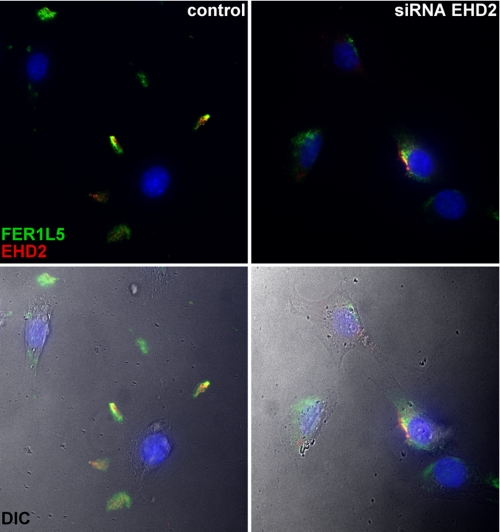
We examined myoblasts with reduced EHD2 immunoreactivity, and we found that Fer1L5 was no longer localized into foci at the plasma membrane (Fig. 9). Instead of discrete structures at the plasma membrane, the majority of Fer1L5 immunostaining was in perinuclear aggregates, most likely in the proximal secretory system. Therefore, normal EHD2 expression is required for normal translocation of Fer1L5 to the plasma membrane.
FIGURE 9.
Reduction of EHD2 inhibits Fer1L5 translocation to the plasma membrane. The left panels show C2C12 myoblasts with normal expression of EHD2. Fer1L5 co-localizes with EHD2 at the cell periphery; note the distance of the immunostaining from the nuclei. The right panels show siRNA-transfected C2C12 cells with reduced expression of EHD2. The majority of Fer1L5 immunostaining was now found localized in a perinuclear pattern, indicating that Fer1L5 requires EHD2 for normal translocation to the peripheral discrete structures. The color images have been merged with the DIC images in the lower two panels.
DISCUSSION
Vesicular fusion to the plasma membrane contributes an additional membrane lipid bilayer, releases contents to the extracellular space through exocytosis, and inserts newly translated or recycled signaling receptors to the cell surface. This process is critical for many cellular functions including myoblast fusion, in which two apposing cellular membranes must recognize the signals of the other cell and coalesce into one cell. Recently, we identified molecules that regulate both endocytic recycling/trafficking and myoblast fusion, suggesting that these processes are molecularly related (14). An increase in intracellular calcium is required for vesicle-plasma membrane fusion. It has been suggested that the disruption of the lipid bilayer by the angled insertion of calcium-sensing proteins induces membrane fusion (37). In a parallel system of vesicular fusion, the role of calcium binding of the synaptotagmin C2 domains has been well studied. In the presence of calcium, the C2A domain of synaptotagmin I binds syntaxin with a high affinity (38) and also binds negatively charged phospholipids with increased affinity (39, 40). The calcium-binding loops of the C2A and C2B domains of synaptotagmin I insert into the lipid bilayer (41). The C2A domains of myoferlin and dysferlin are related to the C2A domain of synaptotagmin I, and the ferlin C2A domains similarly bind negatively charged phospholipids in the presence of calcium (10, 42). The second C2 domain of myoferlin and, as we show now, Fer1L5 protein bind directly to EHD1 and EHD2, and this interaction may be critical for other aspects of the fusion process such as cytoskeletal reorganization required to permit normal membrane fusion. Thus, ferlins are calcium-sensing proteins and are candidate mediators of vesicle-plasma membrane fusion during myoblast fusion.
Reduction of EHD1 or EHD2 Limits Myoblast Fusion and Results in Abnormal Fer1L5 Intracellular Localization
Myoferlin-null myoblasts do not fuse as efficiently as normal myoblasts leading to smaller myotubes in culture and smaller muscle mass in vivo (10). Also, myoferlin-null myoblasts have delayed recycling of transferrin and accumulation of cytoplasmic aggregates, demonstrating the failed exit of vesicles from the endocytic recycling compartment (14). Most recently, we found that cargo within recycling vesicles was aberrantly recycled. Specifically, we found that the insulin-like growth factor was not normally returned to the plasma membrane after internalization (13). Instead, it was redirected toward lysosomes. We hypothesize that this pathway relies on the interaction of ferlin proteins with EHD proteins.
We now show that reduction of EHD1 or EHD2 inhibits myoblast fusion, recapitulating the defect observed in myoferlin-null myoblasts. Although inefficient myoblast fusion may arise from perturbing any number of processes, the co-localization of EHD proteins with Fer1L5 strongly supports a model where multiple ferlin proteins and EHD proteins interact to mediate intracellular trafficking events that are essential for normal myoblast fusion. Both myoferlin and Fer1L5 directly interact with EHD2. Our findings show that reducing EHD2 inhibits the normal translocation of Fer1L5 to the plasma membrane. Similarly, reduction of EHD1 and EHD2 leads to transferrin accumulation in the ERC (17). We hypothesize that it is the absence of Fer1L5 from the plasma membrane that, in part, contributes to reduced myoblast fusion. However, given the number of potential interacting proteins that may participate in this process, it is likely other proteins may be similarly mislocalized from reduction of EHD proteins. Taking these results together, we can conclude that a loss of EHD2 function exerts its effect through multiple ferlin family members.
The successful fusion of two apposed membranes or a vesicle fusing to the plasma membrane requires reorganization of the actin cytoskeleton near the plasma membrane. Discrete structures containing the ferlins and the EHD proteins are devoid of polymerized actin within myoblasts. The interaction between the ferlins and the EHD proteins may lead to the disassembly of the cytoskeleton to permit exiting vesicles to fuse to the plasma membrane. EHD1 and EHD2 interact with an actin-binding protein, EHBP1, through the NPF motif found within EHBP1 (43). Our model suggests that the ferlins and EHBP1 may compete for EHD binding to regulate the complex process of exit from the endocytic recycling compartment, translocation to the plasma membrane, and reorganization of the actin cytoskeleton to facilitate the fusion of exocytosed vesicles to the membrane (14). In addition to EHBP1, EHD1 interacts with MICAL-L1 (44), a member of the molecule interacting with CasL (MICAL) family of proteins. MICALs localize to tubular endosomal membranes, contain a calponin-homology domain, can directly regulate the assembly of actin, and are responsible for myofilament organization and synaptic structure (44–47). Although the functions of EHBP1 and the MICAL family of proteins have not been fully characterized, the interactions of these actin-binding molecules with EHD proteins provide a link between the exit from the endocytic recycling compartment, disassembly of the actin cytoskeleton, and vesicular fusion with the plasma membrane.
Differential Expression Patterns of Ferlin Proteins Suggest Unique Machinery at Different Stages of Myoblast Fusion
Myoferlin is expressed very highly in mononucleated myoblasts just as they begin to differentiate (42). These prefusion myoblasts are elongated and have already begun to express genes consistent with differentiated muscle. In contrast, we find that Fer1L5 protein is more abundant in nascent microtubes, which are myotubes that contain two to four nuclei and represent the fusion of a small number of myoblasts. The largest myotubes in culture, those containing five or more nuclei, expressed high levels of dysferlin. It should be emphasized that these data are derived from a cell culture model of muscle development where the cells are not synchronized, and each culture is a mixture of myoblasts, small myotubes, and large myotubes. Nonetheless, these data support a model where myoferlin expression dominates in prefusion mononucleated myotubes and dysferlin in the mature myotube, whereas Fer1L5 is characteristic of the smaller myotubes that can likely undergo additional fusion steps to grow larger.
It is likely that these processes are not unique to myoblasts or muscle cells. Muscle cells and myoblasts may be more dependent on these processes than other cell types. However, the pathway of endocytic recycling and the broad expression pattern of ferlin proteins and EHD proteins suggests that these mechanisms occur in many if not all cell types, where they may be essential for normal growth and development.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants NS047726 (to E. M. M.), CA105489 (to H. B.), CA99163 (to H. B.), CA87986 (to H. B.), CA116552 (to H. B.), and T32 HL7381. This work was also supported by the Muscular Dystrophy Association and the Jain Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- SMART
- Simple Modular Architecture Research Tool
- EH
- Eps15 homology
- DIC
- differential interference contrast.
REFERENCES
- 1. Achanzar W. E., Ward S. (1997) J. Cell Sci. 110, 1073–1081 [DOI] [PubMed] [Google Scholar]
- 2. Washington N. L., Ward S. (2006) J. Cell Sci. 119, 2552–2562 [DOI] [PubMed] [Google Scholar]
- 3. Bai J., Earles C. A., Lewis J. L., Chapman E. R. (2000) J. Biol. Chem. 275, 25427–25435 [DOI] [PubMed] [Google Scholar]
- 4. Davis A. F., Bai J., Fasshauer D., Wolowick M. J., Lewis J. L., Chapman E. R. (1999) Neuron 24, 363–376 [DOI] [PubMed] [Google Scholar]
- 5. Davletov B. A., Südhof T. C. (1993) J. Biol. Chem. 268, 26386–26390 [PubMed] [Google Scholar]
- 6. Bashir R., Britton S., Strachan T., Keers S., Vafiadaki E., Lako M., Richard I., Marchand S., Bourg N., Argov Z., Sadeh M., Mahjneh I., Marconi G., Passos-Bueno M. R., Moreira Ede S., Zatz M., Beckmann J. S., Bushby K. (1998) Nat. Genet. 20, 37–42 [DOI] [PubMed] [Google Scholar]
- 7. Liu J., Aoki M., Illa I., Wu C., Fardeau M., Angelini C., Serrano C., Urtizberea J. A., Hentati F., Hamida M. B., Bohlega S., Culper E. J., Amato A. A., Bossie K., Oeltjen J., Bejaoui K., McKenna-Yasek D., Hosler B. A., Schurr E., Arahata K., de Jong P. J., Brown R. H., Jr. (1998) Nat. Genet. 20, 31–36 [DOI] [PubMed] [Google Scholar]
- 8. Yasunaga S., Grati M., Cohen-Salmon M., El-Amraoui A., Mustapha M., Salem N., El-Zir E., Loiselet J., Petit C. (1999) Nat. Genet. 21, 363–369 [DOI] [PubMed] [Google Scholar]
- 9. Bansal D., Miyake K., Vogel S. S., Groh S., Chen C. C., Williamson R., McNeil P. L., Campbell K. P. (2003) Nature 423, 168–172 [DOI] [PubMed] [Google Scholar]
- 10. Doherty K. R., Cave A., Davis D. B., Delmonte A. J., Posey A., Earley J. U., Hadhazy M., McNally E. M. (2005) Development 132, 5565–5575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Doberstein S. K., Fetter R. D., Mehta A. Y., Goodman C. S. (1997) J. Cell Biol. 136, 1249–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kalderon N., Gilula N. B. (1979) J. Cell Biol. 81, 411–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Demonbreun A. R., Posey A. D., Heretis K., Swaggart K. A., Earley J. U., Pytel P., McNally E. M. (2010) FASEB J. 24, 1284–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Doherty K. R., Demonbreun A. R., Wallace G. Q., Cave A., Posey A. D., Heretis K., Pytel P., McNally E. M. (2008) J. Biol. Chem. 283, 20252–20260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pohl U., Smith J. S., Tachibana I., Ueki K., Lee H. K., Ramaswamy S., Wu Q., Mohrenweiser H. W., Jenkins R. B., Louis D. N. (2000) Genomics 63, 255–262 [DOI] [PubMed] [Google Scholar]
- 16. Caplan S., Naslavsky N., Hartnell L. M., Lodge R., Polishchuk R. S., Donaldson J. G., Bonifacino J. S. (2002) EMBO J. 21, 2557–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. George M., Ying G., Rainey M. A., Solomon A., Parikh P. T., Gao Q., Band V., Band H. (2007) BMC Cell Biol. 8, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Grant B. D., Caplan S. (2008) Traffic 9, 2043–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guilherme A., Soriano N. A., Furcinitti P. S., Czech M. P. (2004) J. Biol. Chem. 279, 40062–40075 [DOI] [PubMed] [Google Scholar]
- 20. Park M., Penick E. C., Edwards J. G., Kauer J. A., Ehlers M. D. (2004) Science 305, 1972–1975 [DOI] [PubMed] [Google Scholar]
- 21. Park S. Y., Ha B. G., Choi G. H., Ryu J., Kim B., Jung C. Y., Lee W. (2004) Biochemistry 43, 7552–7562 [DOI] [PubMed] [Google Scholar]
- 22. Rotem-Yehudar R., Galperin E., Horowitz M. (2001) J. Biol. Chem. 276, 33054–33060 [DOI] [PubMed] [Google Scholar]
- 23. Shao Y., Akmentin W., Toledo-Aral J. J., Rosenbaum J., Valdez G., Cabot J. B., Hilbush B. S., Halegoua S. (2002) J. Cell Biol. 157, 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Daumke O., Lundmark R., Vallis Y., Martens S., Butler P. J., McMahon H. T. (2007) Nature 449, 923–927 [DOI] [PubMed] [Google Scholar]
- 25. de Beer T., Carter R. E., Lobel-Rice K. E., Sorkin A., Overduin M. (1998) Science 281, 1357–1360 [DOI] [PubMed] [Google Scholar]
- 26. de Beer T., Hoofnagle A. N., Enmon J. L., Bowers R. C., Yamabhai M., Kay B. K., Overduin M. (2000) Nat. Struct. Biol. 7, 1018–1022 [DOI] [PubMed] [Google Scholar]
- 27. Enmon J. L., de Beer T., Overduin M. (2000) Biochemistry 39, 4309–4319 [DOI] [PubMed] [Google Scholar]
- 28. Naslavsky N., Boehm M., Backlund P. S., Jr., Caplan S. (2004) Mol. Biol. Cell 15, 2410–2422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Letunic I., Copley R. R., Pils B., Pinkert S., Schultz J., Bork P. (2006) Nucleic Acids Res. 34, D257–D260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Davis D. B., Delmonte A. J., Ly C. T., McNally E. M. (2000) Hum. Mol. Genet. 9, 217–226 [DOI] [PubMed] [Google Scholar]
- 31. Patel P., Harris R., Geddes S. M., Strehle E. M., Watson J. D., Bashir R., Bushby K., Driscoll P. C., Keep N. H. (2008) J. Mol. Biol. 379, 981–990 [DOI] [PubMed] [Google Scholar]
- 32. Therrien C., Dodig D., Karpati G., Sinnreich M. (2006) J. Neurol. Sci. 250, 71–78 [DOI] [PubMed] [Google Scholar]
- 33. Yaffe D., Saxel O. (1977) Nature 270, 725–727 [DOI] [PubMed] [Google Scholar]
- 34. Salcini A. E., Confalonieri S., Doria M., Santolini E., Tassi E., Minenkova O., Cesareni G., Pelicci P. G., Di Fiore P. P. (1997) Genes Dev. 11, 2239–2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mintz L., Galperin E., Pasmanik-Chor M., Tulzinsky S., Bromberg Y., Kozak C. A., Joyner A., Fein A., Horowitz M. (1999) Genomics 59, 66–76 [DOI] [PubMed] [Google Scholar]
- 36. Kieken F., Jović M., Tonelli M., Naslavsky N., Caplan S., Sorgen P. L. (2009) Protein. Sci. 18, 2471–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Peuvot J., Schanck A., Lins L., Brasseur R. (1999) J. Theor. Biol. 198, 173–181 [DOI] [PubMed] [Google Scholar]
- 38. Chapman E. R., Hanson P. I., An S., Jahn R. (1995) J. Biol. Chem. 270, 23667–23671 [DOI] [PubMed] [Google Scholar]
- 39. Brose N., Petrenko A. G., Südhof T. C., Jahn R. (1992) Science 256, 1021–1025 [DOI] [PubMed] [Google Scholar]
- 40. Perin M. S., Fried V. A., Mignery G. A., Jahn R., Südhof T. C. (1990) Nature 345, 260–263 [DOI] [PubMed] [Google Scholar]
- 41. Bai J., Wang P., Chapman E. R. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 1665–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Davis D. B., Doherty K. R., Delmonte A. J., McNally E. M. (2002) J. Biol. Chem. 277, 22883–22888 [DOI] [PubMed] [Google Scholar]
- 43. Guilherme A., Soriano N. A., Bose S., Holik J., Bose A., Pomerleau D. P., Furcinitti P., Leszyk J., Corvera S., Czech M. P. (2004) J. Biol. Chem. 279, 10593–10605 [DOI] [PubMed] [Google Scholar]
- 44. Sharma M., Giridharan S. S., Rahajeng J., Naslavsky N., Caplan S. (2009) Mol. Biol. Cell 20, 5181–5194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hung R. J., Yazdani U., Yoon J., Wu H., Yang T., Gupta N., Huang Z., van Berkel W. J., Terman J. R. (2010) Nature 463, 823–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Beuchle D., Schwarz H., Langegger M., Koch I., Aberle H. (2007) Mech. Dev. 124, 390–406 [DOI] [PubMed] [Google Scholar]
- 47. Fischer J., Weide T., Barnekow A. (2005) Biochem. Biophys. Res. Commun. 328, 415–423 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.