Abstract
The plastids of cryptophytes, haptophytes, and heterokontophytes (stramenopiles) (together once known as chromists) are surrounded by four membranes, reflecting the origin of these plastids through secondary endosymbiosis. They share this trait with apicomplexans, which are alveolates, the plastids of which have been suggested to stem from the same secondary symbiotic event and therefore form a phylogenetic clade, the chromalveolates. The chromists are quantitatively the most important eukaryotic contributors to primary production in marine ecosystems. The mechanisms of protein import across their four plastid membranes are still poorly understood. Components of an endoplasmic reticulum-associated degradation (ERAD) machinery in cryptophytes, partially encoded by the reduced genome of the secondary symbiont (the nucleomorph), are implicated in protein transport across the second outermost plastid membrane. Here, we show that the haptophyte Emiliania huxleyi, like cryptophytes, stramenopiles, and apicomplexans, possesses a nuclear-encoded symbiont-specific ERAD machinery (SELMA, symbiont-specific ERAD-like machinery) in addition to the host ERAD system, with targeting signals that are able to direct green fluorescent protein or yellow fluorescent protein to the predicted cellular localization in transformed cells of the stramenopile Phaeodactylum tricornutum. Phylogenies of the duplicated ERAD factors reveal that all SELMA components trace back to a red algal origin. In contrast, the host copies of cryptophytes and haptophytes associate with the green lineage to the exclusion of stramenopiles and alveolates. Although all chromalveolates with four membrane-bound plastids possess the SELMA system, this has apparently not arisen in a single endosymbiotic event. Thus, our data do not support the chromalveolate hypothesis.
Keywords: Emiliania huxleyi, secondary endosymbiosis, chromalveolate hypothesis, complex plastid, plastid protein import, algal evolution
Introduction
The chromalveolate hypothesis (for details see, Cavalier-Smith 1999) posits that chromists and alveolates are a monophyletic group descended from a single secondary symbiosis involving a red algal endosymbiont and a heterotrophic host. The single symbiotic origin of the chromalveolates remains debated because of conflicting and equivocal data (Frommolt et al. 2008; Sanchez-Puerta and Delwiche 2008; Baurain et al. 2010). Other recent findings point to a specific relationship between cryptophytes and haptophytes to the exclusion of stramenopiles and alveolates (Harper et al. 2005; Rice and Palmer 2006; Hackett et al. 2007), pointing in turn to a more complex history of organisms with a secondary red algal symbiont, with newer data suggesting that the Rhizaria might be the sisters of the chromalveolate supergroup, more precisely to the alveolates and stramenopiles (Lane and Archibald 2008; Elias and Archibald 2009). Hence, the distribution of secondary plastids among the chromalveolates might have entailed a larger number of symbioses than the chromalveolate hypothesis suggests (Becker et al. 2008; Sanchez-Puerta and Delwiche 2008; Moustafa et al. 2009; Nozaki et al. 2009).
For the plastids of chromalveolates to function, hundreds of nuclear-encoded plastid proteins have to be imported across the surrounding membranes in a highly coordinated fashion, and the components of those protein transport machineries involved should, in turn, reflect the history of these complex cells. Progress is being made in understanding the principles and components underlying protein import in complex plastids and indeed plastid protein import seems to follow very similar principles in all members of the chromalveolates analyzed so far (Hempel et al. 2007). First, all nucleus-encoded plastid proteins are equipped with N-terminal bipartite signal sequences (bipartite targeting signal [BTS]) consisting of a classical endoplasmic reticulum (ER) signal peptide (SP) followed by a stretch of amino acids called transit peptide (TP) (Patron and Waller 2007). Second, in phototrophic chromists, where the outermost of the four surrounding membranes is continuous with the rough ER (rER) and the nuclear envelope (fig. 1), host-encoded proteins destined for the symbiont pass the outermost plastid membrane in a Sec61-dependent manner. This is in contrast to peridinin-containing dinoflagellates (whose plastids are surrounded by three membranes only) and apicomplexa, where plastid proteins enter the classical secretory pathway first (reviewed in Harper and Keeling 2003; Bolte et al. 2009). Thereafter, the protein transport in peridinin-containing dinoflagellates and in the apicomplexa (in the case that the apicoplast is not located in the ER lumen, see Tonkin et al. 2008) is dependent upon vesicles, which bud either from the ER or Golgi, respectively, and fuse with the outermost membrane of the plastid of dinoflagellates or the apicoplast (Nassoury et al. 2003; Tonkin et al. 2006). From an evolutionary standpoint, protein import across the first (rER or epiplastid) membrane does not appear to require too much in the way of molecular inventions, as preexisting components for ER import are used.
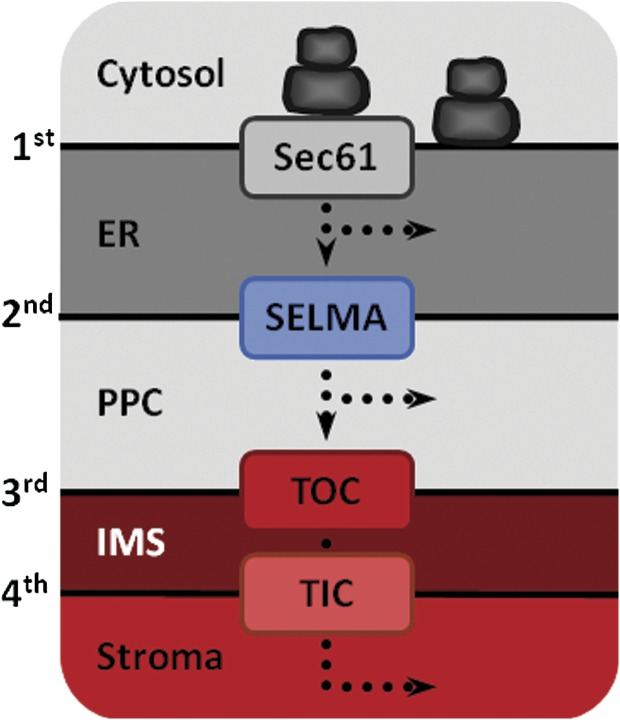
FIG. 1.—
Plastid architecture of stramenopiles and haptophytes. The secondary plastids of, for example, Phaeodactylum tricornutum and Emiliania huxleyi both have the same membrane organization. They are surrounded by four membranes with the outermost one being contiguous to the ER. Plastid proteins presumably cross the 1st, 2nd, 3rd, and 4th membrane by Sec61, SELMA (investigated in this study), TOC, and TIC translocons, respectively. IMS, intermembrane space.
However, the subsequent steps of protein import, in particular, across the second outermost (or periplastid) membrane, have remained far more elusive, and a number of suggestions regarding the nature of this process have been put forth. These include vesicle-shuttling models (Gibbs 1979; Kilian and Kroth 2005) and various different translocator-based mechanisms (Cavalier-Smith 2003; Bodyl 2004). Data on the issue has been lacking. Recently, Sommer et al. (2007) identified components of the ER-associated degradation (ERAD) machinery that are specific to the symbiont and hence are termed symbiont-specific ERAD-like machinery (SELMA) components (Hempel et al. 2009). These are expressed in cryptomonads, diatoms, and apicomplexa (Sommer et al. 2007). This machinery coexists with the host ER-associated degradation (hERAD) machinery and is proposed to constitute the translocon for crossing the second outermost membrane in those organisms investigated. The current working hypothesis posits that the SELMA system is located in the second outermost membrane of the four membrane–bound plastids and facilitates the passage of symbiont-targeted proteins across this membrane (Hempel et al. 2009, 2010). Although the sequence of the remnant nucleomorph genome in cryptophytes has been known for some time (Douglas et al. 2001), only recently some of the genes encoding SELMA components were identified in that genome (Sommer et al. 2007). In stramenopiles and apicomplexa, these factors are encoded as preproteins in the nuclear genomes, where they are equipped with an N-terminal bipartite signal sequence to ensure their localization within the second outermost membrane or the periplastid compartment (PPC), respectively (Hempel et al. 2007; Sommer et al. 2007; Bolte et al. 2009).
Finally, the transport of proteins across the innermost two membranes—homologous to the plastid envelope of the red algal endosymbiont—is poorly understood. Although theory predicts that homologues of the Toc and Tic machinery of primary plastids would be the most likely candidates (Cavalier-Smith 2003; Tonkin et al. 2008), the growing number of sequenced chromalveolate genomes generally appear to lack readily identifiable homologues of the corresponding genes, with the exception of some homologs of Tic subunits (McFadden and van Dooren 2004; Hempel et al. 2007), which recently were shown to be indeed involved in protein transport in apicomplexa (van Dooren et al. 2008). However, we recently identified an Omp85 protein and localized it to the third outermost membrane in Phaeodactylum tricornutum, where it likely functions as the translocon (i.e., as a Toc75 homologue) of the respective membrane (Bullmann et al. 2010).
Despite such progress, few lineages within the chromalveolates have yet been studied with respect to protein import into the complex plastid. For the haptophytes, which harbor plastids surrounded by four membranes, no data are currently available. If the SELMA pathway is used by all chromalveolates, it should be present in the haptophytes as well. We have screened the available genomic data from the haptophyte Emiliania huxleyi for these genes and studied the encoded proteins. Here, we report that in addition to a host-specific ERAD system, components for SELMA are present in E. huxleyi, the targeting signals of which are able to direct green fluorescent protein (GFP) to the predicted cellular localization in heterologous expression experiments.
Materials and Methods
In silico Analyses
Data mining for symbiont and host-specific ERAD components of E. huxleyi (strain CCMP1516) was performed as described in Sommer et al. (2007) based on publicly available single-read raw sequences of the E. huxleyi genome project (http://genome.jgi-psf.org/Emihu1/Emihu1.home.html) downloaded from the NCBI Trace File Archive (ftp://ftp.ncbi.nih.gov/pub/TraceDB). Blast against a local database was done using P. tricornutum and Guillardia theta gene models as baits. The cryptophyte raw sequences of Uba1 and of the host version of Cdc48 were produced by the Joint Genome Institute’s Community Sequencing Program (http://www.jgi.doe.gov/).
Emiliania huxleyi Cell Culture and cDNA
Emiliania huxleyi was grown in f/2 medium at 21 °C in a 14:10 h light:dark cycle. Total mRNA was extracted using the TRIzol reagent (Invitrogen). cDNA synthesis was carried out with the Qiagen OneStep RT-PCR Kit using gene-specific oligonucleotides.
Cloning and In vivo GFP/YFP Localization Studies
Each presequence was inserted into the plasmid pPhaT1 together with the gene encoding enhanced green fluorescent protein (eGFP) via EcoRI and HindIII restriction sites. For localization of the hERAD components, the first 300 N-terminal amino acids were fused to yellow fluorescent protein (YFP) using the same restriction sites. The leader sequences and the reporters were ligated at NcoI sites. The presequence of AtpG together with the first 32 amino acids of the predicted mature protein was amplified using the Failsafe PCR System (EPICENTRE Biotechnologies). Further sequences encoding targeting signals have been synthesized at the GeneArt Company with optimized codon usage for expression in P. tricornutum.
The transformation of P. tricornutum was carried out as described (Apt et al. 1996). Analysis of recombinant cells was performed with a Leica TCS SP2 confocal laser-scanning microscope using the settings as described (Gould et al. 2006b) for the GFP and chlorophyll fluorescence. YFP images were obtained by excitation of the chromophore at 514 nm and the usage of a DD 458/514 beam splitter. YFP fluorescence was detected at a bandwidth of 550–600 nm.
Phylogenetic Reconstruction
For the alignments of Uba1 and Cdc48, the 38 and 39 sequences for each protein were aligned using fast statistical alignment (FSA) (Bradley et al. 2009). Due to several large insertions in some sequences, the alignments contained 7,999 and 2,812 sites, respectively. The highly divergent regions in the alignments have been excluded from the phylogenetic analysis using Gblocks (Talavera and Castresana 2007). The remaining 438 and 646 sites were used to estimate distances with PROTDIST (Felsenstein 1989) using the Jones, Taylor, and Thorton (JTT) substitution matrix. With these distances, NNet splits graphs (Bryant and Moulton 2004) were constructed and visualized with SplitsTree4 (Huson and Bryant 2006). Both alignments were also used to construct bootstrapped maximum likelihood (ML) trees using PhyML (Guindon and Gascuel 2003) under the LG + I + G model with four substitution rate categories, estimated gamma distribution parameter, and number of invariable sites employing 100 bootstrap replicates. The starting tree topology—a BioNJ tree—was optimized using both simultaneous nearest neighbor interchange and subtree pruning and regrafting (SPR). Branch lengths and rate parameters were optimized as well. Because PhyML uses an “uphill-climbing” algorithm, it can get stuck in local optima without finding the ML tree despite SPR (Guindon et al. 2010). Hence, we computed two additional trees using RAxML (Stamatakis 2006; Stamatakis et al. 2008) under the JTT + G model with four substitution rate categories, estimated gamma distribution parameter, optimized topology, and optimized branch lengths employing 1,000 bootstrap replicates. In an effort to avoid local optima, every fifth of those bootstrap trees was used as a starting tree in the ML analysis.
Results
Data Mining
By screening the available E. huxleyi genome data for genes encoding components of the ERAD machinery (reviewed in Meusser et al. 2005; Ismail and Ng 2006), we identified, among others, homologs for the 4-fold membrane spanning protein Der1, the ubiquitin-dependent AAA-ATPase Cdc48, its cofactor Ufd1, as well as homologs of the ubiquitin-activating enzyme Uba1 (table 1). As shown for other members of the chromalveolates (Sommer et al. 2007), all of these genes were detected in at least two copies, one encoding the putative copy of the host ERAD machinery (hERAD) and one encoding a preprotein with a N-terminally located bipartite targeting sequence (determined by targetP and signalP) (Emanuelsson and von Heijne 2001; Emanuelsson et al. 2007; Sommer et al. 2007). These genes were tentatively assigned as symbiont-specific ERAD factors and further analyzed.
Table 1.
Identified Host- and Symbiont-specific ERAD(-Like) Components in Emiliania huxleyi
| Gene Model/Protein | Protein IDa | Functionb | Predicted Localization |
| hCdc48 | 439359 | AAA-ATPase | Cytosolic |
| sCdc48 | 421356 | PPC | |
| hUfd1 | 467386 | Cofactor of Cdc48 | Cytosolic |
| sUfd1 | 362690 | PPC | |
| hUba1 | 425780 | Ubiquitin-activating enzyme | Cytosolic |
| sUba1 | 459406 | PPC | |
| hDer1-1 | 100253 | Putative channel component for soluble ERAD substrates | ER membrane |
| hDer1-2 | 420318 | ||
| sDer1-like 1 | 273097 | PPM | |
| sDer1-like 2 | 96871 |
Known in classical ERAD; PPM, periplastidic membrane.
Expression of E. huxleyi Genes in P. tricornutum
For genetic accessibility and cellular localization studies, the diatom P. tricornutum is still a model system of choice for phototrophic chromalveolates. Recently, we have shown that the same subcellular localization as in cryptophytes can be obtained with various constructs expressing cryptophyte targeting sequences fused to GFP (Gould et al. 2006a, 2006b). Because heterologous expression seemed to deliver correct compartmentalization, we expressed the predicted targeting signals from the identified symbiont-specific ERAD components Der1, Uba1, and Cdc48 from E. huxleyi in P. tricornutum as GFP-fusion proteins (see table 1). Unlike Cdc48, where the start codon of the open reading frame was obtained coincidently, three different putative start codons for the SP of the bipartite leader were predicted for sDer1-1 by the various neural network prediction tools available, with the shortest one being most likely the genuine start methionine of the protein and from which we obtained amplification products only by reverse transcriptase-polymerase chain reaction experiments. However, the targeting sequences of Cdc48 as well as all three possible lengths of sDer1-1 (l, long; m, medium length; s, short) were fused to GFP and subsequently expressed. In the case of sUba1, the first out of four in-frame methionines was predicted as the start codon due to the best SP prediction and expressed as a GFP-fusion protein. As controls, we fused the targeting signals from the gene encoding the E. huxleyi plastid protein AtpG as well as from the ER-residential 70-kDa family heat-shock protein (BiP) to GFP to determine their in vivo localization. In addition, the N-termini of the host versions of Uba1 (hUba1) and Cdc48 (hCdc48) were expressed as YFP-fusion proteins.
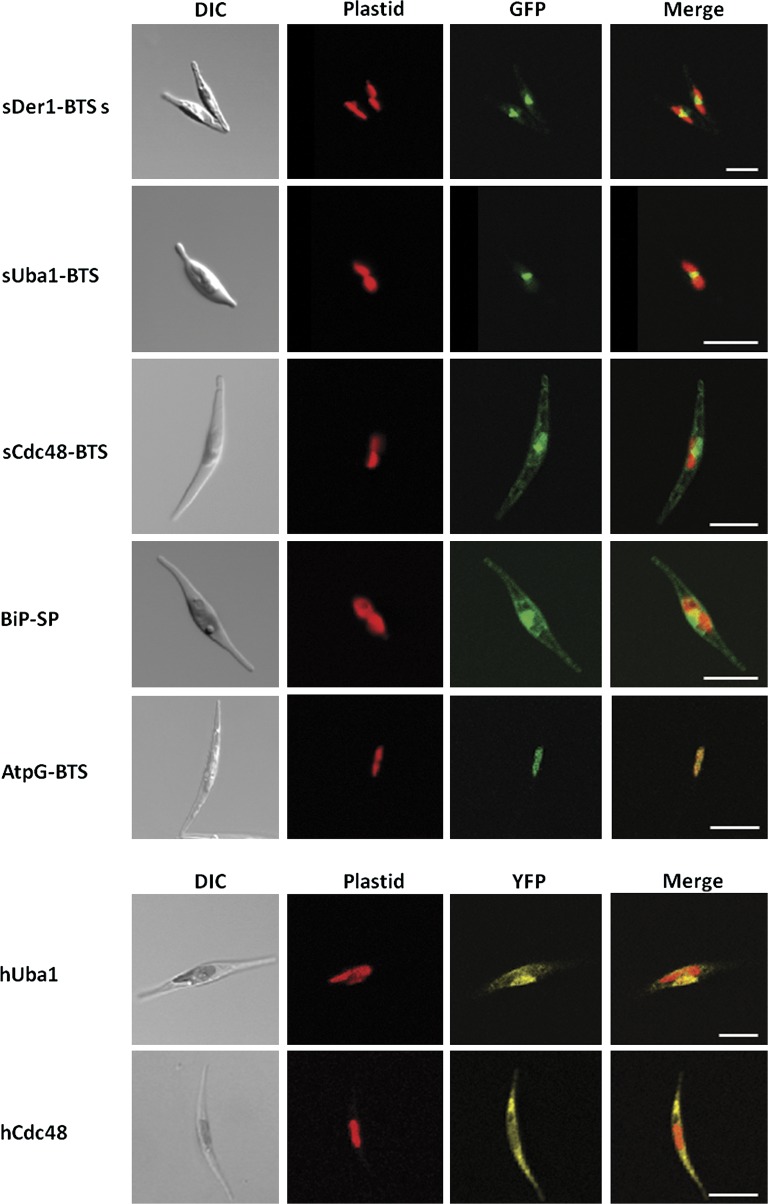
Some of the initially obtained clones showed no fluorescence at all, pointing to some kind of general incompatibility. Emiliania huxleyi has a high G + C content of up to ≥75% in the analyzed presequences, which is far higher than that of P. tricornutum with a G + C content of about 50% in coding regions suggesting a differential codon usage in these algae (Armbrust et al. 2004). We presumed this to be the reason for the missing fluorescence. In order to overcome the problem in expressing the haptophytic genes in the diatom, we customly synthesized the sequences to be tested in the codon bias determined for P. tricornutum. As shown in figure 2, the shortest sDer1-1 and the sUba1 constructs, in which the nucleotides but not the amino acids are converted and expressed as a GFP fusion, showed a fluorescence in a typical “blob-like structure,” recently described as being the region homologous to the symbiont’s remnant cytosol in the diatom (PPC; see Gould et al. 2006a). Moreover, the two longer presequences of the symbiontic Der1-1 led to a PPC localization of the GFP too (data not shown). Unexpectedly, the targeting signal of sCdc48 drove GFP not to the PPC, but showed—compared with the BiP construct, which clearly localizes to the ER-lumen—what appears to be dual localization in the ER and the PPC. An ER localization can appear in different fluorescence patterns. With the sCdc48 clone, one can see a mesh-like structure, whereas with BiP, the cell periphery is marked. As Cdc48 is a nonmembrane protein, located at the plasmatic site of ERAD systems, its localization therefore indicates an incomplete transport of the reporter protein population across only one membrane into the ER-lumen (fig. 2). Finally, the N-termini of the hUba1 and hCdc48 localized YFP to the host cytoplasm (fig. 2).
FIG. 2.—
Import properties of heterologously expressed haptophytic sequences. The topogenic signals of sDer1-1 and sUba1 direct GFP to the PPC. The GFP fusion comprising the targeting signal of sCdc48 predominantly localizes to the ER. The presequences of BiP and AtpG lead to either ER or stromal localization of GFP, respectively. The host versions of Uba1 and Cdc48 localize within the cytosol. Columns from left to right: light microscopic images, chlorophyll autofluorescence, GFP fluorescence and YFP fluorescence, respectively, merged chlorophyll and GFP/YFP fluorescence. Scale bar represents 10 μm.
Phylogenetic Analyses
Together with our previous investigations (Sommer et al. 2007), this work indicates the presence of a symbiont-specific ERAD (-like) system in all four membrane–bound plastid/apicoplast-bearing chromalveolates investigated so far. In order to reconstruct the phylogenetic origin of the plastid-located ERAD systems, we analyzed the available genomic data, searching for further homologous ERAD components. The Der1 sequences were too divergent to reconstruct any reliable phylogeny. Independent data sets were created for Uba1 and Cdc48, for which sequences from vertebrates, invertebrates, fungi, chlorophytes and land plants, a rhodophyte, and several representatives from the chromalveolates (cryptophytes, haptophytes, diatoms, pelagophytes, oomycetes, ciliates, and alveolates) were included.
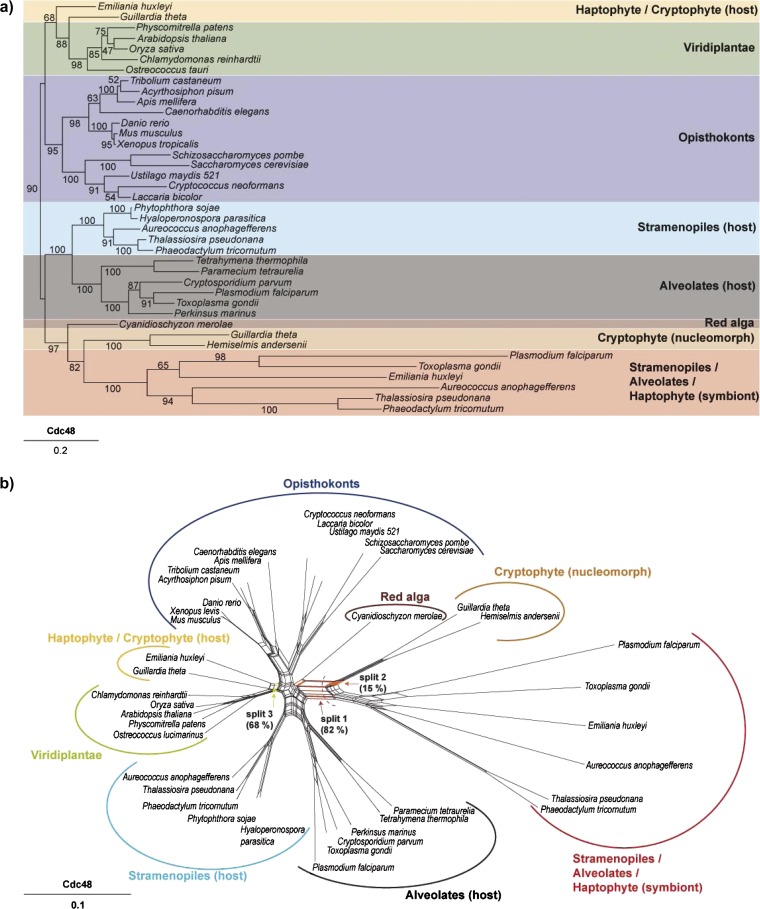
To depict the evolutionary affinities of Cdc48 and Uba1, we generated both phylogenetic networks (fig. 3b and d) and phylogenetic trees (fig. 3a and c). The splits in the network serve the same purpose as the familiar branches in a phylogenetic tree, that is, they separate the sequences into two groups, one either side of the branch or split. Splits are depicted as a series of parallel lines. Networks have the advantage that they can depict conflicting splits (signals) in the data simultaneously. For example, the network constructed with the Cdc48 data set (fig. 3b) shows a strong split (labeled as split 1) that unites the symbiont derived Emiliania Cdc48 with its symbiont derived homologues from stramenopiles and alveolates, all of which are in turn united with their homologues from cryptophyte nucleomorphs. The corresponding branch has a bootstrap value of 82% in the phylogenetic tree constructed with ML (fig. 3a). At the same time, however, nucleomorph-encoded Cdc48 also shares a conflicting split (split 2, fig. 3b) with the red algal homologue from Cyanidioschyzon merolae, but the corresponding branch is not depicted in the ML tree. The branch that unites the symbiont-derived Cdc48 homologues with the homologue from the red alga has a BP of 97%, supporting the view that the symbiont-derived SELMA/ERAD system does indeed stem from the nuclear genome of the secondary red algal endosymbiont. The Emiliania host-derived Cdc48 sequence clusters with its homologues from the cryptophyte nucleus and green plants (split 3, fig. 3b) a position also reflected in the ML tree but with a low BP (68%, fig. 3a).
FIG. 3.—
Phylogenetic bootstrap trees and Neighbor Net (NNet) splits graphs. (a) PHyML bootstrap tree derived from an FSA alignment of Cdc48 sequences from 39 taxa (646 of 2,812 sites were used after curation with Gblocks). Bootstrap analyses were conducted using 100 bootstrap replicates. (b) NNet splits graph derived from an FSA alignment of Cdc48 sequences from 39 taxa (646 of 2812 sites that were left after curation with Gblocks were used for the estimation of distances using PROTDIST and the JTT model). With these distances, a NNet splits graph was constructed which was visualized with Splitstree4. Three splits are highlighted: Split 1 separates the symbiontic sequences from all other taxa. This split also occurs in 82 of 100 constructed bootstrap trees. Split 2 separates the nucleomorph-encoded sequences and the red algal copy from all other taxa, whereas split 3 unites the Emiliania host-derived Cdc48 sequence with its homologues from the cryptophyte nucleus and green plants with a bootstrap support of 68%. (c) PHyML bootstrap tree derived from an FSA alignment of Uba1 sequences from 38 taxa (438 of 7,999 sites were used after curation with Gblocks). Bootstrap analyses were conducted using 100 bootstrap replicates. (d) NNet splits graph derived from an FSA alignment of Uba1 sequences from 38 taxa (438 of 7,999 sites that were left after curation with Gblocks were used for the estimation of distances using PROTDIST and the JTT model). With these distances, a NNet splits graph was constructed which was visualized with Splitstree4. Split 4 separates the green lineage together with the Emiliania huxleyi and the Guillardia theta sequences from all other taxa and has a bootstrap support of 98% in the corresponding ML tree.
The phylogenetic network of the somewhat less well-conserved Uba1 protein (fig. 3d) shows a more complex phylogeny with generally lower bootstrap values in the ML tree. The red algal sequence clusters within the monophyletic group containing the Emiliania symbiont-derived gene, its homologues from stramenopiles, alveolates, and the nucleomorph-encoded Uba1, as expected for an origin of these genes from the secondary endosymbiont genome. The host-derived E. huxleyi Uba1 gene again clusters with the cryptophyte nuclear and green algal homologues (split 4, fig. 3d), also in the ML tree (fig. 3c, BP 98).
Thus, both the symbiont-derived E. huxleyi Cdc48 and Uba1 genes in the SELMA pathway reflect a red algal ancestry, whereas the host-derived E. huxleyi Cdc48 and Uba1 genes trace the host ERAD ancestry to a cryptophyte and green algal signal.
In addition to PhyML, we also computed ML trees with RAxML (supplementary figs. 1 and 2, Supplementary Material online), using every fifth tree (200 in total) that was computed during the bootstrap search as a starting tree for the ML optimization in the tree search. This somewhat more extensive RAxML topology search ultimately delivered trees that were identical in topolgy to those obtained with PhyML, only the bootstrap values were slightly different. As the most extreme example, the branch in the Cdc48 tree dividing the ophistokonts, viridiplantae, E. huxleyi, and G. theta from the other species has a BP value of 90 in the PhyML tree and of 31 in the RAxML tree. Thus, if PhyML is getting stuck in local optima with this data, RAxML is getting stuck in the same local optimum.
Discussion
The chromalveolate hypothesis (Cavalier-Smith 1999, 2003) posits that all chlorophyll c-containing algae and their nonphotosynthetic sister groups are descended from one, and the same secondary symbiotic event entailing a red algal endosymbiont. This concept has been controversially discussed, and more recently, alternative hypotheses have been put forward (Harper et al. 2005; Reyes-Prieto et al. 2007; Gould et al. 2008; Sanchez-Puerta and Delwiche 2008; Elias and Archibald 2009). However, a better understanding of protein import among the chromalveolates should contribute to our understanding of endosymbioses and plastid origins within the group.
A tenet of the single secondary symbiotic origin of the chromalveolates is that the plastid preprotein import machinery evolved once in the progenitor of the group. Recently, several laboratories (Sommer et al. 2007; Agrawal et al. 2009; Hempel et al. 2009; Kalanon et al. 2009; Spork et al. 2009) presented evidence to suggest that the plastid preprotein translocator for the second outermost membrane of the four membrane–bound plastids of chromalveolates is derived from the ERAD system, the original function of which is for directing misfolded proteins out of the ER lumen. Genes encoding components of this putative ERAD-derived translocon machinery have been identified in the genomes of cryptophytes, diatoms, and apicomplexa (Sommer et al. 2007; Agrawal et al. 2009; Kalanon et al. 2009; Spork et al. 2009). In the case of the cryptophytes, some of these genes still reside in the nucleomorph. If the assumption of an ancient import machinery in the progenitor of chromalveolates is correct, the proposed translocator should have attained its functional state before the chromalveolates diverged into their modern groups. Hence, the corresponding genes or traces thereof should be detectable in the genomes of the plastid/apicoplast-harboring chromists and alveolates. Recently, the genomes of several further chromists have been sequenced, among them that of the haptophyte E. huxleyi. Besides their ecological importance, haptophytes are fascinating organisms with a complex cell architecture.
We identified E. huxleyi genes for the postulated ERAD-derived translocation machinery, namely Cdc48—the putative energy providing ubiquitin-dependent AAA-ATPase—its cofactor Ufd1, two Der1-like proteins (discussed as a possible pore protein see Meusser et al. [2005], table 1), and the E1-activity Uba1. All identified proteins exist at least as two copies in parallel, one for the symbiont and one for host-specific ERAD machinery. The encoded SELMA proteins possess BTS sequences necessary to facilitate their proper targeting to or across the second outermost membrane of the plastid, as predicted by neural network tools. Indeed, the customized, heterologously expressed targeting sequences of sDer1-1 and sUba1 fused to GFP as well as the control constructs of BiP and AtpG, hCdc48 and hUba1, did localize to the subcellular compartment expected under our working hypothesis (fig. 2). By contrast, the fluorescence of the bipartite signal sequence of Cdc48 fused to GFP was observed in the ER lumen (with presumably only a minor part in the PPC). Cdc48 is a soluble protein of the cytoplasm (and of the periplastic space in case of sCdc48, respectively), hence the localization within the extraplasmatic lumen between the first and second membrane indicates that, in this case, the targeting signal permits an import across the first of the two membranes but functions less effective for transport across the second outermost membrane. This implies that targeting signals might have so far unidentified characteristics or motifs that contribute to their role in intracellular trafficking, which could differ in diatoms (the heterologous expression system) and haptophytes (the source of the genes expressed). Obvious differences in the composition of targeting signals have been recognized by inspection of the TP sequences of organisms of secondary symbiotic origin, indicating that the canonical phenylalanine residue at the first amino acid position of TPs of plastid-targeted proteins in cryptophytes and diatoms is present in some but not all TPs of haptophytes (Patron and Waller 2007).
Recent reconstructions of the phylogeny of chromalveolates were mainly based either on plastid-derived or host-specific markers. The symbiont/host ERAD factors take advantage of the fact that—at least for the phototrophic members and the apicomplexa—data can be obtained from the eukaryotic (endo-) symbiont and its host in parallel. Hence, findings from the ERAD-based phylogeny might contribute to the phylogenetic issues, in addition to providing needed information about the process of protein import itself. We sampled ERAD-homologs from opisthokonts, chromalveolates (including ciliates and oomycetes), and viridiplantae for single gene-based phylogenies. The sequences of Cdc48 and Uba1 provide an interpretable view of the evolution of the ERAD systems investigated (fig. 3). In both phylogenetic trees, the symbiont versions group together with those of the cryptophyte and of a red alga. This pattern, which was seen in a recent study on Cdc48 as well (Agrawal et al. 2009), might indicate that the ERAD complex for the symbiont is of red algal origin and was introduced once by the possible progenitor of chromalveolates. However, the branching of the host ERAD versions indicates a more sophisticated pattern. Here, the hCdc48 sequences of stramenopiles form a monophyletic group with the sequences of the alveolates with a BP of 100%, whereas in the case of hUba1, these groups neither in the tree nor in the network branch together. Most strikingly, the hCdc48 copy and both copies of hUba1 from cryptophytes and that of the haptophyte group together with the viridiplantae (the green line) reflecting a similar branching as it has been shown for other markers (Harper et al. 2005; Rice and Palmer 2006; Hackett et al. 2007). The grouping of the symbiont versus host components raises the question, why all symbiont-specific factors trace back to rhodophytes in spite of an indicated host polyphyly. The SELMA system might have been established repeatedly in serial secondary endosymbioses (Burki et al. 2007) (meaning individual endosymbiosis events with rhodophytes) due to cellular demands for the conversion of an endosymbiont to a complex plastid. An alternative explanation, to the monophyletic chromalveolate clade, has been offered by Sanchez-Puerta and Delwiche (2008), who argued for tertiary endosymbioses (meaning the internalization of an alga belonging to the haptophyte/cryptophyte clade) at the onset of stramenopiles and alveolates.
Conclusions
Accumulating data, including that presented here, suggest that the evolution of chromists is more complex than formulated in the chromalveolate hypothesis. Accordingly, several new hypotheses have recently been put forth regarding chromist evolution (e.g., Becker et al. 2008; Frommolt et al. 2008; Sanchez-Puerta and Delwiche 2008; Moustafa et al. 2009; Nozaki et al. 2009; Cavalier-Smith 2010). Our present data indicate that an ERAD-related machinery may be used for protein import into the complex plastid in cryptophytes, haptophytes, stramenopiles, and alveolates. Like other recent findings (Harper et al. 2005; Hackett et al. 2007; Becker et al. 2008; Frommolt et al. 2008; Nozaki et al. 2009; Baurain et al. 2010), our data do not support the view that a single, red-algal derived secondary endosymbiosis underlies the origin of chromalveolates. The recruitment of the ERAD machinery for protein targeting into complex plastids might not be restricted to the red lineage, hence the identification and study of its components among chlorarachniophytes is of interest with respect to the principles governing evolutionary integration of eukaryotic genomes during secondary endosymbiosis.
Supplementary Material
Supplementary figures 1–2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank J. M. Archibald, M. W. Gray, P. J. Keeling, G. I. McFadden, and C. E. Lane for access to preliminary genome sequence data from Guillardia theta, which was produced by the Joint Genome Institute’s Community Sequencing Program (http://www.jgi.doe.gov/). This work was supported by grants from the Deutsche Forschungsgemeinschaft SFB593, Graduate School 1216 (to U.G.M.) and SFB TR1 (to W.M.) and by the European Research Council (Networkorigins) (to W.M).
References
- Agrawal S, van Dooren GG, Beatty WL, Striepen B. Genetic evidence that an endosymbiont-derived ERAD system functions in import of apicoplast proteins. J Biol Chem. 2009;284:33683–33691. doi: 10.1074/jbc.M109.044024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apt KE, Kroth-Pancic PG, Grossman AR. Stable nuclear transformation of the diatom Phaeodactylum tricornutum. Mol Gen Genet. 1996;252:572–579. doi: 10.1007/BF02172403. [DOI] [PubMed] [Google Scholar]
- Armbrust EV, et al. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- Baurain D, et al. Phylogenomic evidence for separate acquisition of plastids in cryptophytes, haptophytes, and stramenopiles. Mol Biol Evol. 2010;27:1698–1709. doi: 10.1093/molbev/msq059. [DOI] [PubMed] [Google Scholar]
- Becker B, Hoef-Emden K, Melkonian M. Chlamydial genes shed light on the evolution of photoautotrophic eukaryotes. BMC Evol Biol. 2008;8:203. doi: 10.1186/1471-2148-8-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodyl A. Evolutionary origin of a preprotein translocase in the periplastid membrane of complex plastids: a hypothesis. Plant Biol (Stuttg) 2004;6:513–518. doi: 10.1055/s-2004-821092. [DOI] [PubMed] [Google Scholar]
- Bolte K, et al. Protein targeting into secondary plastids. J Eukaryot Microbiol. 2009;56:9–15. doi: 10.1111/j.1550-7408.2008.00370.x. [DOI] [PubMed] [Google Scholar]
- Bradley RK, et al. Fast statistical alignment. PLoS Comput Biol. 2009;5:e1000392. doi: 10.1371/journal.pcbi.1000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant D, Moulton V. Neighbor-net: an agglomerative method for the construction of phylogenetic networks. Mol Biol Evol. 2004;21:255–265. doi: 10.1093/molbev/msh018. [DOI] [PubMed] [Google Scholar]
- Bullmann L, et al. Filling the gap: the evolutionary conserved Omp85 in plastids of chromalveolates. J Biol Chem. 2010;285:6848–6856. doi: 10.1074/jbc.M109.074807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki F, et al. Phylogenomics reshuffles the eukaryotic supergroups. PLoS One. 2007;2:e790. doi: 10.1371/journal.pone.0000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Principles of protein and lipid targeting in secondary symbiogenesis: euglenoid, dinoflagellate, and sporozoan plastid origins and the eukaryote family tree. J Eukaryot Microbiol. 1999;46:347–366. doi: 10.1111/j.1550-7408.1999.tb04614.x. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Genomic reduction and evolution of novel genetic membranes and protein-targeting machinery in eukaryote-eukaryote chimaeras (meta-algae) Philos Trans R Soc Lond B Biol Sci. 2003;358:109–133. doi: 10.1098/rstb.2002.1194. discussion. 133–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Kingdoms Protozoa and Chromista and the eozoan root of the eukaryotic tree. Biol Lett. 2010;6:342–345. doi: 10.1098/rsbl.2009.0948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas S, et al. The highly reduced genome of an enslaved algal nucleus. Nature. 2001;410:1091–1096. doi: 10.1038/35074092. [DOI] [PubMed] [Google Scholar]
- Elias M, Archibald JM. Sizing up the genomic footprint of endosymbiosis. Bioessays. 2009;31:1273–1279. doi: 10.1002/bies.200900117. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, von Heijne G. Prediction of organellar targeting signals. Biochim Biophys Acta. 2001;1541:114–119. doi: 10.1016/s0167-4889(01)00145-8. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.6. Cladistics. 1989;5:164–166. [Google Scholar]
- Frommolt R, et al. Ancient recruitment by chromists of green algal genes encoding enzymes for carotenoid biosynthesis. Mol Biol Evol. 2008;25:2653–2667. doi: 10.1093/molbev/msn206. [DOI] [PubMed] [Google Scholar]
- Gibbs SP. The route of entry of cytoplasmically synthesized proteins into chloroplasts of algae possessing chloroplast ER. J Cell Sci. 1979;35:253–266. doi: 10.1242/jcs.35.1.253. [DOI] [PubMed] [Google Scholar]
- Gould SB, et al. Nucleus-to-nucleus gene transfer and protein retargeting into a remnant cytoplasm of cryptophytes and diatoms. Mol Biol Evol. 2006a;23:2413–2422. doi: 10.1093/molbev/msl113. [DOI] [PubMed] [Google Scholar]
- Gould SB, et al. Protein targeting into the complex plastid of cryptophytes. J Mol Evol. 2006b;62:674–681. doi: 10.1007/s00239-005-0099-y. [DOI] [PubMed] [Google Scholar]
- Gould SB, Waller RF, McFadden GI. Plastid evolution. Annu Rev Plant Biol. 2008;59:491–517. doi: 10.1146/annurev.arplant.59.032607.092915. [DOI] [PubMed] [Google Scholar]
- Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hackett JD, et al. Phylogenomic analysis supports the monophyly of cryptophytes and haptophytes and the association of Rhizaria with chromalveolates. Mol Biol Evol. 2007;24:1702–1713. doi: 10.1093/molbev/msm089. [DOI] [PubMed] [Google Scholar]
- Harper JT, Keeling PJ. Nucleus-encoded, plastid-targeted glyceraldehyde-3-phosphate dehydrogenase (GAPDH) indicates a single origin for chromalveolate plastids. Mol Biol Evol. 2003;20:1730–1735. doi: 10.1093/molbev/msg195. [DOI] [PubMed] [Google Scholar]
- Harper JT, Waanders E, Keeling PJ. On the monophyly of chromalveolates using a six-protein phylogeny of eukaryotes. Int J Syst Evol Microbiol. 2005;55:487–496. doi: 10.1099/ijs.0.63216-0. [DOI] [PubMed] [Google Scholar]
- Hempel F, et al. Transport of nuclear-encoded proteins into secondarily evolved plastids. Biol Chem. 2007;388:899–906. doi: 10.1515/BC.2007.119. [DOI] [PubMed] [Google Scholar]
- Hempel F, Bullmann L, Lau J, Zauner S, Maier UG. ERAD-derived pre-protein transport across the 2nd outermost plastid membrane of diatoms. Mol Biol Evol. 2009;26:1781–1790. doi: 10.1093/molbev/msp079. [DOI] [PubMed] [Google Scholar]
- Hempel F, Felsner G, Maier UG. New mechanistic insights into pre-protein transport across the second outermost plastid membrane of diatoms. Mol Microbiol. 2010;76:793–801. doi: 10.1111/j.1365-2958.2010.07142.x. [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Ismail N, Ng DT. Have you HRD? Understanding ERAD is DOAble! Cell. 2006;126:237–239. doi: 10.1016/j.cell.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Kalanon M, Tonkin CJ, McFadden GI. Characterization of two putative protein translocation components in the apicoplast of Plasmodium falciparum. Eukaryot Cell. 2009;8:1146–1154. doi: 10.1128/EC.00061-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian O, Kroth PG. Identification and characterization of a new conserved motif within the presequence of proteins targeted into complex diatom plastids. Plant J. 2005;41:175–183. doi: 10.1111/j.1365-313X.2004.02294.x. [DOI] [PubMed] [Google Scholar]
- Lane CE, Archibald JM. The eukaryotic tree of life: endosymbiosis takes its TOL. Trends Ecol Evol. 2008;23:268–275. doi: 10.1016/j.tree.2008.02.004. [DOI] [PubMed] [Google Scholar]
- McFadden GI, van Dooren GG. Evolution: red algal genome affirms a common origin of all plastids. Curr Biol. 2004;14:R514–R516. doi: 10.1016/j.cub.2004.06.041. [DOI] [PubMed] [Google Scholar]
- Meusser B, Hirsch C, Jarosch E, Sommer T. ERAD: the long road to destruction. Nat Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- Moustafa A, et al. Genomic footprints of a cryptic plastid endosymbiosis in diatoms. Science. 2009;324:1724–1726. doi: 10.1126/science.1172983. [DOI] [PubMed] [Google Scholar]
- Nassoury N, Cappadocia M, Morse D. Plastid ultrastructure defines the protein import pathway in dinoflagellates. J Cell Sci. 2003;116:2867–2874. doi: 10.1242/jcs.00517. [DOI] [PubMed] [Google Scholar]
- Nozaki H, et al. Phylogenetic positions of glaucophyta, green plants (Archaeplastida) and haptophyta (Chromalveolata) as deduced from slowly evolving nuclear genes. Mol Phylogenet Evol. 2009;53:872–880. doi: 10.1016/j.ympev.2009.08.015. [DOI] [PubMed] [Google Scholar]
- Patron NJ, Waller RF. Transit peptide diversity and divergence: a global analysis of plastid targeting signals. Bioessays. 2007;29:1048–1058. doi: 10.1002/bies.20638. [DOI] [PubMed] [Google Scholar]
- Reyes-Prieto A, Weber AP, Bhattacharya D. The origin and establishment of the plastid in algae and plants. Annu Rev Genet. 2007;41:147–168. doi: 10.1146/annurev.genet.41.110306.130134. [DOI] [PubMed] [Google Scholar]
- Rice DW, Palmer JD. An exceptional horizontal gene transfer in plastids: gene replacement by a distant bacterial paralog and evidence that haptophyte and cryptophyte plastids are sisters. BMC Biol. 2006;4:31. doi: 10.1186/1741-7007-4-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Puerta MV, Delwiche CF. A hypothesis for plastid evolution in chromalveolates. J Phycol. 2008;44:1097–1107. doi: 10.1111/j.1529-8817.2008.00559.x. [DOI] [PubMed] [Google Scholar]
- Sommer MS, et al. Der1-mediated pre-protein import into the periplastid compartment of chromalveolates? Mol Biol Evol. 2007;24:1–11. doi: 10.1093/molbev/msm008. [DOI] [PubMed] [Google Scholar]
- Spork S, et al. An unusual ERAD-like complex is targeted to the apicoplast of Plasmodium falciparum. Eukaryot Cell. 2009;8:1134–1145. doi: 10.1128/EC.00083-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Tonkin CJ, Kalanon M, McFadden GI. Protein targeting to the malaria parasite plastid. Traffic. 2008;9:166–175. doi: 10.1111/j.1600-0854.2007.00660.x. [DOI] [PubMed] [Google Scholar]
- Tonkin CJ, Struck NS, Mullin KA, Stimmler LM, McFadden GI. Evidence for Golgi-independent transport from the early secretory pathway to the plastid in malaria parasites. Mol Microbiol. 2006;61:614–630. doi: 10.1111/j.1365-2958.2006.05244.x. [DOI] [PubMed] [Google Scholar]
- van Dooren GG, Tomova C, Agrawal S, Humbel BM, Striepen B. Toxoplasma gondii Tic20 is essential for apicoplast protein import. Proc Natl Acad Sci U S A. 2008;105:13574–13579. doi: 10.1073/pnas.0803862105. [DOI] [PMC free article] [PubMed] [Google Scholar]