Abstract
Drosophila Kinesin-73 (Khc-73), which plays a role in mitotic spindle polarity in neuroblasts, is a metazoan-specific member of the Kinesin-3 family of motors, which includes mammalian KIF1A and Caenorhabditis elegans Unc-104. The mechanism of Kinesin-3 motors has been controversial because some studies have reported that they transport cargo as monomers whereas other studies have suggested a dimer mechanism. Here, we have performed single-molecule motility and cell biological studies of Khc-73. We find that constructs containing the motor and the conserved short stretches of putative coiled-coil-forming regions are predominantly monomeric in vitro, but that dimerization allows for fast, processive movement and high force production (7 piconewtons). In Drosophila cell lines, we present evidence that Khc-73 can dimerize in vivo. We also show that Khc-73 is recruited specifically to Rab5-containing endosomes through its “tail” domain. Our results suggest that the N-terminal half of Khc-73 can undergo a monomer-dimer transition to produce a fast processive motor and that its C-terminal half possesses a specific Rab5-vesicle binding domain.
Keywords: Kinesin, Membrane Trafficking, Microscopic Imaging, Single-molecule Biophysics, Vesicles, Khc-73, Rab5
Introduction
Microtubule motors in the kinesin superfamily transport a variety of molecular cargoes in eukaryotic cells, regulate microtubule dynamics, and organize microtubule arrays in the mitotic spindle (for review, see Refs. 1, 2). The best characterized motor in the kinesin superfamily is Kinesin-1 (conventional kinesin), which is involved in organelle and mRNA transport (3–5). Kinesin can take >100 8-nm steps along a microtubule without dissociating, a process known as processive movement (for review, see Ref. 6). The bias for the plus end-directed movement is thought to be driven by the docking of a mechanical element (the neck linker) to the core of the enzyme when ATP binds to the active site (7, 8).
The Kinesin-3 class is another well established and important class of cargo-transporting kinesins. The best studied member, KIF1A (mouse)/Unc104 (Caenorhabditis elegans), is involved in the transport of synaptic vesicles to the nerve terminus (9, 10). Unlike Kinesin-1, KIF1A/Unc104 and other Kinesin-3 members lack the extensive coiled-coil (CC) domain that conventional kinesin uses for dimerization. Indeed, the first characterizations of Kinesin-3 family members, mouse KIF1A and KIF1B, suggested that these motors are monomeric in vitro (10, 11) and gave rise to the hypothesis that these monomeric motors might use a biased diffusion mechanism to move along microtubules in contrast to the hand-over-hand movement of Kinesin-1 (12, 13). However, other studies have suggested that Kinesin-3 motors might dimerize through several short CC motifs when concentrated in solution or on a membrane vesicle and move by a Kinesin-1-like mechanism (14, 15). More recently, a publication has suggested that full-length KIF1A may always be a dimer both in vitro and in vivo (16). Thus, the mechanism by which Kinesin-3 family members move cargo remains an open question.
An intriguing, metazoan-specific member of the Kinesin-3 family is typified by Khc-733 in Drosophila (17) and GAKIN in humans (18). Although there are four Kinesin-3 family members in Drosophila, Khc-73 is unique among Kinesin-3 family members in that it contains a C-terminal CAP-Gly (cytoskeleton-associated protein Glycine-rich) domain, which is found in several microtubule-binding proteins (19, 20). Khc-73 interacts with the Discs Large tumor suppressor in neuroblasts and is necessary for proper mitotic spindle orientation (21). GAKIN also binds the human homolog of Discs Large and is necessary for the enrichment of phosphatidylinositol trisphosphate-containing vesicles at the tips of neurites (18, 22).
Because Khc-73/GAKIN remains poorly characterized and its motile properties have not been fully studied, we sought to investigate its single-molecule motility and to determine its localization when expressed in cells. Here, we show that dimeric Khc-73 motors undergo rapid (∼1.5 μm/s) processive movement and generate forces comparable with conventional Kinesin-1 (∼7 pN). In Drosophila S2 and BG2 cells, Khc-73 forms a specific interaction with Rab5-containing endosomes through its C-terminal domain. Our results also suggest that Khc-73 can dimerize both in vitro and in vivo and that the dimer is likely to be the active form of the motor.
EXPERIMENTAL PROCEDURES
Cloning of Khc-73 and Rab Constructs
All Khc-73 clones were amplified from the full-length Khc-73 construct generously provided by C. Doe (21). The GCN4 leucine zipper (LZ) motif was amplified from a construct provided by K. Slep (23). Shorter Khc-73 constructs used for Drosophila cell line transfection were subcloned into pENTR/D-TOPO (Invitrogen) and then moved into either a Gateway C-terminal GFP or mCherry vector under the control of the copper-inducible metallothionein promoter (pMTWG and pMTWCherry; Drosophila Gateway Collection). For generation of GFP-tagged Rabs, each Rab ORF was amplified from the appropriate full-length cDNA clone (primer sequences available on request) and then subcloned into the pENTR/D-TOPO vector. The Rab ORF was then moved into an N-terminal Gateway GFP vector under the control of the actin promoter (pAGW; Drosophila Gateway Collection).
Expression and Purification of Khc-73 Constructs
Bacterially expressed constructs were cloned into pET17b with a C-terminal GFP followed by His6, expressed, and purified as described previously (14). Khc-73-positive nickel-nitrilotriacetic acid column eluates were dialyzed with BRB 80 buffer (80 mm PIPES (pH 6.8), 1 mm MgCl2, 1 mm EGTA) supplemented with 1 mm ATP and 1 mm DTT, concentrated, aliquotted, and snap frozen in liquid nitrogen. Before use in further assays, aliquots were subjected to microtubule binding and release to purify active Khc-73 as described previously (14).
Analysis of Native Molecular Mass
The molecular mass of Khc-73 proteins in solution was determined by measuring the Stokes radius by gel filtration and the sedimentation coefficient by sucrose gradient centrifugation (24). Gel filtration was performed over a Superdex 200 column connected to an AKTA FPLC (GE Healthcare). All fractions were collected, and peak protein levels were confirmed as Khc-73-positive by SDS-PAGE followed by Coomassie Blue staining. Confirmed peaks were compared with a calibration curve generated by standards of known Stokes radius (thyroglobulin (8.5 nm), ferritin (6.1 nm), aldolase (4.5 nm), BSA (3.6 nm) ovalbumin (3.1 nm), and cytochrome c (1.6 nm)) to determine the radius of each construct. For determination of sedimentation coefficients, constructs were layered over a 12-ml, 5–40% sucrose gradient and centrifuged at 35,000 rpm (∼150,000 × g) for 18 h at 4 °C in an SW 40 rotor. A total of 24 fractions were collected from each sample, and peak fractions were identified by Coomassie-stained SDS-PAGE. Sedimentation coefficients were determined by comparing with a standard curve generated from calibration standards (thyroglobulin, ferritin, catalase, lactate dehydrogenase, and BSA; GE Healthcare) analyzed in the same centrifugation run. The values of Stokes radii and sedimentation coefficients were used to derive the molecular mass (Mr) of each construct according to the following equation,
where N is Avogadro's number, ν is the partial specific volume of the protein (estimated as 0.725 ml/g in this study), s is the sedimentation coefficient, a is the Stokes radius, and η and ρ are the viscosity (1.6 g · m−1 · s−1) and density (1 g/ml) of water at 4 °C, respectively.
Optical Trapping Assay
Stall-force and step-size measurements of single Khc-73 molecules were performed at 25 ± 1 °C with a custom-built force clamp optical trapping microscope as described previously (25). In brief, carboxylated polystyrene beads (0.92-mm diameter; Invitrogen) were sparsely covered with the GFP-tagged motor proteins via affinity-purified anti-GFP antibodies (14). Measurements were performed in a ∼10-μl flow cell constructed by placing two strips of double-sided sticky tape between a standard microscope slide and a 160-μm thick 18 mm × 18 mm coverslip to form a channel. The assay solution consisted of 30 mm HEPES (pH 7.2), 2 mm magnesium acetate, 1 mm EGTA, 1 mm MgATP, 1 mg/ml casein, 10 mm DTT, 4.5 mg/ml glucose, and an oxygen scavenger system (26). Bead displacement was detected by a quadrant photodiode and recorded at 2 kHz. Before each experiment, the trapped bead was scanned along the x axis of the object field across the detection region to obtain the detector's response. A trapped bead was then positioned over a rhodamine-labeled sea urchin sperm flagellar axoneme that was immobilized onto a coverslip and aligned with the x axis of the object field (coincides with the x axis of the position detector). Experiments were performed at dilutions at which the fraction of beads moving was ≤0.3 to ensure measurements on a single-molecule level (27). Trap stiffness was calibrated for each trapped bead from the amplitude of the thermal diffusion. Stall forces were determined by multiplying the trap stiffness by the mean maximum distance reached. Force-feedback was operational in an area of ±200 nm along the x axis of the object field. Motor steps (approximate center-of-mass movement of kinesin) were determined from the bead displacement records using a step-finding algorithm developed by Kerssemakers et al. (28). This algorithm assumes that steps are hidden in normal-distributed noise but makes no assumptions about steps sizes and durations. The resulting data were then visually screened, and only those steps that could be visually separated from noise were included in the step size histograms.
In Vitro Motility Assays
For microtubule gliding assays, flow chambers were made as described above, and 10 μl of a 0.5 mg/ml GFP antibody solution was added and allowed to attach for 5 min. Flow chambers were washed twice with motor buffer (25 mm PIPES (pH 6.8), 75 mm KCl, 1 mm EGTA, 1 mm MgCl2, 1 mm DTT, 20 μm Taxol, 5% sucrose), then incubated for 5 min with motor buffer containing 1 mg/ml casein to block nonspecific binding to the coverglass. Khc-73 motors were diluted to 50 nm in motor buffer with casein and allowed to incubate for 5 min prior to two additional washes in motor buffer. Finally, 20 μl of motility buffer (motor buffer with 1 mg/ml casein, 5 mm ATP, oxygen scavenger, and rhodamine-labeled microtubules) was added. Time-lapse imaging was performed on a Nikon Eclipse TE2000-E microscope equipped with an Andor EM CCD camera driven by Micro-Manager imaging software. To determine the microtubule gliding velocity, the leading edge of a microtubule was tracked through a minimum of 10 successive frames (30 s), and the average instantaneous velocity (sum of distances traveled in each successive frame divided by the number of frames multiplied by the time between each frame) was obtained using ImageJ. The average for each construct was determined by applying a simple one-peak Gaussian fit to a histogram of the velocities using Origin 8 software (OriginLab Corporation). Single-molecule motility assays along rhodamine-labeled sea urchin axonemes were performed using a standard assay (29). Axonemes were identified using wide field fluorescence imaging, and GFP-labeled Khc-73 constructs were imaged at 10 Hz using total internal reflection fluorescence (TIRF) microscopy with a 100×, 1.49 NA TIRF objective and an Andor EMCCD camera. Kymograph analysis in ImageJ was used to determine the velocity and run length for each experiment. The slope of particle movement over time was used for velocity, and total distance traveled corresponded to run length. Movement frequency was generated by adding the total number of motility events and dividing by the number of axonemes in each frame and the total time of observation. Motility events were defined as movement along the length of an individual axoneme that persists through at least five individual acquisition frames. Average velocity was determined by applying a simple one-peak Gaussian fit to a histogram of the velocities for each construct, and the run length was determined by fitting an exponential decay plot to a histogram of the run lengths in Origin 8.
Khc-73 Antibody Production
The 20 amino acids at the C terminus of Khc-73 are not found in the other Kinesin-3 family members in Drosophila, and BLAST searches of the Drosophila data base show no significant homology of this sequence with any other expressed protein. A peptide of the final 20 C-terminal amino acids of Khc-73 was synthesized with an N-terminal cysteine residue that was conjugated to keyhole limpet hemocyanin (Maine Biotechnology). Antigen was used to generate rabbit polyclonal serum against Drosophila Khc-73 (Maine Biotechnology). Serum was tested by Western blotting against a BSA-conjugated antigen peptide, with preimmune serum and BSA used as negative controls. A portion of crude serum was further purified by incubation with protein G-Sepharose beads (Gamma-bind). Use of this total IgG fraction in Western blots of Drosophila S2 cell lysate recognized several bands, but only one prominent band was recognized in the size range of Khc-73, which was subsequently reduced in cells treated with RNAi targeted against Khc-73 (supplemental Fig. 5).
Cell Culture and RNAi
Drosophila Schneider cell line (S2) cells (Invitrogen) and Drosophila BG2 cell line (Drosophila Genomics Resource Center) were cultured as described previously (30, 31). RNAi construct generation and 7-day RNAi treatment were performed as described previously (32). Two separate dsRNA constructs were used in this study. To target the 3′-UTR of the Khc-73, the following primers were used: TGTACCCAAAGTGTTCGCATCAG and CATTTGCGGCATGGGGTGAGAAT. To target the coding region of the Khc-73 transcript, the following primers were used: ATTCACGAGCTTAATGACAACG and CAGCGTGTAAAACTTATGTCCG. Neither construct generates 19 nucleotide sequences with predicted off-target effects, as determined by analysis of the dsRNA sequences generated by the Genome RNAi data base at the German Cancer Research Center. RNAi treatment of S2 cells with either construct led to a reduction of intensity of a single band using the Khc-73 antibody that we generated. For each experimental RNAi treatment, a sample was collected and analyzed by Western blotting to determine the level of Khc-73.
Live Cell Imaging and Analysis
Full-length Khc-73 and shorter Khc-73 constructs were cloned into the pENTR/D-TOPO vector (Invitrogen) and moved into C-terminal Gateway GFP and mCherry vectors under the control of the metallothionein promoter described previously (33). Drosophila S2 and BG2 cells were transfected using the Effectene transfection reagent (Qiagen) using a standard protocol. 16 h before imaging, expression was induced by the addition of 50 μm copper sulfate. 1 h prior to imaging, S2 cells were plated on concanavalin A-coated (Sigma-Aldrich), glass bottom dishes (MatTek) and allowed to spread. For analysis of Rab5-GFP motility rates, Drosophila S2 cells were treated with 7-day RNAi to knock down Khc-73 or mock treated, plated on concanavalin A-coated coverglass and allowed to spread for 1 h, then incubated with 5 μm cytochalasin D to induce cellular projections of bundled microtubules in parallel orientation as described previously (34). Time-lapse images were acquired using wide field fluorescence on the Nikon microscope described above. Images were analyzed visually in the Micro-Manager 5D viewing frame, and representative frames were chosen for documentation. Images shown were imported into Photoshop CS4 (Adobe), then enhanced by applying a linear adjustment of intensity boundaries. Individual images were inserted into corresponding channels of a single RGB image to create the merged images shown. Corresponding QuickTime movies were generated by exporting image stacks from ImageJ.
RESULTS
Analysis of the Biophysical and Biochemical Behavior of Khc-73
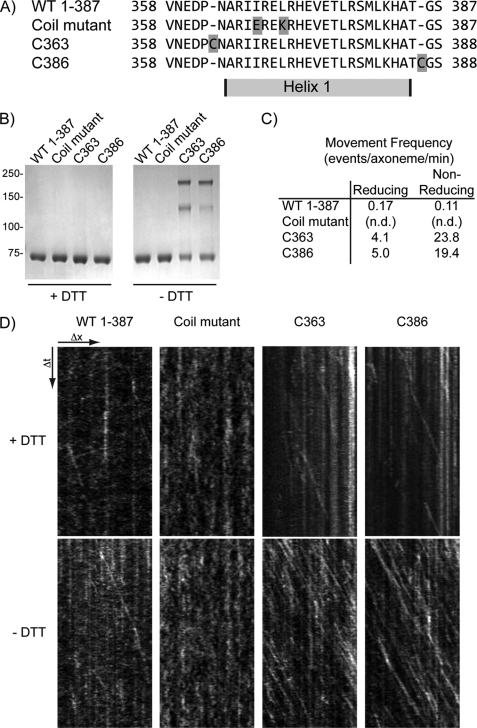
Previous work has shown that Unc104, a C. elegans member of the Kinesin-3 family, undergoes concentration-dependent dimerization in vitro as a result of two short helical domains that are directly C-terminal to the neck linker (14). The homologous region of the Mus musculus Kinesin-3 family member, KIF1A was also predicted to form a CC (10, 16). Alignment of the Khc-73 amino acid sequence with Unc104 and KIF1A identified two potential helical regions in Khc-73 with an intermediate predicted potential for CC formation (Fig. 1A). However, a 38-amino acid stretch distal to the forkhead-associated domain (a domain characteristically found in Kinesin-3 motors) shows high CC-forming probability (Fig. 1B). Thus, we designed motor-containing constructs with varying portions of these regions for further biophysical and biochemical characterization (Fig. 1C).
FIGURE 1.
Khc-73 domain analysis and construct design. A, sequences of D. melanogaster Khc-73, C. elegans Unc104, and M. musculus KIF1A were aligned using the ClustalW multiple sequence alignment tool at EMBL-EBI. Dark green shaded regions indicate sequence identity, whereas light green shaded regions indicate sequence similarity. B, D. melanogaster Khc-73 sequence was analyzed for coil-forming probability by the COILS program. Probabilities shown are the output using windows of 21 amino acids. C, schematic of Khc-73 constructs used in this work is shown. Label numbers refer to the Khc-73 amino acid boundaries of each construct.
C-terminal GFP-tagged constructs of the Khc-73 motor were expressed in bacteria and purified. To ensure that the purified proteins were catalytically active, we performed sequential microtubule binding and release reactions as described previously (14). Analysis of the bulk activity of each construct via microtubule gliding assays showed a similar rate of microtubule motility (Table 1). To determine the processive behavior of Khc-73 motor constructs, we examined single molecules by using TIRF microscopy. Molecules of each of the Khc-73 constructs attached to and moved along sea urchin axonemes with similar velocities in the range of 1.5 μm/s, as determined by the peak of a Gaussian curve fit to a histogram of the velocities (Table 1). A histogram of the distance traveled per microtubule encounter revealed an exponential decay constant (defined as the run length) (Table 1 and supplemental Fig. 1). In addition, because experiments were performed with the same concentration of motor (50 nm), we are able to compare the relative frequency of processive movement events (construct movements along the length of the axoneme that persisted for more than five individual acquisition frames), normalized to axoneme number and imaging time. We noticed that although the velocity of each construct is similar, both the run length and movement frequency increased for the longer constructs that had more potential CC-forming domains. As a positive control for dimer formation, we expressed and purified a “constitutive dimer” in which the shortest motor construct was fused to the GCN4 LZ dimerization motif (1–387-LZ-GFP). In single-molecule motility assays, this construct showed the same velocity, but a longer run length and nearly 100-fold higher frequency of movement compared with the same construct without the LZ. This result suggests that dimerization is needed for efficient processive movement. The finding that the truncated Khc-73 constructs undergo infrequent processive movement compared with the constitutive dimer suggests that they may be in equilibrium between pools of a predominant monomer and a dimer.
TABLE 1.
Motile and hydrodynamic behavior of Khc-73 motor-containing constructs
| Khc-73 construct | Microtubule gliding velocitya | Single molecule motilityb |
Sedimentation coefficient | Stokes radius | Molecular mass |
|||
|---|---|---|---|---|---|---|---|---|
| Velocityc | Run lengthc | Frequency | Calculatedd | Predictede | ||||
| μm/s | μm/s | μm | events/axoneme/min | ×10−13s | Å | kDa | kDa | |
| 1–387-GFP | 1.45 ± 0.07 | 1.57 ± 0.26 | 0.45 ± 0.09 | 0.09 | 3.0 | 42.5 | 84.2 | 70.8 |
| 1–432-GFP | 1.51 ± 0.02 | 1.58 ± 0.12 | 1.30 ± 0.49 | 0.15 | 3.0 | 43.3 | 85.7 | 76.1 |
| 1–547-GFP | 1.62 ± 0.10 | 1.33 ± 0.23 | 0.76 ± 0.11 | 0.13 | 3.0 | 44.0 | 87.1 | 88.8 |
| 1–623-GFP | 1.58 ± 0.04 | 1.54 ± 0.46 | 1.04 ± 0.11 | 0.28 | 3.0 | 45.5 | 90.1 | 97.8 |
| 1–387-LZ-GFP | N.D. | 1.56 ± 0.15 | 1.51 ± 0.06 | 6.77 | 5.1 | 43.3 | 144.8 | 75.0 |
a Mean ± S.D. shown for >50 measurements.
b Each data set comes from multiple experiments using protein from a single preparation.
c Mean ± S.D. shown for >40 measurements.
d Molecular mass based on the calculation described under “Experimental Procedures.”
e Molecular mass based on the sum of amino acids in the Khc-73 construct, linker, GFP, and His6 affinity tag.
To determine the native state of the motor constructs in vitro, we performed gel filtration coupled with sucrose density gradient centrifugation (24) (Table 1). The constructs lacking the LZ motif had a calculated molecular mass close to that predicted for a monomer. In contrast, and as expected, the calculated molecular mass of 1–387-LZ-GFP was close to that predicted for a dimer of two polypeptide chains. This result suggests that the majority of the non-LZ-containing constructs are monomeric in solution, although a small amount of dimer (predicted from our single-molecule motility experiments) would not likely be detected in these hydrodynamic measurements, as the dimer would likely dissociate over the time course of the gel filtration and sucrose gradient runs.
If the putative CC domains in the tail region of Khc-73 facilitate dimer formation and this dimer formation is necessary for the in vitro motility we observe, then interrupting the CC domain should abolish Khc-73 motility. Likewise, stabilization of the CC should increase the frequency of Khc-73 motility. To test this hypothesis, we created a series of mutations in the shortest (1–387) Khc-73 construct. Tomishige et al. (14) showed that two point mutations in the putative CC motif in Unc104 (I362E, L365K) abolished its in vitro motility. Because sequence alignment of Unc104 and Khc-73 (Fig. 1A) shows that these residues are conserved, we made the same mutations in the Khc-73 1–387 molecule (Fig. 2A, coil mutant). At the same concentration as the wild-type 1–387 construct, we observed no in vitro motility (total imaging time of 12.5 min, 30 experiments) of the coil mutant construct (Fig. 2D). To stabilize in vitro dimer formation, we engineered constructs with a single cysteine on either side of the single CC domain in the 1–387 construct (Fig. 2A, C363 and C386). When final purification steps were carried out in the absence of a reducing agent, both the Cys-363 and Cys-386 constructs showed a significant population that shifted to higher molecular mass bands on a nonreducing gel compared with their migration on a reducing gel (Fig. 2B). This behavior was not seen for wild-type 1–387 or the coil mutant purified in the same fashion (Fig. 2D). When their motility was assayed in vitro, both Cys-363 and Cys-386 showed a >100-fold increase in motility frequency compared with wild-type 1–387 (Fig. 2, C and D). These results lend further credence to the model that the Khc-73 dimer is the active form of the molecule and that dimer formation is facilitated by the predicted CC helices adjacent to the Khc-73 motor domain. It also supports the notion that Khc-73 exists in a dynamic equilibrium between monomers and dimers. When diluted to nanomolar concentrations in the motility assay, the prevalent species is likely to be monomers. However, with cysteine-cysteine cross-linking, the dimer population is stable and does not dissociate to monomers upon dilution.
FIGURE 2.
Coiled-coil domain of Khc-73 facilitates processive movement. A, sequences of the regions surrounding helix 1 in wild type, coil mutant (I367E, L370K), and cysteine insertions before (C363) and after (C386) helix 1. B, side-by-side preparations of wild type, coil mutant, C363 and C386 constructs boiled in SDS sample buffer containing or omitting reducing agent (+DTT and −DTT, respectively) and run on the same gel. C, in vitro motility assays performed as described above with the frequency of processive motility calculated by adding the total number of events and dividing by the number of axonemes observed and the total time of imaging. Experiments were performed after incubating each construct in the presence and absence of 5 mm tris(2-carboxyethyl)phosphine for 6 h at 4 °C (reducing and nonreducing conditions, respectively). n.d., not detected (in 12.5 min of observation, 30 experiments). D, sample kymographs of Khc-73 construct motility on individual axonemes. Distance is in the x-coordinate, and time is in the y-coordinate. Diagonal lines indicate processive movement of single molecules along an axoneme.
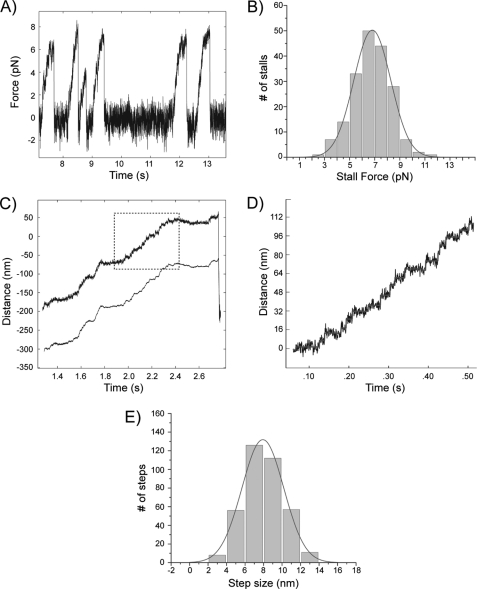
To determine the mechanical properties of Khc-73, we attached single dimeric 1–387-LZ-GFP molecules to latex beads and performed optical trap assays. Because optical trap studies necessitate the use of a uniform population of stable dimer and not a species that is subject to monomer-dimer equilibrium, we utilized the well established GCN4 LZ construct to ensure dimer stability. Using the laser trap to position and hold a motor-bound bead over a coverslip-attached, rhodamine-labeled axoneme, we measured the stall force (the maximal force generated by the motor before it detached from a microtubule) of the Khc-73 motor (Fig. 3A). Fitting a Gaussian curve to a histogram of the individual stall forces revealed an average stall force of 6.8 ± 1.5 pN (mean ± S.D., Fig. 3B). Using a feedback-controlled optical trap to maintain a constant load (6 pN) on the motor, we could visualize clear stepwise movement of the motor (see enlarged region of the motor trace in Fig. 3D). A histogram of the individual step sizes revealed an average step size of 7.92 ± 0.07 nm (mean ± S.E., Fig. 3E), which is approximately the distance between α-β-tubulin dimers and the step size of the Kinesin-1 motor (35).
FIGURE 3.
Force generation and processive movement of single dimeric Khc-73 molecules in the optical trapping microscope. The Khc-73 1–387-LZ construct was used for each of the optical trap experiments. A, displacement of a single kinesin molecule at 1 mm ATP in a fixed (nonfeedback) optical trap showing successive motor detachments and stalling events (trap stiffness: k = 0.057 pN/nm). B, stall force distribution (n = 187). C, processive microtubule plus end-directed motion of Khc-73 under a constant 6 pN opposing load (force-feedback mode) in the presence of 1 mm ATP (trap stiffness: k = 0.057 pN/nm). The bead displacement is shown in the upper trace and the trap position in the lower trace. The outlined portion of the bead displacement trace is enlarged in D. D, enlargement of outlined trace in C. The raw data are shown in black, and the steps detected by the step-finding program are shown in gray. E, histogram of step sizes for microtubule plus end-directed movement under 6 pN opposing load in the presence of 1 mm ATP (n = 370).
Khc-73 Is Enriched at the Ends of Microtubules and Is Recruited to Rab5-containing Vesicles
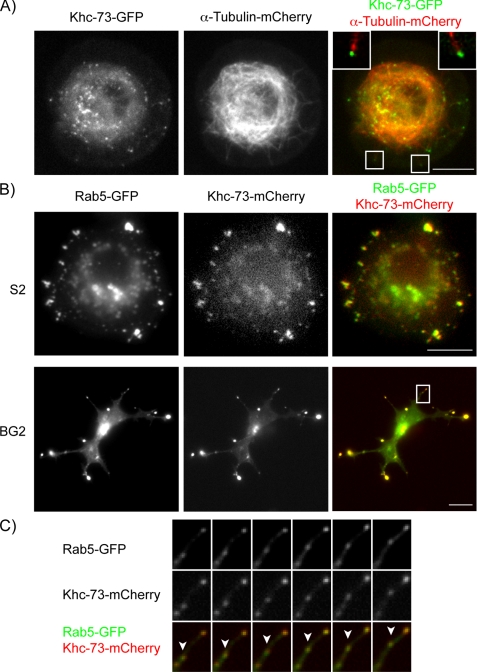
Khc-73 has been shown to have a role in mitotic spindle orientation in Drosophila neuroblasts (21), but its interphase localization has not been explored. We expressed full-length Khc-73 in Drosophila S2 cells and saw two distinct localizations, the first enriched at the peripheral tips of microtubules, and the second on motile puncta in the cytosol (Fig. 4A and supplemental Movie 1). A subset of microtubule-associated proteins known as +TIPs bind to the polymerizing plus ends of microtubules (36). This is particularly relevant to Khc-73 because it contains a C-terminal CAP-Gly domain, which is present in some +TIP proteins and in certain cases has been shown to be sufficient to endow plus end tracking behavior if the CAP-Gly domain is present as a dimer (23, 37). To test whether the Khc-73 CAP-Gly domain might account for its accumulation at microtubule plus ends, we expressed and localized the CAP-Gly domain as a monomer and a dimer (fused to a leucine zipper, CAP-Gly-LZ). The monomeric form (CAP-Gly) exhibited diffuse cellular localization, whereas the dimeric form (CAP-Gly-LZ) bound nonspecifically along the microtubule lattice (supplemental Fig. 2). Thus, the microtubule tip enrichment of Khc-73 is apparently not due to CAP-Gly-dependent plus end tracking.
FIGURE 4.
Khc-73 is enriched at the distal ends of microtubules and co-localizes with Rab5-GFP. A, Drosophila S2 cells were transiently transfected with constructs containing mCherry-tagged α-tubulin and GFP-tagged, full-length Khc-73 under the control of the metallothionein promoter (see “Experimental Procedures”). Images shown are an individual frame of a time-lapse movie provided as supplemental Movie 1. Merged panel shows enlargement of boxed regions in the image with Khc-73 puncta at the tips of microtubules. Top left corner corresponds to the box on the lower left, and top right corner corresponds to the boxed region in the lower right. B, Drosophila S2 and BG2 cells were transiently transfected with constructs containing mCherry-tagged Khc-73 under the control of the metallothionein promoter and GFP-tagged Rab5 under the control of the actin promoter. Images shown are individual frames of time-lapse movies provided as supplemental Movies 2 and 3. C, enlargement of boxed region in the merged image of the BG2 cell from B is shown. Arrowhead points to a motile puncta where both Khc-73-mCherry and Rab5-GFP co-localize.
Full-length Khc-73 might also enrich at microtubule plus ends through its motor activity and might be carrying a cargo to the tip. Because many kinesins associate with membrane vesicles, we examined whether Khc-73 co-localized with distinct Rab GTPases, which are good markers for different intracellular membrane vesicles (38, 39). We created a collection of GFP-tagged Rab GTPases (Rabs 5, 6, 8, 9, 10, 11, 14, 18, 19, 23, 30, 32, and 40) and performed pairwise co-transfections of Drosophila S2 cells with each individual construct and mCherry-labeled full-length Khc-73. Although most co-transfections yielded fluorescence signals in nonoverlapping compartments (supplemental Fig. 3), we found that Khc-73-mCherry exhibited nearly complete overlap with Rab5-GFP, a well characterized marker of early endosomes (40–42) (Fig. 4B and supplemental Movie 2). Because Khc-73 has been previously characterized in Drosophila neuroblasts (21), we also co-expressed Rab5-GFP and Khc-73-mCherry in the Drosophila neuronal BG2 cell line and saw similar co-localization both at the periphery and in motile puncta (Fig. 4B and supplemental Movie 3). Thus, in Drosophila cells lines, Khc-73 exhibits a very specific localization to Rab5-containing early endosomes.
Knockdown of Endogenous Khc-73 Reveals Dimerization and Rab5-containing Vesicle Recruitment Domain
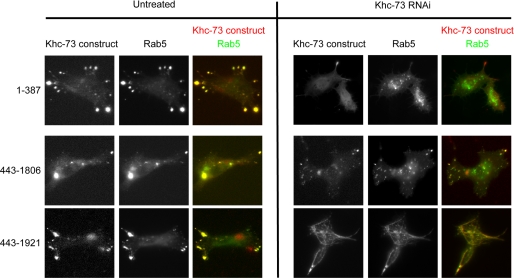
To determine the portion of Khc-73 that is responsible for binding Rab5-containing vesicles, we co-expressed mCherry-tagged constructs of Khc-73 with Rab5-GFP and examined their localization. To our surprise, all constructs (many of which were nonoverlapping) were recruited to Rab5-containing vesicles, with the exception of 1–432 which showed diffuse cytosolic localization (Fig. 5 and supplemental Fig. 4). This result might suggest that the Khc-73 molecule has nonoverlapping N- and C-terminal domains that target the motor to Rab5-containing vesicles. However, another possibility is that some of the expressed constructs were localizing to Rab5 vesicles indirectly through an association (e.g. dimerization) with endogenous Khc-73. To test this hypothesis, we depleted the endogenous Khc-73 by performing RNAi targeted to the 3′-UTR of Khc-73 in BG2 cells (supplemental Fig. 5), then co-expressed Rab5-GFP and the same set of mCherry-tagged Khc-73 constructs. Under these conditions where endogenous Khc-73 was depleted, we found that all of the N-terminal constructs (containing the motor up to the CC motif C-terminal to the forkhead-associated domain) no longer co-localized with Rab5-GFP (Fig. 5 and supplemental Fig. 6). The requirement of endogenous Khc-73 for these N-terminal constructs to localize with early endosomes suggests that these constructs may dimerize with endogenous motor (see “Discussion”). Under conditions of endogenous Khc-73 depletion, N-terminal constructs still accumulated at microtubule-rich regions at the periphery without co-localization with Rab5 (Fig. 5 and supplemental Fig. 6), which might reflect dimerization and processive movement of these constructs toward microtubule plus ends.
FIGURE 5.
RNAi-mediated knockdown of endogenous Khc-73 identifies Rab5 vesicle-targeting domain in Khc-73. Drosophila BG2 cells were subjected to 7-day RNAi treatment with an RNAi construct targeting the 3′-UTR of Khc-73 or mock treated. On day 4, cells were transfected with constructs containing the mCherry-tagged portion of Khc-73 shown under the control of the metallothionein promoter and GFP-tagged Rab5 under the control of the actin promoter. On day 6, cells were incubated with 50 mm copper sulfate for 16 h. On day 7, time-lapse imaging was performed.
In contrast to the N-terminal constructs, all of the C-terminal constructs (containing the portion of the tail between the CC and the CAP-Gly domain) co-localized with Rab5-GFP in the absence of endogenous Khc-73 (Fig. 5 and supplemental Fig. 6). However, there were distinct differences in constructs that contained or did not contain the CAP-Gly domain. A C-terminal construct (443–1806) that lacked the CAP-Gly domain co-localized with Rab5-GFP in its normal punctate pattern (Fig. 5). However, inclusion of the CAP-Gly domain in this construct (443–1921) resulted in strong microtubule binding and altered the normal localization of Rab5-GFP in BG2 cells, recruiting these membranes all along the microtubule lattice (Fig. 5). The fact that the expression of full-length Khc-73 does not show this strong microtubule binding and abnormal Rab-5 localization suggests that the presence of the motor domain in the wild-type protein masks this behavior. These results indicate that the C-terminal half of Khc-73 contains a cargo binding site for Rab5-containing early endosomes and that the CAP-Gly domain is not needed for directing Khc-73 to these membranes.
DISCUSSION
In this study, we have performed the first in vitro characterization of the motility of Khc-73, a member of the Kinesin-3 family. Khc-73 is well conserved in metazoans, but not found in lower organisms, and appears to have important functions in neuronal development (17, 18, 22), although it likely serves additional roles as well. Although it has been controversial whether Kinesin-3 family members are monomeric or dimeric (10, 11, 14, 16), our results suggest that truncated Khc-73 constructs are in an equilibrium between monomers and dimers and that the dimer is a fast and highly processive motor that generates forces comparable in magnitude to Kinesin-1. We also provide evidence that Khc-73 strongly associates with early endosomes in vivo, an interaction mediated through its nonmotor domain. Collectively, these findings provide new insight into the biophysical mechanism and cell biological roles for this poorly understood kinesin.
Evidence that Kinesin-3 Can Form Dimers in Vitro and in Vivo
The Kinesin-3 class of motors was originally considered to be “monomeric” based upon the initial biochemical characterization of KIF1A as a monomer (10) and the finding that Kinesin-3 members have lower propensity of predicted CC formation compared with other kinesin classes. In addition, single-molecule studies suggested that monomeric KIF1A might transport cargo by a biased diffusion mechanism (12). In this work, a series of Khc-73 constructs of the N-terminal, motor domain-containing half of the protein behave predominantly as monomers in hydrodynamic analysis. However, a weak dimer might dissociate to a monomer during these prolonged separation techniques. Indeed, our in vitro motility experiments are most consistent with Khc-73 being in equilibrium between a monomer and dimer. In support of this idea, Khc-73 dimerized constitutively by fusion to a C-terminal LZ shows longer run lengths and a much higher (∼20-fold) frequency of processive movement than a comparable construct without the LZ, but the velocities of the LZ and non-LZ constructs were nearly identical. These results are most easily explained by a model in which the majority of the expressed Khc-73 molecules are monomers that do not exhibit processive movement, but a subpopulation of Khc-73 forms a dimer with identical motility properties to Khc-73-LZ. Interestingly, comparably truncated Unc104 constructs do not exhibit processive motility unless LZ-dimerized (14). Unc104/KIF1A possess a long flexible hinge between two helices next to the motor domain, which is thought to create an autoinhibited conformation through the formation of an intramolecular CC that must be broken to form a processive motor dimer (43). Khc-73 appears to lack this flexible loop (Fig. 1A), which may allow these helices to form intermolecular CCs more readily.
Our transfection data also suggest that Khc-73 can dimerize in cells. In the presence of endogenous Khc-73, a variety of different length Khc-73 constructs all co-localize with Rab5-GFP. However, when cells are incubated with RNAi constructs knocking down the endogenous Khc-73 protein, none of the motor-containing constructs that lack the central domain between the CC and the CAP-Gly domain co-localized with Rab5-GFP. This endogenous Khc-73-dependent localization result suggests that endogenous Khc-73 is responsible for recruiting the expressed motor constructs to the surface of Rab5-containing vesicles. Although we cannot rule out a scenario in which a higher order protein complex dependent on full-length Khc-73 is responsible for this recruitment, it seems most likely that the expressed proteins are recruited to Rab5-containing vesicles by forming heterodimers with the endogenous full-length protein.
Collectively, our data suggest that in vitro and in vivo Khc-73 exists in equilibrium between a monomer and dimer. Our biochemical analysis of purified Khc-73 showing a predominant monomer species is in general agreement with previous results with KIF1A by the Hirokawa laboratory (11, 13). However, the Hirokawa laboratory did not report or suggest the possibility that a subpopulation of this kinesin can self-associate to create a dimer. In a contrasting story, Hammond et al. propose that KIF1A is constitutively dimerized, based upon co-immunoprecipitation from lysate, chemical cross-linking, intracellular FRET, co-migration with conventional kinesin by gradient centrifugation, and single-molecule motility (16). It is important to note, though, that while our current in vitro work with Khc-73 and the previous work on KIF1A used bacteria- and baculovirus-expressed constructs (11, 13), the recent study by Hammond et al. used KIF1A from cell lysate from cell lines expressing KIF1A constructs for in vitro studies. This presents the possibility that there is a cellular component that stabilizes the Kinesin-3 dimer, which is consistent with our in vivo data with Khc-73. Future studies are targeted toward identifying this potential cellular factor.
Khc-73 and Rab5-containing Endosomes
We observed a striking in vivo co-localization of Khc-73 with Rab5-GFP, a marker of early endosomes (38, 39). Khc-73 co-localized with Rab5-GFP at the tips of microtubules as well as on a pool of motile intracellular puncta. By co-expressing various constructs of Khc-73 with Rab5 in vivo, we found the Rab5-containing vesicle targeting domain to be between the CC domain and the CAP-Gly domain in a region of the protein that has no predicted conserved domains. There is precedence of a Kinesin-3 family member binding to Rab5-containing vesicles, as one of the mammalian homologs, KIF16B, transports early endosomes to the plus end of microtubules (44). However, in the case of KIF16B, a C-terminal pleckstrin homology domain (not found in Khc-73) was shown to be important for the recruitment and transport of Rab5-containing endosomes by KIF16B. In addition, although KIF16B was shown to be necessary for the normal localization of early endosomes in HeLa cells (44), we have found that Rab5-GFP-containing endosomes continue to move after RNAi-mediated knockdown of Khc-73 in Drosophila S2 and BG2 cell lines (supplemental Fig. 5). This result might suggest that although Khc-73 is recruited to early endosomes, it does not transport endosomes along microtubules and might serve some other role. However, it also could be that Khc-73 transports early endosomes but that a redundant kinesin also moves early endosomes, thereby accounting for their continued motility after Khc-73 knockdown. Another possibility that cannot be excluded is that residual Khc-73 after RNAi treatment (supplemental Fig. 5) might suffice for early endosome motility.
A critical question that remains is how the biophysical properties and endosomal localization of Khc-73 might be related its biological functions in the living animal. The clearest biological activity ascribed to Khc-73 is its mitotic role in spindle orientation in Drosophila neuroblasts (21). Through its interaction with Discs Large, Khc-73 induces the cortical polarity of the Pins/Gαi complex that is necessary for asymmetric cell division. The Khc-73 homolog in mammals also has been found to bind to membrane scaffolding proteins (18, 45). In our study, Khc-73 also localizes to early endosomes at the tips of microtubules that are very close to the cell cortex. In summary, there are now several results implicating Khc-73 with functions of microtubules/cargo at the cell cortex, but its exact activity and mechanism remain unclear and constitute an interesting topic for future investigation.
Supplementary Material
Acknowledgments
We thank Chris Doe and Kevin Slep for providing constructs, Nico Stuurmann for microscopy assistance, and Andrew Carter for assistance with the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants 38499 (to R. D. V.) and CA094414 (to A. H. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–6 and Movies 1–3.
- Khc-73
- Kinesin-73
- CAP-Gly
- cytoskeleton-associated protein glycine-rich
- CC
- coiled-coil
- LZ
- leucine zipper
- pN
- piconewtons
- TIRF
- total internal reflection fluorescence.
REFERENCES
- 1. Vale R. D. (2003) Cell 112, 467–480 [DOI] [PubMed] [Google Scholar]
- 2. Hirokawa N., Nitta R., Okada Y. (2009) Nat. Rev. Mol. Cell Biol. 10, 877–884 [DOI] [PubMed] [Google Scholar]
- 3. Pfister K. K., Wagner M. C., Stenoien D. L., Brady S. T., Bloom G. S. (1989) J. Cell Biol. 108, 1453–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schnapp B. J., Reese T. S., Bechtold R. (1992) J. Cell Biol. 119, 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brendza R. P., Serbus L. R., Duffy J. B., Saxton W. M. (2000) Science 289, 2120–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gennerich A., Vale R. D. (2009) Curr. Opin. Cell Biol. 21, 59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rice S., Lin A. W., Safer D., Hart C. L., Naber N., Carragher B. O., Cain S. M., Pechatnikova E., Wilson-Kubalek E. M., Whittaker M., Pate E., Cooke R., Taylor E. W., Milligan R. A., Vale R. D. (1999) Nature 402, 778–784 [DOI] [PubMed] [Google Scholar]
- 8. Hwang W., Lang M. J., Karplus M. (2008) Structure 16, 62–71 [DOI] [PubMed] [Google Scholar]
- 9. Hall D. H., Hedgecock E. M. (1991) Cell 65, 837–847 [DOI] [PubMed] [Google Scholar]
- 10. Okada Y., Yamazaki H., Sekine-Aizawa Y., Hirokawa N. (1995) Cell 81, 769–780 [DOI] [PubMed] [Google Scholar]
- 11. Nangaku M., Sato-Yoshitake R., Okada Y., Noda Y., Takemura R., Yamazaki H., Hirokawa N. (1994) Cell 79, 1209–1220 [DOI] [PubMed] [Google Scholar]
- 12. Nitta R., Kikkawa M., Okada Y., Hirokawa N. (2004) Science 305, 678–683 [DOI] [PubMed] [Google Scholar]
- 13. Okada Y., Hirokawa N. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 640–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tomishige M., Klopfenstein D. R., Vale R. D. (2002) Science 297, 2263–2267 [DOI] [PubMed] [Google Scholar]
- 15. Klopfenstein D. R., Tomishige M., Stuurman N., Vale R. D. (2002) Cell 109, 347–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hammond J. W., Cai D., Blasius T. L., Li Z., Jiang Y., Jih G. T., Meyhofer E., Verhey K. J. (2009) PLOS Biol. 7, e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li H. P., Liu Z. M., Nirenberg M. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 1086–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hanada T., Lin L., Tibaldi E. V., Reinherz E. L., Chishti A. H. (2000) J. Biol. Chem. 275, 28774–28784 [DOI] [PubMed] [Google Scholar]
- 19. Weisbrich A., Honnappa S., Jaussi R., Okhrimenko O., Frey D., Jelesarov I., Akhmanova A., Steinmetz M. O. (2007) Nat. Struct. Mol. Biol. 14, 959–967 [DOI] [PubMed] [Google Scholar]
- 20. Steinmetz M. O., Akhmanova A. (2008) Trends Biochem. Sci. 33, 535–545 [DOI] [PubMed] [Google Scholar]
- 21. Siegrist S. E., Doe C. Q. (2005) Cell 123, 1323–1335 [DOI] [PubMed] [Google Scholar]
- 22. Horiguchi K., Hanada T., Fukui Y., Chishti A. H. (2006) J. Cell Biol. 174, 425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Slep K. C., Vale R. D. (2007) Mol. Cell 27, 976–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Siegel L. M., Monty K. J. (1966) Biochim. Biophys. Acta 112, 346–362 [DOI] [PubMed] [Google Scholar]
- 25. Gennerich A., Carter A. P., Reck-Peterson S. L., Vale R. D. (2007) Cell 131, 952–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yildiz A., Forkey J. N., McKinney S. A., Ha T., Goldman Y. E., Selvin P. R. (2003) Science 300, 2061–2065 [DOI] [PubMed] [Google Scholar]
- 27. Svoboda K., Block S. M. (1994) Cell 77, 773–784 [DOI] [PubMed] [Google Scholar]
- 28. Kerssemakers J. W., Munteanu E. L., Laan L., Noetzel T. L., Janson M. E., Dogterom M. (2006) Nature 442, 709–712 [DOI] [PubMed] [Google Scholar]
- 29. Reck-Peterson S. L., Derr N., Stuurman N. (2010) Cold Spring Harbor Protoc. pdb.prot5399 [DOI] [PubMed] [Google Scholar]
- 30. Rogers S. L., Wiedemann U., Stuurman N., Vale R. D. (2003) J. Cell Biol. 162, 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ui-Tei K., Nishihara S., Sakuma M., Matsuda K., Miyake T., Miyata Y. (1994) Neurosci. Lett. 174, 85–88 [DOI] [PubMed] [Google Scholar]
- 32. Goshima G., Vale R. D. (2003) J. Cell Biol. 162, 1003–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Griffis E. R., Stuurman N., Vale R. D. (2007) J. Cell Biol. 177, 1005–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kural C., Serpinskaya A. S., Chou Y. H., Goldman R. D., Gelfand V. I., Selvin P. R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 5378–5382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Svoboda K., Schmidt C. F., Schnapp B. J., Block S. M. (1993) Nature 365, 721–727 [DOI] [PubMed] [Google Scholar]
- 36. Lansbergen G., Akhmanova A. (2006) Traffic 7, 499–507 [DOI] [PubMed] [Google Scholar]
- 37. Perez F., Diamantopoulos G. S., Stalder R., Kreis T. E. (1999) Cell 96, 517–527 [DOI] [PubMed] [Google Scholar]
- 38. Deneka M., Neeft M., van der Sluijs P. (2003) Crit. Rev. Biochem. Mol. Biol. 38, 121–142 [DOI] [PubMed] [Google Scholar]
- 39. Zhang J., Schulze K. L., Hiesinger P. R., Suyama K., Wang S., Fish M., Acar M., Hoskins R. A., Bellen H. J., Scott M. P. (2007) Genetics 176, 1307–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bucci C., Parton R. G., Mather I. H., Stunnenberg H., Simons K., Hoflack B., Zerial M. (1992) Cell 70, 715–728 [DOI] [PubMed] [Google Scholar]
- 41. Horiuchi H., Lippé R., McBride H. M., Rubino M., Woodman P., Stenmark H., Rybin V., Wilm M., Ashman K., Mann M., Zerial M. (1997) Cell 90, 1149–1159 [DOI] [PubMed] [Google Scholar]
- 42. Nielsen E., Severin F., Backer J. M., Hyman A. A., Zerial M. (1999) Nat. Cell Biol. 1, 376–382 [DOI] [PubMed] [Google Scholar]
- 43. Al-Bassam J., Cui Y., Klopfenstein D., Carragher B. O., Vale R. D., Milligan R. A. (2003) J. Cell Biol. 163, 743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hoepfner S., Severin F., Cabezas A., Habermann B., Runge A., Gillooly D., Stenmark H., Zerial M. (2005) Cell 121, 437–450 [DOI] [PubMed] [Google Scholar]
- 45. Asaba N., Hanada T., Takeuchi A., Chishti A. H. (2003) J. Biol. Chem. 278, 8395–8400 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.