Abstract
Prolactin (PRL) is essential for normal reproduction and signals through two types of receptors, the short (PRL-RS) and long (PRL-RL) form. We have previously shown that transgenic mice expressing only PRL-RS (PRLR−/−RS) display abnormal follicular development and premature ovarian failure. Here, we report that MAPK, essential for normal follicular development, is critically inhibited by PRL in reproductive tissues of PRLR−/−RS mice. Consequently, the phosphorylation of MAPK downstream targets are also markedly inhibited by PRL without affecting immediate upstream kinases, suggesting involvement of MAPK specific phosphatase(s) in this inhibition. Similar results are obtained in a PRL-responsive ovary-derived cell line (GG-CL) that expresses only PRL-RS. However, we found the expression/activation of several known MAPK phosphatases not to be affected by PRL, suggesting a role of unidentified phosphatase(s). We detected a 27-kDa protein that binds to the intracellular domain of PRL-RS and identified it as dual specific phosphatase DUPD1. PRL does not induce expression of DUDP1 but represses its phosphorylation on Thr-155. We also show a physical association of this phosphatase with ERK1/2 and p38 MAPK. Using an in vitro phosphatase assay and overexpression studies, we established that DUPD1 is a MAPK phosphatase. Dual specific phosphatase inhibitors as well as siRNA to DUPD1, completely prevent PRL-mediated MAPK inhibition in ovarian cells. Our results strongly suggest that deactivation of MAPK by PRL/PRL-RS contributes to the severe ovarian defect in PRLR−/−RS mice and demonstrate the novel association of PRL-RS with DUPD1 and a role for this phosphatase in MAPK deactivation.
Keywords: Cell Differentiation, Cytokine, ERK, Ovary, p38 MAPK, DUPD1, Decidua, Prolactin
Introduction
PRL,3 a versatile hormone synthesized and secreted principally by the pituitary, plays a key role in normal ovarian development and function (reviewed in Refs. 1–4). It affects the survival and steroidogenic capacity of both the follicles and corpus luteum (1, 2, 5, 6) by activating its receptor (PRLR), a member of the cytokine receptor superfamily that lacks intrinsic tyrosine kinase activity. Two isoforms of the PRLR have been identified in several species (4, 7–10). They are generated by alternative splicing, differ in the length and composition of their cytoplasmic tail, and are referred to as short (PRL-RS) and long (PRL-RL). The most extensively characterized isoform is the PRL-RL, which transduces both mitogenic and differentiative signals (11–14). It is known to activate the Jak/Stat pathway, a prototype signaling pathway used by all cytokines. As to PRL-RS, it has been cloned from several species including humans (15), rat (16), mouse (9), cow, and sheep (17). In rats, only one PRL-RS isoform is generated by alternative splicing, whereas three isoforms have been described in mice (9, 18). Among these, one clone (PR-1) is highly homologous to that of the rat (5) and contains a potential signal transduction motif (3).
Conflicting results on the function of the PRL-RS have been reported. PRL-RS was originally considered as an inactive receptor that acts as a dominant negative to PRL-RL, preventing Jak2 activation and cell proliferation (19, 20). However, a signaling role for this receptor was proposed by Das and Vonderhaar (21), who showed that activation of the mouse PRL-RS in NIH-3T3 fibroblasts induces MAPK activity and suggested that it may be involved in cell proliferation. The human PRL-RS can also activate MAPK in cultured cells, although this activation is delayed and prolonged, and therefore a role in differentiation rather than proliferation was suggested (22). Using a transgenic mouse model, Binart et al. (23) reported that overexpression of PRL-RS in PRLR+/− mice can rescue a mammopoiesis defect in the heterozygote mice. This led to the conclusion that, in mammary glands, PRL acting through PRL-RS may mediate activation of MAPK. Generation of transgenic mice expressing only the PRL-RS in the background of PRLR null mice has recently established that in vivo activation of PRL-RS by PRL elicits profound effects in the ovary, causing a clear defect in follicular development leading to premature ovarian failure and repression of key transcription factors essential for ovarian function (5, 24).
Recent investigations have established a key role for MAPK in the normal development and function of the ovary. Generation of mice with granulosa cell-specific deletion of ERK1 and ERK2 (25) revealed a critical role for these kinases in granulosa cell differentiation and ovulation, whereas expression of a constitutively active K-RAS mutant causes impaired follicular development and premature ovarian failure, presumably because of inappropriate activation of ERK1/2 in granulosa cells of growing follicles (26).
The abnormal follicular development and premature ovarian failure observed in the ovary of transgenic mice expressing only PRL-RS led us to examine whether MAPK signaling is defective in the ovary. Because MAPK also plays an important role in normal formation of the decidua (27–29), another important target tissue of PRL (8, 30, 31), we examined whether PRL activation of PRL-RS impacts MAPK activation in this tissue as well. In addition, using cells expressing only PRL-RS, we examined the mechanism by which PRL regulates MAPK activation. Our results obtained both in vivo and in cell culture show clearly and in complete opposition to previous reports (20, 22), that PRL signaling through PRL-RS deactivates both ERK1/2 and p38 MAPK pathways. We established the novel finding that this deactivation involves the association of a novel phosphatase DUPD1 with PRL-RS and with both ERK1/2 and p38 and established a novel PRL signaling mechanism through PRL-RS.
EXPERIMENTAL PROCEDURES
Animal Model and Tissue Preparation
Generation of transgenic mice expressing PRL-RS in the background of PRLR−/− (PRLR−/−RS) has been described previously (5, 23). The mice were genotyped by PCR using genomic DNA isolated from tail as described previously (5). The mice were kept at 25 °C with a 14-h light/10-h dark cycle and fed a commercial pellet diet ad libitum. All of the experimental procedures were performed in accordance with the principles of the National Institutes of Health Guide for the Care and Use of Laboratory and approved by the Institutional Animal Care and Use Committee. Pseudopregnant PRLR−/−RS mice were prepared as previously described (24). In brief, PRLR−/−RS females were mated with vasectomized males to induce pseudopregnancy and treated with progesterone from the day a vaginal plug was found. Decidualization was induced with an intrauterine oil injection on day 4 of pseudopregnancy. On day 9, the mice were treated with ergocryptine to block endogenous PRL secretion followed, 6 h later, with an intraperitoneal injection of either recombinant ovine PRL (60 μg, purchased from Dr. Arieh Gertler, Protein Laboratories Rehovot Ltd., Rehovot, Israel) or vehicle (0.1% BSA). The mice were sacrificed 15, 30, and 120 min later. Ovaries and decidua were isolated, frozen in liquid nitrogen, and stored at −80 °C until processing for RNA or protein extraction.
Cell Culture
GG-CL cells, a rat ovarian cell line generated in our laboratory (35), were incubated in a humidified atmosphere of 5% CO2 at 33 °C. For transient transfection, the cells are grown at 50–60% confluency in 2% CDT-FBS (Hyclone, Logan, UT) in 6-well plates. The cells were transfected using Lipofectamine 2000 (Invitrogen) or Effectene (Qiagen) according to the manufacturer's protocol. The cells were transfected with rat PRL-RS or PRL-RL expression vectors (24), each at 0.8 μg/well. For overexpression of DUPD1-GFP, GG-CL cells were co-transfected with PRL-RS (0.8 μg) and DUPD1-GFP (catalog number RG214361; OriGene Technologies, Inc., Rockville, MD) at various concentrations, and the total amount of DNA was adjusted with pcDNA. For co-transfection of DUPD1 siRNA (siRNA 1, Santa Cruz Biotechnology, catalog number sc-156160; siRNA 2, Ambion, catalog number 4390771) or control siRNA with PRL-RS expression vector. Lipofectamine 2000 (Invitrogen) or Attractene reagent (Qiagen) were used according to the manufacturer's protocol. After 48 h, the cells were serum-starved for 2 h and treated thereafter with either PRL (1 μg/ml) or vehicle. For inhibitor studies, all of the inhibitors were treated 1 h prior to PRL treatment. The concentrations of inhibitors are protein-tyrosine phosphatase 1B inhibitor (250 nm), NSC87877 (50 μm), okadaic acid (100 nm), AG490 (25 μm), NSC95397 (30 μm), and NSC663284 (10 μm). All of the inhibitors were obtained from Calbiochem and EMD Crop Bioscience, Inc. (Brookfield, WI). At different time points, the cells were rinsed twice with PBS and were frozen at −80 °C until RNA and protein extraction.
RT-PCR and qPCR
Total RNA was extracted from ovary and decidua using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. One microgram of total RNA is reverse transcribed using Superscript Polymerase II kit according to the manufacturer's instructions (Invitrogen). The resulting cDNA was then diluted to a total volume of 100 μl with sterile H2O. Rat PRL-RS and PRL-RL expressions were detected using procedures described previously (24). The primers used were as follows: a common sense primer (5′-AAG TAT CTT GTC CAG ACT CGC TG-3′) was combined with a specific PRL-RS antisense primer (5′-TTG TAT TTG CTT GGA GAG AGC CAG-3′) or a specific PRL-RL antisense primer (5′-AGC AGT TCT TCA GAC TTG CCC TT-3′). The PCR products were then separated by gel electrophoresis on a 0.7% agarose gel, and the intensity was measured using UV transilluminator and a digital camera (Eastman Kodak Co., New Haven, CT).
Each qPCR consisted of 5 μl of diluted RT product, 1× SYBR Green PCR Master Mix (PE Applied Biosystems, Foster City, CA), and 50 nm forward and reverse primers. The reactions were carried out on an ABI PRISM 7700 sequence detection system (PE Applied Biosystems) for 40 cycles (95 °C for 15 s and 60 °C for 1 min) after an initial 10-min incubation at 95 °C. The primer sets used for qPCR are as follows: PRL-RS (5′-CCT GCA TCT TTC CAC CAG TTC-3′ and 5′-GCA CTC AGC AGT TCT TCA GAC TTG-3′), PRL-RL (5′-CCT GCA TCT TTC CAC CAG TTC-3′ and 5′-GAT CCA CCT TGT ATT TGC TTG GAG-3′), MKP1 (5′-AGC ACC CCT CTC TAC GAC CAG-3′ and 5′-AAT TGG CCG AGA CGT TGA TC-3′), MKP3 (5′-GCC AGC TGC TTG ACT TTG-3′ and 5′-GAG GGC GCG GTG AAG TAG A-3′), MKP5 (5′-CTC AGT CTG TCC CCT CCA CC-3′ and 5′-CTG AGC ATC CTG CTC ATT GC-3′), and DUPD1 (5′-TCC TGG TTC ACT GTG CCA TG-3′ and 5′-CGG TTC TTA GCC ACT TGT TGG-3′). The fold change in expression of each gene was calculated using the ΔΔCt method, with the ribosomal protein 36B4 mRNA as an internal control.
Western Blot Analysis
For Western blots, 25 μg of protein was resolved on SDS-PAGE, transferred to PVDF membranes, and blocked for nonspecific binding. The membranes were then incubated with the appropriate primary antibodies overnight at 4 °C. After a series of washes, the blots were incubated with a secondary antibody linked to horseradish peroxidase for 1 h. The bands were detected by using an enhanced chemiluminescence system. Antibodies to phospho-p90RSK, phospho-ATF2, phospho-ELK1, phospho-MEK1/2, phosphp-MKK3/6, total p38, phospho-ERK1/2 (catalog number 9101), and phospho-RXRXX(S/T) (catalog number 9611) were from Cell Signaling, Denvers, MA. Other primary antibodies used were phospho-p38 MAPK (Promega, Madison, WI), total ERK1/2 (Upstate Biotechnology, Lake Placid, NY), β-Actin (Abcam Inc., Cambridge, MA), and DUPD1 (Santa Cruz Biotechnology; sc-79465 and sc-79462).
GST Pulldown Assay
Construction and expression of the fusion protein containing the cytoplasmic domain of PRL-RS tagged with glutathione S-transferase (RS-GST) was described previously (33). Decidual and GG-CL protein extracts were incubated with glutathione-agarose beads for 60 min at room temperature to remove proteins that bound nonspecifically (precleaned). The samples were centrifuged at 1,000 × g, and the supernatant was recovered and adjusted to 1% CHAPS. Fusion protein containing PRL-RS were isolated on glutathione-agarose beads, and the beads were then incubated overnight at 4 °C with 300 μg of precleaned protein lysates. GST alone (C-GST) was used as control. Following incubation, the beads were collected by centrifugation and washed in ice-cold phosphate-buffered saline. PRL-RS fusion proteins or C-GST and their attached proteins were then eluted by boiling for 5 min with SDS-PAGE sample buffer. Eluted proteins were analyzed by SDS-PAGE. After electrophoresis, the proteins were stained directly with silver staining (34) or transferred to PVDF for Western blot analysis with DUPD1 antibody.
Immunoprecipitation
GG-CL cell lysates were solubilized in 1% Nonidet P-40 or 1% CHAPS and 0.25% deoxycholate. 300 μg of solubilized proteins were incubated with polyclonal total ERK1/2 (12.5 μg/ml), DUPD1 (15 μg/ml) polyclonal antibodies, monoclonal antibody to phosphotyrosine (Upstate or Millipore Corp.), or nonspecific rabbit/goat/mouse IgG (Santa Cruz Biotechnology) for overnight at 4 °C with shaking. 30 μl of protein G-agarose (Santa Cruz Biotechnology) beads were added into the immune complexes and incubated at 4 °C or room temperature for 30 min. The beads were collected by centrifugation and washed extensively. The beads were then suspended in 30 μl of SDS-PAGE sample buffer and subjected to SDS-PAGE. The proteins were transferred to PVDF membranes and analyzed by Western blotting as described above.
Immunocytochemistry
GG-CL cells were grown for 24 h in M199 medium supplemented with 2% CDT-FBS on Lab-Tek chamber slides (Nalge Nunc International, Rochester, NY). The cells were transfected with PRL-RS or empty vector using Effectene transfection reagent for 12 h. The cells were washed, serum-starved, and then cultured with recombinant PRL (1 μg/ml) or vehicle for 30 min and processed for immunocytochemistry as described previously (35). Antibody to p-ERK1/2 (1:100) and DUPD1 (1:100) were used as primary antibodies, whereas Cy2-conjugated donkey anti-rabbit IgG and Cy3-conjugated donkey anti-goat IgG (1:800; Jackson ImmunoResearch Laboratories, West Grove, PA) were used as secondary antibodies.
Immunohistochemistry
Paraffin-embedded sections were subjected to the avidin-biotin-peroxidase complex method using a Vectastain ABC kit (Vector Laboratories, Inc., Burlingame, CA) as described previously with some modifications (5). Briefly, formalin-fixed, paraffin-embedded sections (5 μm) were deparaffinized, hydrated, and blocked with 5% normal donkey serum. The slides were then incubated overnight at 4 °C with a polyclonal antibody to DUPD1 (Santa Cruz Biotechnology) at 1:100. The slides were then exposed for 4 h at room temperature to Cy3-conjugated donkey antigoat IgG (Jackson ImmunoResearch Laboratories) at a 1:800 dilution. The slides were washed, and the procedure was repeated with polyclonal antibody to ERK1/2 and Cy2-conjugated donkey anti-rabbit IgG as primary and secondary antibodies, respectively. The slides were mounted in Vectashield medium (Vector Laboratories) containing a counterstain for DAPI and were observed with a Carl Zeiss (Oberkochen, Germany) LSM 510 laser confocal microscope equipped with a 63× water immersion objective lens (NA 1.2).
In Vitro Phosphatase Assay
DUPD1 was immunoprecipitated from GG-CL cells expressing PRL-RS by polyclonal antibody to DUPD1 as described in the previous section. As control, cell lysates were immunoprecipitated with goat IgG. The immunoprecipitates were resuspended in phophatase assay buffer (36) and incubated for 1 h in the presence of recombinant active p38 (Cell Signaling, Danvers, MA). The reaction was stopped by adding SDS-PAGE loading buffer. The samples were boiled for 5 min and resolved by SDS-PAGE and immunoblotted for phospho-p38, DUPD1, and total p38.
Statistical Analysis
The data were examined by one-way analysis of variance followed by the Tukey test using Prism software (GraphPad Software Inc., San Diego, CA). The values were considered statistically significant at p < 0.05 (*) and p < 0.01 (**).
RESULTS
Inhibition of MAPK Activity by PRL Signaling through PRL-RS
We examined whether PRL administered in vivo to pseudopregnant mice expressing only PRL-RS (Fig. 1A) regulates MAPK activity in the ovary and decidua. Surprisingly and in sharp contrast to results reported in cell culture by others (21), we found that PRL administration in vivo induces a rapid decrease in ERK1/2 as well as p38 MAPK phosphorylation in both ovary (Fig. 1B) and decidua (Fig. 1C) of PRLR−/−RS mice.
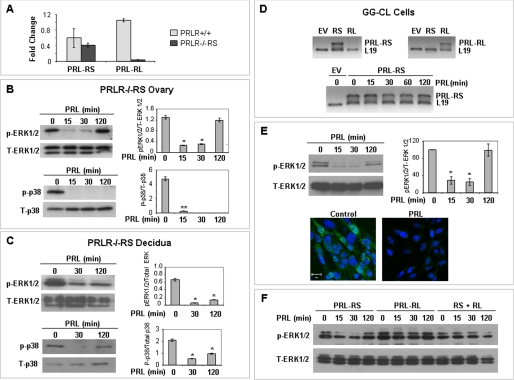
FIGURE 1.
PRL mediated inhibition of MAPK in ovary, decidua, and GG-CL cells expressing only PRL-RS. A, PRL-RS and PRL-RL expression were examined by qPCR in ovaries of wild type (PRLR+/+) and PRLR−/−RS mice. The expression of PRL-RS was not significantly different from wild type, whereas expression of PRL-RL was not detected in PRLR−/−RS ovaries. B and C, pseudopregnant PRLR−/−RS mice were injected with ergocryptine (200 μg, subcutaneously) to inhibit endogenous PRL secretion and were treated 6 h later with a single IP injection of PRL (60 μg). Activation status of ERK1/2 and p38 MAPK were measured by Western blot analysis using phosphospecific ERK1/2 (Thr-202/Tyr-204) and p38 MAPK (Thr-180/Tyr-182) antibodies in either ovaries or decidua. Total ERK1/2 and total p38 were used as loading controls. D, GG-CL cells were transfected with either PRL-RS or PRL-RL expression vectors, and specific expression of either receptor was analyzed by RT-PCR (upper panel). PRL treatment had no effect on PRL-RS expression (lower panel). E, activation status of ERK1/2 was measured by Western blot analysis using phospho-antibodies against ERK1/2 in GG-CL cells. Total ERK1/2 was used as loading control. Density of p-ERK was plotted against T-ERK (upper panel). GG-CL cells transfected with PRL-RS were treated with either PRL (1 μg/ml) or vehicle, and localization of active ERK1/2 was analyzed by immunocytochemistry as described under “Experimental Procedures.” Lower panel, green, p-ERK1/2; blue, DAPI. F, GG-CL cells transfected with PRL-RS, PRL-RL, or both were treated with PRL for different time points as indicated. Activation status of ERK1/2 was measured by Western blot analysis using phosphospecific ERK1/2 (Thr-202/Tyr-204) antibody. The values are expressed as the means ± S.E. (n = 3). *, p < 0.05; **, p < 0.01 versus 0 min.
We further established that PRL represses MAPK phosphorylation in an ovarian cell line (GG-CL) generated in our laboratory (37) and transfected with PRL-RS. As shown in Fig. 1D, PRL-RS is well expressed after transfection. The level of PRL-RS expression is not affected by PRL at any time point examined. However, PRL treatment caused a dramatic inhibition in the state of ERK1/2 phosphorylation as shown by both Western analysis and immunocytochemistry (Fig. 1E). The time course of this inhibition was very similar to the one observed in vivo in PRL-RS-expressing mice (Fig. 1A). Interestingly, this PRL-RS-mediated inhibition of MAPK is totally prevented when cells were co-transfected with PRL-RL (Fig. 1F).
Downstream Targets of MAPK but Not the Upstream Kinases Are Affected by PRL Signaling through PRL-RS in Vivo and in Vitro
The results shown in Fig. 2 indicate that PRL treatment in vivo inhibits markedly the phosphorylation of ERK1/2 downstream targets (p90RSK and ELK-1) as well as that of p38 MAPK (ATF-2) in both ovary (Fig. 2, A and B) and decidua (Fig. 2, C and D). Interestingly, however, the phosphorylation of MEK1/2 and MKK3/6, upstream kinases of ERK1/2 and p38, respectively, are not affected by PRL at any time investigated in either ovary (Fig. 2, A and B) or decidua (Fig. 2, C and D). Similar results are seen in cell culture (Fig. 3) where PRL induces a dephosphorylation of downstream MAPKs targets but not upstream kinases. This strongly suggests that the dephosphorylation of p38 MAPK and ERK1/2 by PRL signaling through PRL-RS is not due to deactivation of upstream kinases but rather due to activation of specific phospahatase(s).
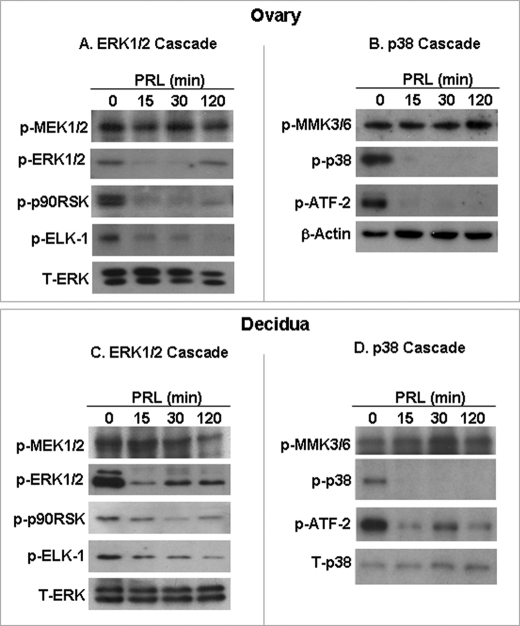
FIGURE 2.
Downstream targets of MAPK are affected by PRL/PRL-RS but not the upstream kinases in PRLR−/−RS mice. Decidualization in pseudopregnant PRLR−/−RS mice was induced as described under “Experimental Procedures.” PRLR−/−RS mice were injected with ergocryptine (200 μg, subcutaneously) for 6 h followed by a single IP injection of PRL (60 μg). ERK1/2 and p38 MAPK cascades were examined by Western blot analysis in the ovary (A and B) and decidua (C and D) using phospho-specific antibodies to ELK1, p90RSK, ATF2, MEK1/2, and MKK3/6. Total ERK1/2 or β-actin was used loading control.
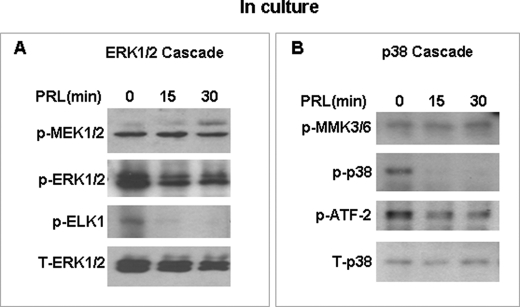
FIGURE 3.
Downstream targets of MAPK are affected by PRL/PRL-RS but not the upstream kinases in GG-CL cells expressing PRL-RS. GG-CL cells were transfected with PRL-RS and treated with PRL as indicated. Total protein extracts were analyzed for activated ERK1/2 (A) or activated p38 MAPK pathway (B) by Western analysis using phospho-specific antibodies to ELK1, ATF2, MEK1/2, and MKK3/6. Total ERK1/2 and total p38 were used as loading controls.
Inhibition of ERK1/2 Activity and Effect of Phosphatase Inhibitors in GG-CL Cells Expressing PRL-RS
To determine the phosphatase(s) responsible for PRL-mediated inhibition of MAPK activity, we treated GG-CL cells expressing PRL-RS with PRL in the presence or absence of protein-tyrosine phosphatase 1B inhibitor, SHP2 inhibitor (NSC 87877), and PP1 and PP2A inhibitor (okadaic acid) and with Jak2 inhibitor (AG490). As shown in Fig. 4A (top and middle panel), these inhibitors do not prevent PRL-mediated inhibition of ERK1/2, although all of the inhibitors used beside SHP2 inhibitors enhance the basal activity of ERK1/2.
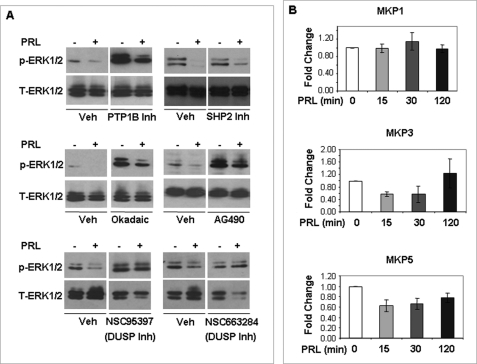
FIGURE 4.
Effect of various phosphatase inhibitors on the PRL-mediated inhibition of ERK1/2 and mRNA expression of MKPs. A, GG-CL cells transfected with PRL-RS were treated with PRL (1 μg/ml) for 15 min in the presence of protein-tyrosine phosphatase 1B (PTP1B) inhibitors, SHP2 inhibitor, okadaic acid, AG-490, NSC95397, and NSC663284 as described under “Experimental Procedures.” Activation status of ERK1/2 was measured by Western blot analysis using phospho-antibodies against ERK1/2. Total ERK1/2 was used as loading control. B, GG-CL cells transfected with PRL-RS were treated with PRL (1 μg/ml) for several time points as indicated. The mRNA levels of MKP1, MKP3, and MKP5 were analyzed by qPCR in these cells, and the expression levels were normalized to L19 or 36B4.
Interestingly, two dual specific phosphatase inhibitors (NSC 95397 and NSC 663284) completely reversed the inhibition of ERK1/2 phosphorylation by PRL signaling through PRL-RS (Fig. 4A, bottom panel). NSC 95397 is known to inhibit the MAPK phosphatases MKP1 and MKP3, two phosphatases whose recognized mode of regulation is by rapid induction in response to activating stimulus. We therefore analyzed the expression levels of MKP1/3 mRNA and also that of MKP5, another inducible MAPK phosphatase, by qPCR. As shown in Fig. 4B, PRL treatment had no significant effect on the expression of any of the MKPs examined.
Identification of DUPD1 as PRL-RS Associated Protein
To find out whether phosphatase(s) associates with PRL-RS, fusion proteins with glutathione S-transferase containing the cytoplasmic domain of PRL-RS (RS-GST) were produced, purified, and applied to glutathione-Sepharose columns. Cell extracts, obtained from either PRLR−/−RS mice decidua (Fig. 5A) or GG-CL cells (Fig. 5B) treated with PRL, were applied to the columns and eluted with SDS-PAGE sample buffer. Eluted proteins were analyzed by SDS-PAGE and silver staining. RS-GST pulls down a band approximately at 27 kDa both in decidual and GG-CL extracts (Fig. 5, A and B). PRL treatment had no effect on the association of this protein with PRL-RS. RS-GST alone (without cell lysates, RS) was eluted at ∼25 kDa.
FIGURE 5.
Identification of DUPD1 as PRL-RS associated protein. PRL-RS-GST (RS-GST) fusion proteins were prepared as described under “Experimental Procedures.” RS-GST interacting proteins were pulled down using decidual cell extracts from PRLR−/−RS mice treated with PRL (60 μg/animal) or vehicle (A) or GG-CL cells transfected with PRL-RS and treated with PRL (1 μg/ml) for different time points (B). The bands were detected by silver staining. The molecular mass markers (lanes M) were as indicated to determine the size of the bands. As a control, a well of pre-pulldown PRL-RS-GST (RS) was loaded showing a band of 25 kDa. The band (27–30 kDa) pulldown from decidual and GG-CL cell lysates by RS-GST fusion protein is indicated by arrow. C, RS-GST pulldown samples of GG-CL cell lysates were analyzed by Western blot using a polyclonal antibody to DUPD1. To show the specific interaction of DUPD1 with RS-GST, GST alone (C-GST) was used to pull down GG-CL cell lysates as a control. GG-CL cells were transfected with PRL-RS and treated with PRL (1 μg/ml) for different time points. Expression of DUPD1 was measured by qPCR (D) or Western analysis (E). *, p < 0.05 versus 0 min.
A dual specific phosphatase (DUSP) of similar molecular mass was recently cloned, and an antibody against it was generated (38, 39). The 220-amino acid active human DUSP (human chromosome 10) was named DUSP27 by Friedberg et al. (38). However, throughout the text we refer to this protein by its official name, DUPD1.4 We examined the possibility that DUPD1 associates with PRL-RS using RS-GST pulldown assay followed by Western analysis with DUPD1 antibody. As shown in Fig. 5C, DUPD1 was clearly detected in association with the RS-GST but not with the control GST. The association of DUPD1 with the PRL-RS occurred in the absence of PRL, suggesting a constitutive association irrespective of ligand activation. Moreover, PRL had no effect on the localization of this protein in GG-CL cells (supplemental Fig. S1).
We examined whether PRL affects the expression and/or the state of phosphorylation of DUPD1 in GG-CL cells. The results shown in Fig. 5 (D and E) indicate that PRL does not induce rapidly the expression of DUPD1 mRNA and protein levels to account for early ERK1/2 and p38 dephosphorylation (Figs. 1 and 2). Although a delayed stimulation was observed, this stimulation occurred long after MAPKs were dephosphorylated.
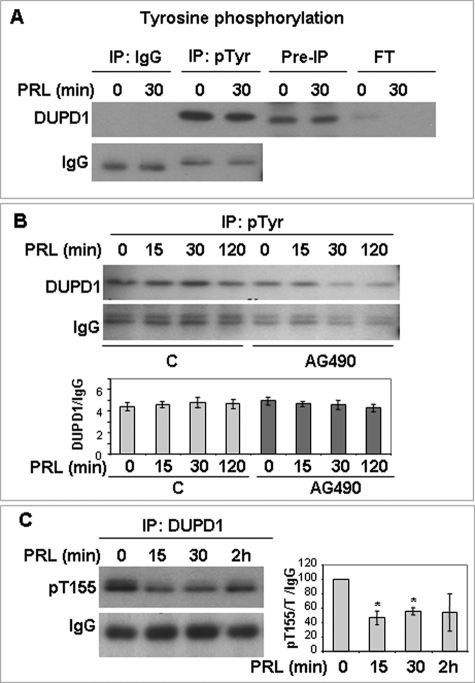
Motif scan analysis (40) of the rat DUPD1 sequence (supplemental Fig. S2) revealed two putative tyrosine phosphorylation sites at Tyr-32 and Tyr-100. As shown in Fig. 6 (A and B), DUPD1 is phosphorylated on tyrosine; however, this phosphorylation is not affected by PRL treatment. Furthermore, treatment with AG490, a Jak2 inhibitor, had no apparent effect on tyrosine phosphorylation of DUPD1 at any time point of PRL treatment (Fig. 6B).
FIGURE 6.
DUPD1 is a phosphoprotein. A, GG-CL cells were transfected with PRL-RS and treated with PRL for different time points. Total cell lysates were immunoprecipitated with anti-phosphotyrosine antibody or control IgG and immunoblotted for DUPD1. B, GG-CL cells were transfected with PRL-RS and treated with PRL in the presence or absence of AG490. Total cell lysates were immunoprecipitated with anti-phophotyrosine antibody and immunoblotted for DUPD1. Density of DUPD1 was plotted against IgG. C, GG-CL cells were transfected with PRL-RS and treated with PRL. Total cell lysates were immunoprecipitated with DUPD1 and immunoblotted with an anti-phospho-RXRXX(S/T) antibody. The density of phospho-DUPD1 was plotted against IgG. *, p < 0.05 versus 0 min.
Motif scan analysis of the rat DUPD1 also revealed two putative serine/threonine phosphorylation sites at Ser-153 and Thr-155 (supplemental Fig. S3). GG-CL extracts immunoprecipitated with DUPD1 antibody and immunoblotted with a targeted antibody against the Thr-155 site, which lies within the RXRXXT motif, indicates that DUPD1 is phosphorylated on threonine and that this phosphorylation is markedly inhibited by PRL (Fig. 6C).
DUPD1 Physically Associates with ERK1/2 and p38 MAPK
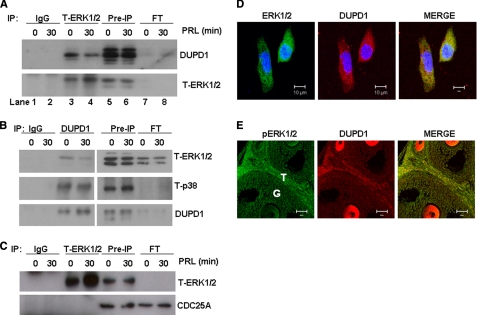
We examined whether DUPD1 associates with MAPK by immunoprecipitating total protein extracts from GG-CL cells with antibody against either ERK1/2 or DUPD1. The ERK1/2 immunoprecipitates were analyzed either by gel silver staining or by immunoblotting with DUPD1 antibody. Whereas silver-stained gels showed association of ERK1/2 with a band ∼27 kDa (supplemental Fig. S4), results shown in Fig. 7A clearly depict that DUPD1 co-immunoprecipitates with ERK1/2. Fig. 7 also shows efficient depletion of DUPD1 in the flow through as compared with pre-immunoprecipitation (IP) samples and immunoprecipitation with ERK1/2 antibody but not with IgG. Conversely, immunoprecipitation with DUPD1 antibodies shows (Fig. 7B) that ERK1/2 as well as p38 MAPK co-precipitate with DUPD1, but not with control IgG. These results taken together demonstrate that DUPD1 physically associates with both ERK1/2 and p38 MAPK.
FIGURE 7.
DUPD1 physically associates with ERK1/2 and p38 MAPK. GG-CL cells were transfected with PRL-RS and treated with PRL (1 μg/ml) for 30 min. A, total protein extracts were subjected to IP with T-ERK1/2 or rabbit IgG, and immunocomplexes were resolved on SDS-PAGE, transferred onto PVDF membrane, and immunoblotted against DUPD1 or T-ERK1/2s. Lanes 1 and 2, IP with IgG (0 and 30 min PRL-treated samples); lanes 3 and 4, IP with T-ERK1/2 (0 and 30 min PRL-treated samples); lanes 5 and 6, pre-IP samples (total protein extracts); lanes 7 and 8, flow through after IP (unbound supernatants). B, cells were IP with DUPD1 antibody or goat IgG, and immunocomplexes were resolved and immunoblotted as described for A. C, cells were immunoprecipitated with T-ERK1/2 antibody or goat IgG, and immunocomplexes were resolved and immunoblotted against CDC25A and T-ERK1/2. D, GG-CL cells were transfected with PRL-RS and treated with PRL for 30 min. Co-localization of T-ERK1/2 (green) and DUPD1 (red) was examined by immunocytochemistry using specific antibodies. Blue, DAPI. E, paraffin-embedded ovary sections from PRLR−/−RS mice (2.5 days pregnant) were analyzed for co-localization of active ERK1/2 (green) and DUPD1 (red).
To determine the specificity of ERK1/2 association with DUPD1, we examined whether CDC25A also associates with this kinase. This DUSP phosphatase was selected because of its sensitivity to NSC 95397 and NSC 663284, which prevent PRL-mediated ERK1/2 inhibition (Fig. 4). The results (Fig. 7C) indicate no association of ERK1/2 with CDC25A. This was also confirmed by the fact that an equal amount of proteins was eluted in the flow through as that of pre-IP samples.
The immunofluorescence studies shown in Fig. 7, D and E, further confirm the association of ERK1/2 or phospho-ERK1/2 with DUPD1. They co-localize in GG-CL cells, and in the ovary of PRLR−/−RS mice, a strong coexpression of this kinase and the phosphatase is apparent in theca cells as well as in oocytes. Co-localization is also observed in granulosa cells (Fig. 7E).
DUPD1 Is Responsible for PRL-mediated MAPK Inhibition
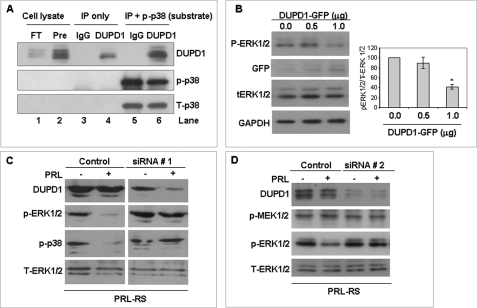
We examined whether DUPD1 is a MAPK phosphatase using an in vitro phosphatase assay. As shown in Fig. 8A (lanes 3 and 4), DUPD1 was efficiently and specifically immunoprecipitated from GG-CL cells by DUPD1 antibody. The phosphatase assay was carried out using excess recombinant phosphorylated p38 MAPK. As shown in Fig. 8A (lanes 5 and 6), incubation of p38 substrate with DUPD1 substantially reduced its state of phosphorylation. We also examined whether DUPD1 causes the dephosphorylation of ERK1/2. However, because an ERK1/2 phosphorylated substrate that can be differentiated from the endogeneous ERK1/2 (by molecular mass) is not available for the in vitro phosphatase assay, we transfected GG-CL cells with DUPD1-GFP expression vector and examined the state of phosphorylation of the endogenous ERK1/2. We found (Fig. 8B) a dose-dependant inhibition in ERK1/2 phosphorylation by overexpression of DUPD1-GFP.
FIGURE 8.
DUPD1 is a MAPK phosphatase. A, total protein lysates of GG-CL cells expressing PRL-RS were subjected to IP with a polyclonal antibody to DUPD1 or control IgG. In vitro phosphatase assay was carried out using these immunoprecipitates and a recombinant active p38 (rp-p38) as substrate. Finally, the proteins were resolved on SDS-PAGE and immunoblotted against p-p38 to examine the remaining substrate. The blot was also reprobed for DUPD1 and T-p38. Lane 1, flow through after the IP; lane 2, pre-IP; lane 3, IP with IgG alone; lane 4, IP with DUPD1 alone; lane 5, IP with IgG and rp-p38 (after the assay); lane 6, IP with DUPD1 and rp-p38 (after the assay). B, GG-CL cells were co-transfected with PRL-RS and increasing doses of DUPD1-GFP expression vectors or empty vectors. Phosphorylation status of ERK1/2 was examined using phospho-specific antibody. Expression of exogenous DUPD1 was measured by Western blot against GFP antibody. Total ERK1/2 and GAPDH were used as loading controls. The density of p-ERK1/2 was plotted against T-ERK1/2. *, p < 0.05 versus 0 μg. C, GG-CL cells were co-transfected with PRL-RS and DUPD1 siRNA (siRNA # 1) or a control siRNA. After 48 h, the cells were treated with or without PRL for 15 min. DUPD1, p-ERK1/2, and p-p38 were measured in the total cell lysates by Western blotting. Total ERK1/2 was used as loading control. D, GG-CL cells were co-transfected with PRL-RS and DUPD1 siRNA (siRNA # 2) or a control siRNA. After 48 h, the cells were treated with or without PRL for 15 min. DUPD1, p-MEK1/2 and p-ERK1/2 were measured in the total cell lysates by Western blotting. Total ERK1/2 was used as a loading control.
We further established, using siRNA to silence DUPD1, that this phosphatase is involved in PRL-mediated dephosphorylation of MAPK. As shown in Fig. 8C (top panels) DUPD1 siRNA 1, but not control siRNA, represses the expression of DUPD1 protein levels in GG-CL cells. PRL induces down-regulation of ERK1/2 as well as p38 phosphorylation in GG-CL transfected with control siRNA (left panels). This PRL effect is totally reversed in the DUPD1 siRNA transfected group (right panels).
We further examined whether DUPD1 knockdown affects ERK1/2 upstream kinase. As shown in Fig. 8D, DUPD1 siRNA significantly down-regulates endogeneous DUPD1 in GG-CL cells without affecting the state of MEK1/2 phosphorylation. Here also, PRL-mediated inhibition of ERK1/2 is totally prevented by the second set of DUPD1 siRNA.
DISCUSSION
The results of the present investigation reveal the original finding that PRL acting through PRL-RS inhibits ERK1/2 and p38 MAPK activity as well as their downstream targets in two PRL reproductive target tissues: the ovary and decidua. This PRL/PRL-RS-mediated inhibition is clearly detectable in whole animal as well as in cultured cells expressing PRL-RS. The results also demonstrate that deactivation of the MAPKs is not due to dephosphorylation of upstream kinases but is rather due to PRL/PRL-RS activation of a novel member of the DUSP phosphatase family, DUPD1. In addition, our results show that DUPD1 binds to the intracellular domain of PRL-RS and also associates with ERK1/2 and p38. Finally, we have shown that PRL activation of PRL-RS induces the dephophorylation of DUPD1 and have established an as yet undiscovered role for DUPD1 in the dephosphorylation of both MAPKs.
The present finding of MAPK inhibition sharply contrast with the previous report indicating activation of MAPK by PRL in NIH3T3 cells expressing PRL-RS (21). Another study (41) has shown that PRL can activate ERK phosphorylation in PC3 or DU145 cells expressing human PRL-RS isoforms. However, this activation did not occur with PRL but only in response to S179D PRL, a molecular mimic of the phosphorylated form of PRL. Activation of ERK1/2 by PRL/PRL-RS may be unique to these cell lines, or alternatively PRL may have different signaling mechanisms in reproductive tissues.
The MAPK family has a central role in various cellular responses including cell proliferation, differentiation, survival, and apoptosis (42, 43). Recently, it was shown that MAPK critically regulate the fate of ovarian granulosa cells (25, 42). Activation of p38 MAPK by FSH leads to phosphorylation of HSP-27 in granulosa cells, resulting in cell rounding and aggregation promoting microfilament reorganization and stabilization (44), suggesting a role in granulosa cell survival. Attenuation of Raf-1-MEK-ERK signaling pathway has also been shown to correlate with granulosa cell apoptosis (45). Deletion of ERK1/2 at relatively advanced stages of follicular development prevents follicular differentiation and ovulation (25). Our present results demonstrate that the PRL/PRL-RS elicits repression of ERK1/2 and p38 MAPK as well as that of their downstream targets; these repressions may well be associated with the recognized severe defects in follicular development and premature ovarian failure that occur in response to exclusive PRL-RS stimulation (5). We have previously shown that PRL-RL prevents the deteriorative effect of PRL signaling through PRL-RS (5). This finding is further supported by our present results indicating that PRL-mediated inhibition of MAPK is totally prevented when PRL-RL is coexpressed with PRL-RS.
We have examined the mechanism by which PRL activation of PRL-RS leads to MAPK inhibition as well as their downstream substrates. Our finding that MEK1/2 and MKK3/6, the upstream dual specific kinases that phosphorylate tyrosine and threonine residues in the ERK1/2 or p38 activation loops, remain unchanged in response to PRL suggested to us that PRL/PRL-RS activates a MAPK specific phosphatase and that this phosphatase most likely has affinity for both ERK1/2 and p38. We examined the role of PRL on known inducible MAPK phosphatases in PRLR−/−RS mice and found no PRL-mediated stimulation. Because not many phosphatase inhibitors can be used in vivo, we transfected an ovarian cell line generated in our laboratory with PRL-RS and established that PRL causes similar inhibition to the one seen in vivo in terms of MAPK signaling. Treatment with an array of phosphatase inhibitors revealed that only the DUSP inhibitors (NSC 95397 and NSC6 663284) could prevent PRL-mediated ERK1/2 dephosphorylation, suggesting involvement of a DUSP in this process. There are several known dual specificity MKPs that dephosphorylate and inactivate MAPK isoforms in mammalian cells (46). Among these MKPs, MKP1 and MKP3 are known to be induced rapidly in granulosa cells, are inhibited by NSC 95397, and regulate ERK1/2 activity negatively (26, 47). This led us to examine whether the rapid increase in the expression levels of these phosphatases could play a role in PRL-mediated inhibition of ERK1/2; however, we found no such PRL-mediated stimulation. NSC 663284, the other DUSP inhibitor that prevents PRL-mediated down-regulation of ERK1/2 phosphorylation, is known to inhibit CDC25 (48), a DUSP known to be involved in the cell cycle and to inhibit the ERK1/2 pathway upstream of the MEK kinase level (49). We examined whether PRL causes a physical association of CDC25 family with ERK1/2. We found no such association in either the presence or absence of PRL, ruling out the involvement of CDC25 in the PRL-mediated inhibition of ERK1/2 activity. All of these results suggest that the phosphatase is a DUSP but not a known MPK phosphatase.
Luckily, we found a dual specific phosphatase DUPD1 to be physically associated with PRL-RS. We discovered that DUPD1 not only interacts with PRL-RS but also co-localizes and associates with ERK1/2 and p38 MAPK in both ovaries and an ovarian cell line expressing only the PRL-RS. We have established, using in vitro phosphatase assay and DUPD1 overexpression, that ERK1/2 and p38 are dephosphorylated by DUPD1. Most importantly we have shown that knockdown of endogenous DUPD1 can completely prevent PRL-mediated MAPK inhibition without affecting upstream kinase, establishing a key role for this phosphatase in PRL-mediated dephosphorylation of MAPK.
A previous study predicted DUPD1 to have 446 amino acid residues with a pro-isomerase domain and a cyclophilin-like domain in addition to its catalytic phosphatase domain (38). When it was expressed, it was found to have an open reading frame that translates into a 220-amino acid protein with a calculated molecular mass of 25.33 kDa. However, the recombinant protein runs slightly higher than this size (38), suggesting post-translational modifications. Indeed, our pulldown as well as Western analysis detected DUPD1 at ∼27 kDa. Other members of the DUSP family such as DUSP15 have been shown to be myristoylated and can be targeted to plasma membrane. Whether such post-translational modification of DUPD1 occurs and whether this facilitates interaction with PRL-RS are not known. Previously, our lab has shown that another microsomal protein, PRAP, is associated with PRL-RS (33). However, very little is known about how molecules associate with PRL-RS. One possibility is that ligand binding causes Jak2 activation, which allows recruitment of these factors. However, in the case of DUPD1, this is unlikely because DUPD1 can efficiently bind to the receptor without PRL stimulation. Moreover, Jak2 kinase inhibitor had no effect on PRL-mediated ERK1/2 inhibition. Therefore, it appears that Jak2 is not involved in either association or activation of DUPD1 and that both Jak2 and DUPD1 are constitutively associated with PRL-RS. Because other DUSPs have been shown to be early inducible genes (50), we examined whether DUPD1 is induced by PRL. We found no increase in DUPD1 expression related to MAPK dephosphorylation. Post-translation modifications of DUSPs are known to affect their phosphatase activity. Acetylation of MKP-1 promotes the interaction of MKP-1 with its substrate p38 MAPK, which results in dephosphorylation of p38 MAPK (51). However, PRL/PRL-RS-mediated activation of DUPD1 does not involve acetylation because no acetyl modification was detected on DUPD1 in response to PRL treatment (results not shown). Phosphorylation of DUSPs has been shown to regulate their activity (52). Whether DUPD1 function is modified by phosphorylation is not yet known. However, our results indicate that PRL activation of PRL-RS causes a significant inhibition in DUPD1 phosphorylation at threonine 155. Interestingly, the targeted threonine, threonine 155, lies just three residues C-terminal to the absolute required arginine at residue 152 within the catalytic core of the phosphatase. Therefore, it is highly possible that phosphorylation on threonine 155 prevents its catalytic activity and PRL induces its activity by causing dephosphorylation on this site. Further studies are necessary to examine the intriguing possibility that inhibition of this phosphorylation site may be involved in the stability or activation of this phosphatase.
Supplementary Material
Acknowledgment
We acknowledge Moe EL-Ahmad for technical help.
This work was supported, in whole or in part, by National Institutes of Health Grants HD11119 and HD 12356 (to G. G.) and T32 HL007692 (to J. L. and A. M. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
The phosphatase cloned, characterized, and renamed (DUSP27) by Friedberg et al. (38) is currently known as DUPD1, consistent with the official human gene symbol indicated by HUGO Gene Nomenclature Committee (DUPD1; HGNC:23481), the official mouse gene symbol according to Mouse Genome Database (Dupd1; MGI:3647127), and the official rat nomenclature according to the Rat Genome Database (Dupd1; RGD:1310229). The reason we use its official name (DUPD1) is to avoid confusion with the large 1100-amino acid inactive human phosphatase (human chromosome 1), which is now designated officially as DUSP27 (HGNC:25034).
- PRL
- prolactin
- PRLR
- PRL receptor
- qPCR
- quantitative PCR
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- DUSP
- dual specific phosphatase.
REFERENCES
- 1. Risk M., Gibori G. (2001) in Prolactin (Horseman N. D. ed) pp. 265–295, Kluwer Academic, Boston, MA [Google Scholar]
- 2. Stocco C., Telleria C., Gibori G. (2007) Endocr. Rev. 28, 117–149 [DOI] [PubMed] [Google Scholar]
- 3. Bachelot A., Binart N. (2007) Reproduction 133, 361–369 [DOI] [PubMed] [Google Scholar]
- 4. Ben-Jonathan N., LaPensee C. R., LaPensee E. W. (2008) Endocr. Rev. 29, 1–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Halperin J., Devi S. Y., Elizur S., Stocco C., Shehu A., Rebourcet D., Unterman T. G., Leslie N. D., Le J., Binart N., Gibori G. (2008) Mol. Endocrinol. 22, 513–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Russell D. L., Richards J. S. (1999) Mol. Endocrinol. 13, 2049–2064 [DOI] [PubMed] [Google Scholar]
- 7. Frasor J., Gaspar C. A., Donnelly K. M., Gibori G., Fazleabas A. T. (1999) J. Clin. Endocrinol. Metab. 84, 3344–3350 [DOI] [PubMed] [Google Scholar]
- 8. Gu Y., Srivastava R. K., Clarke D. L., Linzer D. I., Gibori G. (1996) Endocrinology 137, 4878–4885 [DOI] [PubMed] [Google Scholar]
- 9. Davis J. A., Linzer D. I. (1989) Mol. Endocrinol. 3, 674–680 [DOI] [PubMed] [Google Scholar]
- 10. Telleria C. M., Parmer T. G., Zhong L., Clarke D. L., Albarracin C. T., Duan W. R., Linzer D. I., Gibori G. (1997) Endocrinology 138, 4812–4820 [DOI] [PubMed] [Google Scholar]
- 11. Lesueur L., Edery M., Ali S., Paly J., Kelly P. A., Djiane J. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 824–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. O'Neal K. D., Yu-Lee L. Y. (1994) J. Biol. Chem. 269, 26076–26082 [PubMed] [Google Scholar]
- 13. Llovera M., Touraine P., Kelly P. A., Goffin V. (2000) Exp. Gerontol. 35, 41–51 [DOI] [PubMed] [Google Scholar]
- 14. Jabbour H. N., Critchley H. O., Boddy S. C. (1998) J. Clin. Endocrinol. Metab. 83, 2545–2553 [DOI] [PubMed] [Google Scholar]
- 15. Hu Z. Z., Meng J., Dufau M. L. (2001) J. Biol. Chem. 276, 41086–41094 [DOI] [PubMed] [Google Scholar]
- 16. Boutin J. M., Jolicoeur C., Okamura H., Gagnon J., Edery M., Shirota M., Banville D., Dusanter-Fourt I., Djiane J., Kelly P. A. (1988) Cell 53, 69–77 [DOI] [PubMed] [Google Scholar]
- 17. Bignon C., Binart N., Ormandy C., Schuler L. A., Kelly P. A., Djiane J. (1997) J. Mol. Endocrinol. 19, 109–120 [DOI] [PubMed] [Google Scholar]
- 18. Clarke D. L., Linzer D. I. (1993) Endocrinology 133, 224–232 [DOI] [PubMed] [Google Scholar]
- 19. Berlanga J. J., Garcia-Ruiz J. P., Perrot-Applanat M., Kelly P. A., Edery M. (1997) Mol. Endocrinol. 11, 1449–1457 [DOI] [PubMed] [Google Scholar]
- 20. Chang W. P., Ye Y., Clevenger C. V. (1998) Mol. Cell Biol. 18, 896–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Das R., Vonderhaar B. K. (1995) Mol. Endocrinol. 9, 1750–1759 [DOI] [PubMed] [Google Scholar]
- 22. Huang K., Ueda E., Chen Y., Walker A. M. (2008) J. Mammary Gland Biol. Neoplasia 13, 69–79 [DOI] [PubMed] [Google Scholar]
- 23. Binart N., Imbert-Bollore P., Baran N., Viglietta C., Kelly P. A. (2003) Mol. Endocrinol. 17, 1066–1074 [DOI] [PubMed] [Google Scholar]
- 24. Devi Y. S., Shehu A., Stocco C., Halperin J., Le J., Seibold A. M., Lahav M., Binart N., Gibori G. (2009) Endocrinology 150, 3327–3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fan H. Y., Liu Z., Shimada M., Sterneck E., Johnson P. F., Hedrick S. M., Richards J. S. (2009) Science 324, 938–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fan H. Y., Shimada M., Liu Z., Cahill N., Noma N., Wu Y., Gossen J., Richards J. S. (2008) Development 135, 2127–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thienel T., Chwalisz K., Winterhager E. (2002) Mol. Hum. Reprod. 8, 465–474 [DOI] [PubMed] [Google Scholar]
- 28. Shoji T., Yoshida S., Mitsunari M., Miyake N., Tsukihara S., Iwabe T., Harada T., Terakawa N. (2007) J. Reprod. Immunol. 75, 82–90 [DOI] [PubMed] [Google Scholar]
- 29. Yoshino O., Osuga Y., Hirota Y., Koga K., Hirata T., Yano T., Ayabe T., Tsutsumi O., Taketani Y. (2003) J. Clin. Endocrinol. Metab. 88, 2236–2241 [DOI] [PubMed] [Google Scholar]
- 30. Bao L., Tessier C., Prigent-Tessier A., Li F., Buzzio O. L., Callegari E. A., Horseman N. D., Gibori G. (2007) Endocrinology 148, 2326–2334 [DOI] [PubMed] [Google Scholar]
- 31. Gu Y., Jayatilak P. G., Parmer T. G., Gauldie J., Fey G. H., Gibori G. (1992) Endocrinology 131, 1321–1328 [DOI] [PubMed] [Google Scholar]
- 32. Deleted in proof.
- 33. Duan W. R., Linzer D. I., Gibori G. (1996) J. Biol. Chem. 271, 15602–15607 [DOI] [PubMed] [Google Scholar]
- 34. Yan J. X., Wait R., Berkelman T., Harry R. A., Westbrook J. A., Wheeler C. H., Dunn M. J. (2000) Electrophoresis 21, 3666–3672 [DOI] [PubMed] [Google Scholar]
- 35. Bao L., Devi S., Bowen-Shauver J., Ferguson-Gottschall S., Robb L., Gibori G. (2006) Mol. Endocrinol. 20, 3240–3250 [DOI] [PubMed] [Google Scholar]
- 36. Wang P. Y., Liu P., Weng J., Sontag E., Anderson R. G. (2003) EMBO J. 22, 2658–2667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sugino N., Zilberstein M., Srivastava R. K., Telleria C. M., Nelson S. E., Risk M., Chou J. Y., Gibori G. (1998) Endocrinology 139, 1936–1942 [DOI] [PubMed] [Google Scholar]
- 38. Friedberg I., Nika K., Tautz L., Saito K., Cerignoli F., Godzik A., Mustelin T. (2007) FEBS Lett. 581, 2527–2533 [DOI] [PubMed] [Google Scholar]
- 39. Alonso A., Sasin J., Bottini N., Friedberg I., Friedberg I., Osterman A., Godzik A., Hunter T., Dixon J., Mustelin T. (2004) Cell 117, 699–711 [DOI] [PubMed] [Google Scholar]
- 40. Obenauer J. C., Cantley L. C., Yaffe M. B. (2003) Nucleic Acids Res. 31, 3635–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wu W., Ginsburg E., Vonderhaar B. K., Walker A. M. (2005) Cancer Res. 65, 7509–7515 [DOI] [PubMed] [Google Scholar]
- 42. Tamura M., Nakagawa Y., Shimizu H., Yamada N., Miyano T., Miyazaki H. (2004) J. Reprod. Dev. 50, 47–55 [DOI] [PubMed] [Google Scholar]
- 43. Zhang W., Liu H. T. (2002) Cell Res. 12, 9–18 [DOI] [PubMed] [Google Scholar]
- 44. Hunzicker-Dunn M., Maizels E. T. (2006) Cell Signal. 18, 1351–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gebauer G., Peter A. T., Onesime D., Dhanasekaran N. (1999) J. Cell Biochem. 75, 547–554 [PubMed] [Google Scholar]
- 46. Owens D. M., Keyse S. M. (2007) Oncogene 26, 3203–3213 [DOI] [PubMed] [Google Scholar]
- 47. Shepherd E. G., Zhao Q., Welty S. E., Hansen T. N., Smith C. V., Liu Y. (2004) J. Biol. Chem. 279, 54023–54031 [DOI] [PubMed] [Google Scholar]
- 48. Guo J., Parise R. A., Joseph E., Lan J., Pan S. S., Joo B., Egorin M. J., Wipf P., Lazo J. S., Eiseman J. L. (2007) Anticancer Res. 27, 3067–3073 [PubMed] [Google Scholar]
- 49. Nemoto K., Vogt A., Oguri T., Lazo J. S. (2004) Prostate 58, 95–102 [DOI] [PubMed] [Google Scholar]
- 50. Keyse S. M. (2008) Cancer Metastasis Rev. 27, 253–261 [DOI] [PubMed] [Google Scholar]
- 51. Chi H., Flavell R. A. (2008) Sci. Signal 1, pe44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jeffrey K. L., Camps M., Rommel C., Mackay C. R. (2007) Nat. Rev. Drug. Discov. 6, 391–403 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.