Abstract
The LHR has an essential role in sexual development and reproductive function, and its transcription is subjected to several modes of regulation. In this study, we investigated PC4 coactivator function in the control of LHR transcription. Knockdown of PC4 by siRNA inhibited the LHR basal promoter activity and trichostatin A (TSA)-induced gene transcriptional activation and expression in MCF-7 cells. While overexpression of PC4 alone had no effect on the LHR gene, it significantly enhanced Sp1- but not Sp3-mediated LHR transcriptional activity. PC4 directly interacts with Sp1 at the LHR promoter, and this interaction is negatively regulated by PC4 phosphorylation. The coactivator domain (22–91 aa) of PC4 and DNA binding domain of Sp1 are essential for PC4/Sp1 interaction. ChIP assay revealed significant occupancy of PC4 at the LHR promoter that increased upon TSA treatment. Disruption of PC4 expression significantly reduced TSA-induced recruitment of TFIIB and RNAP II, at the promoter. PC4 functions are beyond TSA-induced phosphatase release, PI3K-mediated Sp1 phosphorylation, and HDAC1/2/mSin3A co-repressor release indicating its role as linker coactivator of Sp1 and the transcriptional machinery. These findings demonstrated a critical aspect of LHR modulation whereby PC4 acts as a coactivator of Sp1 to contribute to the human of LHR transcription.
Keywords: Coregulator Transcription, Gene Regulation, Gene Transcription, Sp1, Transcription Coactivators
Introduction
The luteinizing hormone receptor (LHR)2 is a member of the G protein-coupled receptor family and is essential for sexual development and reproduction in mammals. The LHR is predominantly located on the plasma membrane of gonadal cells, where it mediates the luteinizing hormone signals that regulate ovarian granulosa/luteal and testicular Leydig cell development and function. It is also found in non-gonadal cells, tumoral tissues, and cancer cells (1–2), and these have provided a convenient model to study modalities of LHR transcriptional regulation.
The TATA-less LHR promoter contains two activating Sp1 binding domains and an upstream inhibitory motif that binds nuclear orphan receptors (3–8). Characterization of LHR transcriptional mechanisms revealed that the LHR gene is subject to repression/derepression through complex and diverse networks that include an epigenetic modulation at the promoter and association/dissociation of multiple effectors centered at the proximal Sp1 site of the promoter. Local chromatin changes resulting from histone acetylation and cell-specific CpG island methylation/demethylation within the promoter are critical for silencing and reactivation of the LHR gene in cancer cells (9). Sp1 acts as an anchor to recruit histone deacetylases (HDAC)1/2/mSin3A corepressor complex and p107 repressor protein. This results in promoter localized hypo-acetylation that contributes to the silencing of LHR transcriptional expression. The participation of the PI3K/PKCζ was found to be essential for histone deacetylase inhibitor TSA-induced LHR activation in cancer cells (10). PKCζ directly associates with Sp1 and phosphorylates Sp1 at Ser-641, which triggers dissociation of p107 repressor from Sp1, recruitment of TFII B and Pol II and LHR gene activation. TSA-induced chromatin changes cause cell-specific release of phosphatases which associate directly through Sp1, or indirectly through HDAC1/2 at the promoter. This serves as an on switch for Sp1 phosphorylation that triggers release of p107 repressor from Sp1 at the promoter and marked transcriptional activation of the LHR gene (11). Maximal derepression of the LHR gene upon TSA treatment is dependent on complete demethylation of the promoter, in conjunction with histone hyperacetylation and release of repressors (p107 and HDAC/mSin3A).
Whereas the role of repressor/corepressors (e.g. HDAC1/2, mSin3A, p107) and their association/dissociation with Sp1 in TSA-induced repression/derepression of the LHR gene have been well characterized, the involvement of transcriptional coactivators in this process has not been elucidated. Histone acetylase transferases p300 and CBP have been reported to participate in transcriptional activation of many genes in response to the HDAC inhibitors (12–13). However, the absence of participation of these coactivators in LHR activation induced by TSA (10) suggested the participation of other Sp1-associated coactivator(s). Positive cofactor 4 (PC4) is a highly abundant and multifunctional nuclear protein that has important roles in transcription, replication and DNA-repair (14). As a transcriptional coactivator, PC4 is proposed to facilitate activator-dependent class II gene transcription through providing bridge interactions between components of the general transcription machinery and transcriptional activators such as GAL4-Sp1, GAL4-VP16, GAL4-BRCAL1, GAL4-OCA-B (15–19) as demonstrated in studies using reconstituted systems. PC4 was also found to stimulate the function of several activators in vivo including activator protein 2, AP2 in ras-transformed PA-1 cells (20–21), HIV transactivator TAT (22) and more recently p53-mediated transactivation (23–24). While PC4 was shown to increase transactivation of GAL-Sp1 in an in vitro reconstituted cell- free transcription system (15–16), no functional in vivo evidence was provided for PC4-mediated Sp1 activation. In this study, we investigated the role of PC4 in basal LHR promoter activity and TSA-induced LHR gene transcription/expression. These studies have demonstrated a crucial aspect of LHR gene modulation, whereby PC4 acts as an essential coactivator of Sp1 to facilitate LH receptor transcription.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Trichostatin A (TSA) was purchased from Calbiochem. The antibodies against PC4, Sp1, TFIIB, p107, HDAC1, HDAC2, mSin3A, PP1, MED17 antibody, and actin were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). The Pol II antibody was from Upstate Biotechnology (Lake Placid, NY). V5 and Flag antibodies were purchased from Invitrogen (Carlsbad, CA) and Sigma-Aldrich. Calf intestine phosphatase (CIP) and casein kinase II (CKII) were obtained from New England Biolabs Inc. (Ipswich, MA). Purified recombinant PC4 was purchased from Protein One (Rockville, MD).
Reporter Gene Constructs and Expression Vectors
The reporter gene construct for LHR promoter was generated by cloning the human LHR (hLHR) gene promoter region (−176 to +1) into the SacI/BglII sites of pGL2 basic vector (25). The full-length human p21 promoter-luciferase reporter construct pWWP-luc was provided by Addgene Inc. (Cambridge, MA). The probasin promoter construct was generated by inserting the region (+11 to −256 bp) into the KpnI/BglII site of pGL2 basic vector. The pCMV6-PC4 and pCMV-Sp1 were purchased from Origene (Rockville, MD), and used as PCR template for generation of constructs expressing PC4 or Sp1. The pcDNA 1.1-Sp3 vector was described previously (26). 3×Flag PC4 vector was created by inserting PCR-amplified PC4 cDNA into the EcoRI and KpnI sites of the p3×FLAG-CMV-7.1 vector (Sigma). The V5-tagged PC4 1–127, PC4 22–127, PC4 43–127, PC4 1–91, PC4 1–62, PC4 1–42, and Sp1 1–778, Sp1 1–617, Sp1 618–778, Sp1 83–778, Sp1 83–714, 83–684, 83–655 vectors were constructed by subcloning corresponding PCR-amplified PC4 or Sp1 fragments into EcoRI and XhoI sites of the pcDNA3.1 V5/His A vector (Invitrogen). The GST vectors carrying full length PC4 or Sp1 were generated by inserting PCR amplified cDNA fragments corresponding to full-length of PC4 or Sp1 into the sites of EcoRI and XhoI of pET-41a (+) vector (Novagen).
Cell Culture, Transfection, and Reporter Gene Assay
MCF-7 A2 cells (MCF-7), kindly provided by Dr. Erica Berleth (C. Roswell Park Cancer Institute, Buffalo, NY) (27) were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum and 1% antibiotic/antimycotic solution (Invitrogen). Transfections were performed using Lipofectamine and Plus Reagent according to the manufacturer's instruction (Invitrogen). The DNA amount in each well was equalized with empty vector DNA. The luciferase activity was normalized to light units per microgram protein. All experiments were performed for at least three times in triplicate, and results were expressed as the mean ± standard errors.
RNA Isolation and Real-time RT-PCR
Total RNA was extracted with RNeasy Mini Kit (Qiagen, Valencia, CA) followed by treatment with DNase I (Invitrogen) for 15 min at room temperature. 4 μg of total RNA was then reverse-transcribed with random primer for synthesis of the first strand cDNA using the High Capacity cDNA Kit (Applied Biosystems, Foster City, CA). The relative levels of LHR mRNA was then determined with real-time PCR using SYBR-Green Master Mix in an ABI 7500 sequence detection system (Applied Biosystems) as described previously (28).
Preparation of Whole Cell Lysate and Nuclear Protein and Western Blot Analyses
Whole cell lysates were extracted with the M-PER Mammalian Protein Extraction reagents (Pierce). The cytosolic and nuclear proteins were isolated with NE-PER Nuclear and Cytoplasmic Extraction reagents (Pierce). All of the preparations were performed in the presence of 1× protease inhibitor (Roche). Protein concentration was determined by the colorimetric method of Bradford using Bio-Rad Protein Assay reagents (Bio-Rad). 50 μg of whole cell lysates or 15 μg of nuclear proteins for each sample were loaded into 4–20% Tris-glycine gel (Invitrogen) and analyzed by Western blot, which was performed as described previously (7).
Immunodepletion
Immunodepletion was performed as previously described (29) with modifications. Briefly, 10 μg of Sp1 antibody or IgG (Santa Cruz Biotechnology) were incubated with 200 μl of protein A-agarose beads (Santa Cruz Biotechnology) in 1 ml of binding buffer (20 mm Tris-HCl, pH 8.0, 100 mm NaCL, 1 mm EDTA, 0.1% Nonidet P-40) for 2 h at 4 °C. The antibody-bound beads were washed three times with phosphate-buffered saline (PBS), and subsequently incubated with 200 μg nuclear proteins for 6 h at 4 °C. The supernatant was collected by centrifugation, and subjected to one more round of depletion. Immunodepletion of Sp1 was confirmed by Western blot with Sp1 antibody.
DNA Affinity Precipitation Assay (DAPA)
DAPA was performed as previously described (30). Briefly, the sense and antisense oligonucleotides corresponding to the wild-type or mutant Sp1–1-binding site of the hLHR promoter were biotin-labeled at 5′-end and annealed at 1:1 molar ratio at 75 °C for 10 min in the presence of 100 mm NaCl. 50 μg of nuclear extracts of MCF-7 cells were incubated with 0.4 μg of wild-type or mutant biotinylated probe in binding buffer (60 mm KCl, 12 mm HEPES pH 7.9, 4 mm Tris-HCl, 5% glycerol, 0.5 mm EDTA, 1 mm dithiothreitol, 1× protease inhibitor) on ice for 45 min. The DNA-protein complexes were then incubated with 36 μl of streptavidin beads (Invitrogen). The incubation was continued for 1 h at 4 °C with gentle rotation. DNA-protein complexes were then washed five times with the binding buffer, and resuspended in 36 μl of 2× protein sample buffer (Invitrogen). After 5 min boiling, the supernatant proteins were analyzed by SDS-PAGE and Western blot detection with specific antibodies. For DAPA of CIP-treated nuclear extracts, proteins were incubated with CIP at 37 °C for 15 min in the reaction buffer (100 mm NaCl, 50 mm Tris-HCl, 10 mm MgCl2, 1 mm dithiothreitol, pH 7.9) prior to incubation with the corresponding probes.
GST Pull-down Assay
GST, GST-Sp1, and GST- PC4 were expressed in Escherichia coli strain RosettaTM 2 (DE3) (Novagen). After overnight culture at 37 °C, cells were diluted at 1:50 and cultured at 30 °C for 2–3 h (A600 of 0.6–0.7), and subsequently incubated with 0.1 mm IPTG for additional 3 h at 30 °C. Cultures were then harvested and lysed in ice-cold 1× GST·bind/wash buffer (4.3 mm Na2HPO4, 1.47 mm KH2PO4, 137 mm NaCl, 2.7 mm KCl, pH 7.3, 1× protease inhibitor) followed by sonication. Supernatant was subjected to purification with GST·BindTM Resin (Novagen). The specificity and purity of expressed protein was validated by SDS-PAGE and Coomassie Blue staining. In vitro coupled transcription and translation system was used to synthesize V5-tagged full-length Sp1 and its deletion mutants with T7 promoter upstream of the cloned gene in pcDNA 3.1/V5/His A. Various V5-tagged full length and deletion mutants of PC4 was synthesized with in vitro TNT coupled wheat germ extract systems (Promega). The expression of in vitro translated proteins was confirmed by Western blot with V5-antibody. For GST-pull down assay, bacterially expressed GST or GST-Sp1, GST-PC4 protein was immobilized to GST·BindTM resin (Novagen), and incubated with in vitro translated V5-Sp1 or V5-PC4 proteins in GST·bind/wash buffer which contains 0.1% Nonidet P-40 overnight. The beads bound with proteins were then washed four times with GST·bind/wash buffer to remove unbound proteins, and resuspended in 2× sample loading buffer (Invitrogen). The samples were then boiled for 5 min and resolved on SDS-PAGE for Western blot analysis with V5 antibody.
siRNA Analyses
Validated siRNAs designed to knock-down the endogenous expression of PC4, MED17, and scrambled siRNA as negative control (NTC) were purchased from Ambion (Austin, TX). Transfections of siRNA to MCF-7 were carried out with siPORTNeoFX reagent (Ambion) as previously described (10, 28). At 24 h post-transfection, cells were replaced with normal growth medium and cultured for additional 40 h before harvest. For reporter gene assays, the LHR gene promoter/reporter gene was introduced by Lipofectamine and Plus Reagent at 24 h post-transfection of siRNA, and the luciferase activity was determined 40 h later. For cells treated with TSA, 500 ng/ml or vehicle was added to cells 16 h prior to termination.
Chromatin Immunoprecipitation (ChIP)
ChIP assays were performed with MAGnifyTM Chromatin Immunoprecipitation system from Invitrogen following the manufacturer's instruction. Briefly, MCF-7 cells were transfected with siRNA of PC4 or scrambled NTC siRNA, and treated with or without 500 ng/ml TSA for 16 h. Cells were then crosslinked with 1% formaldehyde for 10 min at 37 °C followed by incubation with 125 mm glycine for 5 min at room temperature. Chromatin DNA was sheared to the size of 500–2000 bp with 2.5 in diameter cup horn using Misonix Sonicator S-3000 (Misonix, Farmingdale, NY). The sheared chromatin was immunoprecipitated overnight with 1–2 μg of various antibodies of interest or normal IgG, which were pre-coupled with Dynabeads. After extensive washes with IP buffer 1 and IP buffer 2, the antibody-Dynabeads complexes were reverse-crosslinked and DNA was purified with DNA purification magnetic beads. The relative binding of proteins of interest to the human LHR promoter was quantitatively analyzed by real-time PCR assay of the precipitated DNA and input DNA using SYBER Green Master Mix in an ABI 7500 sequence detection system. The primers utilized for amplification of the hLHR gene promoter region were described previously (11, 28). For detection of Flag-tagged PC4 recruitment, cells were transfected with 3xFlag PC4 and treated with or without TSA (500 ng/ml, 16 h), and Flag antibody was used to assess the association of Flag-PC4 at the LHR promoter.
Statistical Analysis
Statistical significance was evaluated by various test analyses with computer programs Statview (Abacus Concepts, Berkeley, CA) and Superanova (Abacus Concepts).
RESULTS
PC4 Activates the LHR Promoter Activity in a Sp1-dependent Manner
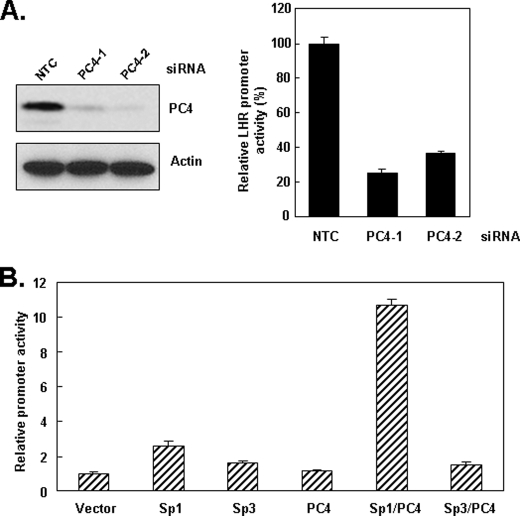
Following the exclusion of a number of coactivators in the Sp1-directed basal and TSA-induced transcriptional activity including p300, CBP, and PCAF and others, we investigated the participation of PC4, a potential coactivator of Sp1, in the transcriptional control of LHR (Ref. 10).3 For these studies, we knocked-down PC4 expression with siRNA to assess its impact on luciferase reporter activity driven by the minimal promoter of human LHR gene. To rule out off-target effects, two siRNAs targeting different regions of PC4 were used. As shown in Fig. 1A, left, the expression of endogenous PC4 in MCF-7 cells was effectively knocked down by both PC4 siRNAs. Consequently, the LHR promoter activity was largely inhibited by PC4 knockdown by either of siRNAs compared with the activity in NTC-transfected cells (Fig. 1A, right). These results indicated that PC4 is required for the basal LHR promoter activity and also suggested its potential coactivator function on Sp1/Sp3 transcriptional activation of LHR gene.
FIGURE 1.
Effect of PC4 knockdown and overexpression on the LHR promoter activity. A, left, whole cell lysates of MCF-7 cells transfected with two specific siRNAs (PC4–1, PC4–2) against PC4 or negative control scrambled siRNA (NTC) were analyzed for expression of PC4 by Western blot with PC4 antibody. The expression of actin served as control. Right, reporter gene analyses of LHR promoter activity in MCF-7 cells transfected with PC4–1, PC4–2 siRNA, and NTC siRNA. Relative luciferase activity was expressed as the percentage over the basal promoter activity in cells transfected with NTC siRNA, which was set as 100. B, LHR promoter was cotransfected with pcDNA3.1 vector, pCMV-Sp1, pcDNA 1.1-Sp3 and pCMV-PC4 expression vectors, or both Sp1/PC4, Sp3/PC4 vectors in MCF-7 cells followed by luciferase reporter gene assay. The amount of transfected DNA in each well was equalized by addition of empty vector (pcDNA3.1). Relative luciferase activity was expressed as fold-induction over the promoter activity with transfection of empty vector only.
Subsequent studies were designed to examine whether PC4 exerts its stimulatory effect on the LHR gene by cooperating with Sp1 or Sp3, both of which bind the LHR promoter and are of central importance for LHR basal promoter activity (3). To this end, the luciferase reporter construct carrying the LHR promoter was co-transfected with the constructs expressing PC4, Sp1, or Sp3 alone, or together. Transfection of Sp1, or Sp3 alone significantly activated the promoter activity confirming their activating role on the LHR promoter activity (Fig. 1B). While transfection of PC4 per se showed no significant effect on the LHR basal promoter activity, cotransfection of PC4 with Sp1 significantly enhanced the Sp1-dependent activity of LHR gene (∼2.6-fold versus ∼10.7-fold). In contrast, cotransfection of PC4 with Sp3 was unable to further augment the Sp3-dependent activity. These data demonstrated that PC4 functionally interacts with Sp1 to activate the Sp1-dependent LHR promoter activity.
PC4 Is Involved in TSA-induced LHR Activation
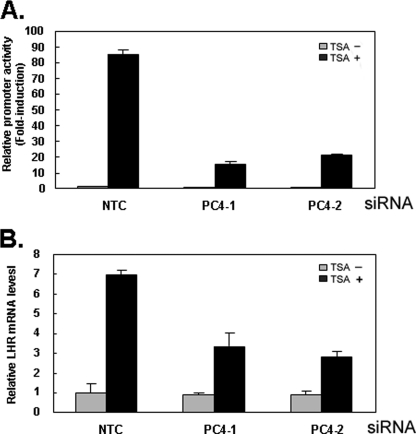
Having established that PC4 plays a Sp1-dependent stimulatory role for the promoter activity of LHR gene, it was of interest to determine whether PC4 participates in the TSA-induced LHR gene activation, which is mediated by Sp1 bound to the proximal Sp1 (I) site of the promoter (10). As expected, TSA treatment caused a marked induction of the LHR promoter activity in the cells transfected with NTC siRNA (Fig. 2A). However, this induction was largely inhibited upon depletion of PC4 expression with either of PC4 siRNAs (Fig. 2A) indicating an involvement of PC4 in the Sp1-dependent LHR activation in response to TSA. In agreement with this result, disruption of PC4 expression with two siRNAs substantially reduced TSA-stimulated LHR gene expression at the mRNA level (Fig. 2B). These data demonstrated that PC4 participates in TSA-induced LHR gene expression and suggested an interaction between with PC4 and Sp1 at the LHR promoter. We further investigated whether the effect resulting from PC4 knockdown is specific to the LHR gene. To this end, the effect of PC4 on a Sp1-dependent p21 promoter and the Sp1-independent probasin promoter was tested in the presence and absence of TSA treatment. Consistent with previous reports (31–32), TSA treatment markedly induced the promoter activity of p21 in MCF-7 cells (data not shown). However, transfection of PC4 siRNAs did not exert a significant inhibition on both p21 basal and TSA-mediated promoter activity. Similarly, no significant inhibitory role of PC4 was observed on the activity of the probasin promoter which does not contain Sp1 binding site and was not stimulated by TSA treatment (data not shown). Taken together, these data demonstrated that PC4 plays a specific role in the transcriptional control of LHR gene expression in MCF-7 cells.
FIGURE 2.
PC4 is involved in TSA-induced activation of LHR gene. A and B, reporter gene analyses of LHR gene promoter activity (A) and real-time PCR analyses of hLHR expression (B) in MCF-7 cells transfected with two PC4 siRNAs (PC4–1, PC4–2) or scrambled siRNA (NTC). Cells were treated with or without TSA (500 ng/ml) for 16 h. Luciferase activity were normalized to light units/μg of protein and expressed as the means ± S.E. of three independent experiments in triplicate wells. Relative luciferase activity or mRNA level was expressed as fold-induction over the promoter activity or mRNA level in the absence of treatment in NTC-transfected cells (1-fold).
PC4 Associates with Sp1 at the LHR Promoter
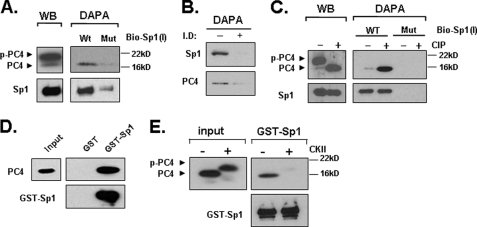
The observation that PC4 stimulates the LHR promoter in a Sp1-dependent manner and its requirement for Sp1-dependent LHR stimulation in response to TSA prompted us to investigate whether PC4 associates with Sp1 at the LHR promoter. DAPAs were performed by incubation of nuclear extracts from MCF-7 cells and 5′-end biotin-labeled probes corresponding to the wild type (WT) Sp1 (I) site and its mutant (Mut) which is devoid of Sp1 binding. As shown in Fig. 3A, right, the expected binding of Sp1 was shown to the WT but not the Mut Sp1 (I) probe. PC4 was specifically pulled-down by the WT Sp1 (I), whereas no detectable PC4 was pulled-down by the Mut probe. We further tested whether the association of PC4 with LHR promoter is dependent on recruitment of Sp1 by DAPA using nuclear proteins that were immunodepleted with an Sp1 antibody. As shown in Fig. 3B, the binding of PC4 to Sp1 (I) binding site (−, control) was substantially reduced upon immunodepletion of Sp1 (+, nuclear extracts immunodepleted of Sp1). Western blot analyses of nuclear extracts from MCF-7 cells showed expression of two distinct forms of PC4 protein, a slower migrating phosphorylated form, which accounted for at least 90% of total PC4, and a fasting migrating non-phosphorylated form of PC4 (<10%) (Fig. 3A, left). Despite of the large abundance of phosphorylated PC4 in the extracts, this form did not bind to the promoter since this species was not pulled-down by the WT probe (Fig. 3A, right). In contrast, a significant band of non-phosphorylated PC4 was pulled-down by the WT probe. This result indicated that PC4 binds to Sp1 at the LHR promoter and that the phosphorylation of PC4 has a negative effect in the association of two proteins. This was confirmed by treatment of nuclear extracts with calf intestinal phosphatase (CIP), which efficiently converted the phospho-PC4 form into the non-phospho-PC4 form (Fig. 3C, left). CIP-induced shift of phospho-PC4 to non-phospho-PC4 substantially increased the binding of PC4 to the promoter, whereas the specific binding of Sp1 remained unchanged (Fig. 3C, right).
FIGURE 3.
PC4 associates with Sp1 at the LHR promoter and phosphorylation of PC4 prevents this association. A, nuclear proteins isolated from MCF-7 cells were analyzed by Western blot (left) for expression of PC4 and Sp1, or by DAPA using 5′-biotin-labeled wild type Sp1–1 probe (WT) or its mutant oligomer devoid of Sp1 binding activity (Mut) (right). The avidin-precipitated protein complexes were analyzed by Western blots with PC4 and Sp1 antibodies. p-PC4 represents the phosphorylated form of PC4. B, Sp1-immunopleted nuclear proteins from MCF-7 cells and control samples were analyzed by DAPA using 5′-biotin-labeled wild type Sp1–1 probe followed by Western blot with Sp1 and PC4 antibodies. I.D, immunodepleted; −, control; +, nuclear extracts immunodepleted of Sp1 by Sp1 antibody (see “Experimental Procedures”). C, nuclear extracts from MCF-7 cells were treated with or without calf intestinal phosphatase (CIP) for 15 min at room temperature, and analyzed by Western blot (WB) for expression of PC4 and Sp1 (left), or precipitated by the WT or Mut Sp1–1 probe with DAPA (right). D, GST and GST-Sp1 was immobilized to GST·BindTM-agarose beads and incubated with 100 units of recombinant PC4 protein. The pull-down samples were analyzed with immunoblotting with anti-PC4 antibody. The blot was also probed with anti-Sp1 antibody. E, GST-pull down assay of non-phosphorylated and CKII-phosphorylated PC4 with GST-Sp1. The pull-down samples were analyzed by immunoblotting with anti-PC4 and anti-Sp1 antibody. Input represents 5% of the total PC4 protein used in each assay.
To determine whether PC4 directly interacts with Sp1, GST pull-down assays were carried out with bacterial expressed GST-fused Sp1 immobilized on glutathione-Sepharose resin and purified recombinant PC4. Immunoblots of pull-down samples with PC4 antibody showed that PC4 was significantly bound with GST-Sp1, but not with GST alone indicating a specific direct interaction between PC4 and Sp1 (Fig. 3D). We further tested if this interaction is affected by PC4 phosphorylation that regulates its coactivator function. Recombinant PC4 was efficiently phosphorylated by CKII as manifested by complete shift of the non-phosphorylated lower mobility species to the high mobility phosphorylated species (Fig. 3E, left). When we incubated immobilized GST-Sp1 with nonphosphorylated PC4, a significant amount of PC4 was retained. In contrast, no detectable binding of phosphorylated PC4 was observed (Fig. 3E, right). Thus, the unphosphorylated PC4 but not phosphorylated-PC4 interacted with Sp1. Together these data demonstrated that PC4 associates with the LHR promoter through a direct interaction with Sp1, which is negatively regulated by PC4 phosphorylation. PC4 might act as a coactivator of Sp1 to participate in the transcriptional activation of LHR gene.
The Coactivator Domain (22–91 aa) of PC4 Interacts with Sp1
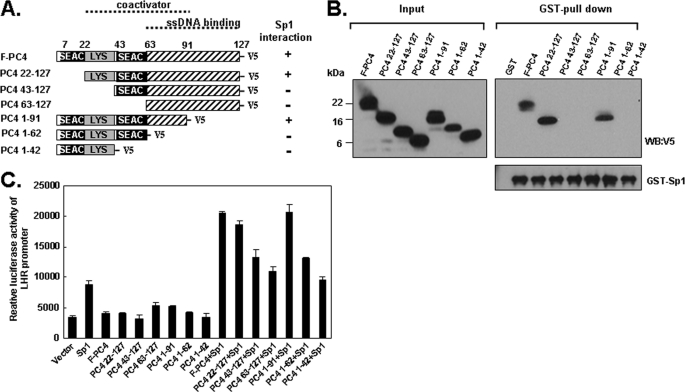
We next sought to characterize the domain of PC4 that interacts with Sp1. As depicted in Fig. 4A, full-length PC4 is composed of several distinct domains (15). Two serine-rich regions, termed SEAC (residue 7–22, and residue 43–63) and one lysine-rich domain (residue 22–43) are present at the N-terminal. SEAC domains are subjected to phosphorylation by CKII, which negatively regulates its coactivator function. The C-terminal region is localized between residues 63 and 127, which is important for binding single-stranded DNA. The region between residue 22 and 91 was shown to be a domain necessary and sufficient for the coactivator activity of PC4 (17). To determine the region of PC4 for Sp1 interaction, we generated a series of N-terminal, C-terminal deletion forms of PC4, which were expressed as V5-tagged proteins using TNT wheat germ cell lysate (Fig. 4A). These in vitro translated proteins were incubated with GST and GST-Sp1 that were immobilized onto the glutathione-Sepharose resin for the interaction assay. After precipitation and extensive washing, the precipitants were analyzed via Western blot with V5 antibody. As shown in Fig. 4B, and also summarized in Fig. 4A, both full-length PC4 and N-terminal deletion PC4-(22–127) showed strong interaction with GST-Sp1, whereas other N-terminal deletions of PC4-(43–127), PC4-(63–127) did not interact with GST-Sp1. This indicated that the region between residue 1 and 22 that contains the first SAEC is not required for the interaction to Sp1. When C-terminal deletions of PC4-(1–91), PC4-(1–62), and PC4-(1–41) were tested in the binding assay, only PC4-(1–91) but not PC4-(1–62), and PC4-(1–41) interacted with GST-Sp1, which suggested that the PC4/Sp1 interaction does not require the region between residue 91 and 127. The detected interaction between GST-Sp1 and PC4 is specific since GST only did not show interaction with PC4 protein (Fig. 4B, GST). Furthermore, the reprobed blot with Sp1 antibody confirmed the presence of equal amount of GST-Sp1 among reactions indicating that the observed differences of Sp1/PC4 interactions were not related to GST-Sp1 concentration. We further examined the effect of PC4 and its truncated mutants on Sp1-mediated LHR promoter activity. Reporter gene assay was performed by cotransfection of LHR promoter with Sp1 and PC4/PC4 mutant constructs. Consistent with pull-down results, transfection of full-length PC4, the truncation mutants PC4-(22–127) and PC4-(1–91) could induce Sp1-mediated LHR activity, whereas other mutants PC4-(43–127), PC4-(63–127), PC4-(1–62) and PC4-(1–42) could not or minimally induce the promoter activity driven by Sp1 (Fig. 4C). Together these data indicated that the middle region of PC4 (amino acid residues 22–91), which is the coactivator domain of this protein is important for the Sp1 binding.
FIGURE 4.
Coactivator domain (22–91 aa) of PC4 interacts with Sp1. A, schematic representation of the structure of full-length PC4 (F-PC4) and its deletion mutants. Two serine-rich domains (residues 7–22 and residues 43–63), which are termed SEAC, and one lysine-rich region (LYS) are shown. The coactivator domain (residues 22–91) and the domain for binding single-stranded DNA (residues 63–127) are indicated with dotted line. Interaction and noninteraction of tested PC4 with Sp1 are indicated by + and −, respectively. B, GST pull-down assay of PC4/Sp1 interaction with GST (control) and GST-Sp1 and in vitro translated full-length PC4 and its deletion mutants. The pull-down assay was analyzed by immunoblotting with antibodies against V5 and Sp1. C, LHR promoter was cotransfected with pCMV-Sp1, and various pcDNA3.1 V5/His-PC4 expression vectors alone, or both in MCF-7 cells followed by luciferase reporter gene assay. The amount of transfected DNA in each well was equalized by addition of empty vector (pcDNA3.1). Relative luciferase activity was normalized to light units/μg of protein and expressed as the means ± S.E. of three independent experiments in triplicate wells.
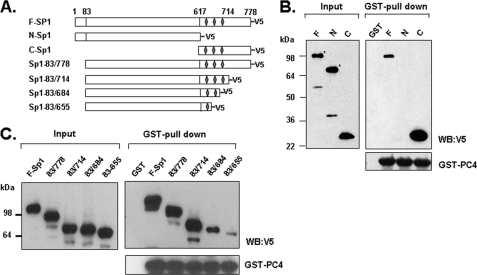
The DNA Binding Domain of Sp1 Is Involved in the Interaction with PC4
Although in vitro reconstitution studies have revealed that PC4 facilitates transcription in response to the Sp1-activating domain, which was fused to the minimal DNA-binding domain of GAL4 (16), and suggested an interaction of PC4 and Sp1, the essence of such interaction remained to be elucidated. To further investigate the direct interaction between PC4 and Sp1, we mapped the interacting domain of Sp1. In vitro translated full-length, N-terminal, and C-terminal of Sp1 were subjected to GST pull down assay with bacterial expressed GST or GST-PC4 which were immobilized onto the glutathione-Sepharose resin. While N-terminal activating domain of Sp1-(1–617), and C-terminal region containing DNA binding domain (618–778) of Sp1 were equally expressed in vitro as shown by Western blot, only C-terminal of Sp1 but not N-terminal of Sp1 was observed in the complex pulled-down by GST-PC4 (Fig. 5B). This result indicated that the C-terminal region which contains DNA binding domain is involved in its interaction with PC4.
FIGURE 5.
C-terminal domain of Sp1 is required for PC4 interaction. A, schematic representation of the structure of full-length Sp1 (F-Sp1) and its deletion mutants. Three zinc finger domains of Sp1 are shown as diamonds. B and C, GST pull-down assays of PC4/Sp1 interaction with bacterial GST/GST-PC4 and in vitro translated full-Sp1, N-terminal Sp1, C-terminal Sp1, and a series of its deletion mutants including Sp1-(83–778), Sp1-(83–714), Sp1-(83–684), and Sp1-(83–655). The pull-down samples were analyzed by immunoblotting with antibodies against V5 and PC4. (F-Sp1, full-length Sp1 (1–778 aa); N-Sp1, N-terminal Sp1 (1–617 aa); C-Sp1, C-terminal Sp1 (617–778 aa)).
The C-terminal Sp1 comprises of the DNA binding domain (amino acids 622–720) containing three Cys2His2 zinc fingers which are required for sequence-specific DNA binding to GC-rich promoter sequence, and the D domain (amino acids 721–788), which is required for synergistic activation along with its activation domains (33). To determine the region of Sp1 C-terminal involved in the interaction with PC4, we constructed a panel of Sp1 constructs that express Sp1 with deletions of zinc finger (s) or/and D-domain (Fig. 5A), and tested their interactions with GST-PC4. Western blot with V5-antibody showed similar levels of expression for in vitro translated Sp1 proteins (Fig. 5C). GST pull-down assays revealed that the Sp1 mutant (83–714) lacking the D-domain retained a comparable interaction of PC4 with the Sp1-(83–778) that contains intact C-terminal of Sp1 indicating that the D-domain of Sp1 is not required for interaction with PC4. Notably, the Sp1-(83–684) protein with deletion of the third zinc finger displayed substantially reduced association with PC4. Additional deletion of the second zinc finger further reduced the PC4/Sp1 interaction (Fig. 5C, Sp1-(83–655)). As in previous binding assays, all the blots were reprobed with PC4 antibody to confirm the presence of equal amount of GST-PC4 protein in each assay (Fig. 5C). Accordingly, our data demonstrated that the DNA binding domain of Sp1 is responsible for PC4/Sp1 interaction, with each zinc finger contributing to this interaction.
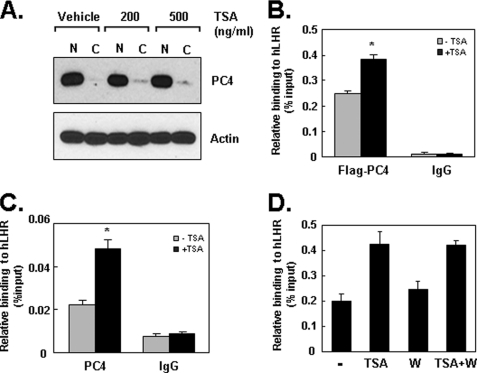
PC4 Is Recruited to the LHR Promoter in Response to TSA
Having established that PC4 directly associated with Sp1 at the LHR promoter and its participation in TSA-induced LHR activation, we next investigated whether the involvement of PC4 in TSA action is associated with a change of PC4 protein expression or its translocation within cellular compartments. Western blot showed that treatment with increasing concentration of TSA did not significantly alter the pattern of nuclear and cytoplasmic distribution of PC4 indicating that TSA-mediated LHR expression is not attributable to the changes on PC4 expression (Fig. 6A). However, PC4 could be recruited by Sp1 to participate in the TSA-mediated LHR transcription. To assess the association of PC4 at the LHR promoter, ChIPs were initially performed on cells transfected with 3×Flag-tagged version of PC4 in the absence and presence of TSA treatment. Significant binding of PC4–3×Flag at the promoter was detected under basal non-TSA treated condition (Fig. 6B) which is consistent with our observation that PC4 is required for basal promoter activity of the LHR gene (Fig. 1A). Moreover, TSA treatment induced a further increase of PC4–3×Flag association at the promoter (Fig. 6B). To confirm the PC4 recruitment to the LHR promoter in response to TSA, the binding of endogenous PC4 was further examined by ChIP. Consistent with the above findings, a significant increase of endogenous PC4 recruitment was observed in the cells treated with TSA (Fig. 6C). We then examined if this PC4 recruitment is dependent on PI3K/PKCζ-mediated Sp1 phosphorylation which has previously shown to induce the TSA-stimulated LHR activation (10). Inhibition of PI3K/PKCζ with its specific inhibitor (Wortmannin) did not affect the association of PC4 at the promoter that is trigged by TSA (Fig. 6D). These data indicated that PC4 is recruited to the LHR promoter during TSA-elicited gene expression in a manner independent of PI3K/PKCζ-mediated Sp1 phosphorylation.
FIGURE 6.
PC4 is recruited to the LHR promoter in response to TSA. A, cytosolic (C) and nuclear (N) proteins prepared from MCF-7 cells that were treated with vehicle or two doses of TSA (200 ng/ml and 500 ng/ml, 16 h) were analyzed by immunoblotting with antibodies against PC4 and actin. B, MCF-7 cells transfected with 3×Flag PC4 were subjected to quantitative ChIP analyses of the association of Flag-PC4 at the LHR promoter using Flag antibody in the absence and presence of TSA (500 ng/ml, 16 h). Binding of IgG antibody served as negative control. Results are expressed as percentages of the total input DNA (*, p < 0.01). C, quantitative ChIP analyses of the binding of endogenous PC4 in MCF-7 cells treated with or without TSA (500 ng/ml, 16 h). D, quantitative ChIP analyses of the association of 3xFlag PC4 to LHR promoter in MCF-7 cells treated with or without TSA (500 ng/ml, 16 h), PKCζ inhibitor wortmannin (W), or in combination.
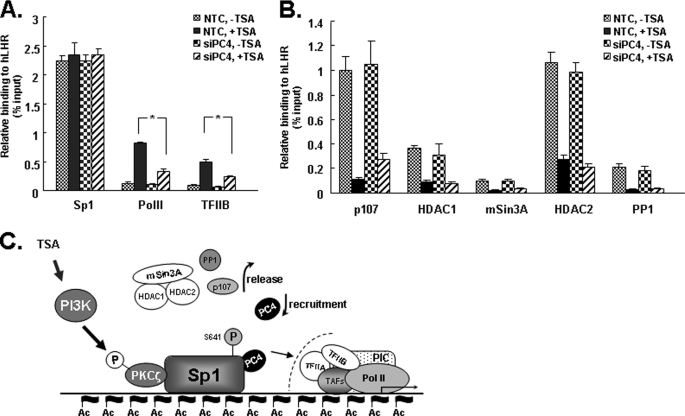
Knockdown of PC4 Impairs RNAP II and TFIIB Recruitment in Response to TSA
We next explored possible mechanism(s) by which promoter-associated PC4 exerts its function in TSA-induced LHR transcription. ChIP assays were performed to test whether PC4 knock-down affects the recruitment of RNA polymerase II (Pol II) and TFIIB at the promoter. TSA treatment greatly enhanced the recruitment of both Pol II and TFIIB at the region of LHR promoter in NTC cells (Fig. 7A). In contrast, TSA-mediated recruitment of Pol II and TFIIB was significantly inhibited upon depletion of PC4 (Fig. 7A). This is consistent with our observations showing that PC4 knockdown markedly reduced TSA-mediated activation of LHR promoter activity and mRNA (Fig. 2). In agreement with our previous observation, Sp1 binding at the promoter remained unchanged regardless of TSA treatment (9–10), and this Sp1 binding pattern was also unchanged when PC4 expression was knocked down (Fig. 7A). Our previous studies have demonstrated that the release of corepressor p107, HDCA1/2/mSin3A repressor complex and phosphatase PP1 from the promoter are critical for TSA-mediated de-repression (9–11). We therefore examined whether PC4 knockdown could influence the dissociation of those factors from the promoter following TSA treatment. Our ChIP assays showed that TSA caused the expected release of p107, HDAC1, HDAC2, mSin3A, and PP1 from promoter in NTC cells (Fig. 7B). PC4 knockdown had no significant effect on the dissociation of those factors except for a minor reduction of the dissociation of p107 observed in cells transfected with PC4 siRNA. Taking together, these data demonstrated that PC4 played a critical role for recruitment of Pol II and TFIIB to the promoter in response to TSA stimulation. These findings also indicated that PC4 is recruited to the promoter by Sp1 to facilitate or stabilize the formation of preinitiation complex at the LHR promoter during TSA-induced gene activation.
FIGURE 7.
Effect of PC4 knockdown on TSA-mediated recruitment of Pol II, TFIIB, and other promoter-associated factors at LHR promoter. A and B, quantitative ChIP analyses of the association of Sp1, PolII, TFIIB, p107, HDAC1, mSin3A, HDAC2, and PP1 with the LHR promoter in NTC and PC4 siRNA-transfected cells. Cells were treated with or without TSA (500 ng/ml) for 16 h. C, proposed role of PC4 in TSA-induced LHR activation. TSA-induced LHR activation is achieved by the combined actions of chromatin changes, release of inhibitory factors (p107, HDAC1/2/mSin3A, and phosphatase PP1), and recruitment of PC4, which is independent to PI3K/PKCζ/Sp1 phosphorylation. PC4 might function as a linker to bridge Sp1 and one or more components of the PIC.
DISCUSSION
These studies have investigated the role of PC4 in the silencing and re-activation of LHR gene transcription. We found that PC4 markedly activates the Sp1- but not Sp3-dependent LHR promoter activity. This activation is associated with direct interaction between the coactivation domain of PC4 and Zn finger DNA binding domain of Sp1. Furthermore, PC4 is involved in Sp1-regulated LHR activation in response to TSA, and PC4 recruitment at the LHR promoter is significantly increased by TSA stimulation. Depletion of PC4 impaired TSA-induced recruitment of Pol II and TFIIB at the LHR promoter. Taken together, these findings have revealed a critical aspect of LHR modulation in which PC4 is a necessary coactivator in Sp1-dependent LHR derepression induced by TSA.
In this study, we have demonstrated the coactivator function of PC4 with Sp1 in MCF-7 cells. PC4 markedly enhanced Sp1-dependent LHR promoter activity, but did not display an augmenting effect on the activity that is driven by Sp3 though both Sp1 and Sp3 function as activators of basal LHR gene expression (Fig. 1B). This result revealed a specific coactivator function of PC4 for Sp1-activated LHR gene transcription. This finding was supported by the association of PC4 with Sp1 at the Sp1 (I) site of LHR promoter (Fig. 3A). Moreover, the physical interaction between PC4 and Sp1 as determined by GST pull-down assay provides further evidence to support the coactivator role of PC4 for Sp1 activity in vivo (Fig. 3D). The identification of PC4 as a coactivator of LHR transcription in MCF-7 cells has revealed its crucial role in the physiological control of LHR gene expression.
PC4 interacts with distinct domains of activators to mediate their functions. While the activation domains of many activators such as VP16, GAL4, and AP2 are responsible for PC4 interaction (16–17, 20–21), the basic RNA binding domain of HIV-TAT rather than its activating domain is found to interact with PC4 (22). In contrast, both the DNA binding domain and the highly flexible C-terminal regulatory domain of p53 bind PC4 to mediate the interaction with PC4 (24). The current study has demonstrated that PC4 interacts with the DNA binding domain (DBD) of Sp1 in vitro, and three Zn fingers in the DBD contribute to this interaction (Fig. 5). The differential functional regions of activators required for their interactions with PC4 suggests a diverse mechanism underlying its coactivator function to mediate the transcriptional response to distinct types of activators. The Zn finger DBD of Sp1 has also been shown to be involved in interaction with other cofactors, such as human TAFII55, p300, the ATP-dependent nucleosomal remodeling enzyme SWI/SNF, and corepressor SMRT, NCoR, and BcoR (34–38). Those interactions are crucial in the regulation of many cellular functions by controlling gene expression at the transcriptional level. On the other hand, GST pull-down assay using the bacterial expressed Sp1 and in vitro translated PC4 proteins showed that the Sp1-interacting region of PC4 is localized to the region between residue 22 and 91, which is its coactivator domain, that further indicated the coactivator function of PC4 on Sp1 activity (Fig. 4, A and B). The relevance of this region for Sp1 interaction was also supported by functional analyses (Fig. 4C) in which the Sp1-mediated LHR promoter activity could be substantially activated by transfection of full-length PC4, deletion mutant PC4-(22–127), or PC4-(1–91) but not by the mutants that are devoid of the intact 22–91 region. Although we were unable to detect an interaction between Sp1 and PC4-(43–127) and PC4-(1–62) in GST pull-down assay, transfection of those two constructs could marginally stimulate the Sp1-driven LHR activity. This implies that a weak interaction might exist between Sp1 and the PC4 region of residue 43–91 or region of residue 22–63, which could ultimately contribute to the minimal activation of Sp1-mediated promoter activity observed.
We have previously established that Sp1 is not only important for basal LHR promoter activity, but also has a key role in mediation of activated-LHR expression in cancer cells that are stimulated by TSA (3–4, 10–11). At the silenced LHR promoter, Sp1 interacts directly or indirectly with a variety of repressor/corepressor factors (e.g. HDAC1/2/mSin3A, p107, PP1), and the release of such inhibitory complex/molecules is dependent on Sp1 phosphorylation and local chromatin changes (9–11). These are essential regulatory mechanisms that lead to re-activation of LHR gene expression. This study reveals an additional regulatory component that involves recruitment of the coactivator PC4 of Sp1 in the control of LHR gene transcription. We found that PC4 directly associated with Sp1 at the promoter. Depletion of PC4 inhibited the basal LHR promoter activity and largely blocked TSA-induced LHR activation and mRNA expression. The residual promoter activity/transcription is indicative of the presence of other coactivator (s) that might be also involved in the TSA action. This coactivator activity could work independently or cooperatively with PC4 to mediate the overall activation of the LHR gene.
PC4 has been reported to be regulated by post-translational modifications including phosphorylation and acetylation (15, 24, 39–40). CKII phosphorylates PC4 in vivo and such phosphorylation negatively regulates its coactivator functions, its interactions with the activator and TATA box-binding protein/TFIIA (15, 39). In our studies, phosphorylation of PC4 exhibited a negative effect on the interaction between PC4 and Sp1 (Fig. 3). Sp1 interacted with the bacterial recombinant PC4 that is unphosphorylated, but not with PC4 that was phosphorylated by CKII. Conversely, nuclear PC4 that was treated with CIP largely increases the association of the two proteins at the promoter in DAPA experiments. Therefore, these results indicated that PC4 phosphorylation may play an important role in the TSA-induced activation. On the other hand, PC4 acetylation could also affect the function of PC4 with Sp1. PC4 is known to be acetylated by p300, and this acetylation stimulates its double stranded DNA binding activity, which is correlated with its coactivator function (40). Thus, it is conceivable that the inhibition of HDAC with TSA could lead to recruitment of a yet unknown protein with HAT activity that could mediate PC4 acetylation, which would enhance its interaction with Sp1 and thereby LHR gene activation.
This study indicates that PC4 has an important role in the formation/assembly of general transcriptional machinery in the TSA-mediated LHR transcription. PC4 was recruited by Sp1 to the LHR promoter following TSA treatment. Loss of PC4 significantly blocked TSA-stimulated recruitment of Pol II and TFIIB (Fig. 7), which is correlated with a substantial reduction of TSA-mediated LHR activation. Similarly, the yeast PC4 homolog SUB1 facilitates osmoresponse gene transcription by promoting preinitiation complex formation (41). Although PC4 is also believed to be implicated in chromatin organization (42), PC4 does not seem in this case to participate in TSA-induced chromatin structural alteration, because silencing of PC4 did not change the release of HDAC1/2/Sin3A and phosphatase release from Sp1, two events that are dependent on TSA-elicited DNA methylation and chromatin decompaction resulting from histone acetylation at local promoter (9–11). In addition, while TSA-induced TFIIB recruitment is largely dependent on PC4 recruitment, lack of interaction between PC4 and TFIIB in vitro and in vivo (data not shown) has ruled out the possibility that TFIIB is the potential target of PC4 to link Sp1 to the basal transcriptional machinery in TSA-induced LHR transcription. This is consistent with the report that in an in vitro system PC4 is unable to interact with free TFIIB, while it interacts with free or DNA-bound TFIIA and TBP components of the basal transcription machinery (17). Moreover, knockdown of Med17/Trap80 did not block TSA-induced LHR activation in MCF-7 cells and it was not found to be recruited to the LHR promoter upon TSA treatment (data not shown). Thus, interaction with other members of this initiation complex and/or mediator complex is anticipated.
PC4 binds the LHR promoter in vivo, and TSA treatment further induces its recruitment. These data, along with our previous observations, have revealed complex regulation that releases inhibitory factors, and recruitment of positive PC4 cofactor cooperatively contributes to maximum activation of this gene. However, while the release of repressor p107 is triggered by PI3K/PKCζ-mediated Sp1 phosphorylation (10), recruitment of PC4 to LHR promoter is independent of such phosphorylation since the inhibition of PI3K/PKCζ with its specific inhibitor (Wortmannin) did not block TSA-induced PC4 recruitment (Fig. 6D). A mechanism other than Sp1 phosphorylation might engage to cause its recruitment. In addition, TSA-mediated dissociation of p107, HDAC/mSin3A complex and phosphatase from the promoter was not significantly affected by knockdown of PC4 (Fig. 7B). Together these findings led us to propose that the critical role of PC4 in TSA-induced LHR gene expression is independent of Sp1 phosphorylation and release of corepressors, and it may function as a coactivator linker between Sp1 and one or more components of the preinitiation complex (Fig. 7C).
This work was supported, in whole or in part, by the National Institutes of Health Intramural Research Program through the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
M. L. Dufau, unpublished data.
- LHR
- luteinizing hormone receptor
- DAPA
- DNA affinity precipitation assay
- TSA
- trichostatin A
- PC4
- positive cofactor 4
- HDAC
- histone deacetylase
- aa
- amino acids.
REFERENCES
- 1. Dufau M. L. (1998) Annu. Rev. Physiol. 60, 461–496 [DOI] [PubMed] [Google Scholar]
- 2. Dufau M. L., Tsai-Morris C.-H. (2007) in Contemporary Endocrinology (Payne A. H., Hardy M. P. eds) pp. 227–252, Humana Press Inc., Totowa [Google Scholar]
- 3. Tsai-Morris C. H., Geng Y., Xie X. Z., Buczko E., Dufau M. L. (1994) J. Biol. Chem. 269, 15868–15875 [PubMed] [Google Scholar]
- 4. Tsai-Morris C. H., Xie X., Wang W., Buczko E., Dufau M. L. (1993) J. Biol. Chem. 268, 4447–4452 [PubMed] [Google Scholar]
- 5. Tsai-Morris C. H., Geng Y., Buczko E., Dufau M. L. (1998) J. Clin. Endocrinol. Metab. 83, 288–291 [DOI] [PubMed] [Google Scholar]
- 6. Zhang Y., Dufau M. L. (2000) J. Biol. Chem. 275, 2763–2770 [DOI] [PubMed] [Google Scholar]
- 7. Zhang Y., Dufau M. L. (2001) Mol. Endocrinol. 15, 1891–1905 [DOI] [PubMed] [Google Scholar]
- 8. Zhang Y., Dufau M. L. (2003) Mol. Cell. Biol. 23, 6958–6972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang Y., Fatima N., Dufau M. L. (2005) Mol. Cell. Biol. 25, 7929–7939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang Y., Liao M., Dufau M. L. (2006) Mol. Cell. Biol. 26, 6748–6761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang Y., Liao M., Dufau M. L. (2008) J. Biol. Chem. 283, 24039–24046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang X., Pan L., Feng Y., Wang Y., Han Q., Han L., Han S., Guo J., Huang B., Lu J. (2008) Oncogene 27, 1894–1904 [DOI] [PubMed] [Google Scholar]
- 13. Kim S. N., Kim N. H., Lee W., Seo D. W., Kim Y. K. (2009) Mol. Cancer Res. 7, 735–744 [DOI] [PubMed] [Google Scholar]
- 14. Conesa C., Acker J. (2010) RNA Biol. 7, 287–290 [DOI] [PubMed] [Google Scholar]
- 15. Kretzschmar M., Kaiser K., Lottspeich F., Meisterernst M. (1994) Cell 78, 525–534 [DOI] [PubMed] [Google Scholar]
- 16. Ge H., Roeder R. G. (1994) Cell 78, 513–523 [DOI] [PubMed] [Google Scholar]
- 17. Kaiser K., Stelzer G., Meisterernst M. (1995) EMBO J. 14, 3520–3527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haile D. T., Parvin J. D. (1999) J. Biol. Chem. 274, 2113–2117 [DOI] [PubMed] [Google Scholar]
- 19. Luo Y., Ge H., Stevens S., Xiao H., Roeder R. G. (1998) Mol. Cell. Biol. 18, 3803–3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kannan P., Tainsky M. A. (1999) Mol. Cell. Biol. 19, 899–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhong L., Wang Y., Kannan P., Tainsky M. A. (2003) Gene 320, 155–164 [DOI] [PubMed] [Google Scholar]
- 22. Holloway A. F., Occhiodoro F., Mittler G., Meisterernst M., Shannon M. F. (2000) J. Biol. Chem. 275, 21668–21677 [DOI] [PubMed] [Google Scholar]
- 23. Banerjee S., Kumar B. R., Kundu T. K. (2004) Mol. Cell. Biol. 24, 2052–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Batta K., Kundu T. K. (2007) Mol. Cell. Biol. 27, 7603–7614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Geng Y., Tsai-Morris C. H., Zhang Y., Dufau M. L. (1999) Biochem. Biophys. Res. Commun. 263, 366–371 [DOI] [PubMed] [Google Scholar]
- 26. Hu Z. Z., Zhuang L., Meng J., Dufau M. L. (1998) J. Biol. Chem. 273, 26225–26235 [DOI] [PubMed] [Google Scholar]
- 27. Ujházy P., Klobusická M., Babusíková O., Strausbauch P., Mihich E., Ehrke M. J. (1994) Int. J. Cancer 59, 83–93 [DOI] [PubMed] [Google Scholar]
- 28. Liao M., Zhang Y., Dufau M. L. (2008) Mol. Endocrinol. 22, 1449–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dong J., Tsai-Morris C. H., Dufau M. L. (2006) J. Biol. Chem. 281, 18825–18836 [DOI] [PubMed] [Google Scholar]
- 30. Zhang Y., Dufau M. L. (2002) J. Biol. Chem. 277, 33431–33438 [DOI] [PubMed] [Google Scholar]
- 31. Sowa Y., Orita T., Minamikawa S., Nakano K., Mizuno T., Nomura H., Sakai T. (1997) Biochem. Biophys. Res. Commun. 241, 142–150 [DOI] [PubMed] [Google Scholar]
- 32. Huang L., Sowa Y., Sakai T., Pardee A. B. (2000) Oncogene 19, 5712–5719 [DOI] [PubMed] [Google Scholar]
- 33. Suske G. (1999) Gene 238, 291–300 [DOI] [PubMed] [Google Scholar]
- 34. Chiang C. M., Roeder R. G. (1995) Science 267, 531–536 [DOI] [PubMed] [Google Scholar]
- 35. Suzuki T., Kimura A., Nagai R., Horikoshi M. (2000) Genes Cells 5, 29–41 [DOI] [PubMed] [Google Scholar]
- 36. Lee J. A., Suh D. C., Kang J. E., Kim M. H., Park H., Lee M. N., Kim J. M., Jeon B. N., Roh H. E., Yu M. Y., Choi K. Y., Kim K. Y., Hur M. W. (2005) J. Biol. Chem. 280, 28061–28071 [DOI] [PubMed] [Google Scholar]
- 37. Doetzlhofer A., Rotheneder H., Lagger G., Koranda M., Kurtev V., Brosch G., Wintersberger E., Seiser C. (1999) Mol. Cell. Biol. 19, 5504–5511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kadam S., McAlpine G. S., Phelan M. L., Kingston R. E., Jones K. A., Emerson B. M. (2000) Genes Dev. 14, 2441–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ge H., Zhao Y., Chait B. T., Roeder R. G. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 12691–12695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kumar B. R., Swaminathan V., Banerjee S., Kundu T. K. (2001) J. Biol. Chem. 276, 16804–16809 [DOI] [PubMed] [Google Scholar]
- 41. Rosonina E., Willis I. M., Manley J. L. (2009) Mol. Cell. Biol. 29, 2308–2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Das C., Gadad S. S., Kundu T. K. (2010) J. Mol. Biol. 397, 1–12 [DOI] [PubMed] [Google Scholar]