Abstract
ATP7B is a P-type ATPase involved in copper transport and homeostasis. In experiments with microsomes isolated from COS-1 cells or HepG2 hepatocytes sustaining ATP7B heterologous expression, we found that ATP7B utilization of ATP includes autophosphorylation of an aspartyl residue serving as ATPase catalytic intermediate as well as phosphorylation of serine residues by protein kinase D (PKD). The latter was abolished by specific PKD inhibition with CID755673. The presence of PKD protein in the microsomal fraction was demonstrated by Western blotting. PKD is a serine/threonine kinase that associates with the trans-Golgi network, regulating fission of transport carriers destined to the cell surface. Parallel studies on cultured cells showed that nascent WT ATP7B transits to the Golgi complex where it undergoes serine phosphorylation by PKD. Misfolded ATP7B protein (especially if subjected to deletions) underwent proteasome-mediated degradation, which provides effective quality control. Inhibition of proteasome-mediated degradation with MG132 yielded additional, but nonfunctional protein. On the other hand, serine phosphorylation protected WT ATP7B from degradation. Protection was enhanced by PKD activation with phorbol esters and limited by PKD inhibition with CID75673. As a final step, phosphorylated ATP7B was transferred from the Golgi complex to cytosolic trafficking vesicles. Phosphorylation and trafficking were completely prevented by mutations of critical copper binding sites, demonstrating copper dependence of both PKD-assisted phosphorylation and trafficking. ATP7B trafficking was markedly reduced by the Ser-478/481/1121/1453 to Ala mutation. We conclude that PKD plays a key role in copper-dependent serine phosphorylation, permitting high levels of ATP7B protein expression and trafficking.
Keywords: ATPases, Phosphorylation Enzymes, Protein Kinases, Serine/Threonine Protein Kinase, Trafficking
Introduction
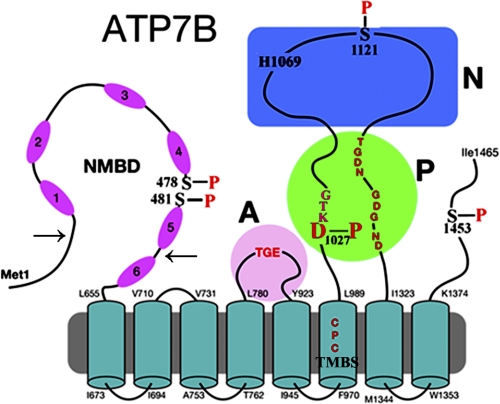
ATP7B is a member of the P1B-ATPase subfamily, playing important roles in delivery of copper to nascent metalloproteins and export of excessive copper from the cell (1). ATP7B is involved in the etiology of Wilson disease (2). The ATP7B protein comprises a transmembrane region with eight membrane-spanning segments and a putative transmembrane copper binding site (TMBS)2 in a location corresponding to the cation binding/transport site of other P-ATPases (see Fig. 1). In addition to the A (“actuator”), N (ATP binding), and P (intermediate catalytic phosphorylation) domains that are common to other P-type ATPases, the ATP7B protein includes an N metal binding domain (N terminus extension) (NMBD) with six additional copper binding sites (see Fig. 1). These sites are referred to with numbers from 1 to 6, starting from the N terminus. Furthermore, the presence of serine residues undergoing phosphorylation appears to be relevant in regulating expression and trafficking (3, 4). We identified (5) by mass spectrometry serine residues undergoing phosphorylation as Ser-478 and Ser-481 (NMBD), Ser-1121 (N domain), and Ser-1453 (C-terminal tail).
FIGURE 1.
Two-dimensional folding model of ATP7B sequence. The diagram shows eight transmembrane segments, including a copper binding site (TMBS). The extramembranous region comprises a nucleotide binding domain (N) with the His-1069 residue whose mutation is frequently found in Wilson disease; the P domain with several residues (in red) conserved in P-type ATPases where Asp-1027 undergoes phosphorylation to form the catalytic phosphoenzyme intermediate (EP), the A domain with the TGE conserved sequence involved in catalytic assistance of EP hydrolytic cleavage, the NMBD with six copper binding sites, and a C-terminal chain. Note that serines shown to be phosphorylated (5) reside within flexible loops of the protein (see “Discussion”). These are Ser-478 and Ser-481 (NMBD), Ser-1121 (N domain), and Ser-1453 (C-terminal tail). The arrows on the NMBD delimit the segment that was deleted to obtain the Δ5 construct.
We report here experiments with COS-1 cells or HepG2 hepatocytes sustaining heterologous expression of ATP7B. Using the microsomal fraction derived from these cells, we obtained kinetic and stoichiometric resolution of ATP7B phosphorylation, distinguishing alkali-labile (i.e. aspartyl phosphate catalytic intermediate) and alkali-stable (i.e. serine residues) components and allowing quantitative analysis of the two components. We demonstrate that alkali stable phosphorylation is specifically dependent on protein kinase D (PKD) activity. Furthermore, observing the cells in culture, we found that the level of ATP7B protein and its trafficking are regulated by copper and PKD-dependent phosphorylation of serine residues. PKD is a serine/threonine kinase additional to and distinct from other second message-stimulated kinases, such as protein kinases A (PKA), B, and C (PKC) and Ca2+/calmodulin-dependent kinases (6–8). An important feature of PKD is its association with the trans-Golgi network and its influence on fission of vesicular carriers destined to the cell surface (9–11). In this study, we demonstrate the involvement of PKD in phosphorylation of ATP7B, its determining influence on expression of functional ATP7B, and its association with cytosolic trafficking vesicles (CTV). We also demonstrate the importance of proteasome-mediated degradation in quality control of the nascent ATP7B protein.
MATERIALS AND METHODS
Adenoviral Vector Construction
Recombinant adenovirus vector (rAdATP7Bmyc) containing CMV promoter driven WT human ATP7B cDNA (obtained from Origene) and rAdSERCA1 encoding rabbit skeletal muscle SERCA1 (12) fused with a 3′ c-myc tag were constructed as explained previously (5). Site-directed mutations in ATP7B included D1027N (in the DKTG motif of the N domain); C983A and C985A (in the TMBS); C575A and C578A (in the sixth copper site of the NMBD); and S478A, S481A, S1121A, and S1453A (specific phosphorylation sites observed by phosphoproteomics) and were produced in pShuttleCMV-ATP7Bmyc plasmid using the QuikChange II site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The mutations were confirmed by sequencing, and related plasmids were used for construction of rAdATP7Bmyc mutants. The primers used for the mutation or deletion are given in Table 1. A construct (Δ5) with deletion of the NMBD segment intervening between amino acid residues 69–574, including the first five copper sites (Fig. 1), was also produced.
TABLE 1.
Primers used for mutation or deletion constructs of ATP7B
| Primer name | Sequence |
|---|---|
| D1027N forward | 5′-CAAGATAAAGACTGTGATGTTTAACAAGACTGGCACCATTACCC-3′ |
| D1027N reverse | 5′-GGGTAATGGTGCCAGTCTTGTTAAACATCACAGTCTTTATCTTG-3′ |
| C983A and C985A forward | 5′-TGCTGTGCATTGCCGCCCCCGCCTCCCTGGGGCTGG-3′ |
| C983A and C985A reverse | 5′-CCAGCCCCAGGGAGGCGGGGGCGGCAATGCACAGCA-3′ |
| C575A and C578A forward | 5′-CAATCACAGGGATGACCGCCGCGTCCGCTGTCCACAACATAGAGT-3′ |
| C575A and C578A reverse | 5′-ACTCTATGTTGTGGACAGCGGACGCGGCGGTCATCCCTGTGATTG-3′ |
| S478/481A forward | 5′-GGACATCTTGGCAAAGGCCCCACAAGCAACCAGAGCAGTGGCAC-3′ |
| S478/481A reverse | 5′-GTGCCACTGCTCTGGTTGCTTGTGGGGCCTTTGCCAAGATGTCC-3′ |
| S1121A forward | 5′-CACAGTGAGCGCCCTTTGGCTGCACCGGCCAGTCACCTGA-3′ |
| S1121A reverse | 5′-TCAGGTGACTGGCCGGTGCAGCCAAAGGGCGCTCACTGTG-3′ |
| S1453A forward | 5′-ACGATGATGGGGACAAGTGGGCTCTGCTCCTGAATGGCAGGG-3′ |
| S1453A reverse | 5′-CCCTGCCATTCAGGAGCAGAGCCCACTTGTCCCCATCATCGT-3′ |
| Δ5 forward | 5′-GTCAGGATCTTGGGCATGACTGTGTCTAACATAGAAAG-3′ |
| Δ5 reverse | 5′-CTTTCTATGTTAGACACAGTCATGCCCAAGATCCTGAC-3′ |
Microsomal Preparation and Immunostaining of Recombinant ATP7B or Mutants
Microsomes were prepared from COS-1 cells or HepG2 cells infected with adenoviral vector containing WT ATP7B or mutant ATP7B cDNA or SERCA1 cDNA. The protein expression was evaluated by SDS gel electrophoresis or Western blotting as explained previously (5). Western blotting chemiluminescence was quantified using an Alpha Innotech Fluorchem 8900 Imager and AlphaEaseFC Software version 3.2.3.
For immunostaining, COS-1 cells were grown to 60–70% confluence in DMEM containing 10% FBS in a CO2 incubator at 37 °C on a sterile coverslip that was placed in a 35-mm culture dish. The cells were infected with optimal rAdATP7Bmyc viral titers as determined by preliminary cytotoxicity and expression titrations. Following incubation for 2 h in a CO2 incubator, the cells were treated with 200 μm CuCl2 overnight or kept untreated and then were fixed with 3.7% (v/v) paraformaldehyde in PBS for 20 min followed by permeabilization with 0.1% Triton X-100 in PBS for 15 min at room temperature. The permeabilized COS-1 cells were blocked with 10% horse serum in PBS for 1 h at room temperature followed by incubation with specific primary antibodies (mouse monoclonal antibody 9E10 to c-myc tag and rabbit polyclonal antibody to Giantin (Golgi marker, ab24586)) at 4 °C overnight in block solution. The primary antibodies were detected by incubating the cells with secondary antibodies (goat anti-mouse Alexa Fluor 488 and donkey anti-rabbit Cy5) in the block solution for 2 h at room temperature. The nuclei of the infected cells were stained with propidium iodide (10 μg/ml in PBS) for 10 min. Each step was followed by rinsing three times with PBS. Finally, the stained cells were evaluated for ATP7Bmyc expression using a confocal laser scanning microscope (Nikon, Eclipse TE2000-U).
32P-Phosphoenzyme Formation by Incubation of Microsomes with [γ-32P]ATP
Microsomes derived from COS-1 cells expressing ATP7B (50 μg of microsomal protein/ml) were incubated with 50 μm [γ-32P]ATP (100 μCi/μmol) at 30 °C in a reaction mixture containing 50 mm MES triethanolamine, pH 6.0, 300 mm KCl, 10 mm DTT, 3 mm MgCl2, and 5 μm CuCl2. Microsomes derived from COS-1 cells expressing SERCA1 (50 μg of microsomal protein/ml) were incubated with 50 μm [γ-32P]ATP (100 μCi/μmol) at 5 °C in a reaction mixture containing 50 mm MOPS, pH 7, 80 mm KCl, 3 mm MgCl2, and 5 μm CaCl2. Samples were quenched at serial times with 5% trichloroacetic acid. The quenched samples were pelleted by centrifugation at 5000 rpm for 5 min, washed with 0.125 n perchloric acid, and finally resuspended in pH 8.3 (half of the sample) or pH 6.3 loading buffer (remaining half of the sample) and separated by Laemmli (13) (alkaline buffer) or Weber-Osborn (14) (acid buffer) gel electrophoresis. The gels were dried and exposed to a phosphor screen followed by scanning on a Typhoon scanner (Amersham Biosciences) for stoichiometric determination of phosphoprotein relative to three [γ-32P]ATP standards placed on the gels. Comparative experiments were performed with microsomes derived from COS-1 cells infected with rAdATP7Bmyc (WT and mutants) and cells infected with rAdGFP (“sham”).
Alternatively, 32P-phosphoenzyme formation by utilization of 32P-phosphoenzyme was analyzed in the absence or presence of 20 μm H89 (PKA inhibitor), 250 nm GF10930X (PKC inhibitor), 20 μm PNU 112455A (cyclin-dependent kinase (cdK2/5) inhibitor), 20 μm TDZD8 (glycogen synthase kinase 3β (GSK-3β) inhibitor), or 20 μm CID755673 (PKD inhibitor). 32P-Phosphoenzyme formation was carried out for 30 s at 30 °C.
Ex Vivo Analysis of ATP7B Expression Level in COS-1 Cells Infected with Adenoviral Vector
COS-1 cells were grown to 80% confluence in DMEM containing 10% FBS in a CO2 incubator at 37 °C in a 35-mm culture dish. The cells were infected with adenovirus vector (∼50 multiplicity of infection) expressing WT ATP7B or sixth NMBD copper site, TMBS copper site, or S478A/S481A/S1121A/S1453A mutant. Following a 16-h incubation, the infected cells were treated with 50 μm protein kinase D inhibitor CID755673 (Tocris Biosciences) or 150 nm protein kinase D activator phorbol 12-myristate 13-acetate (PMA) (Sigma) or were kept untreated (see Fig. 6) for an additional 24 h. The cells were then harvested and lysed in cell resuspension buffer (50 mm K+-MOPS, pH 7.0, 1 mm Na-EDTA, 10 mm NaF, and 300 mm sucrose) containing 0.2% SDS and protease inhibitors (Roche Applied Science) by sonication. The ATP7B expression level in the lysate was evaluated by Western blot using monoclonal antibody (9E10) against c-myc tag. Simultaneously, actin was stained using rabbit anti-actin antibody (Sigma) as a loading control. Western blotting chemiluminescence was quantified using an Alpha Innotech Fluorchem 8900 Imager and AlphaEaseFC Software version 3.2.3.
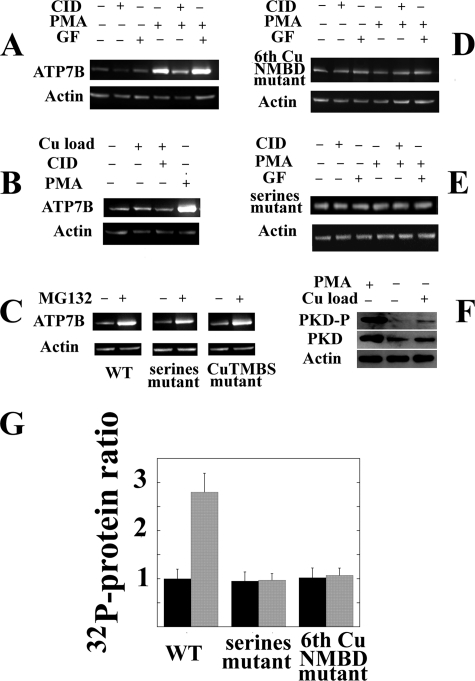
FIGURE 6.
Levels of heterologously expressed ATP7B and endogenous PKD in total lysates of COS-1 cells following experimental variables in vivo. Sixteen hours following delivery of WT ATP7B (A, B, E, and F), 6thCuNMBD mutant (C), Ser-478, Ser-481, Ser-1121, and Ser-1453 mutant (D and E), or CuTMBS mutant (E) cDNA with adenovirus vector, 50 μm CID755673 (CID; PKD inhibitor), 150 nm PMA (PKC activator), 250 nm GF10930X (GF; PKC inhibitor), 20 μm MG132 (proteasome-mediated degradation inhibitor), or 200 μm CuCl2 was added to the culture medium. The cells were harvested 24 h later, and ATP7B expression was evaluated by Western blots of total cell lysates using c-myc tag antibodies (A, B, C, D, and E). Alternatively (F), the levels of endogenous PKD in its phosphorylated (“activated”) form or total PKD protein were evaluated with specific antibodies. Actin expression was also evaluated in the same samples. G, ratios (means and S.D. obtained from four experiments) of ATP7B protein levels in lysates of COS-1 cells expressing WT ATP7B, serine mutant, and sixth copper site NMBD mutant in the absence (−) or presence (+) of 150 nm PMA (PKD activator). Note that PMA increases the level of WT but not the levels of mutants. Error bars represent standard deviations derived from five separate determinations.
Immunodetection of Protein Kinase D in COS-1 Cell Lysate and in Microsomes
Microsomal protein prepared from COS-1 cells infected with adenoviral vector encoding ATP7B or uninfected COS-1 cells were separated by SDS gel electrophoresis. The SDS-PAGE-separated protein was subjected to Western blotting, and the presence of protein kinase D was detected using rabbit anti-PKD antibody (Cell Signaling Technology). The presence of PKD was also evaluated in uninfected COS-1 cell lysate. The cell lysate was prepared as detailed above.
RESULTS
32P-Phosphoprotein Formation by Utilization of [32P]ATP in Vitro: Experiments with Microsomes Obtained from Cells Sustaining Expression of ATP7B
Electrophoretic analysis of microsomes obtained from cells sustaining expression of WT ATP7B showed that, under favorable conditions, the recombinant protein accounts for ∼5% of the total microsomal protein (Fig. 2). Addition of [γ-32P]ATP yielded phosphorylation of the ATP7B protein within the second time scale at 30 °C. The resulting 32P-phosphoprotein included alkali-labile and alkali-stable components that can be clearly distinguished by electrophoresis in acid or alkaline medium (Fig. 3A). The alkali-labile component reached maximal levels within 5 s, whereas the alkali-stable phosphorylation was slower and reached higher levels with prolonged incubation. It should be pointed out that microsomes obtained from COS-1 cells infected with empty (no ATP7B cDNA) viral vectors did not exhibit any phosphoprotein following incubation with [γ-32P]ATP in control experiments.
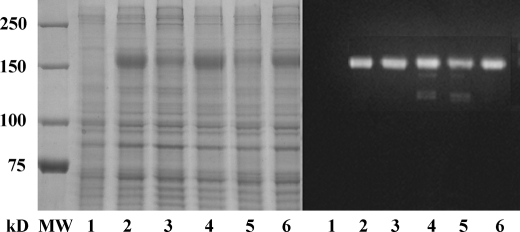
FIGURE 2.
Electrophoretic analysis (Laemmli) of microsomal fractions derived from COS-1 cells expressing WT and mutant ATP7B. The protein bands were stained with Coomassie Blue (left) or evidenced by Western blots obtained with antibodies against the c-myc tag in the expressed protein (right). Lane 1 was obtained with fractions derived from COS-1 cells infected with adenovirus vector carrying GFP cDNA (sham), lane 2 corresponds to WT ATP7B, lane 3 corresponds to the S478A/S481A/S1121A/S1453A mutant, lane 4 corresponds to the 6thCuNMBD mutant, lane 5 corresponds to the D1027N mutant, and lane 6 corresponds to the CuTMBS mutant. Note the different levels of expression, which were determined by chemiluminescence and taken into account to compare the stoichiometry of phosphorylation.
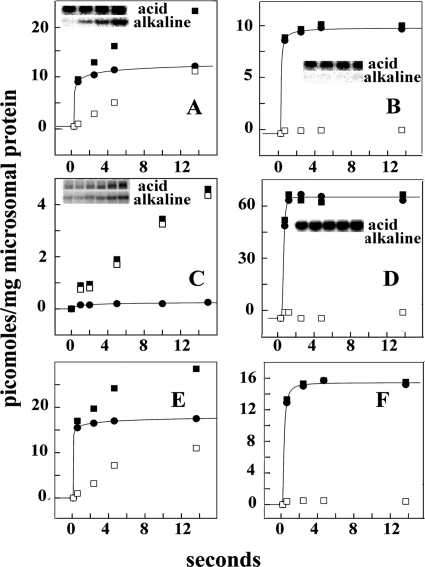
FIGURE 3.
Phosphorylation of WT ATP7B (A, B, E, and F), ATP7B D1027N mutant (C), and SERCA1 (D) in absence (A, C, D, and E) and presence (B and F) of PKD inhibitor. Reactions performed with microsomes obtained from COS-1 cells (A, B, C, and D) or HepG2 hepatocytes (E and F) sustaining heterologous expression of WT ATP7B (A, B, E, and F), ATP7B D1027N mutant (C), or SERCA1 (D) were incubated with 50 μm [γ-32P]ATP at 30 °C in the absence (A, C, D, and E) or presence (B and F) of 20 μm CID755673 (PKD inhibitor) as explained under “Materials and Methods.” Electrophoresis in acid buffer or alkaline buffer was then performed to distinguish total 32P-phosphoprotein (■) from alkali-resistant 32P-phosphoprotein (serine and/or threonine; □). The difference is considered alkali-labile 32P-phosphoprotein (aspartate; ●) and attributed to formation of phosphorylated enzyme intermediate. “Acid” and “alkaline” refer to the media used for resuspension of samples and electrophoresis. The stoichiometry of phosphoprotein refers to total microsomal protein. A, WT ATP7B from COS-1 cells. B, WT ATP7B from COS-1 cells with 20 μm CID755673. C, D1027N ATP7B from COS-1 cells. D, SERCA1 from COS-1 cells. E, WT ATP7B from HepG2 hepatocytes. F, WT ATP7B from HepG2 hepatocytes with 20 μm CID755673. Note the different stoichiometric scale of the vertical axis in the left and right panels. Electrophoresis gel images correspond to sequential samples obtained within the time scale shown in the horizontal axis.
We found that alkali-stable phosphorylation was totally prevented by specific PKD inhibition with 20 μm CID755673, whereas alkali-labile phosphorylation was not affected (Fig. 3B). In fact, the alkali-labile component is related to phosphorylation of a catalytic aspartyl residue conserved in all P-type ATPases and was not obtained with ATP7B subjected to a single D1027 mutation (Fig. 3C). It is noteworthy that only alkali-labile (i.e. catalytic phosphoenzyme intermediate), and no alkali-stable phosphorylation was formed by other P-type ATPases, such as SERCA1 (15, 16), even when obtained by the same recombinant method used for ATP7B (Fig. 3D). On the other hand, both alkali-labile and alkali-stable components were also obtained with ATP7B expressed in cells other than COS-1, such as HepG2 hepatocytes in which formation of the alkali-stable component was again PKD-dependent (Fig. 3, E and F).
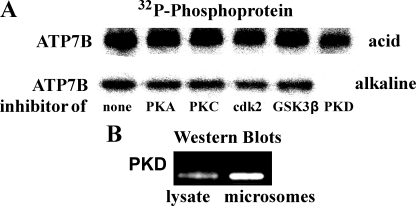
The effects of various protein kinase inhibitors (added in vitro to microsomes derived from COS-1 cells) on ATP7B phosphorylation by [γ-32P]ATP are shown in Fig. 4. It is clear that only the PKD inhibitor CID755673 prevented formation of alkali-stable phosphoprotein (Fig. 4A, lower lane). When electrophoresis was run at acid pH, the radioactive band intensity was proportionally reduced because the alkali-stable phosphorylation was prevented by PKD inhibition, whereas alkali-labile phosphoprotein was still present (Fig. 4A, upper lane). Known inhibitors of other kinases, such as PKA, PKC, cdK2/5, and GSK-3β, were not effective. Most importantly, the presence of PKD protein in the total lysate and microsomal fraction of COS-1 cells could be demonstrated by Western blotting (Fig. 4B). PKD is associated with the trans-Golgi network (9–11), and trans-Golgi network fragments are evidently associated with the microsomal fraction of COS-1 cells.
FIGURE 4.
Demonstration of PKD activity and presence in microsomal fraction and total lysate of COS-1 cells. A, microsomes obtained from COS-1 cells sustaining heterologous ATP7B expression were incubated at 30 °C for 30 s with 50 μm [γ-32P]ATP in the absence or presence of 20 μm H89 (PKA inhibitor), 250 nm GF10930X (PKC inhibitor), 20 μm PNU 112455A (cdK2/5 inhibitor), 20 μm TDZD8 (GSK-3β inhibitor), or 20 μm CID755673 (PKD inhibitor). Phosphorylation and electrophoresis were as described for Fig. 3. B, Western blots obtained with specific antibodies reveal the presence of PKD in total cell lysate and in the microsomal fraction obtained from COS-1 cells. The same amount (20 μg) of protein from total lysate or microsomal fraction was subjected to electrophoresis by the Laemmli method (13), and immunodetection was as described under “Materials and Methods.” Actin standards are not shown because of the absence of actin in the microsomal fraction.
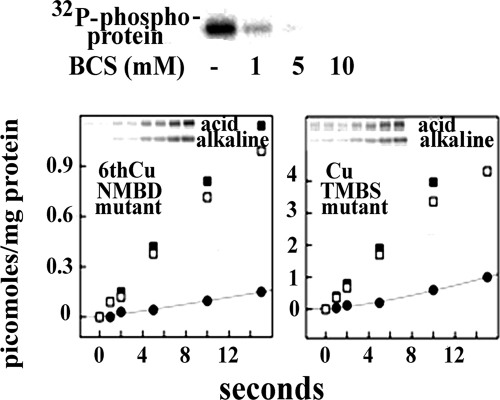
Addition of CuCl2 (up to 200 μm in the presence of 10 mm DTT) did not increase significantly the level of ATP7B phosphorylation obtained by incubation of microsomes with [γ-32P]ATP. On the other hand, chelation of contaminant copper with bathocuproine disulfonate prevented phosphorylation of ATP7B (Fig. 5). To distinguish the effect of copper binding to NMBD or TMBS, we studied the effects of mutations expected to prevent copper binding. We found that, even in the presence of copper (μm range as in the experiments shown in Fig. 3), mutation of the sixth NMBD copper site interfered with alkali-labile as well as alkali-stable phosphorylation (Fig. 5). On the other hand, mutation of the TMBS copper site interfered with alkali-labile phosphorylation (i.e. catalytic intermediate) but not with alkali-stable phosphorylation (Fig. 5; note the different stoichiometric scale of the vertical axis in the left and right panels), demonstrating that copper binding at the TMBS site has a specific effect on catalytic activation.
FIGURE 5.
Copper dependence of ATP7B phosphorylation following addition of [γ-32P]ATP to microsomal fraction containing ATP7B: effects of site-directed mutations. Top, microsomal protein was subjected to phosphorylation and acid electrophoresis as described for Fig. 3 for determination of total (aspartate and serine) phosphorylation, which is strongly inhibited by bathocuproine disulfonate (BCS). Lower quadrants, following phosphorylation of microsomes obtained from COS-1 cells expressing ATP7B mutants (as described for Fig. 1), electrophoretic analysis was performed in acid or alkaline medium to detect total phosphoprotein (■) and distinguish alkali-labile (●; aspartyl phosphate) and alkali-stable (□; phosphorylated serines) fractions. 6thCuNMBD mutant, C575A/C578A mutation at the sixth NMBD copper site; CuTMBS mutant, C983A and C985A mutation of the TMBS. Note the different stoichiometric scale on the vertical axis of the two panels, indicating inhibition of aspartate phosphorylation in both mutants but inhibition of serine phosphorylation only in the sixth NMBD copper site mutant.
Effects of Experimental Variables in Vivo on Cells Sustaining Expression of ATP7B
In a parallel series of experiments, we tested the effects of kinase inhibition or activation by adding specific compounds to cell culture media (in vivo) and then determining the levels of expressed ATP7B by Western blotting on whole cell homogenates. Fig. 6A shows that the expression level of ATP7B was slightly reduced by PKD inhibition with CID755673 but was markedly increased by protein kinase activation with PMA, which we demonstrated directly by detection of the phosphorylated PKD “active form” (Fig. 6F). Most importantly, this increase was totally prevented by PKD inhibition with CID755673. It is apparent that activation of PKD, either directly or through a kinase cascade (8), is responsible for the observed increase in the level of expressed ATP7B. GF10930X (a PKC inhibitor) did not interfere with the increase in ATP7B level produced by the kinase activator PMA (Fig. 6A).
It should be pointed out that endogenous copper was present in all these experiments, and addition of CuCl2 (up to 200 μm) to the culture media did not increase significantly the expression level even though the increase was still produced by PMA (Fig. 6B). Therefore, endogenous copper present in culture media was sufficient to sustain the effect of PKD on the ATP7B expression level.
We then tested whether PKD activation has any effect on expression of an ATP7B construct subjected to a mutation of the sixth NMBD copper site that interferes with phosphorylation (Fig. 1). In fact, we found that the sixth NMBD copper mutation prevented (Fig. 6D) the increase in the level of expressed protein produced by PKD activation. This interference was also produced (Fig. 6E) by mutation of serine residues (Ser-478/481/1121/1453 to Ala). A quantitative determination of the effect of PKD activation on the level of expressed ATP7B is shown in Fig. 6G. It is also shown in Fig. 6G that PKD activation was not effective on the sixth copper site NMBD mutant and on the serines mutant. An interesting finding is that a significantly higher level of expressed ATP7B was obtained if N-[(phenylmethoxy)carbonyl]-l-leucyl-N-[(1S)-1-formyl-3-methylbutyl]-l-leucinamide (MG132), a known inhibitor of proteasome-mediated degradation (17–21), was added to the culture medium of cells sustaining ATP7B expression (Fig. 6C). The MG132 effect was also obtained when the CuTMBS or the serines mutants were used (Fig. 6C).
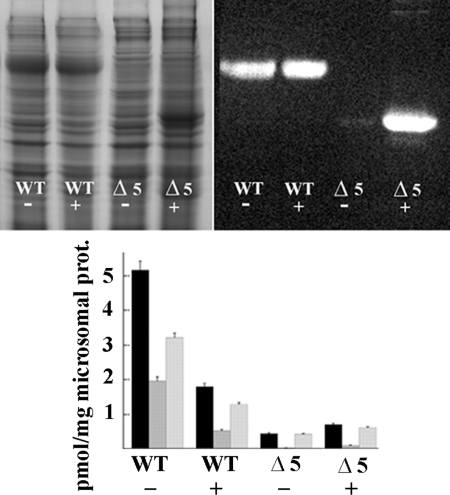
In Fig. 7, we show the ATP7B protein content and phosphorylation levels observed in microsomes (as opposed to total cells homogenates as in Fig. 6) obtained from COS-1 cells sustaining heterologous expression in the absence or presence of MG132. In this case, it is possible that some misfolded and aggregated protein was lost during microsomal preparation by multiple centrifugations. At any rate, it is apparent that MG132 had a much higher effect on the expression levels of an ATP7B construct subjected to deletion of an NMBD segment including the first five copper binding sites (Δ5). Although the quantity of Δ5 protein associated with microsomes of COS-1 cells infected with vector carrying Δ5 cDNA was negligible in control experiments, the amount of Δ5 protein recovered from cells exposed to MG132 was much higher and reached the same level as that of WT ATP7B (Fig. 7). Interestingly, the additional Δ5 protein recovered from cells exposed to MG132 was nonfunctional, and no additional phosphorylation by ATP was observed (Fig. 7). Furthermore, in the case of WT ATP7B, the level of protein recovered in the presence of MG132 was approximately the same as in the absence of MG132 but was evidently diluted with inactive protein that normally would be degraded by quality control. These experiments indicate that proteasome-mediated degradation of misfolded protein provides a very powerful quality control for ATP7B. Only protein exhibiting acceptable folding and function is allowed to remain.
FIGURE 7.
Protein levels and phosphorylation of WT and Δ5 ATP7B in microsomal fractions of COS-1 cells sustaining heterologous expression. The upper panels show gels (Laemmli) stained with Coomassie Blue for detection of all proteins (left) or evidenced by Western blots obtained with antibodies against the c-myc tag in the expressed protein (right). Note that Δ5 protein migrates faster due to its smaller size. The lower panel shows levels of phosphorylation (means and S.D. obtained from three experiments) detected by acid (black columns) or alkaline (second columns in each group) electrophoresis and net alkali-labile phosphoenzyme intermediate (third columns in each group). The presence of 5 μm MG132 (proteasome-mediated degradation inhibitor) in the culture media is denoted by the + sign. Note the large increase in Δ5 protein level in the presence of MG132. The additional WT and Δ5 proteins obtained in the presence of MG132 do not undergo phosphorylation. prot, protein. Error bars represent standard deviations derived from five separate determinations.
Cellular Trafficking of ATP7B
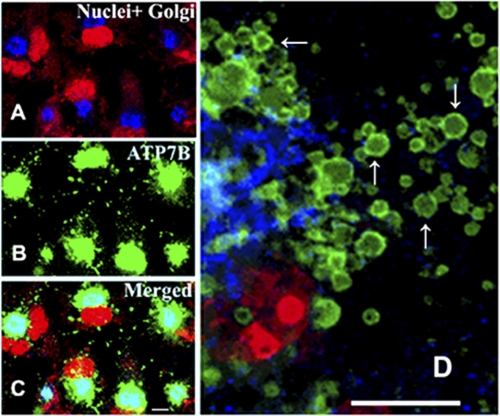
Trafficking of ATP7B is involved in copper homeostasis, yielding CTV that were identified as late endosomes by Harada et al. (22). In our experiments with COS-1 cells, the expressed ATP7B was initially transferred from an initial perinuclear location (i.e. endoplasmic reticulum) to the Golgi complex (Fig. 8A, blue stain, corner of the red stained nuclei). Immunostaining with specific antibodies demonstrated that in fact ATP7B (Fig. 8B, green stain) and Golgi complex are superimposed (Fig. 8C). Under basal conditions (i.e. endogenous copper present), formation of CTV including ATP7B protein was then observed. Addition of CuCl2 (200 μm) to the culture media produced more intense trafficking, and numerous CTV were noted exiting the Golgi complex (Fig. 8D). As the ATP7B expression level was not markedly increased by the 200 μm CuCl2 load (Fig. 6), it is apparent that high copper increases the association of ATP7B with membrane vesicles (CTV) in the cytoplasm.
FIGURE 8.
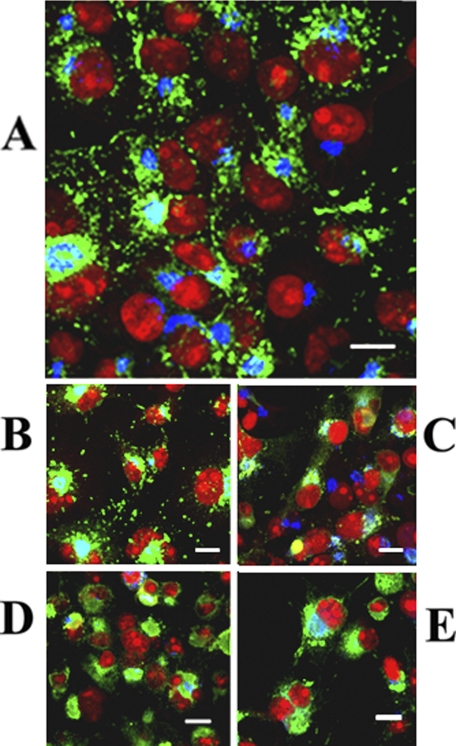
Immunostaining of COS-1 cells sustaining expression of WT ATP7B. The cells were stained simultaneously with antibodies specific for the Golgi marker protein Giantin (most evident in A) and ATP7B c-myc tag (most evident in B) 24 h following infection with adenovirus vector carrying WT ATP7B cDNA. Secondary antibodies were goat anti-mouse Alexa Fluor 488 for the ATP7B c-myc tag (green) and donkey anti-rabbit Cy5 for Golgi (blue). Red color indicating nuclei stained with propidium iodide (with nucleoli undergoing denser staining) is most evident in A and C. The lower panel (C) shows the merging of the stains, demonstrating colocalization of ATP7B with the Golgi marker in a distinct and polarized location relative to the nuclei. The right panel (D) shows an enlarged view of CTV (some indicated by arrows) containing ATP7B (green), budding from the Golgi region (blue), and trafficking throughout the cytoplasm of COS-1 cells exposed overnight to 200 μm CuCl2 1 day after infection with adenovirus vector for delivery of WT ATP7B cDNA. Scale bars, 15 μm.
We also studied whether specific mutations interfere with formation of cytosolic vesicles in cells exposed to a copper load (200 μm CuCl2 added to culture media 2 h following infection with adenovirus vector). We found that mutation of Asp-1027 (involved in formation of phosphoenzyme intermediate upon utilization of ATP) did not interfere with formation of CTV (Fig. 9, compare A and B). On the other hand, mutation of serines (Ser-478/481/1121/1453 to Ala) whose involvement in phosphorylation was previously demonstrated by mass spectrometry (5) interfered with formation of CTV (Fig. 9C). We also found that mutation of the transmembrane (TMBS) copper site, which was required for catalytic activation but not for PKD-assisted phosphorylation (Fig. 5), interfered with formation of CTV (Fig. 9D). Interference was also produced (Fig. 9E) by mutation (C575A/C578A) of the sixth NMBD copper site, which interferes with both PKD-assisted phosphorylation and catalytic activation (Fig. 5).
FIGURE 9.
Intracellular distribution of ATP7B in COS-1 cells expressing WT ATP7B (A) or ATP7B subjected to mutations at Asp-1027 (B), at Ser-478, Ser-481, Ser-1121, and Ser-1453 (C), at the transmembrane domain (TMBS) copper site (D), or at the sixth NMBD copper site (E). All panels present different fields of cells treated identically. A copper load (200 μm) was added to the culture media 2 h following infection with adenovirus vector for delivery of ATP7B cDNA (WT or mutant). The cells were fixed 24 h later. Secondary antibodies were goat anti-mouse Alexa Fluor 488 for the ATP7B c-myc tag (green) and donkey anti-rabbit Cy5 for Golgi (blue). Red color indicates nuclei stained with propidium iodide. Scale bar, 10 μm.
It is then apparent that lack of serine phosphorylation interferes with the effect of PKD activation on the level of expressed ATP7B as well as with ATP7B trafficking and formation of CTV. In addition, the different effects of Asp-1027 and TMBS copper site mutations indicate that the cation-activated state (i.e. E1Cu) but not necessarily the state obtained following utilization of ATP (i.e. E1Cu-P) is required for formation of CTV.
DISCUSSION
We obtained kinetic and stoichiometric resolution of alkali-labile and alkali-stable ATP7B phosphorylation upon addition of [γ-32P]ATP to the microsomal fraction of COS-1 cells or HepG2 hepatocytes infected with adenovirus vector and sustaining heterologous expression of ATP7B. These findings were rendered possible by a relatively high yield (Fig. 2) of recombinant protein expressed in COS-1 cells or HepG2 hepatocytes. The high yield is due to efficient delivery of cDNA to all cells in culture by infection with adenovirus vector and not to excessive expression in each single cell. Furthermore, proteasome-mediated degradation (Fig. 7) provides a very efficient quality control whereby only functional protein is recovered as demonstrated by phosphorylation (Fig. 3) and active displacement of bound copper upon ATP utilization (23).
Alkali-labile phosphorylation can be unambiguously attributed to the conserved aspartate serving as catalytic intermediate in all P-type ATPases. On the contrary, alkali-stable phosphorylation, which does not commonly occur in other P-type ATPases, involves serine residues (Fig. 1) that were identified (5) by mass spectrometry.
Alkali-labile phosphorylation (catalytic intermediate) and alkali-stable phosphorylation (serines) were both copper-dependent (Fig. 5) but through different mechanisms. Formation of phosphoenzyme intermediate was inhibited by mutation of the TMBS copper site, a location analogous to those required for specific cation binding and catalytic activation in other P-type ATPases. On the other hand, mutation of the sixth NMBD copper site inhibited both alkali-labile and alkali-stable phosphorylation.
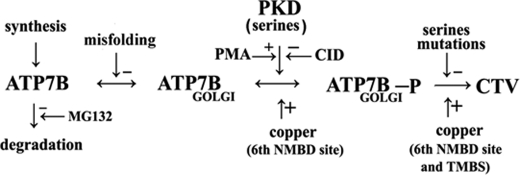
The alkali-stable phosphorylation of serines is specifically catalyzed by PKD as demonstrated by total interference by the PKD inhibitor CID755673 (Figs. 3 and 4) when incubation with [γ-32P]ATP was performed in vitro with microsomes derived from cells sustaining ATP7B expression. Interestingly, PKD activation with PMA or conversely PKD inhibition with CID755673 in the cultured cells (in vivo) resulted in increased or decreased levels of expressed ATP7B (Fig. 6), respectively. In fact, an important feature of PKD is its association with the trans-Golgi network where it favors fission of transport carriers specifically destined to the cell surface (6–11). Our experiments show that the nascent ATP7B protein migrates first to the Golgi network and then undergoes trafficking with CTV. This steady state trafficking, favored by phosphorylation, removes the nascent ATP7B from proteasome-mediated degradation (17–21), which we found to be more prominent when ATP7B was subjected to NMBD deletions, providing a very effective quality control. Sequential distribution of the ATP7B species, as shown in Scheme 1, includes transfer of nascent ATP7B to the Golgi complex where PKD activation (favored by PMA and inhibited by CID755673) produces phosphorylation of serines, thereby reducing the species that undergoes proteasome-mediated degradation (inhibited by MG132).
SCHEME 1.
Sequential reactions for quality control and serine phosphorylation for ATP7B expressed in COS-1 cells. CID, CID755673.
Transfer to the Golgi network requires proper folding, and very effective degradation was observed when the Δ5 (large NMBD segment deleted) construct was used (Fig. 7). Even discrete mutations within the NMBD sequence were shown to interfere with targeting and Golgi retention of nascent ATP7B (24). In fact, the rather extended NMBD sequence is flexible and undergoes functionally relevant conformational interactions as shown by its structural resolution by NMR spectroscopy (25, 26), its propensity to interact with chaperone proteins (27), and the influence of its copper site redox status on overall conformation (28). Finally, we found that mutation of the serine residues undergoing phosphorylation interfered strongly with formation of CTV, underscoring the role of PKD-sustained phosphorylation in the final trafficking step. These serines reside within flexible loops relatively distant to each other, suggesting a requirement for approximation and proper folding for interaction with partner proteins.
A most important observation is that mutation of the sixth NMBD copper site interfered with PKD-assisted phosphorylation and formation of trafficking vesicles as well as catalytic activity (Figs. 5 and 9). Therefore, this copper site has a key role in overall ATP7B function. Its occupancy by copper may optimize folding and interactions of NMBD with other ATP7B domains and/or other proteins. It is of interest that mutation of the TMBS copper site involved in catalytic activation interfered with formation of CTV (Fig. 9) even though it did not interfere with phosphorylation of serines (Fig. 5). In this light, lack of interference by the Asp-1027 mutation with formation of CTV suggests that the cation-activated state (i.e. E1Cu) but not necessarily the state obtained following utilization of ATP (i.e. E1Cu-P) is required for formation of CTV. On the other hand, in the presence of ATP, it is clear that association of WT ATP7B with CTV would be instrumental in transfer and dissociation of bound copper.
It is clear that the involvement of protein kinase D in phosphorylation and trafficking of ATP7B demonstrates a unique feature of this copper ATPase that is not shared at all by other cation ATPases. Most cation (Ca2+ and Na+/K+) transport ATPases function from a stable location on a membrane, creating a 3–4-order of magnitude concentration gradient. On the other hand, ATP7B delivers copper by trafficking as well as ATP-dependent displacement of bound copper onto acceptor sites. Perhaps an appropriate name for ATP7B would be “transfer” rather than “transport” ATPase. We demonstrated here that trafficking is dependent on phosphorylation of specific serines, and protein kinase D plays a crucial role in this regard.
This work was supported, in whole or in part, by National Institutes of Health Grant RO301-69830 from the NHLBI.
- TMBS
- transmembrane copper binding site
- CID755673
- PKD inhibitor
- CTV
- cytosolic trafficking vesicles
- NMBD
- N metal binding domain (N terminus extension)
- PKD
- protein kinase D
- PMA
- phorbol 12-myristate 13-acetate.
REFERENCES
- 1. Lutsenko S., Barnes N. L., Bartee M. Y., Dmitriev O. Y. (2007) Physiol. Rev. 87, 1011–1046 [DOI] [PubMed] [Google Scholar]
- 2. Forbes J. R., Cox D. W. (1998) Am. J. Hum. Genet. 63, 1663–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vanderwerf S. M., Cooper M. J., Stetsenko I. V., Lutsenko S. (2001) J. Biol. Chem. 276, 36289–36294 [DOI] [PubMed] [Google Scholar]
- 4. Bartee M. Y., Ralle M., Lutsenko S. (2009) Biochemistry 48, 5573–5581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pilankatta R., Lewis D., Adams C. M., Inesi G. (2009) J. Biol. Chem. 284, 21307–21316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Valverde A. M., Sinnett-Smith J., Van Lint J., Rozengurt E. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 8572–8576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Van Lint J., Rykx A., Maeda Y., Vantus T., Sturany S., Malhotra V., Vandenheede J. R., Seufferlein T. (2002) Trends Cell Biol. 12, 193–200 [DOI] [PubMed] [Google Scholar]
- 8. Rozengurt E., Rey O., Waldron R. T. (2005) J. Biol. Chem. 280, 13205–13208 [DOI] [PubMed] [Google Scholar]
- 9. Maeda Y., Beznoussenko G. V., Van Lint J., Mironov A. A., Malhotra V. (2001) EMBO J. 20, 5982–5990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jamora C., Yamanouye N., Van Lint J., Laudenslager J., Vandenheede J. R., Faulkner D. J., Malhotra V. G. (1999) Cell 98, 59–68 [DOI] [PubMed] [Google Scholar]
- 11. Liljedahl M., Maeda Y., Colanzi A., Ayala I., Van Lint J., Malhotra V. (2001) Cell 104, 409–420 [DOI] [PubMed] [Google Scholar]
- 12. Liu Y., Pilankatta R., Lewis D., Inesi G., Tadini-Buoninsegni F., Bartolommei G., Moncelli M. R. (2009) J. Mol. Biol. 391, 858–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 14. Weber K., Osborn M. (1969) J. Biol. Chem. 244, 4406–4412 [PubMed] [Google Scholar]
- 15. MacLennan D. H., Brandl C. J., Korczak B., Green N. M. (1985) Nature 316, 696–700 [DOI] [PubMed] [Google Scholar]
- 16. Toyoshima C., Nakasako M., Nomura H., Ogawa H. (2000) Nature 405, 647–655 [DOI] [PubMed] [Google Scholar]
- 17. Meusser B., Hirsch C., Jarosch E., Sommer T. (2005) Nat. Cell Biol. 7, 766–772 [DOI] [PubMed] [Google Scholar]
- 18. Ding W. X., Yin X. M. (2008) Autophagy 4, 141–150 [DOI] [PubMed] [Google Scholar]
- 19. Vashist S., Ng D. T. (2004) J. Cell Biol. 165, 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ruddock L. W., Molinari M. (2006) J. Cell Sci. 119, 4373–4380 [DOI] [PubMed] [Google Scholar]
- 21. Vembar S. S., Brodsky J. L. (2008) Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Harada M., Kawaguchi T., Kumemura H., Terada K., Ninomiya H., Taniguchi E., Hanada S., Baba S., Maeyama M., Koga H., Ueno T., Furuta K., Suganuma T., Sugiyama T., Sata M. (2005) Am. J. Pathol. 166, 499–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tadini-Buoninsegni F., Bartolommei G., Moncelli M. R., Pilankatta R., Lewis D., Inesi G. (2010) FEBS Lett. 584, 4619–4622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Braiterman L., Nyasae L., Guo Y., Bustos R., Lutsenko S., Hubbard A. (2009) Am. J. Physiol. Gastrointest. Liver Physiol. 296, G433–G444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dmitriev O., Tsivkovskii R., Abildgaard F., Morgan C. T., Markley J. L., Lutsenko S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 5302–5307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Banci L., Bertini I., Cantini F., Inagaki S., Migliardi M., Rosato A. (2010) J. Biol. Chem. 285, 2537–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Banci L., Bertini I., Cantini F., Massagni C., Migliardi M., Rosato A. (2009) J. Biol. Chem. 284, 9354–9360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. LeShane E. S., Shinde U., Walker J. M., Barry A. N., Blackburn N. J., Ralle M., Lutsenko S. (2010) J. Biol. Chem. 285, 6327–6336 [DOI] [PMC free article] [PubMed] [Google Scholar]