Abstract
Niemann-Pick C1-like 1 (NPC1L1) is an essential protein for dietary cholesterol absorption. Nonsynonymous (NS) variants of NPC1L1 in humans have been suggested to associate with cholesterol absorption variations. However, information concerning the characteristics and mechanism of these variants in cholesterol uptake is limited. In this study, we analyzed the cholesterol uptake ability of the 19 reported NS variants of NPC1L1 identified from cholesterol low absorbers. Among these variants, L110F, R306C, A395V, G402S, T413M, R693C, R1214H, and R1268H could partially mediate cellular cholesterol uptake and were categorized as partially dysfunctional variants. The other 11 variants including T61M, N132S, D398G, R417W, G434R, T499M, S620C, I647N, G672R, S881L, and R1108W could barely facilitate cholesterol uptake, and were classified into the severely dysfunctional group. The partially dysfunctional variants showed mild defects in one or multiple aspects of cholesterol-regulated recycling, subcellular localization, glycosylation, and protein stability. The severely dysfunctional ones displayed remarkable defects in all these aspects and were rapidly degraded through the ER-associated degradation (ERAD) pathway. In vivo analyses using adenovirus-mediated expression in mouse liver confirmed that the S881L variant failed to localize to liver canalicular membrane, and the mice showed defects in biliary cholesterol re-absorption, while the G402S variant appeared to be similar to wild-type NPC1L1 in mouse liver. This study suggests that the dysfunction of the 19 variants on cholesterol absorption is due to the impairment of recycling, subcellular localization, glycosylation, or stability of NPC1L1.
Keywords: Cholesterol, Endocytosis, Genetic polymorphism, Glycosylation, Membrane Trafficking, NPC1L1, Cholesterol Absorption, Ezetimibe
Introduction
Exogenous cholesterol absorption is a major way for humans to obtain cholesterol and accounts for more than 50% of total renewed cholesterol in the human every day (1). The cholesterol absorption ability is genetically inherited and the absorption efficiency for dietary cholesterol varies from 29 to 80% among humans (2, 3).
Niemann-Pick C1-like 1 (NPC1L1)2 has been identified as a critical protein in the exogenous cholesterol absorption. In mice, NPC1L1 is selectively expressed in intestine, whereas in humans, NPC1L1 is highly expressed in both intestine and liver (4, 5). NPC1L1-deficient mice show dramatic reductions in dietary cholesterol absorption (5). Overexpression of NPC1L1 in mouse liver significantly decreases biliary cholesterol concentration and increases plasma cholesterol level, indicating that NPC1L1 in liver mediates cholesterol re-absorption from bile (6).
Our previous studies have revealed that NPC1L1 mediates cholesterol uptake through vesicular endocytosis, and it recycles between the endocytic recycling compartment (ERC) and plasma membrane (PM) in responding to alterations in cholesterol concentration (7). Depletion of cholesterol causes the transport of NPC1L1 from ERC to PM in a myosin Vb·Rab11a·Rab11-FIP2-dependent manner (8). Once reaching the PM, it can mediate cholesterol uptake through clathrin-AP2-mediated endocytosis (7). Ezetimibe, a hypocholesterolemic drug, binds NPC1L1 and blocks its endocytosis, thereby inhibiting cholesterol uptake (7, 9–11).
Human NPC1L1 protein contains 1,332 amino acid residues with 13 transmembrane domains (12). Recently, multiple rare sequence variants in NPC1L1 were found to be associated with variations in cholesterol absorption and plasma levels of LDL-Cholesterol (LDL-C) (13). Cohen et al. (13) used the campesterol: lathosterol ratio as a surrogate marker of cholesterol absorption and identified a series of nonsynonymous (NS) sequence variations of NPC1L1 associating with high or low cholesterol absorption. Further studies indicated that some sequence variations associated with low cholesterol absorption destabilized NPC1L1, resulting in lower steady-state levels of NPC1L1 in cells (14). Individuals with variations like I647N and R693C showed significant reductions in cholesterol absorption compared with unaffected family members (14). Four of the low cholesterol absorption-associated variants (D398G, T413M, R417W, and G434R) were also reported to have reduced expression levels, altered subcellular localization, or lower transport activity of cholesterol in Caco-2 cells (15). However, systematic examination and comparison of all these NPC1L1 variants are still lacking.
Here, we analyzed the cholesterol uptake ability, cholesterol-regulated recycling, intracellular localization, glycosylation, and protein stability of all the 19 reported NS variants of NPC1L1 in vitro. In addition, in vivo analyses in mouse liver were also performed to study their localization and the influence on biliary and hepatocytic cholesterol levels.
EXPERIMENTAL PROCEDURES
Animals, Materials, Primary Antibodies, and Plasmids
Animal protocols were approved by the institutional animal care and use committee of the Shanghai Institutes for Biological Sciences (Chinese Academy of Sciences). Experiments were performed with age-matched 8–10-week-old ICR strain mice (3–6 mice for each experimental group) housed in a specific pathogen-free animal facility in plastic cages in a temperature-controlled room (22 °C) with a 12-hour light/12-hour dark cycle. The mice were fed ad libitum a cereal-based rodent chow diet.
As for materials, we obtained horseradish peroxidase-conjugated donkey anti-mouse and anti-rabbit IgG from Jackson ImmunoResearch Laboratories; filipin, cycloheximide solution (CHX), monoclonal anti-MRP2 antibody (M3692) from Sigma; endoglycosidase H (Endo H), and peptide N-glycosidase F (PNGase F) from New England Biolabs, Inc.; MG-132 from Calbiochem; Alexa Fluor 555 donkey anti-mouse IgG from Invitrogen; mouse anti-calnexin antibody from upstate; methyl-β-cyclodextrin (CDX) from Cyclodextrin Technologies Development, Inc., and other reagents from previously described sources (7, 16, 17). Lipoprotein-deficient serum (LPDS, density >1.215 g/ml) was prepared from newborn calf serum by ultracentrifugation (18).
The coding region of human NPC1L1 was amplified from human liver cDNA by standard PCR and cloned into the pEGFP-N1 vector. The resulting plasmid encodes full-length human NPC1L1 fused with EGFP at the COOH terminus. The NPC1L1 variants and the S881L(K0) constructs, in which all the 13 lysines located in the intracellular part of NPC1L1(S881L) were substituted by arginines were generated using the QuikChange® Site-directed Mutagenesis kit (Stratagene). The coding region of Rab11a was amplified from HEK293 cell cDNA and cloned into the pRFP-C1 vector. The pRFP-C1, a vector encoding monomeric red fluorescent protein (RFP), was inserted in place of GFP in a pEGFP-C1 vector (Clontech). The resulting construct encodes RFP-Rab11a. The plasmid VCP-DN (K251Q/K524Q), ATPase-deficient forms of human VCP, were generated from pCMV-VCP-Myc as described previously (16, 19). The coding region of human EHD1 was obtained by RT-PCR from total RNA of HeLa cells as described previously (20). For colocalization studies, it was integrated into the linearized pDsRed-Monomer-N1 vector (Clontech) with the restriction enzymes XhoI and HindIII.
Cell Culture and Transfection
CRL-1601 (a McArdle RH7777 rat hepatoma cell line) cells were grown in monolayer at 37 °C in 5% CO2. The cells were maintained in medium A (Dulbecco's modified Eagle's medium containing 100 units/ml penicillin and 100 μg/ml streptomycin sulfate) supplemented with 10% FBS. Cholesterol-depleting medium was medium A supplemented with 5% LPDS, 10 μm compactin, 50 μm mevalonate, and 1.5% CDX. Cholesterol-replenishing medium contained medium A supplemented with 5% LPDS, 10 μm compactin, 50 μm mevalonate, and 15 μg/ml of cholesterol-CDX. The cholesterol-CDX inclusion complexes were prepared as described previously (19). Transient expression of NPC1L1 variants in CRL-1601 cells was accomplished using the FUGEN HD reagent (Roche) according to the manufacturer's manual.
In Vitro Glycosylation Assay
Cells were harvested and lysed in 0.5 ml of Buffer A (10 mm Hepes-KOH, pH 7.6, 1.5 mm MgCl2, 10 mm KCl, 5 mm EDTA, 5 mm EGTA, 250 mm sucrose, 5 μg/ml pepstatin A, 10 μg/ml leupeptin, 5 μm MG-132, 1 mm PMSF). The cell suspension was passed through a 22-gauge needle 30 times and centrifuged at 1,000 × g for 7 min at 4 °C. The supernatant from this spin was used to prepare the membrane fraction by centrifugation at 16,000 × g for 15 min at 4 °C. Each membrane pellet was resuspended in 0.2 ml of SDS-lysis buffer (10 mm Tris-HCl, pH 6.8, 1% (w/v) SDS, 100 mm NaCl, 1 mm EDTA, and 1 mm EGTA) and incubated at 37 °C for 30 min. Then the samples were treated with or without PNGase F or Endo H by following instructions. Immunoblot analysis was carried out with anti-EGFP antibody as previously described (17).
The Cycloheximide (CHX) Chase Assay
Plasmids encoding human wild-type and mutant NPC1L1 were transfected into CRL-1601 cells. After 24 h, cells were treated with 100 μm CHX. After different time durations (0, 1.5, 4.5 h), cells were harvested, and membrane fractions were prepared as described above. Western blot was carried out with anti-EGFP antibody.
Immunofluorescence Microscopy and Filipin Staining
Cells were fixed with 4% paraformaldehyde (PFA) in PBS and permeabilized with 0.2% Triton X-100 in PBS. Then they were blocked with 1% BSA in PBS and labeled with primary antibodies and fluorescent secondary antibodies. Leica TCS SP5 laser scanning microscope was used for detecting immunofluorescence. In each experiment, images of the same channel were acquired at identical laser parameters.
For filipin staining, a fresh 5 mg/ml stock solution of filipin was prepared in ethanol. Cells were fixed as described earlier, washed twice with PBS, and incubated with 50 μg/ml filipin in the dark for 30 min at room temperature following three washes with PBS. Cellular filipin signals were analyzed with a Leica TCS SP5 laser scanning microscope equipped with a two-photon laser using an excitatory wavelength of 720 nm. Red pseudo-color was assigned to show the filipin signal. In each experiment, images were acquired at identical laser parameters.
Fluorescence Quantification
For quantification of NPC1L1-EGFP on PM, two circles, one outlining the whole cell and the other beneath the plasma membrane, were drawn manually. After subtraction of the background, the fluorescence intensities in each circle were measured by Image-Pro Plus 5.02 and regarded as whole-cell and intracellular fluorescence intensity, respectively. The intensity of each cell was arbitrarily defined as 1, against which the intracellular intensity was normalized. Plasma membrane intensity was obtained using the whole-cell intensity minus the intracellular intensity (7). For each variant, 50 cells were randomly selected and calculated. The data shown in the figures are representative of three or more independent experiments.
Adenovirus-mediated Gene Expression
The AdEasyTM adenovirial vector system was utilized to construct the adenovirus expression vectors (21). For EGFP, NPC1L1-EGFP, NPC1L1(G402S)-EGFP, and NPC1L1(S881L)-EGFP expression, the encoding sequences were subcloned into pShuttle-CMV vector and recombined with pAdEasy vector. The adenoviruses were packaged in HEK293A cells and purified with CsCl ultracentrifugation. The viruses were titered and administrated via caudal vein injection (109 pfu viruses per mouse). At 4 days later, mouse tissues and bile were collected as described previously (6). Total lipids from liver lysates or bile were extracted and subjected to examination of cholesterol (Wako, Cholesterol E) and phospholipids (Wako, Phospholipids C).
Liver Immunohistochemistry
After 4 days injection with indicated adenoviruses, the mice were sacrificed, and similar sizes of liver tissues were collected. After fixation in 4% PFA and deaquation in 20% sucrose, the tissues were embedded in Tissue Freezing Medium (OCT, Leica) and frozen at −80 °C. Finally, 10-μm frozen sections were obtained and processed for immunohistochemistry with anti-MRP2 antibody as described previously (6).
RESULTS
Cellular Cholesterol Uptake Ability of Different NPC1L1 Variants
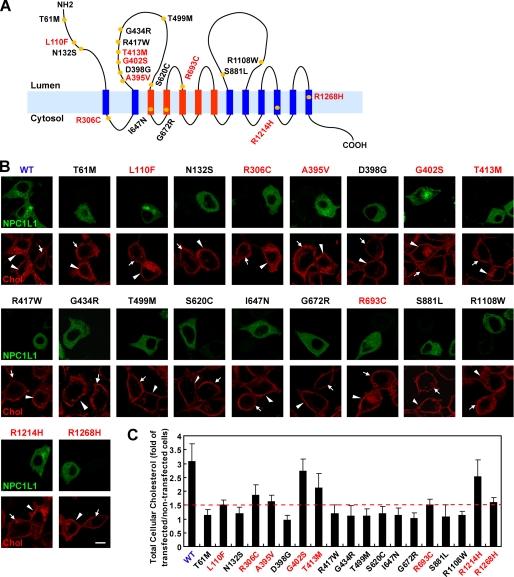
Cohen et al. (13) identified 20 NS NPC1L1 variants associated with low cholesterol absorption from humans. To further investigate the mechanism(s) by which these variants influence the function of NPC1L1, we chose 19 NS variants in our study, with the exception of W1014X because this truncated NPC1L1 completely loses function (data no shown). For measuring the cellular cholesterol uptake mediated by these NPC1L1 variants, expression vectors for different NPC1L1 variants were transiently transfected into rat hepatic CRL-1601 cells. All vectors were constructed with the EGFP tag, so the localization of these variants could be easily detected using fluorescent imaging. Meanwhile, the cellular cholesterol uptake mediated by these variants was analyzed by filipin staining (22). Consistent with our previous study (7), transient expression of wild-type NPC1L1 increased cholesterol uptake by about 2-fold compared with the control cells (non-transfected cells) as shown by the enhanced filipin signal (Fig. 1B, WT, red signal, compare arrowheads pointing to cells to arrows pointing to cells; Fig. 1C, WT). The NPC1L1 variants showed different capabilities for cholesterol uptake. Among these variants, L110F, R306C, A395V, G402S, T413M, R693C, R1214H, and R1268H enhanced cellular cholesterol uptake to 1.5∼2.8-fold of the non-transfected control cells, which was less than that of wild-type NPC1L1 (Fig. 1, B and C, indicated with red letters). We categorized these variants as partially dysfunctional variants. The other 11 low cholesterol absorption-related variants could not enhance the cholesterol uptake of the transfected cells, in which the filipin signals was similar to that of the control cells (Fig. 1, B and C, indicated with black letters). These variants were classified as the severely dysfunctional variants.
FIGURE 1.
Cellular cholesterol uptake mediated by different NPC1L1 variants. A, model of membrane topology of human NPC1L1. The 19 NS variants of NPC1L1 identified from low cholesterol absorbers are shown with yellow circles. The blue and red strips indicate the transmembrane domains of NPC1L1, and the red ones denote the sterol-sensing domain. B, cellular cholesterol uptake mediated by NPC1L1. Equal amounts of plasmids encoding human wild-type and the 19 NS variants of NPC1L1 were transfected into CRL-1601 cells, respectively. 24 h after transfection, cells were incubated in cholesterol-depleting medium to reduce cellular cholesterol, and then were refed with cholesterol-replenishing medium to deliver cholesterol. Then the cells were fixed, stained with filipin, and examined by confocal microscopy. Arrows indicate the cells without expression of the transfected plasmids. Arrowheads show the cells expressing the indicated NPC1L1 variants. Scale bar, 10 μm. C, quantification of cellular cholesterol uptake mediated by different NPC1L1 variants in B. For each variant, the filipin fluorescence intensity was measured by Image-Pro Plus 5.02 as described under “Experimental Procedures.” The dashed red line denotes the 1.5-fold of total cellular cholesterol of non-transfected control cells. Error bars represent standard deviations.
Taken together, these data showed that the 19 NS variants of NPC1L1 had defects in mediating cellular cholesterol uptake to different extents, even though they were all identified from cholesterol low absorbers. The mechanism(s) by which these NS variants impair NPC1L1 function may not be the same.
Intracellular Trafficking and Subcellular Localization of NPC1L1 Variants
It has been revealed that NPC1L1 mediates cholesterol uptake through vesicular endocytosis, and NPC1L1 recycles between ERC and PM in responding to cellular cholesterol alterations (7).
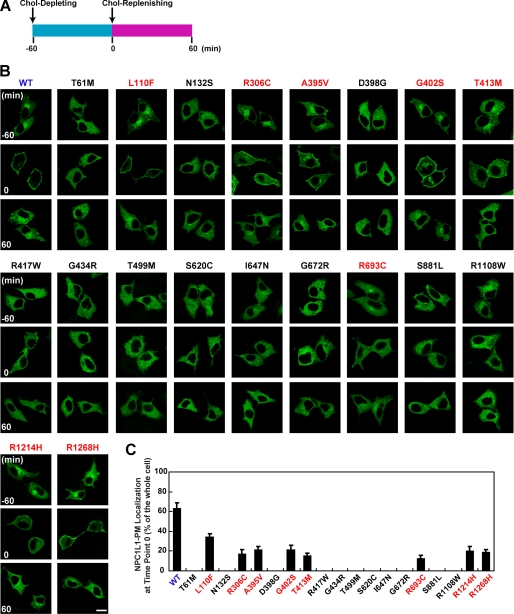
To explore whether these NPC1L1 variants were subjected to cholesterol-regulated recycling, they were transiently expressed in CRL-1601 cells. At steady state, wild-type NPC1L1 protein was mainly present in a perinuclear compartment area (Fig. 2B, WT, time point −60 min). After cells were refed with cholesterol-depleting medium, NPC1L1 gradually moved to the PM (Fig. 2B, WT, time point 0 min). About 60–70% of total NPC1L1 showed PM localization after 1 h of cholesterol depletion (Fig. 2C, WT). Then, cholesterol replenishment induced the internalization of NPC1L1 (Fig. 2B, WT, time point 60 min).
FIGURE 2.
The cholesterol-regulated recycling of NPC1L1variants. A, diagram showing the procedure used to treat cells. Briefly, 24 h after transfection, cells were incubated in cholesterol-depleting medium for 60 min to deplete cellular cholesterol, and then refed with cholesterol-replenishing medium for another 60 min to deliver cholesterol. B, intracellular localization of wild-type and different variants of NPC1L1 at different time points. CRL-1601 cells transfected with indicated plasmids were treated as shown in A. Cells were fixed at indicated time points (−60, 0, 60 min) and imaged with confocal microscopy. Scale bar, 10 μm. C, quantification of the plasma membrane (PM) localization of wild-type and different variants of NPC1L1 at time point 0 shown in B. Error bars represent standard deviations.
Similarly, for the 8 partially dysfunctional variants, only a small fraction (< 40%) of the total proteins moved to the PM when depleting cellular cholesterol (Fig. 2, B and C, time point 0 min, indicated with red letters). When refed with cholesterol-replenishing medium, these variants were also internalized (Fig. 2B, time point 60 min, indicated with red letters). In contrast, the 11 severely dysfunctional variants were diffusely localized in the cells and did not respond to cellular cholesterol alterations (Fig. 2, B and C, indicated with black letters). These results are in agreement with their cholesterol absorption abilities, further confirming that NPC1L1 facilitates cholesterol uptake via vesicular endocytosis.
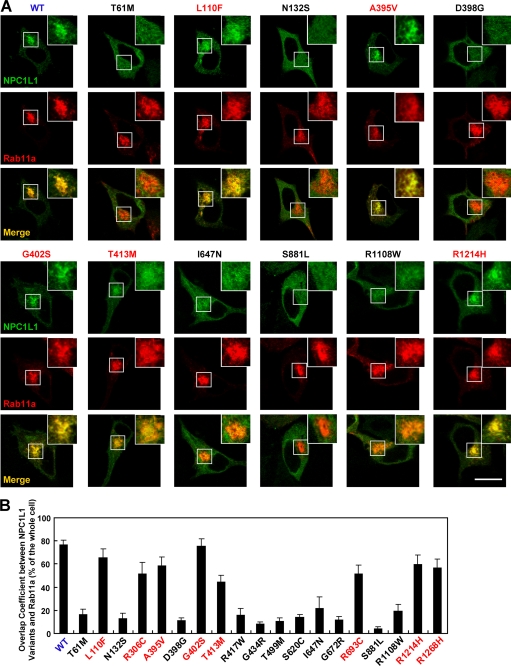
To further validate the subcellular localization of these NPC1L1 variants, the colocalization of NPC1L1 with two ERC markers, Rab11a (23) and the Eps15 homology domain (EHD) 1 (24), together with calnexin, an ER marker, was examined. Wild-type NPC1L1 mainly localized in the ERC area where it co-localized with Rab11a and EHD1 but not calnexin at steady state (Fig. 3 and supplemental Figs. S1–S3, WT). Similar results were found in cells expressing the 8 partially dysfunctional NPC1L1 variants (Fig. 3 and supplemental Figs. S1–S3, indicated with red letters). But the cellular localization of the other 11 variants was completely different. They co-localized with calnexin very well but not with either Rab11a or EHD1 (Fig. 3 and supplemental Figs. S1–S3, indicated with black letters), suggesting that the 11 variants were mostly retained in the ER.
FIGURE 3.
Colocalization of NPC1L1 variants with Rab11a, an endocytic recycling compartment marker. A, EGFP-tagged wild-type or different variants of NPC1L1 were co-expressed with RFP-Rab11a in CRL-1601 cells, respectively. 24 h after transfection, cells were fixed and examined by confocal microscopy. Scale bar, 10 μm. B, quantification of the overlap coefficient between NPC1L1 variants and RFP-Rab11a shown in A. For each variant, the intracellular green fluorescence intensity that overlapped with the red fluorescence was measured by Image-Pro Plus 5.02 and normalized by the total cellular green fluorescence intensity. Error bars represent standard deviations.
NPC1L1 Variants Identified from Low Absorbers Are Immaturely Glycosylated
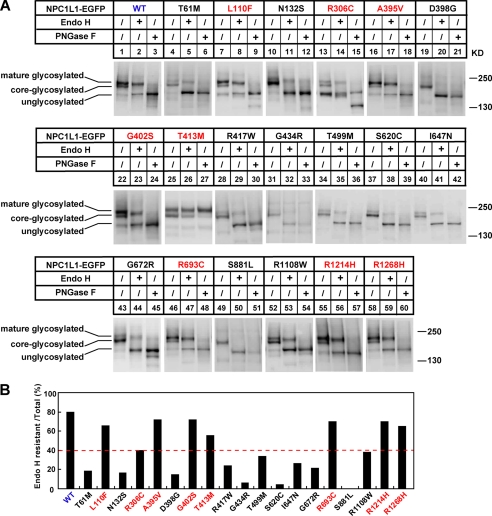
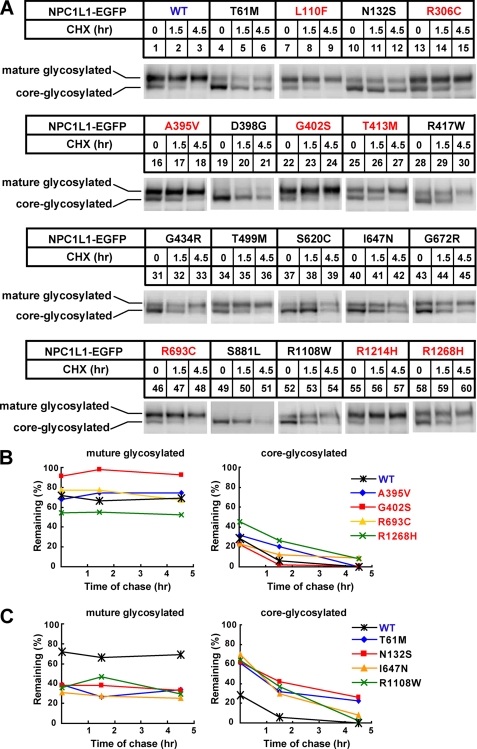
NPC1L1 is a 13-transmembrane protein with multiple potential N-glycosylation sites. Protein glycosylation is usually essential for folding and secretion. N-Linked oligosaccharides on proteins residing in the ER are sensitive to endoglycosidase H (Endo H) digestion. When glycoproteins are transported to the Golgi, their sugar chains are successively modified and are resistant to Endo H digestion (25). The observation of abnormal cellular localization of some NPC1L1 variants (Figs. 2–3, supplemental Figs. S1–S3) prompted us to analyze the glycosylation of NPC1L1. Wild-type NPC1L1 showed three different glycosylated forms: mature/high glycosylated form, core-/low glycosylated form, and un/non-glycosylated form. With Endo H treatment, the core-glycosylated NPC1L1, but not the mature glycosylated protein, was cleaved into the unglycosylated form. However, PNGase F treatment could remove all N-linked carbohydrates and convert the glycosylated form to the unglycosylated form (26). For wild-type NPC1L1, the mature/high glycosylated protein is the major form (∼80%) (supplemental Fig. S4, Fig. 4, A and B, WT), indicating that most of the wild-type NPC1L1 present in the cells have exited the ER and traversed the Golgi.
FIGURE 4.
The glycosylation of NPC1L1 variants. A, glycosidase treatment of wild-type and different variants of NPC1L1. 24 h after transfection, the cell membrane fractions were prepared as described under “Experimental Procedures” and treated with Endo H or PNGase F. Finally, the samples were analyzed by Western blot with anti-EGFP antibody. B, quantification of the Endo H-resistant NPC1L1 shown in A. The ratio was calculated as described in supplemental Fig. S2. The dashed red line denotes the 40% Endo H-resistant ratio.
We then analyzed the glycosylation status of the 19 NPC1L1 variants. As shown in Fig. 4A, the glycosylation of the 8 partially dysfunctional variants was similar to that of wild-type NPC1L1 (Fig. 4A, indicated with red letters). They expressed a high percentage of mature glycosylated protein (40–70%) that was resistant to Endo H treatment (Fig. 4B, indicated with red letters). In contrast, the 11 severely dysfunctional variants were mainly core-glycosylated and little mature glycosylated (0–40%) (Fig. 4, A and B, indicated with black letters). When treated with Endo H, most of the variants were converted to the unglycosylated form (Fig. 4A, indicated with black letters). These findings suggest that the amino acid changes impair NPC1L1 glycosylation and maturation, which may in the end result in ER retention and dysfunction of NPC1L1.
The Stability of the NPC1L1 Variants
The accumulation of immature or incorrectly folded proteins in the ER usually leads to the accelerated protein degradation. Because some NPC1L1 variants displayed a high percentage of the core-glycosylated form (Fig. 4, supplemental Fig. S5A), we then examined the stability of these NPC1L1 variants with cycloheximide (CHX) chasing experiments (Fig. 5A). For wild-type NPC1L1, the mature glycosylated form was stable and remained almost unchanged after 4.5 h of CHX treatment, whereas the core-glycosylated form was gradually disappeared with the chase (Fig. 5, A and B, WT). This indicated that the core-glycosylated form of NPC1L1 was mostly degraded. The conversion of the core-glycosylated protein to the mature form during the period of chasing is small, because no obvious increase of the mature glycosylated proteins was observed. Similar to wild-type NPC1L1, the expression of the 8 partially dysfunctional NPC1L1 variants mainly existed in the mature glycosylated form and the core-glycosylated protein (∼30% of total protein) was degraded over time (Fig. 5, A and B, indicated with red letters; supplemental Fig. S5B). In contrast, the 11 severely dysfunctional variants displayed remarkably high percentages of core-glycosylated protein and were rapidly degraded (Fig. 5, A and C, indicated with black letters and supplemental Fig. S5, C and D). These data suggest that the dysfunctional NPC1L1 variants are unstable, which may be attributed to incorrect glycosylation and ER retention.
FIGURE 5.
The protein stability of NPC1L1 variants. A, after transfection of indicated plasmids for 24 h, the cells were treated with 100 μm CHX for different time durations (0, 1.5, 4.5 h). Then cells were harvested, and membrane fractions were prepared and analyzed by Western blot with anti-EGFP antibody. B, and C, amount of mature glycosylated and core-glycosylated form of NPC1L1 in A at each time point was quantified using densitometry, expressed as a percentage of the total NPC1L1 at time 0.
The S881L Variant Is Rapidly Degraded through the ERAD Pathway
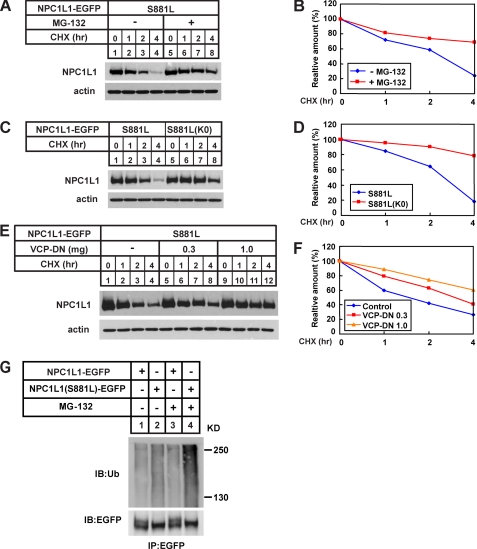
Because most immature or misfolded proteins in the ER are degraded through the ERAD pathway (27), these NPC1L1 severe dysfunctional variants are likely to be degraded via this pathway. To test this hypothesis, the S881L variant was chosen as an example for our further studies, as NPC1L1 (S881L) existed primarily in the core-glycosylated form and was rapidly degraded (Figs. 4 and 5A, S881L).
As shown in Fig. 6, A and B, the addition of MG-132, an inhibitor of proteasome, remarkably blocked the degradation of the S881L variant (compare lanes 5–8 to lanes 1–4), indicating that the ubiquitin-proteasome pathway was involved in its degradation. To confirm this, the K0 mutant form of the S881L variant was constructed, of which all the 13 lysines located in its intracellular part were substituted by arginines to abolish its ubiquitination. In contrast to the S881L variant, this K0 mutation could markedly stabilize the protein (Fig. 6C, compare lanes 5–8 to lanes 1–4; Fig. 6D), which further confirmed that the S881L variant underwent rapid degradation through the ubiquitin-proteasome pathway.
FIGURE 6.
The NPC1L1(S881L) is degraded through the ERAD pathway. A, proteasome inhibitor, MG-132, blocked the degradation of NPC1L1(S881L). 24 h after transfection, cells were treated with 100 μm CHX in the presence or absence of 10 μm MG-132. Then the cell membrane fractions were prepared at indicated time points and analyzed by Western blot with anti-EGFP antibody. C, K0 mutant on the S881L variant significantly stabilized the protein. Cells expressing the S881L variant or NPC1L1(S881L)-K0 were treated with 100 μm CHX for different time durations before harvest. Membrane fractions were prepared and analyzed by Western blot. E, dominant negative VCP blocks degradation of the S881L variant. Different amount of VCP-DN plasmids were co-transfected with the S881L variant. 24 h after transfection, the cells were treated with CHX and analyzed by Western blot. β-Actin was also probed to indicate the equal loading of lysates. B, D, and F, amount of NPC1L1 at each time point was quantified using densitometry and normalized against the protein level at time point 0. G, ubiquitination of wild-type and the S881L variant of NPC1L1. The EGFP-tagged wild-type and the S881L variant of NPC1L1 were transiently expressed in CRL-1601 cells. After 2 h of MG-132 treatment, immunoprecipitation was performed using anti-EGFP beads followed by anti-ubiquitin or anti-EGFP immunoblotting.
The valosin-containing protein (VCP, also known as p97 or Cdc48) is an essential factor in the ERAD pathway. It forms a complex with Ufd1 and Npl4, which binds the ubiquitinated proteins and escorts misfolded proteins to proteasome (28). To further elucidate the involvement of the ERAD pathway in the degradation of NPC1L1 variants, a dominant-negative VCP (VCP-DN) was constructed and co-transfected with the S881L variant. VCP-DN blocked the degradation of the S881L variant in a dose-dependent manner (Fig. 6, E and F). We next examined the ubiquitination of wild-type and the S881L variant of NPC1L1. As shown in Fig. 6G, the S881L variant was more heavily ubiquitinated (compare lane 2 and lane 1), especially when the proteasomal degradation was blocked by MG-132 (compare lanes 3–4 and lanes 1–2). Taken together, these data suggest that the amino acid alteration caused the accumulation of immature or misfolded NPC1L1 protein in the ER, which was eventually degraded through the ERAD pathway.
In Vivo Function of NPC1L1 Variants on Cholesterol Re-absorption in Mouse Liver
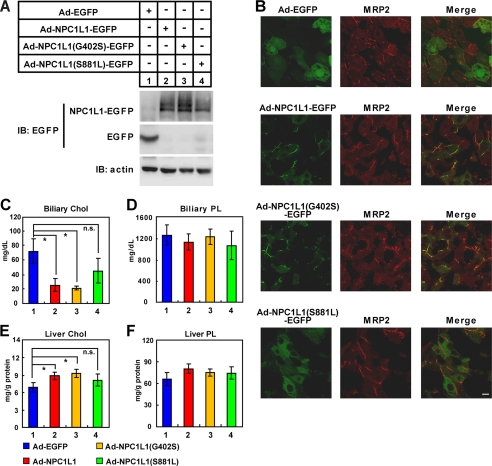
To study the function of NPC1L1 variants in vivo, we performed liver-specific expression of wild-type, a partially dysfunctional variant (G420S), and a severely dysfunctional variant (S881L) of NPC1L1 in mice using the adenovirus system (29, 30). Consistent with previous in vitro results (Fig. 4), the wild-type and G402S variant in mice mainly displayed the mature glycosylated form (Fig. 7A, lanes 2 and 3), whereas the S881L variant was mainly core-glycosylated (Fig. 7A, lane 4). The expression levels of wild-type NPC1L1 and the G402S variant were both higher than that of the S881L variant, which is consistent with their protein stabilities (Fig. 5).
FIGURE 7.
Liver-specific expression of NPC1L1 variants in mice. The adenoviruses expressing EGFP, NPC1L1-EGFP, NPC1L1(G402S)-EGFP, or NPC1L1(S881L)-EGFP were generated and administrated to mice via vena caudalis injection (109 pfu viruses per mouse, three mice for each group). After 4 days, liver tissues and bile were collected and subjected to various analyses as follows. A, adenovirus-mediated expression of EGFP, NPC1L1-EGFP, NPC1L1(G402S)-EGFP, and NPC1L1(S881L)-EGFP in mouse liver. B, localization of EGFP, NPC1L1-EGFP, NPC1L1(G402S)-EGFP, and NPC1L1(S881L)-EGFP in mouse liver. Mouse liver samples were fixed in 4% PFA and processed for frozen section and fluorescence immunohistochemistry with anti-MRP2 antibody. C–F, measurement of bile and hepatic lipid levels in wild-type (WT), G402S and S881L variants of NPC1L1-expressed mice. Total lipids from liver lysates or bile were extracted and subjected to examination of cholesterol (Wako, Cholesterol E) and phospholipids (Wako, Phospholipids C) levels. Chol, cholesterol; PL, phospholipids; *, p < 0.5; n.s., not significant. Error bars represent standard deviations.
Immunofluorescence studies showed that wild-type NPC1L1 localized to the canalicular membrane as it co-localized with the Multidrug Resistance Protein 2 (MRP2) (Fig. 7B, second row), a marker of the canalicular membrane (31). The G402S variant showed a similar localization pattern with wild-type NPC1L1 (Fig. 7B, third row). In sharp contrast, the S881L variant was dispersed in the whole cell (Fig. 7B, fourth row), consistent with its ER localization in cultured cells (Fig. 3 and supplemental Figs. S2–S3, S881L).
The bile and liver samples were also collected and analyzed. Expression of wild-type NPC1L1 or the G402S variant in mouse liver significantly decreased biliary cholesterol concentration (Fig. 7C, compare 2–3 with 1) and increased liver cholesterol (Fig. 7E, compare 2–3 with 1), but did not change biliary and liver phospholipid (PL) levels (Fig. 7, D and F). Expression of the S881L variant had no effect on these parameters (Fig. 7, C--F, compare 4 with 1–3). These results suggest that wild-type NPC1L1 and the G402S variant can specifically absorb cholesterol from bile into hepatocytes, but the S881L variant is not functional in vivo.
DISCUSSION
In this study, we investigated the molecular mechanisms of 19 reported NS NPC1L1 variants identified from cholesterol low absorbers. Based on our results, these variants were classified into two categories: the partially dysfunctional group and the severely dysfunctional group. The partially dysfunctional variants can mediate cellular cholesterol uptake to some extent and recycles between ERC and PM. The severely dysfunctional ones cannot facilitate cholesterol internalization, and they are not properly folded or recycling in a cholesterol-regulated manner. These observations further confirm that NPC1L1 mediates cholesterol uptake via vesicular endocytosis.
A previous study has identified 13 NS NPC1L1 variants in the lowest cholesterol absorbers group, and 10 NS variants in the next lowest group from the Dallas Heart Study (13). Three variants (R306C, I647N, and R693C) appeared in both groups (13). Interestingly, 10 of the 11 severely dysfunctional variants (except G672R) characterized in our study are present in the lowest cholesterol absorbers group, and all of the 8 partially dysfunctional variants are present in the next lowest group. The previous study mainly revealed the correlation between these NS NPC1L1 variants and the cholesterol absorption abilities. We have demonstrated that the NS NPC1L1 variants are dysfunctional in cholesterol uptake to different extents in vitro and in mouse liver. The good consistency between ours and the Cohen et al. study strongly suggests that the NPC1L1 variants may be the direct cause for the low cholesterol absorption phenotype in those humans.
N-Glycosylation is one of the most prevalent post-translational modifications. It occurs during protein synthesis in the ER and has a pivotal role in the folding, targeting, and function of numerous proteins (32, 33). N-Glycosylation usually happens on the Asn-X-Ser/Thr sequence motif. Because NPC1L1 is multiply glycosylated, we used the online NetNglyc server to predict the N-glycosylation sites in human NPC1L1 protein. As a result, 14 potential N-glycosylation sites were found in NPC1L1 with great potential, including N54VSC, N132QSL, N138VTR, N244ESQ, N416RSS, N431FSG, N464ISL, N479TSL, N497RTL, N506QTL, N909FSS, N927FSF, N1037LTS, and N1075ITA. Among the 19 NS NPC1L1 variants, N132S, R417W, G434R, and T499M are the ones with changes in the potential N-glycosylation motifs. In our study, these four variants showed very low amounts of mature glycosylated form and were all classified into the severely dysfunctional group (Fig. 4 and supplemental Fig. S5A). These results indicated that the four amino acid substitutions may directly affect the glycosylation of NPC1L1 and further impair its maturation, stability, and function. For other severely dysfunctional variants, the amino acid changes may disrupt the correct folding of NPC1L1, which would also influence glycosylation or maturation (Fig. 4). However, the glycosylation of the 8 partially dysfunctional variants (except R306C) were more than that of the severely dysfunctional ones but less than that of wild-type NPC1L1 (Figs. 4–5; supplemental Fig. S5A), which suggests that these amino acid substitutions may modestly affect the efficiency of glycosylation, subcellular localization, and thereby the cholesterol uptake of NPC1L1.
It has been reported that the non-coding region and synonymous variations in NPC1L1 are associated with the plasma LDL-C level alterations in different populations, such as −762T>C, −133A>G, −18A>C, 1735 C>G (exon 2), 872 C>G (L272L), 3953 C>T (V1296V), and U3_28650A>G (34–37). Of these variations, NPC1L1 (−133A>G) is reported to associate with hypercholesterolemia, which is associated with enhanced promoter activity and lower binding of transcriptional repressors in the NPC1L1 gene (38). How other polymorphisms in NPC1L1 are related to the plasma LDL-C level variations is unknown and may attract more research attention in near future.
NPC1L1 is the direct target of ezetimibe (11). Several synonymous and nonsynonymous polymorphisms of NPC1L1 were found associating with altered sensitivity to ezetimibe, including hyperresponders (R174H, 872 C>G (L272L), 3929 G>A (Y1291Y)), and nonresponders (the V55L/I1233N heterozygote) (11, 34, 39–42). It is interesting that these hyperresponder or nonresponder variations are not all localized to the putative extracellular loop C of NPC1L1, which was reported to bind ezetimibe (11). In our previous studies, we found that ezetimibe inhibits cholesterol absorption by blocking the sterol-induced endocytosis of NPC1L1 (7). Thus, experiments similar to this study can be carried out to examine the ezetimibe responses of these polymorphisms of NPC1L1, which may provide a basis for selecting therapeutic regimens or drugs for clinical test. In conclusion, this study demonstrates that some naturally occurring amino acid substitutions can impair the recycling, subcellular localization, glycosylation, or stability of NPC1L1, which account for low cholesterol absorption in humans.
Supplementary Material
Acknowledgments
We thank Yu-Xiu Qu and Su-Zhe Pan for technical assistance.
This work was supported by grants from the Ministry of Science and Technology of China (2009CB919000 and 2011CB910900), National Natural Science Foundation of China (30925012 and 90713025), and Shanghai Science and Technology Committee (10QH1402900).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- NPC1L1
- Niemann-Pick C1-like 1
- ER
- endoplasmic reticulum
- ERC
- endocytic recycling compartment
- PM
- plasma membrane
- LDL-C
- low density lipoprotein cholesterol
- NTD
- N-terminal domain
- CDX
- methyl-β-cyclodextrin
- NS
- nonsynonymous
- CHX
- cycloheximide
- EHD1
- Eps15 homology domain 1
- VCP
- valosin-containing protein
- DN
- dominant negative
- EGFP
- enhanced green fluorescent protein
- RFP
- red fluorescent protein.
REFERENCES
- 1. Grundy S. M. (1983) Annu. Rev. Nutr. 3, 71–96 [DOI] [PubMed] [Google Scholar]
- 2. Bosner M. S., Lange L. G., Stenson W. F., Ostlund R. E., Jr. (1999) J. Lipid Res. 40, 302–308 [PubMed] [Google Scholar]
- 3. Davis H. R., Jr., Altmann S. W. (2009) Biochim. Biophys. Acta 1791, 679–683 [DOI] [PubMed] [Google Scholar]
- 4. Davies J. P., Scott C., Oishi K., Liapis A., Ioannou Y. A. (2005) J. Biol. Chem. 280, 12710–12720 [DOI] [PubMed] [Google Scholar]
- 5. Altmann S. W., Davis H. R., Jr., Zhu L. J., Yao X., Hoos L. M., Tetzloff G., Iyer S. P., Maguire M., Golovko A., Zeng M., Wang L., Murgolo N., Graziano M. P. (2004) Science 303, 1201–1204 [DOI] [PubMed] [Google Scholar]
- 6. Temel R. E., Tang W., Ma Y., Rudel L. L., Willingham M. C., Ioannou Y. A., Davies J. P., Nilsson L. M., Yu L. (2007) J. Clin. Invest. 117, 1968–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ge L., Wang J., Qi W., Miao H. H., Cao J., Qu Y. X., Li B. L., Song B. L. (2008) Cell Metab. 7, 508–519 [DOI] [PubMed] [Google Scholar]
- 8. Chu B. B., Ge L., Xie C., Zhao Y., Miao H. H., Wang J., Li B. L., Song B. L. (2009) J. Biol. Chem. 284, 22481–22490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Davis H. R., Veltri E. P. (2007) J. Atheroscler. Thromb. 14, 99–108 [DOI] [PubMed] [Google Scholar]
- 10. Garcia-Calvo M., Lisnock J., Bull H. G., Hawes B. E., Burnett D. A., Braun M. P., Crona J. H., Davis H. R., Jr., Dean D. C., Detmers P. A., Graziano M. P., Hughes M., Macintyre D. E., Ogawa A., O'neill K. A., Iyer S. P., Shevell D. E., Smith M. M., Tang Y. S., Makarewicz A. M., Ujjainwalla F., Altmann S. W., Chapman K. T., Thornberry N. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 8132–8137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Weinglass A. B., Kohler M., Schulte U., Liu J., Nketiah E. O., Thomas A., Schmalhofer W., Williams B., Bildl W., McMasters D. R., Dai K., Beers L., McCann M. E., Kaczorowski G. J., Garcia M. L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 11140–11145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang J., Chu B. B., Ge L., Li B. L., Yan Y., Song B. L. (2009) J. Lipid Res. 50, 1653–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cohen J. C., Pertsemlidis A., Fahmi S., Esmail S., Vega G. L., Grundy S. M., Hobbs H. H. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1810–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fahmi S., Yang C., Esmail S., Hobbs H. H., Cohen J. C. (2008) Hum. Mol. Genet. 17, 2101–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yamanashi Y., Takada T., Suzuki H. (2009) Pharmacogenet. Genomics 19, 884–892 [DOI] [PubMed] [Google Scholar]
- 16. Cao J., Wang J., Qi W., Miao H. H., Wang J., Ge L., DeBose-Boyd R. A., Tang J. J., Li B. L., Song B. L. (2007) Cell Metab. 6, 115–128 [DOI] [PubMed] [Google Scholar]
- 17. Sever N., Song B. L., Yabe D., Goldstein J. L., Brown M. S., DeBose-Boyd R. A. (2003) J. Biol. Chem. 278, 52479–52490 [DOI] [PubMed] [Google Scholar]
- 18. Goldstein J. L., Basu S. K., Brown M. S. (1983) Methods Enzymol. 98, 241–260 [DOI] [PubMed] [Google Scholar]
- 19. Brown A. J., Sun L., Feramisco J. D., Brown M. S., Goldstein J. L. (2002) Mol. Cell 10, 237–245 [DOI] [PubMed] [Google Scholar]
- 20. Caplan S., Naslavsky N., Hartnell L. M., Lodge R., Polishchuk R. S., Donaldson J. G., Bonifacino J. S. (2002) EMBO J. 21, 2557–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Luo J., Deng Z. L., Luo X., Tang N., Song W. X., Chen J., Sharff K. A., Luu H. H., Haydon R. C., Kinzler K. W., Vogelstein B., He T. C. (2007) Nat. Protoc. 2, 1236–1247 [DOI] [PubMed] [Google Scholar]
- 22. Severs N. J. (1997) Subcell. Biochem. 28, 477–505 [DOI] [PubMed] [Google Scholar]
- 23. Ullrich O., Reinsch S., Urbé S., Zerial M., Parton R. G. (1996) J. Cell Biol. 135, 913–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grant B. D., Caplan S. (2008) Traffic 9, 2043–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maley F., Trimble R. B., Tarentino A. L., Plummer T. H., Jr. (1989) Anal. Biochem. 180, 195–204 [DOI] [PubMed] [Google Scholar]
- 26. Plummer T. H., Jr., Tarentino A. L. (1991) Glycobiology 1, 257–263 [DOI] [PubMed] [Google Scholar]
- 27. Hampton R. Y. (2002) Curr. Opin. Cell Biol. 14, 476–482 [DOI] [PubMed] [Google Scholar]
- 28. Meyer H. H., Wang Y., Warren G. (2002) EMBO J. 21, 5645–5652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chakravarthy M. V., Lodhi I. J., Yin L., Malapaka R. R., Xu H. E., Turk J., Semenkovich C. F. (2009) Cell 138, 476–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. He T. C., Zhou S., da Costa L. T., Yu J., Kinzler K. W., Vogelstein B. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Paulusma C. C., van Geer M. A., Evers R., Heijn M., Ottenhoff R., Borst P., Oude Elferink R. P. (1999) Biochem. J. 338, 393–401 [PMC free article] [PubMed] [Google Scholar]
- 32. Varki A., Freeze H. H., Manzi A. E. (2009) in Current Protocols in Protein Science (Coligan J. E., Dunn B. M., Speicher D. W., Wingfield P. T. eds) pp. 57:12.1.1–12.1.10, John Wiley & Sons, Hoboken, NJ [Google Scholar]
- 33. Glozman R., Okiyoneda T., Mulvihill C. M., Rini J. M., Barriere H., Lukacs G. L. (2009) J. Cell Biol. 184, 847–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Simon J. S., Karnoub M. C., Devlin D. J., Arreaza M. G., Qiu P., Monks S. A., Severino M. E., Deutsch P., Palmisano J., Sachs A. B., Bayne M. L., Plump A. S., Schadt E. E. (2005) Genomics 86, 648–656 [DOI] [PubMed] [Google Scholar]
- 35. Maeda T., Honda A., Ishikawa T., Kinoshita M., Mashimo Y., Takeoka Y., Yasuda D., Kusano J., Tsukamoto K., Matsuzaki Y., Teramoto T. (2010) J. Atheroscler. Thromb. 17, 356–360 [DOI] [PubMed] [Google Scholar]
- 36. Chen C. W., Hwang J. J., Tsai C. T., Su Y. N., Hsueh C. H., Shen M. J., Lai L. P. (2009) J. Hum. Genet. 54, 242–247 [DOI] [PubMed] [Google Scholar]
- 37. Polisecki E., Peter I., Simon J. S., Hegele R. A., Robertson M., Ford I., Shepherd J., Packard C., Jukema J. W., de Craen A. J., Westendorp R. G., Buckley B. M., Schaefer E. J. (2010) J. Lipid Res. 51, 1201–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Martín B., Solanas-Barca M., Garcia-Otín A. L., Pampín S., Cofán M., Ros E., Rodríguez-Rey J. C., Pocoví M., Civeira F. (2010) Nutr. Metab Cardiovasc. Dis. 20, 236–242 [DOI] [PubMed] [Google Scholar]
- 39. Hegele R. A., Guy J., Ban M. R., Wang J. (2005) Lipids Health Dis. 4, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhao H. L., Houweling A. H., Vanstone C. A., Jew S., Trautwein E. A., Duchateau G. S., Jones P. J. (2008) Lipids 43, 1155–1164 [DOI] [PubMed] [Google Scholar]
- 41. Pisciotta L., Fasano T., Bellocchio A., Bocchi L., Sallo R., Fresa R., Colangeli I., Cantafora A., Calandra S., Bertolini S. (2007) Atherosclerosis 194, e116–e122 [DOI] [PubMed] [Google Scholar]
- 42. Wang J., Williams C. M., Hegele R. A. (2005) Clin. Genet. 67, 175–177 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.