Abstract
The basement membrane protein laminin-332 (laminin-5) mediates both stable cell adhesion and rapid cell migration and thus has the potential to either restrain or promote tumor cell metastasis. The major cellular receptors for laminin-332 are integrin α3β1, which mediates rapid tumor cell migration, and integrin α6β4, which often mediates stable cell attachment. Tetraspanin protein CD151 interacts directly with both α3β1 and α6β4 integrins and with other tetraspanins, thereby promoting α3β1 and α6β4 association with tetraspanin-enriched microdomains on the cell surface. To explore the possibility of selectively modulating tumor cell responses to laminin-332, we re-expressed a series of CD151 mutants in epidermoid carcinoma cells with near total, RNAi-mediated silencing of endogenous CD151. The interactions of CD151 with its integrin partners or its interactions with other tetraspanins were selectively disrupted by specific mutations in the CD151 large extracellular loop (EC2 domain) or in intracellular CD151 palmitoylation sites, respectively. CD151-integrin association and CD151-tetraspanin association were both important for α3β1 integrin-dependent initial adhesion and rapid migration on laminin-332. Remarkably, however, only CD151-integrin association was required for stable, α6β4 integrin-dependent cell attachment on laminin-332. In addition, we found that a QRD amino acid motif in the CD151 EC2 domain, which had been thought to be crucial for CD151-integrin interaction, is not essential for CD151-integrin association or for the ability of CD151 to promote several different integrin functions. These new data suggest potential strategies for selectively modulating migratory cell responses to laminin-332, while leaving stable cell attachment on laminin-332 intact.
Keywords: Cell Adhesion, Cell Motility, Integrin, Laminin, Protein Palmitoylation, Tetraspanin
Introduction
CD151 is a member of the tetraspanin family of proteins, which possess four transmembrane domains, intracellular amino and carboxyl termini, and one small and one large extracellular domain (named EC1 and EC2, respectively). Tetraspanins share a unique cysteine motif in their EC2 domains that sets them apart from other four-pass proteins. Genetic deletion of CD151 results in defects in the maintenance of kidney and skin epithelial integrity, wound healing, platelet aggregation, and thrombus growth and stability, as well as reduced pathological angiogenesis (1–7). Analysis of clinical samples has revealed potential roles for CD151 in progression and metastasis of colon, prostate, lung, hepatocellular, and breast carcinomas (8–14).
Although CD151 function in platelets has been linked to the platelet integrin αIIbβ3 (3), the basis of CD151 functions in carcinomas and in normal epithelia may be its association with its major integrin partners α3β1, α6β1, and α6β4. CD151 binds directly to these laminin-binding integrins in an interaction involving the CD151 EC2 domain and a membrane proximal region of the α integrin subunit ectodomain. CD151 also associates with other tetraspanins, via a mechanism that depends on palmitoylation of membrane proximal intracellular cysteine residues found on CD151 and other tetraspanins. CD151 thus promotes the association of its integrin partners with other tetraspanins and tetraspanin-associated proteins in cell surface microdomains referred to as tetraspanin-enriched microdomains (TEMs)2 (15). Loss of CD151, by either genetic deletion or RNAi-mediated silencing, dramatically reduces α3 and α6 integrin association with TEMs (5, 11, 16, 17). Concordantly, laminin-dependent adhesion, spreading, motility, morphology, and signaling are all impaired or altered by CD151 ablation (5, 11, 16–20). The ability of CD151 to promote the function of laminin-binding integrins may help to explain why loss of CD151 reduces tumorigenesis, metastatic colonization, growth, and scattering in three-dimensional matrices, and laminin-332 (LM-332)-dependent chemoresistance in experimental systems (11, 12, 14, 21–23).
Although there are compelling reasons to believe that many of the physiological and pathological functions of CD151 stem from its ability to promote laminin-binding integrin association with TEMs, direct evidence in support of that hypothesis is still relatively scarce. Much of the earlier CD151 structure/function analysis was performed in a cellular background of wild type CD151. This, coupled with the fact that CD151 likely exists as a homodimer, has made it difficult to discern whether CD151-tetraspanin and CD151-integrin interactions are both always essential for specific CD151 functions.
To begin to clarify the roles of specific CD151 interactions in CD151 function, we have established epidermoid carcinoma cells with stable, near total, RNAi-mediated silencing of CD151 expression (16, 24). Loss of CD151 in this system impaired cell adhesion and migration on LM-332, phenotypes that could be reversed by re-expressing wild type CD151 (16). Here, we have taken advantage of our system to assess the function of CD151 mutants with little to no interference from endogenous wild type CD151. Our results reveal essential contributions of both CD151-integrin and CD151-tetraspanin interactions for specific functions, as well as important differences between domains required to support certain α3- and α6-dependent cell behaviors. Surprisingly, our data also reveal that a well studied integrin interaction site in the CD151 EC2 domain is not by itself essential for integrin association and is dispensable for at least some CD151-supported integrin functions.
EXPERIMENTAL PROCEDURES
Antibodies and Extracellular Matrix Proteins
Anti-integrin mAbs used in this study were anti-α3, A3-X8, and A3-IIF5 (25); anti-α6, A6-ELE (26); and GoH3 (GeneTex, San Antonio, TX). A polyclonal anti-α3 integrin antibody, A3-CYT, was raised against the synthetic Cys-oligopeptide, CYEAKRQKAEMKSQPSETERLTDDY, derived from the human α3 integrin cytoplasmic tail, similar to the method used in a previous study (27). The anti-α6 integrin polyclonal antibody, AA6NT, was the generous gift of Anne Cress (28). Anti-tetraspanin mAbs used were anti-CD9, ALB6 (Chemicon International, Temecula, CA), and MM2/57 (GeneTex, San Antonio, TX); anti-CD81, mAb M38 (29); and anti-CD151, 5C11 (30). Polyclonal rabbit anti-FLAG epitope tag was from Sigma-Aldrich. For some FLAG immunoprecipitations, monoclonal anti-FLAG M2 affinity gel was used (Sigma-Aldrich). Anti-LM-332 mAb, 6F12 (also called K140; (31)), was used for LM-332 affinity purification. Secondary reagents were Alexa Fluor 680-conjugated goat anti-rabbit and goat anti-mouse antibodies (Invitrogen), Cy2 goat anti-mouse antibody (Jackson ImmunoResearch Laboratories), and IRDye800 goat anti-mouse (Rockland, Gilbertsville, PA). Rat tail collagen I and human fibronectin were purchased from BD Biosciences. Human LM-332 was purified from SCC-25 squamous cell carcinoma-conditioned medium as described previously (16).
Cell Culture, RNAi, and Retroviral Transduction
A431 epithelial carcinoma cells, HaCaT immortalized keratinocyte cells, and retroviral packaging lines, GP2–293 and PT67 (BD Biosciences), were cultured in high glucose DME. SCC-25 squamous carcinoma cells were cultured in a 1:1 mixture of DMEM and F12 media supplemented with 0.4 μg/ml hydrocortisone. All cultures were supplemented with 10% fetal bovine serum, 2 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin (all from Invitrogen). For RNAi, double-stranded oligonucleotides encoding shRNAs targeting the human CD151, α3 integrin, or α6 integrin mRNAs were annealed and then cloned into the pSIREN-RetroQ retroviral vector (BD Biosciences) as described (16). The shRNA targeting sequences were as follows: CD151, 5′-AGTACCTGCTGTTTACCTACA-3′; α3 integrin, 5′-GGATGACTGTGAGCGGATGAA-3′; and α6 integrin, 5′-GTATGTAACAGCAACCTTAAA-3′. After retroviral transduction, stable α3- and α6-silenced cells were obtained by selection and cell sorting, as described previously for CD151-silenced cells (16).
For CD151 re-expression experiments in A431 or HaCaT cells, recombinant PCR was used to construct CD151 cDNAs with (i) silent mutations in the CD151 shRNA targeting sequence, to allow re-expression in cells harboring the CD151 shRNA construct, (ii) an amino-terminal DYKDDDK (FLAG) epitope tag, and (iii) additional mutations in the second extracellular loop (EC2) or in membrane-proximal, intracellular cysteine residues (palmitoylation sites). Three EC2 mutants were created. For the CD151INF mutant, the amino acids QRD at positions 194–196 were changed to INF, which are the amino acids at the analogous positions in tetraspanin CD63 (32). For the CD151VR and CD151EC2 mutants, the variable region (amino acids Ser158 to Gly207) or the entire EC2 domain (amino acids Tyr114 to Arg221), respectively, was replaced with the corresponding regions from tetraspanin TSPAN7 (A15/TM4FS2). To create CD151Palm, six critical intracellular cysteine residues, which are modified by palmitoylation (33, 34), were changed to serines. A wild type FLAG-tagged CD151 re-expression cDNA was also constructed for comparison to the CD151 mutants.
The CD151 constructs, cloned into the LXIZ retroviral vector, were cotransfected with a pVSV-G retroviral coat protein vector into GP2–293 packaging cells using the calcium phosphate method. At 48 h and again at 96 h after transfection, virus-containing medium was collected, 0.45 μm filtered, supplemented with 4 μg/ml polybrene (Sigma-Aldrich), and then used to transduce PT67 packaging cells. Stable packaging lines were selected, and supernatant from these lines was used to transduce CD151-silenced A431 or HaCaT sh3 cells. Stable transductants were selected with 0.5 mg/ml Zeocin (Invitrogen) while maintaining selection with 0.1 μg/ml puromycin (Sigma-Aldrich) to retain the sh3 shRNA retroviral vector. Restored expression of CD151 in these cells was confirmed by flow cytometry using antibodies to CD151 and/or the FLAG epitope tag.
Immunoprecipitation and Immunoblotting
Cells were lysed by scraping into 20 mm HEPES, pH 7.5, 150 mm NaCl, 5 mm MgCl2 (HBSM) supplemented with 1% detergent, and protease inhibitors (2 mm PMSF, 10 μg/ml aprotinin, 5 μg/ml leupeptin, and 5 μg/ml E-64; Roche Diagnostics). Detergents used were Brij 96V, a 1:1 mixture of Brij 96V and Brij 99, or Triton X-100 (all from Sigma-Aldrich). Cells were biotinylated in some experiments using 0.1 mg/ml sulfo-NHS-LC biotin (Thermo Scientific) in HBSM for 1 h at room temperature followed by three rinses with HBSM prior to lysis. Proteins were extracted by rocking at 4 °C for 1 h, and insoluble material was removed by centrifugation for 15 min at 16,000 × g. Lysates were precleared for 1 h at 4 °C with protein G-Sepharose (Pierce Biotechnology) and centrifuged as described previously. Protein concentrations from different cell types were normalized according to the results of a BCA assay (Pierce Biotechnology), and specific antibodies and protein G-Sepharose were added. Immune complexes were collected overnight at 4 °C in most cases. For some experiments using Brij 96V, immune complexes were collected for 1 h at room temperature. After four rinses with lysis buffer, immune complexes were eluted by boiling in sample buffer, resolved by SDS-PAGE, and transferred to nitrocellulose. Blots were blocked in either 5% nonfat milk in TBS or a 1:5 mixture of Aqua-Block (East Coast Bio) and PBS followed by probing with specific primary antibodies diluted in blocking buffer. Blots were developed using Alexa Fluor 680 or IRDye 800-conjugated secondary antibodies diluted 1:5,000 in blocking buffer. DyLight 800 NeutrAvidin (Pierce) was used at 1:20,000 to develop blots with biotin-labeled proteins. Blots were analyzed with a LI-COR Odyssey blot imager (LI-COR Biotechnology).
Flow Cytometry
Cells were stained on ice for 1 h with negative control or specific primary antibodies at 5 μg/ml in blocking buffer (PBS with 10% heat-inactivated goat serum and 0.02% sodium azide). After three rinses in cold PBS, cells were stained on ice for 45 min with a Cy2-conjugated goat anti-mouse secondary antibody (Jackson ImmunoResearch Laboratories) diluted 1:200 in blocking buffer. After three additional rinses, cells were analyzed on a Cell Lab Quanta flow cytometer (Beckman Coulter).
Immunostaining
A431 cells cultured on sterile glass coverslips were fixed with 10% formalin in HEPES-buffered saline with 4% sucrose and 1 mm MgCl2, rinsed twice with TBS, and blocked with 10% goat serum in PBS. For staining the intracellular FLAG epitope, cells were permeabilized with 0.2% Brij 99 (Sigma-Aldrich) during the blocking step. For staining substrate LM-332, cells were extracted with 1% Triton X-100 prior to fixation. Cells or substrates were stained for 1 h with primary antibodies in blocking buffer and then for 45 min with Cy2 goat anti-mouse antibody. Cells were mounted in Prolong Gold (Invitrogen) and analyzed by fluorescence microscopy as described previously (16, 24).
Time-lapse Motility Assays
A431 cells were plated at 5 × 105 cells per 25 cm2 flask on day 1 and starved overnight on day 2 in SFM (DME with 5 mg/ml cell culture grade BSA (Sigma-Aldrich) and 25 mm HEPES, pH 7.2). On day 3, cells were harvested by treating with trypsin-EDTA for 3 min at 37 °C and collected in SFM supplemented with 0.1 mg/ml soybean trypsin inhibitor and 20 μg/ml DNase I (Worthington Biochemical, Lakewood, NJ). After centrifugation, cells were resuspended in SFM, and 2.3 × 105 cells were plated to 35-mm dishes coated overnight with 1 μg/ml LM-332. Cultures were maintained on a Leica DMIRE2 inverted microscope (Deerfield, IL) in a stage incubator (20/20 Technology, Wilmington, NC) providing a humidified 5% CO2, 37 °C atmosphere. OpenLab software (Improvision, Lexington, MA) running on an Apple iMac computer controlled illumination and image acquisition (Cupertino). After allowing 30 min for cells to attach, images were acquired at a rate of 1 frame/min for 3 h using a Hamamatsu ORCA-285 CCD camera (Bridgewater, NJ) and a 20× C Plan phase objective. ImageJ software (49) was used to record the XY position of cell centroids in the frames 3 min apart. Custom Java software (code available upon request) was then used to calculate the distance traveled in each 3-min interval for each cell. Every cell in each field was followed for as long as it remained in view. All cells that could be tracked for at least 1 h were included in the final calculation of velocity (typically 40–60 cells per experiment).
Adhesion Assays
Substrates for adhesion assays were 1 μg/ml LM-332, 20 μg/ml collagen I, 20 μg/ml fibronectin, 100 μg/ml poly-l-lysine (positive control), or 10 mg/ml heat-inactivated BSA (negative control). After overnight coating, wells were rinsed and blocked with 10 mg/ml heat-inactivated BSA. Cells were starved overnight, harvested as for time-lapse motility assays, and resuspended at 3 × 105 cells/ml in SFM. 100 μl of cell suspension was plated to each of four substrate-coated wells per condition in a 96-well plate. After 30 min at 37 °C with 5% CO2, wells were rinsed three times with warm DME, 40 mm HEPES, pH 7.2, using a multichannel pipette and flicking to remove unattached cells. Positive control poly-l-lysine wells were gently rinsed once. Cells remaining after rinses were fixed for 30 min at room temperature with 10% formalin fixative containing 2 mm MgCl2, followed by a 20-min stain with freshly filtered 0.1% crystal violet in double distilled H2O. Cells were destained by rinsing three times with tap water, air dried 10 min, and solubilized overnight with 100 μl/well 1% Triton X-100 in double distilled H2O. Absorbance at 595 nm was determined with a SPECTRAmax PLUS microplate spectrophotometer (Molecular Devices, Sunnyvale, CA). For each cell type, adhesion was expressed as fraction of cells input using absorbances in the poly-l-lysine wells to determine total input.
Proliferation Assays
Cells were plated in replicate 96-well plates, four wells per cell type. On successive days, cells were refed with fresh medium containing WST-1 proliferation reagent (Roche). After 1 h, absorbance at 440 nm was measured in a plate reader.
Resistance to Detachment
A431 or HaCaT cells were plated at 5 × 105 cells per well on a 48-well plate in standard media and allowed to grow for 24–48 h. Cells were rinsed once with DMEM with 40 mm HEPES and then treated with 0.25% trypsin/0.53 mm EDTA for 6 min at 37 °C, 5% CO2. Wells were then rinsed with standard media, followed by two rinses with DMEM with 40 mm HEPES. Medium for rinses was removed by flicking the plates; flicking also helped remove unattached cells. Cells were then fixed, stained with crystal violet, destained, and analyzed as described above for adhesion assays, except that total cells input was determined using wells that were not treated with trypsin-EDTA.
RESULTS
Expression of CD151 Mutants in CD151-silenced Cells
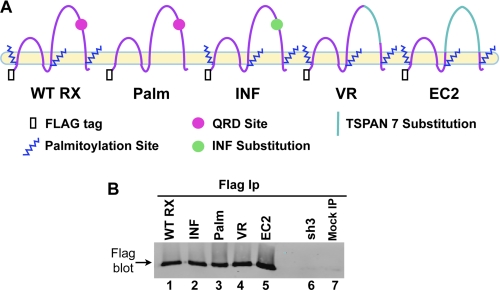
To define the structural motifs of CD151 required for its ability to promote α3β1 and α6β4 integrin function in migration and adhesion, we re-expressed wild type CD151 (CD151WT RX) and a panel of CD151 mutants (Fig. 1A) in A431 cells that had been stably silenced for CD151 (A431 sh3 cells) (16). In the CD151Palm mutant, six membrane-proximal intracellular cysteine residues, which are modified by palmitoylation (33, 34), were changed to serine residues. CD151 palmitoylation is important for its association with other tetraspanins but is not required for its strong association with α3 integrin (33). The CD151INF mutant bears a three amino acid substitution of Ile-Asn-Phe (INF) in place of the amino acids Gln-Arg-Asp (QRD) in the large extracellular loop (EC2) domain. These QRD residues are important for the strong interaction of CD151 with a site in the ectodomain of the α3 integrin subunit (32). In the CD151EC2 mutant, the entire EC2 domain was replaced with the EC2 domain of TSPAN7 (TM4SF2, A15), a tetraspanin that we found in preliminary experiments to associate weakly, if at all, with α3β1 integrin. The CD151VR mutant contains a more limited domain swap with TSPAN7 between Ser158 and Gly207, a part of the EC2 domain that has been termed the variable region (35). All of our CD151 re-expression constructs were labeled at the N terminus with a FLAG epitope tag.
FIGURE 1.
Expression of CD151 constructs in A431 cells. A, schematic representation of CD151 mutant proteins. All constructs had N-DYKDDDDK-C (FLAG) sequences attached to their N termini. B, A431 cells were lysed in a 50/50 mixture of 1% Brij 99 and Brij 96V lysis buffer. Cell lysates were immunoprecipitated (IP) with anti-FLAG M2 agarose and blotted with a polyclonal antibody against the FLAG tag. The mock IP was performed with lysis buffer only instead of cell lysate.
FLAG immunoprecipitation followed by FLAG immunoblotting confirmed that wild type and mutant CD151 constructs were all well expressed upon transduction of CD151-silenced A431 sh3 cells (Fig. 1B, lanes 1–5). No band was detected in the sh3 cells, or in mock immunoprecipitations, as expected (Fig. 1B, lanes 6 and 7). Flow cytometry confirmed that cell surface expression levels of wild type CD151, CD151Palm, and CD151INF were all similar (Table 1). The CD151EC2 and VR mutants could not be directly compared with the other mutants by flow cytometry due to their loss of epitopes for available CD151 antibodies.
TABLE 1.
Flow cytometry of CD151 mutant constructs
Mean fluorescence intensity after subtraction of background fluorescence from a nonimmune negative control antibody.
| Cell type | CD9 | α3 Integrin | α6 Integrin | CD151 |
|---|---|---|---|---|
| Wild type | 1646 | 980 | 534 | 262 |
| CD151 sh3 | 1133 | 788 | 569 | 9 |
| CD151 WTRX | 1539 | 1014 | 600 | 368 |
| CD151 INF | 1507 | 866 | 410 | 473 |
| CD151 Palm | 1668 | 921 | 485 | 212 |
| CD151 VR | 1424 | 943 | 646 | * |
| CD151 EC2 | 1419 | 909 | 717 | * |
* The antibody for CD151 does not recognize these mutants.
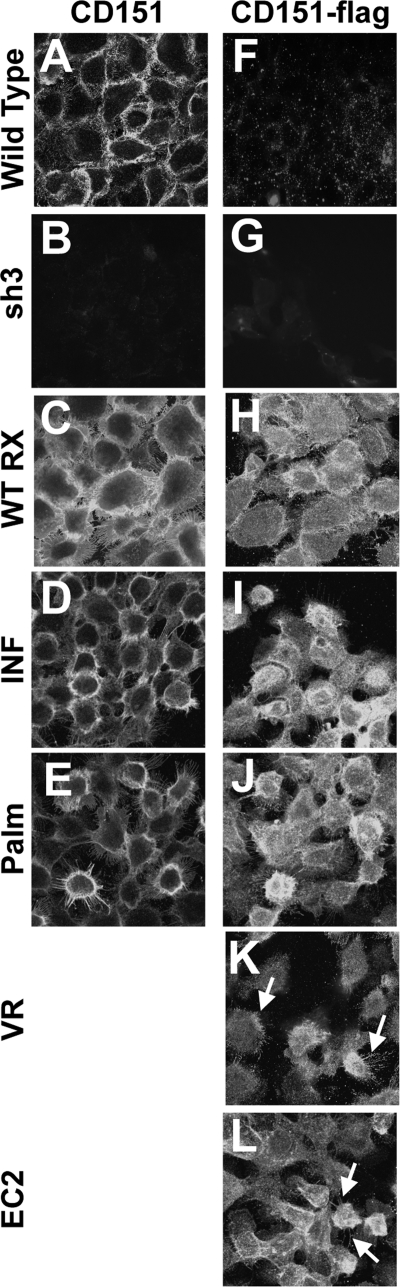
Staining for cell surface CD151 on unpermeabilized cells revealed a similar expression level in wild type parental cells, CD151WT RX, CD151INF, and CD151Palm cells (Fig. 2, A, C, D, and E). Virtually no CD151 was detected in the CD151-silenced sh3 cells (Fig. 2B). Immunostaining for the intracellular FLAG epitope revealed an additional intracellular pool of CD151, as previously described in other cell types (50) (Fig. 2, H–L). There were no obvious differences between the staining pattern or expression levels of wild type CD151 and the various CD151 mutants. Close examination revealed localization of the CD151VR and EC2 mutants on retraction fibers and filopodia, indicative of expression on the cell surface (Fig. 2, K and L, arrowheads). In addition, we have shown in a previous study that the CD151VR mutant is expressed on the A431 cell surface at wild type levels (24). Very little FLAG staining was observed in wild type parental cells or in sh3 cells, as expected (Fig. 2, F and G). Collectively, these data indicated that our CD151 mutants are expressed at similar levels as wild type CD151, with no obvious changes in cellular localization.
FIGURE 2.
CD151 mutants display normal cellular localization and expression. A–E, cells grown on glass coverslips overnight were formalin fixed and incubated with CD151 mAb 5C11 followed by Alexa Fluor 488 secondary antibody. F–L, A431 cells grown overnight on glass coverslips were formalin fixed, permeabilized with 0.2% Brij 99 (Sigma-Aldrich), and then incubated with polyclonal anti-FLAG tag antibody followed by Alexa Fluor 488 secondary antibody. Arrowheads in K and L indicate staining of filopodia or retraction fibers indicative of plasma membrane localization.
Analysis of Integrin and Tetraspanin Interactions in CD151 Mutant Cell Lines
FLAG epitope immunoprecipitation followed by immunoblotting for α3 integrin confirmed that wild type CD151 and the CD151Palm mutant both remain associated with α3β1 integrin in Triton X-100 lysates, whereas the CD151INF mutant does not (Fig. 3A, lanes 1–3), as expected from previous studies (32). Surprisingly, in moderately stringent Brij 96V detergent, the α3β1 association with the CD151INF mutant appeared completely intact, indicating that the QRD→INF mutation weakens but does not abolish the CD151-α3β1 association (Fig. 3A, lanes 4–6). Because the QRD→INF mutation failed to abolish α3β1-CD151 association, we next tested the more extensive CD151VR and EC2 domain swap mutants. Both mutants displayed dramatically impaired α3β1 association in relatively mild Brij 99/96V lysates (Fig. 3B, compare lanes 3 and 4 with lane 1). No α3 was detected in FLAG immunoprecipitations from CD151-silenced sh3 cells (Fig. 3B, lane 2).
FIGURE 3.
The effect of CD151 mutations on associations with other TEM proteins. A, cells were lysed in either 1% Brij 96V or the more stringent Triton X-100 followed by immunoprecipitation (IP) with anti-FLAG M2 agarose and blotting with the anti-α3 integrin polyclonal antibody, A3-CYT. B, cells were lysed in 1% Brij 99/96V followed by anti-FLAG M2 agarose IP and α3 blot with A3-CYT. C, cells were lysed in 1% Brij 99/96V, and then CD9 mAb ALB-6 was used for immunoprecipitation followed by blotting with anti-FLAG polyclonal antibody. D, lanes 1–8 are immunoprecipitations with CD9 mAb ALB-6. Lanes 9–12 are immunoprecipitations with A3-IIF5 anti-α3 mAb. All lanes were lysed in 1% Brij 99/96V and blotted for α3 integrin using polyclonal antibody, A3-CYT. E, cells were lysed in 1% Brij 99/96V and immunoprecipitated using anti-CD81 mAb M38, followed by blotting for CD9 with MM2/57 mAb.
Immunoprecipitation of tetraspanin CD9 followed by FLAG immunoblotting revealed that the various CD151 mutants all retained wild type association with CD9, with the exception CD151Palm mutant (Fig. 3C, lane 4), indicating that the loss of palmitoylation sites successfully disrupted CD151-tetraspanin association, as expected (33, 34). In sum, these data indicated that the VR and EC2 mutations selectively disrupted CD151-α3β1 integrin association, whereas the loss of palmitoylation selectively disrupted CD151-tetraspanin association.
Loss of CD151 dramatically impairs α3β1 association with other tetraspanins (5, 11, 16, 17), indicating that CD151 plays a crucial role in promoting association of α3β1 with TEMs. To test for α3β1-TEM association in our CD151 mutant cells, we immunoprecipitated tetraspanin CD9 and blotted for α3 integrin. Only a low residual level of α3-CD9 association could be detected in cells bearing the CD151Palm, VR, and EC2 mutants (Fig. 3D, lanes 3, 7, and 8), similar to what was observed in CD151-silenced cells (lanes 4 and 6). In contrast, α3-CD9 association remained at wild type levels in CD151INF cells (Fig. 3D, lane 2). Thus, disrupting either CD151-α3 association (VR and EC2 mutants) or CD151-tetraspanin association (Palm mutant) disrupted α3-TEM association, strongly supporting the view that CD151 physically links α3β1 integrin to TEMs. The QRD→INF mutation, while weakening α3β1-CD151 association in more stringent detergent lysates, was not sufficient to abolish this CD151 linker function.
Flow cytometry confirmed that cell surface expression of α3 and α6 integrin and tetraspanin CD9 was not significantly altered in any of the CD151 re-expression mutants or in the CD151-silenced cells (Table 1). In addition, CD9 association with tetraspanin CD81 was also not significantly altered in CD151-silenced cells or in any of the CD151 mutant cell lines, indicating that other interactions within TEMs were not grossly perturbed by the loss of CD151 or the presence of CD151 mutants (Fig. 3E).
α3β1 Integrin Association with TEMs Is Important for Motility and Initial Adhesion on LM-332
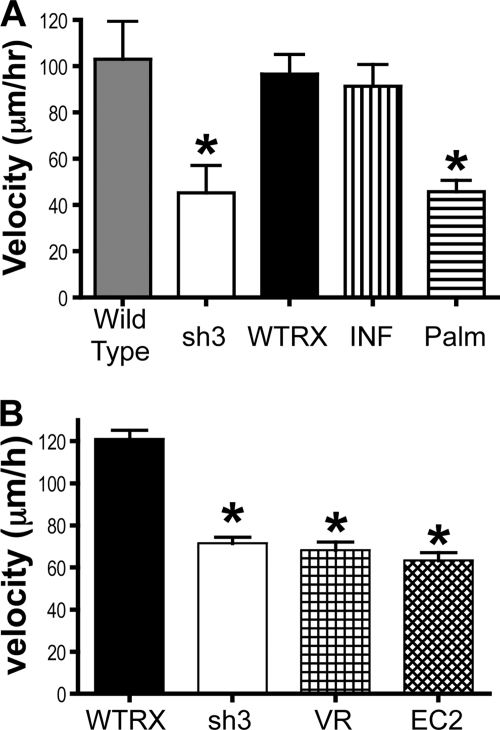
Wild type A431 cells move rapidly and spontaneously on purified LM-332, a behavior that is strongly dependent on α3 integrin (16). When tetraspanin CD151 is silenced, motility is reduced by >50%, and this decrease can be rescued by re-expressing wild type CD151 (16). The biochemical experiments described above revealed that α3β1 integrin association with TEMs could be disrupted by blocking the ability of CD151 to associate with either other tetraspanins (the CD151Palm mutant) or with α3 integrin (the CD151VR and EC2 mutants). In motility experiments on LM-332, CD151-silenced sh3 cells again displayed an ∼60% reduction in migration velocity as compared with either wild type parental cells or CD151WT RX cells (Fig. 4A). The CD151Palm mutant completely failed to rescue the motility defect in sh3 cells. Remarkably, however, the CD151INF mutant supported wild type migration velocity on LM-332 (Fig. 4A). As the CD151INF mutation had no effect on α3β1-dependent motility in our system, we next examined the CD151VR and EC2 mutant cells, in which the CD151-α3 association is more completely disrupted. As shown in Fig. 4B, both of these CD151 mutants completely failed to rescue the reduced motility in sh3 cells. Thus, neither CD151-α3β1 association nor CD151-tetraspanin association was sufficient, on its own, to support wild type α3β1-dependent motility. Only CD151 constructs capable of associating with both α3β1 integrin and tetraspanins were able to rescue the motility defect of the CD151-silenced sh3 cells.
FIGURE 4.
The effect of CD151 mutations on migration on laminin-332. A, motility of wild type and mutant cells on laminin-332 was monitored for 3 h by time-lapse microscopy. Each column represents the average of at least three trials, 40–60 cells per trial. Bars show S.E. *, p < 0.01 compared with CD151WTRX cells (ANOVA with post hoc t test). B, similarly performed motility assays with the CD151VR and CD151EC2 cells. CD151WTRX and CD151sh3 are the average of two trials, whereas CD151VR and CD151EC2 are the average of three trials (40–60 cells/trial). Bars show S.E. *, p < 0.01; **, p < 0.001 compared with CD151WTRX (ANOVA with post hoc t test).
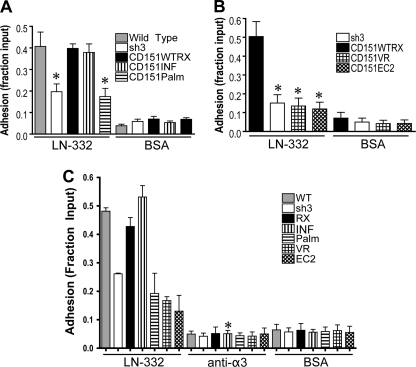
We have previously reported a defect in early adhesion to LM-332 in CD151-silenced cells. The early adhesion defect is rescued upon re-expression of wild type CD151 (16). Similar adhesion defects have been reported in several other studies in which CD151 has been silenced or genetically deleted (16–20). Therefore, we used our CD151 mutants to investigate the requirement for CD151-α3β1 integrin association and CD151-tetraspanin association for initial adhesion to LM-332. Wild type parental cells, CD151WT RX, and CD151INF cells all displayed similar adhesion to LM-332, demonstrating that both wild type and INF constructs could rescue the defect seen in CD151-silenced cells (Fig. 5A). However, CD151Palm, CD151VR, and CD151EC2 cells all performed as poorly as the CD151-silenced cells (Fig. 5, A and B). Thus, as with motility on LM-332, initial adhesion is impaired by mutations that disrupt either CD151-α3β1 association or CD151-tetraspanin association. The reduced adhesion and migration of the CD151-silenced, and the CD151Palm, VR, and EC2 mutant cells was not due to reduced overall viability because growth assays revealed similar proliferation rates for all cell types (supplemental Fig. S1).
FIGURE 5.
The effect of CD151 mutations on initial adhesion to laminin-332. A and B, wild type parental cells, CD151-silenced cells, and cells re-expressing wild type or CD151 mutants were compared in adhesion assays where cells were given 30 min to attach to laminin-332 or BSA-coated wells in a 96-well plate. Nonadherent cells were removed by rinsing, and attached cells were fixed, stained with crystal violet, solubilized, and quantified using a plate reader. Values are reported as the fraction of cells input, measured by cells attached to control wells coated with poly-l-lysine. Each column represents the average of at least three experiments, four wells per condition. Bars show S.E. *, p < 0.01 compared with CD151WTRX cells (ANOVA with post hoc t test). C, adhesion assays comparing wild type, CD151-silenced, and CD151 mutant cells where plated in wells coated with BSA or laminin-332 with or without the addition of 10μg/ml α3-function blocking antibodies. Columns represent the means ± S.E. of three separate trials of four wells per condition per trial. *, p < 0.001 compared with CD151INF cells without α3 function-blocking antibody (ANOVA with post hoc t test).
A431 cells have two LM-332 receptors, α3β1 and α6β4 integrin. However, initial adhesion and motility on LM-332 are predominantly α3β1-dependent processes, with little apparent contribution from α6β4 (16). Given that several previous studies have indicated that the CD151 QRD motif is functionally important in some settings (12, 23, 32, 36), we were surprised by the wild type behavior of our CD151INF cells in adhesion and motility assays. We therefore considered the possibility that α3β1 functions were in fact impaired in CD151INF cells but that a loss of CD151-α3β1 functional interaction in CD151INF cells might actually enhance the ability of CD151 to promote α6β4 function, allowing α6β4 to compensate for reduced α3β1 function. However, an α3 integrin function-blocking antibody virtually abolished adhesion for all of our cell types (Fig. 5C), indicating that initial adhesion to LM-332 is a strongly α3-dependent process in wild type A431 cells, as well as in all the CD151 mutant cells, including CD151INF cells. Thus, the strong, detergent-resistant CD151-α3 integrin association that is provided by the CD151 QRD motif appears to be dispensable for α3β1 functions in some settings.
CD151 Palmitoylation Is Not Required for α6β4 Integrin-dependent Resistance to Detachment
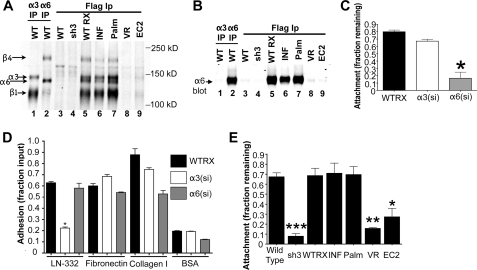
To begin to explore the domains of CD151 required to support α6β4 integrin-dependent functions, we first examined the association of our CD151 mutants with α6β4 in lysates of cell surface-biotinylated cells. Direct immunoprecipitation of α3 integrin yielded bands corresponding to the α3 and β1 subunits (Fig. 6A, lane 1), whereas an α6 immunoprecipitation yielded bands corresponding to the α6 and β4 integrin subunits (lane 2). FLAG epitope immunoprecipitations revealed that α3β1 and α6β4 integrins each associated with the CD151WT RX, CD151INF, and CD151Palm constructs to similar extents (Fig. 6A, lanes 5–7). In contrast, very little α3β1 or α6β4 co-precipitated with the CD151VR or EC2 constructs (Fig. 6A, lanes 8 and 9). No integrin-like bands were co-precipitated by the anti-FLAG epitope antibody from lysates of wild type parental or CD151-silenced sh3 cells, neither of which contain any FLAG-tagged proteins (Fig. 6A, lanes 3 and 4).
FIGURE 6.
Loss of CD151 palmitoylation does not affect α6 integrin-dependent resistance to detachment. A, the indicated cell types were cell surface-labeled with biotin and lysed with 1% Brij 96V. In lanes 1 and 2, anti-α3 integrin mAb, A3X8, and anti-α6 antibody, mAb GoH3, were used for immunoprecipitations (IP) to show where the α3β1 and α6β4 bands, respectively, could be expected in the lanes that follow. The remaining lanes were FLAG immunoprecipitations using anti-FLAG M2 agarose. Proteins were visualized by blotting with DyLight 800 NeutrAvadin. B, immunoprecipitates prepared as in A were analyzed by immunoblotting with the polyclonal anti-α6 integrin antibody, AA6NT. C, de-adhesion assay using CD151WTRX A431 cells and A431 cells silenced for either α3 or α6 integrin. Cells were grown to full confluence in the wells of a 24-well plate for 48–72 h before being exposed to 0.25% trypsin with 0.53 mm EDTA for 6 min at 37 °C. Cells that were detached by trypsin/EDTA were rinsed away, and the remaining cells were fixed, stained with crystal violet, solubilized, and quantified using a plate reader. Values are reported as the fraction of cell input, measured by cells attached to control wells that were not exposed to trypsin. Each column represents the average of three experiments, four wells per condition. Bars show S.E. *, p < 0.001 compared with CD151WTRX cells (ANOVA with post hoc t test). D, adhesion assay of CD151WTRX, α3-silenced, and α6-silenced given 30 min to attach to LN-332, fibronectin, collagen I, or BSA. Attached cells were quantified as described for Fig. 5. This figure represents one of three experiments, each with four replicates per cell type and condition. Bars show S.E. *, p < 0.001 compared with CD151WTRX cells (ANOVA with post hoc t test). E, De-adhesion assay as described for C using wild type, CD151-silenced cells, and CD151 wild type rescue or mutant cells. Each column represents the average of three experiments, four wells per condition. Bars show S.E. *, p < 0.05; **, p < 0.01; ***, p < 0.001 compared with CD151WTRX cells (ANOVA with post hoc t test).
To confirm this pattern of association for α6β4 integrin, we immunoprecipitated α3 or α6 integrin, or the various FLAG-tagged CD151 constructs and immunoblotted for α6 integrin (Fig. 6B). Compared with the robust α6 band in the direct α6 immunoprecipitation, no α6 integrin was detected in the α3 immunoprecipitation (Fig. 6B, lanes 1 and 2). As observed with surface-labeled material, α6 integrin co-precipitated with the FLAG-tagged CD151WT RX, CD151INF, and CD151Palm constructs (Fig. 6B, lanes 5–7) but not with the CD151VR or EC2 constructs (lanes 8 and 9). No α6 bands appeared in control lysates lacking FLAG-tagged proteins (Fig. 6B, lanes 3 and 4). Together, the data in Fig. 6, A and B, showed that the same CD151 mutants that lose association with α3β1 integrin (CD151VR and EC2) also lose association with α6β4 integrin.
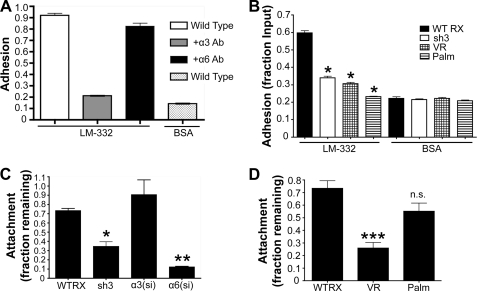
Because α6β4 integrin makes minimal contributions to initial adhesion and migration on LM-332 in our cells, we sought to establish an assay that might capture α6β4 function in stable cell attachment. Previous studies have shown that loss of α6β4 function in keratinocytes results in loss of resistance to detachment upon treatment with trypsin/EDTA (37). Because, like normal keratinocytes, A431 cells deposit their own LM-332 and form hemidesmosome-like structures (38), we tested the relative contributions of α3 and α6 integrin in detachment assays using mature, confluent cell monolayers. Wild type, α3 integrin-silenced, and α6 integrin-silenced A431 cells were plated in 48-well plates and cultured to confluence for 2 days. Cells were then treated with trypsin/EDTA for 6 min, and resistance to detachment was quantified as described in “Experimental Procedures.” Although silencing α3 integrin had little or no effect in this assay, silencing α6 integrin profoundly reduced resistance to detachment (Fig. 6C). In contrast, initial adhesion on LM-332 was significantly impaired for the α3 integrin-silenced cells but not for the α6 integrin-silenced cells (Fig. 6D), as expected from antibody blockade experiments in our previous study (16). Both cell types displayed normal adhesion on fibronectin. On collagen I, adhesion of α6-silenced cells appeared somewhat reduced, but this reduction was variable and was not observed in all trials (data not shown). Thus, silencing α3 but not α6 integrin specifically blocked initial adhesion on LM-332. Flow cytometry confirmed that expression of α3 and α6 integrin was reduced by >90% and >95% in the α3 and α6-silenced cells, respectively (data not shown). Together, these data established resistance to detachment as an assay that would enable us to explore CD151 contributions to a cell behavior that is strongly dependent on α6β4 integrin and largely independent of α3β1 function.
When we performed detachment assays using wild type, CD151-silenced cells, and the CD151 mutant cell lines, we found that between 60–70% of the wild type cells resisted detachment under these conditions (Fig. 6E). In contrast, only ∼10% of the CD151-silenced cells remained attached. Re-expressing wild type CD151 (WT RX) or the CD151INF mutant restored resistance to detachment to normal levels. Remarkably, the CD151Palm mutant was also able to restore normal resistance to detachment, but the CD151VR and EC2 mutants were not (Fig. 6E). Thus, our data demonstrate an important role for CD151-integrin association, but not CD151-tetraspanin association, in α6β4-dependent resistance to detachment. This contrasts with the requirement for both CD151-integrin and CD151-tetraspanin association for α3β1-dependent initial adhesion and rapid motility on LM-332. The reduced resistance to detachment of the CD151-silenced cells and the CD151VR and EC2 mutant cells was not due to reduced LM-332 deposition because immunostaining of cell substrates revealed similar levels of staining for all cell types (supplemental Fig. S2).
To compare results from A431 cells to another cell type, we used HaCaT immortalized keratinocyte cells. HaCaT cells were selected because, like A431 cells, they deposit a LM-332-rich matrix and express α3β1 and α6β4 integrins. An antibody blockade experiment confirmed that initial cell adhesion of HaCaT cells on exogenous LM-332 strongly depends on α3β1 integrin but not α6β4 integrin (Fig. 7A). This α3 integrin-dependent function was impaired in CD151-silenced sh3 cells and in CD151VR and CD151Palm mutant cells (Fig. 7B). Thus, as in A431 cells, mutations that disrupt either CD151-integrin or CD151-tetraspanin association blocked the ability of CD151 to promote α3β1-dependent adhesion. Detachment assays using HaCaT cells silenced for α3 or α6 integrin revealed that resistance to detachment depends strongly on α6 integrin, but not on α3 integrin, and that silencing CD151 significantly impaired this α6-dependent function (Fig. 7C). Re-expressing the CD151VR mutant in CD151-silenced cells completely failed to restore resistance to detachment (Fig. 7D). In contrast, re-expressing the CD151Palm mutant restored resistance to detachment to near wild type levels (Fig. 7D). Overall, results with HaCaT cells were similar to results with A431 cells, with CD151 palmitoylation being crucial for α3β1-dependent initial cell adhesion, but not for α6β4-dependent resistance to detachment.
FIGURE 7.
CD151 mutant constructs behave similarly in HaCaT cells. A, adhesion assay using wild type HaCaT cells plated in wells coated with BSA or laminin-332 with or without the addition of 10 μg/ml α3 or α6 function-blocking antibodies A3-IIF5 and GoH3. Columns represent the means of at least two biological replicates. B, CD151-silenced HaCaT cells and cells re-expressing wild type or CD151Palm or CD151VR mutant constructs were compared in adhesion assays. Attached cells were quantified as described for Fig. 5. This figure is representative of two similar experiments, each with four replicates. Bars show S.E. *, p < 0.001 compared with CD151WTRX cells (ANOVA with post hoc t test). C, de-adhesion assay as described for Fig. 6C with CD151WTRX HaCaT cells and HaCaT cells silenced for CD151 (sh3), or α3 or α6 integrin. Each column represents the average of two experiments, four wells per condition. Bars show S.E. *, p < 0.05; **, p < 0.01 compared with CD151WTRX cells (ANOVA with post hoc t test). D, de-adhesion assay using HaCaT cells expressing wild type CD151 (WTRX) or the CD151VR or CD151Palm mutants. Each column represents four wells per condition. Bars show S.E. ***, p < 0.001; n.s., not significant compared with CD151WTRX cells (ANOVA with post hoc t test).
DISCUSSION
Mounting evidence indicates that tetraspanin CD151 can promote tumor cell motility, invasion, metastatic colonization, and resistance to chemotherapeutics (11, 16, 21, 23, 39). Thus, CD151 has been advanced as a potentially attractive target of anti-cancer strategies (15, 40). However, genetic evidence from mouse models and human patients indicates that tetraspanin CD151 also plays important roles in the maintenance of epithelial integrity and wound healing (2, 4, 6, 41). CD151 makes direct physical contact with laminin-binding integrins, and CD151 also associates with other tetraspanins, thereby localizing its integrin partners to tetraspanin-enriched microdomains on the cell surface (15). However, which of the physiological and pathological functions of CD151 depend on CD151-integrin interactions, CD151-tetraspanin interactions, or both has not yet been completely defined. Therefore, the prospects for selectively targeting specific CD151 functions are at present unclear. In this study, we took advantage of a cell system we previously established, in which endogenous CD151 expression has been almost completely extinguished, to uncover specific roles for different CD151 interaction domains in promoting α3β1 and α6β4 integrin-dependent functions.
CD151 Domains Critical for α3β1 Integrin-dependent Tumor Cell Adhesion and Migration
It had been widely assumed that CD151 promotes α3β1 integrin function by physically linking α3β1 to other tetraspanins and tetraspanin partners in TEMs. However, other models had not been strictly ruled out. For example, although CD151 can associate with both α3β1 and other tetraspanins, it was still conceivable that CD151 association with other tetraspanins might allosterically promote their association with α3β1 integrin, without CD151 actually acting as a physical linker. Our new data strongly support the physical linker model for CD151 function by showing that CD151 must be able to associate with both α3β1 and other tetraspanins to link α3β1 to TEMs and promote α3β1 function in initial adhesion and migration.
Previous studies had revealed that forced expression of a CD151 palmitoylation mutant reduced CD151 and integrin association with other tetraspanins, but the impact on α3 or α6 integrin function was unclear (33, 34). A CD151 palmitoylation mutant expressed in α3-null kidney epithelial cells promoted a more epithelial morphology (33), whereas a similar mutant expressed in Rat1 fibroblasts enhanced Akt activation and focal adhesion formation (34). More recently, siRNA-mediated knockdown of DHHC2, an enzyme that palmitoylates CD9 and CD151, resulted in a more dispersed phenotype in A431 cells, with disrupted cell-cell contacts. Whether these various phenotypes reflect altered function of laminin-binding integrins is not entirely clear. By expressing our CD151Palm mutant in a cell environment with virtually no wild type CD151, we have now revealed an essential role for CD151 palmitoylation in the ability of CD151 to promote α3β1 integrin function. Interestingly, ablation of β4 integrin palmitoylation sites diminished association of the α6β4-CD151 complex with other tetraspanins, resulting in impaired p130Cas phosphorylation and reduced cell spreading on laminin-111 or LM-332 (42). Thus, palmitoylation-dependent targeting of laminin-binding integrins to TEMs may be crucial for dynamic, motility-related behaviors mediated by these integrins in a wide variety of settings.
A surprising outcome of our study was the lack of an obvious phenotype in A431 cells harboring the CD151 QRD→INF mutation in the CD151 large extracellular loop (EC2 domain). Our CD151INF mutant behaved as expected in that it failed to associate with α3β1 integrin in Triton X-100 detergent lysates as reported previously (32). However, because of the lack of interference from endogenous wild type CD151 in our system, our studies revealed that association of the CD151INF mutant with α3β1 in milder Brij detergent conditions does not depend on the presence of wild type CD151, as supposed previously, but instead appears to be an intrinsic feature of the mutant. Thus, we conclude that the QRD→INF mutation may weaken, but does not disrupt, the interaction of CD151 with its laminin-binding integrin partners. Concordantly, in our system, CD151INF was able to mediate wild type levels of α3β1 association with other tetraspanins and wild type α3β1-dependent initial adhesion and motility on LM-332. Our data are consistent with a recent epitope mapping study showing that amino acids just upstream of the QRD motif are also involved in the Triton X-100-resistant CD151-α3β1 integrin interaction (43).
Our data contrast with the original report that over-expression of the CD151INF mutant in Cos7 or NIH3T3 cells inhibited α6/α3 integrin-dependent cable formation in 3D Matrigel as well as cell spreading on thin-coated two-dimensional Matrigel (32). Our data also stand in contrast to recent reports that CD151 mutants lacking the QRD motif were unable to promote α3/α6 integrin-dependent MDA-MB-231 breast carcinoma cell scattering in response to TGF-β (23), or endothelial cell proliferation, transwell migration, and cable formation in Matrigel (36). However, another recent study reported that the QRD motif is dispensable for the ability of CD151 to promote α3β1 integrin-dependent growth in three-dimensional Matrigel by HB2 cells, which resemble mammary epithelial ductal carcinoma in situ (22). Thus, the extent to which CD151 QRD motif is essential for the ability of CD151 to promote the function of laminin-binding integrins remains unclear.
In future studies, it will be important to determine the association of CD151 QRD mutants with laminin-binding integrins in each system, rather than assuming that the QRD mutation dramatically disrupts integrin association. Our data also suggest that in some cases, the effect of the QRD mutation may be more subtle than simple disruption of adhesion or migration on laminin isoforms. For example, it is possible that strong, QRD-dependent association of CD151 with its integrin partners may be more crucial in circumstances such as cable formation or scattering in three-dimensional matrices, where cells might develop strong pulling forces against extracellular substrates. Consistent with this possibility, the difference in the ability of mutant versus wild type CD151 to promote α6β1 integrin-dependent adhesion strengthening to laminin-coated beads was most obvious at higher detachment forces, above 0.9 nN (44).
CD151 Palmitoylation Is Dispensable for α6β4 Integrin-dependent Stable Cell Attachment
A431 and HaCaT cells deposit a LM-332-rich matrix and develop a strong α6β4 integrin-dependent resistance to detachment over time, as we show here. Our new data show that although CD151-α6 association is important for this α6β4-dependent function, CD151 palmitoylation is not. Thus, the presence of CD151, but not linkage to other tetraspanins, appears to be crucial for α6β4-mediated resistance to detachment. These findings are consistent with an earlier report that, although α3β1 and CD151 are both present in pre-hemidesmosomal structures, α3β1 and other tetraspanins are excluded from mature hemidesmosomes containing α6β4 and CD151 (45). Interestingly, CD151 palmitoylation was also not required for CD151 to promote carcinoma cell scattering in response to TGF-β, a process that depended on both α3 and α6 integrins (23). However, β4 integrin palmitoylation, which influences α6β4 association with tetraspanin CD9, is important for MDA-MB-435 cell spreading on laminin (42). Thus, association of α6β4 with tetraspanins other than CD151 appears likely to be dispensable for some α6β4 functions but may be important for others. One possibility is that α6β4-TEM association may be more important in cases where α6β4 is contributing to dynamic, motility-related cell behaviors of the type that are also often mediated by α3β1 integrin.
Implications for Selective Disruption of CD151 Functions in Tumor Cells
Of potential utility, our new data show that it is possible to selectively interfere with the α3β1 integrin-dependent functions of CD151 while leaving at least some of the α6β4 integrin-dependent functions of CD151 intact. The observation that CD151 palmitoylation may not be essential for the ability of CD151 to promote α6β4 integrin-dependent resistance to detachment raises the possibility that targeting CD151 palmitoylation might have relatively specific effects on the promigratory functions of CD151. The recent discovery of DHHC2 as an enzyme responsible for palmitoylation of CD151 and CD9 highlights the potential for developing small molecule inhibitors to block palmitoylation of specific tetraspanins (46). On the other hand, CD151 may also promote the stability of carcinoma cell-cell junctions through a mechanism that depends on α3β1 integrin (24). In addition, the role of CD151 in maintaining kidney epithelial integrity may also involve an α3β1 integrin-dependent mechanism in kidney podocyte foot processes (6, 41). In future studies, it will be important to define the role of CD151 palmitoylation in these and other α3β1 integrin-dependent functions.
In addition to the selective effects of ablating CD151 palmitoylation sites, we also observed that interfering with strong, QRD-dependent CD151-integrin association has no obvious effect on several different α3β1 and α6β4 functions. Together with prior reports that the CD151 QRD motif is important for certain cell behaviors such as cell scattering and cable formation in three-dimensional matrices (23, 32, 36), our data raise the possibility that targeting the CD151 QRD might selectively block functions that may be more important in tumor cell invasion or matrix remodeling. Of potential relevance here is the identification of a monoclonal antibody, Ts151r, that binds to a CD151 epitope that requires the QRD motif (32, 47) and modulates integrin-dependent cell migration and cell-cell interactions (48). However, saturating amounts of Ts151r eventually dissociate CD151 from its integrin partners, disrupt cell-cell contacts, promote cytoskeletal remodeling, and enable cell migration (48), suggesting that Ts151r binding has a more disruptive effect on CD151-integrin interaction and function than does the precise removal of the QRD site by mutation. Another CD151 antibody, which inhibits tumor cell intravasation, has also been identified (39), but the precise epitope on CD151 to which it binds has not been defined.
In conclusion, by expressing CD151 mutants in a background virtually devoid of wild type CD151, we have uncovered selective functions for specific CD151 domains. Our data help to lay the groundwork for future studies aimed at defining the CD151 domains required for the functions of CD151 in tumorigenesis, metastatic colonization, and chemoresistance (11, 12, 21, 23). Such studies may create the possibility of specifically targeting CD151-integrin functions in tumor cells, while leaving important physiological functions relatively undisturbed.
Supplementary Material
Acknowledgments
We thank Marit Meland for creating the CD151 mutants, Steven Gerdes for assisting with immunostaining experiments, Anne Cress for providing anti-α6 integrin polyclonal antiserum, and Mary Herndon for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant CA136664. This work was also supported by American Cancer Society Research Scholar Award RSG-07-043-01-CSM.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- TEM
- tetraspanin-enriched microdomain
- LM-332
- laminin-332
- VR
- variable region
- ANOVA
- analysis of variance.
REFERENCES
- 1. Wright M. D., Geary S. M., Fitter S., Moseley G. W., Lau L. M., Sheng K. C., Apostolopoulos V., Stanley E. G., Jackson D. E., Ashman L. K. (2004) Mol. Cell Biol. 24, 5978–5988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Karamatic Crew V., Burton N., Kagan A., Green C. A., Levene C., Flinter F., Brady R. L., Daniels G., Anstee D. J. (2004) Blood 104, 2217–2223 [DOI] [PubMed] [Google Scholar]
- 3. Lau L. M., Wee J. L., Wright M. D., Moseley G. W., Hogarth P. M., Ashman L. K., Jackson D. E. (2004) Blood 104, 2368–2375 [DOI] [PubMed] [Google Scholar]
- 4. Cowin A. J., Adams D., Geary S. M., Wright M. D., Jones J. C., Ashman L. K. (2006) J. Invest. Dermatol. 126, 680–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Takeda Y., Kazarov A. R., Butterfield C. E., Hopkins B. D., Benjamin L. E., Kaipainen A., Hemler M. E. (2007) Blood 109, 1524–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baleato R. M., Guthrie P. L., Gubler M. C., Ashman L. K., Roselli S. (2008) Am. J. Pathol. 173, 927–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Orlowski E., Chand R., Yip J., Wong C., Goschnick M. W., Wright M. D., Ashman L. K., Jackson D. E. (2009) J. Thromb. Haemost. 7, 2074–2084 [DOI] [PubMed] [Google Scholar]
- 8. Tokuhara T., Hasegawa H., Hattori N., Ishida H., Taki T., Tachibana S., Sasaki S., Miyake M. (2001) Clin. Cancer Res. 7, 4109–4114 [PubMed] [Google Scholar]
- 9. Hashida H., Takabayashi A., Tokuhara T., Hattori N., Taki T., Hasegawa H., Satoh S., Kobayashi N., Yamaoka Y., Miyake M. (2003) Br. J. Cancer 89, 158–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ang J., Lijovic M., Ashman L. K., Kan K., Frauman A. G. (2004) Cancer Epidemiol. Biomarkers Prev. 13, 1717–1721 [PubMed] [Google Scholar]
- 11. Yang X. H., Richardson A. L., Torres-Arzayus M. I., Zhou P., Sharma C., Kazarov A. R., Andzelm M. M., Strominger J. L., Brown M., Hemler M. E. (2008) Cancer Res. 68, 3204–3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sadej R., Romanska H., Baldwin G., Gkirtzimanaki K., Novitskaya V., Filer A. D., Krcova Z., Kusinska R., Ehrmann J., Buckley C. D., Kordek R., Potemski P., Eliopoulos A.. G., Lalani el-N., Berditchevski F. (2009) Mol. Cancer Res. 7, 787–798 [DOI] [PubMed] [Google Scholar]
- 13. Ke A. W., Shi G. M., Zhou J., Wu F. Z., Ding Z. B., Hu M. Y., Xu Y., Song Z. J., Wang Z. J., Wu J. C., Bai D. S., Li J. C., Liu K. D., Fan J. (2009) Hepatology 49, 491–503 [DOI] [PubMed] [Google Scholar]
- 14. Shi G. M., Ke A. W., Zhou J., Wang X. Y., Xu Y., Ding Z. B., Devbhandari R. P., Huang X. Y., Qiu S. J., Shi Y. H., Dai Z., Yang X. R., Yang G. H., Fan J. (2010) Hepatology 52, 183–196 [DOI] [PubMed] [Google Scholar]
- 15. Stipp C. S. (2010) Expert Rev. Mol. Med. 12, e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Winterwood N. E., Varzavand A., Meland M. N., Ashman L. K., Stipp C. S. (2006) Mol. Biol. Cell 17, 2707–2721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yamada M., Sumida Y., Fujibayashi A., Fukaguchi K., Sanzen N., Nishiuchi R., Sekiguchi K. (2008) FEBS J. 275, 3335–3351 [DOI] [PubMed] [Google Scholar]
- 18. Nishiuchi R., Sanzen N., Nada S., Sumida Y., Wada Y., Okada M., Takagi J., Hasegawa H., Sekiguchi K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 1939–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hasegawa M., Furuya M., Kasuya Y., Nishiyama M., Sugiura T., Nikaido T., Momota Y., Ichinose M., Kimura S. (2007) Lab. Invest. 87, 882–892 [DOI] [PubMed] [Google Scholar]
- 20. Geary S. M., Cowin A. J., Copeland B., Baleato R. M., Miyazaki K., Ashman L. K. (2008) Exp. Cell Res. 314, 2165–2175 [DOI] [PubMed] [Google Scholar]
- 21. Yang X. H., Flores L. M., Li Q., Zhou P., Xu F., Krop I. E., Hemler M. E. (2010) Cancer Res. 70, 2256–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Novitskaya V., Romanska H., Dawoud M., Jones J. L., Berditchevski F. (2010) Cancer Res. 70, 4698–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sadej R., Romanska H., Kavanagh D., Baldwin G., Takahashi T., Kalia N., Berditchevski F. (2010) Cancer Res. 70, 6059–6070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Johnson J. L., Winterwood N., DeMali K. A., Stipp C. S. (2009) J. Cell Sci. 122, 2263–2273 [DOI] [PubMed] [Google Scholar]
- 25. Weitzman J. B., Pasqualini R., Takada Y., Hemler M. E. (1993) J. Biol. Chem. 268, 8651–8657 [PubMed] [Google Scholar]
- 26. Lee R. T., Berditchevski F., Cheng G. C., Hemler M. E. (1995) Circ. Res. 76, 209–214 [DOI] [PubMed] [Google Scholar]
- 27. DiPersio C. M., Shah S., Hynes R. O. (1995) J. Cell Sci. 108, 2321–2336 [DOI] [PubMed] [Google Scholar]
- 28. Ports M. O., Nagle R. B., Pond G. D., Cress A. E. (2009) Cancer Res. 69, 5007–5014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fukudome K., Furuse M., Imai T., Nishimura M., Takagi S., Hinuma Y., Yoshie O. (1992) J. Virol. 66, 1394–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yauch R. L., Berditchevski F., Harler M. B., Reichner J., Hemler M. E. (1998) Mol. Biol. Cell 9, 2751–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Marinkovich M. P., Lunstrum G. P., Burgeson R. E. (1992) J. Biol. Chem. 267, 17900–17906 [PubMed] [Google Scholar]
- 32. Kazarov A. R., Yang X., Stipp C. S., Sehgal B., Hemler M. E. (2002) J. Cell Biol. 158, 1299–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yang X., Claas C., Kraeft S. K., Chen L. B., Wang Z., Kreidberg J. A., Hemler M. E. (2002) Mol. Biol. Cell 13, 767–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Berditchevski F., Odintsova E., Sawada S., Gilbert E. (2002) J. Biol. Chem. 277, 36991–37000 [DOI] [PubMed] [Google Scholar]
- 35. Stipp C. S., Kolesnikova T. V., Hemler M. E. (2003) Trends Biochem. Sci. 28, 106–112 [DOI] [PubMed] [Google Scholar]
- 36. Zuo H. J., Lin J. Y., Liu Z. Y., Liu W. F., Liu T., Yang J., Liu Y., Wang D. W., Liu Z. X. (2010) Acta Pharmacol. Sin. 31, 805–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Raymond K., Kreft M., Janssen H., Calafat J., Sonnenberg A. (2005) J. Cell Sci. 118, 1045–1060 [DOI] [PubMed] [Google Scholar]
- 38. Rabinovitz I., Toker A., Mercurio A. M. (1999) J. Cell Biol. 146, 1147–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zijlstra A., Lewis J., Degryse B., Stuhlmann H., Quigley J. P. (2008) Cancer Cell 13, 221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hemler M. E. (2008) Nat. Rev. Drug Discov. 7, 747–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sachs N., Kreft M., van den Bergh Weerman M. A., Beynon A. J., Peters T. A., Weening J. J., Sonnenberg A. (2006) J. Cell Biol. 175, 33–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang X., Kovalenko O. V., Tang W., Claas C., Stipp C. S., Hemler M. E. (2004) J. Cell Biol. 167, 1231–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yamada M., Tamura Y., Sanzen N., Sato-Nishiuchi R., Hasegawa H., Ashman L. K., Rubinstein E., Yáñez-Mó M., Sánchez-Madrid F., Sekiguchi K. (2008) Biochem. J. 415, 417–427 [DOI] [PubMed] [Google Scholar]
- 44. Lammerding J., Kazarov A. R., Huang H., Lee R. T., Hemler M. E. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 7616–7621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sterk L. M., Geuijen C. A., Oomen L. C., Calafat J., Janssen H., Sonnenberg A. (2000) J. Cell Biol. 149, 969–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sharma C., Yang X. H., Hemler M. E. (2008) Mol. Biol. Cell 19, 3415–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Serru V., Le Naour F., Billard M., Azorsa D. O., Lanza F., Boucheix C., Rubinstein E. (1999) Biochem. J. 340, 103–111 [PMC free article] [PubMed] [Google Scholar]
- 48. Chometon G., Zhang Z. G., Rubinstein E., Boucheix C., Mauch C., Aumailley M. (2006) Exp. Cell Res. 312, 983–995 [DOI] [PubMed] [Google Scholar]
- 49. Rasband W. S. (1997–2009) ImageJ, U.S. National Institutes of Health, Bethesda, MD [Google Scholar]
- 50. Sincock P. M., Fitter S., Parton R. G., Berndt M. C., Gamble J. R., Ashman L. K. (1999) J. Cell Sci. 112, 833–844 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.