Abstract
This Technical Note describes a novel modular framework for development and interlaboratory distribution and validation of 3D tractography algorithms based on in vivo diffusion tensor imaging (DTI) measurements. The proposed framework allows individual MRI research centers to benefit from new tractography algorithms developed at other independent centers by “plugging” new tractography modules directly into their own custom DTI software tools, such as existing graphical user interfaces (GUI) for visualizing brain white matter pathways. The proposed framework is based on the Java 3D programming platform, which provides an object-oriented programming (OOP) model and independence of computer hardware configuration and operating system. To demonstrate the utility of the proposed approach, a complete GUI for interactive DTI tractography was developed, along with two separate and interchangeable modules that implement two different tractography algorithms. Although the application discussed here relates to DTI tractography, the programming concepts presented here should be of interest to anyone who wishes to develop platform-independent GUI applications for interactive 3D visualization.
Key Words: Diffusion tensor imaging, white matter, tractography
Background
Diffusion tensor imaging (DTI) is an emergingclinical magnetic resonance imaging (MRI) modality capable of detecting subtle microstructural tissue changes and providing measurements of the location and directionality of white matter fiber “bundles” in the human brain.1 Diffusion tensor imaging has its basis in diffusion-weighted imaging, which was shown more than a decade ago to be an effective tool for early detection of ischemic brain injury.2 Recently, computer algorithms based on the full diffusion tensor have been employed to reconstruct complex three-dimensional white matter structures,3 enabling for the first time studies of the relationship between the development and integrity of white matter pathways and cognitive processes observed by functional imaging modalities such as functional MRI (fMRI).
However, despite the proven worth of DTI for studies of white matter structure, penetration of this technology into clinical practice (and even research) has been held back by the lack of available software tools for DTI processing and visualization, and by the lack of interlaboratory validation of the numerous DTI visualization (“tractography”) algorithms that have been proposed. The tendency to date has been for each clinical MRI research laboratory to spend significant resources to implement its own custom software for DTI processing and 3D tractography, which is often only made available internally or to immediate scientific collaborators. This practice has produced a wide variety of 3D tractography algorithms3 that escape the detailed (code-level) scrutiny by the biomedical engineering community, making clinical and scientific contributions based on such algorithms difficult or impossible to reproduce and validate.
The purpose of this Technical Note is to describe the design and implementation of a modular and portable programming environment for the development, dissemination, and validation of experimental fiber tractography algorithms. The proposed framework relies on object-oriented programming (OOP) concepts and computing-platform independence to produce portable modules (or “objects”) that completely encapsulate the creation and visual representation of individual fibers from DTI data, and that can be shared among clinical research centers for informal peer-review and clinical validation. The Java 3D programming platform (freely obtainable from http://java.sun.com) is ideal for this purpose, and the discussion that follows is devoted to describing the essential components of the Java 3D API that support the proposed modular framework.
Methods
The Java 3D API
The Java 3D API allows the creation of a 3D “universe” that can be manipulated interactively by the end user. In the simplest implementation of the Java 3D API, which is the one used in the examples presented here, individual 3D objects that inherit from the javax.media.j3d.Shape3D class are added to a javax.media.j3d.BranchGroup object, which in turn is added to a built-in universe object (com.sun.j3d.utils.universe.SimpleUniverse). Here, one or more fibers will be added to a single BranchGroup object, and one or more BranchGroup objects will be added to a SimpleUniverse object. The SimpleUniverse object can be added directly to a GUI application based on the Java Swing API. The Java Swing API provides a complete set of GUI components (such as windows, buttons, and other widgets) that are created and manipulated using a consistent and relatively easy-to-use programming interface. The Java Swing API, which replaces the older AWT API, is described at http://java.sun.com/docs/books/tutorial/uiswing. For useful information on integrating Java 3D components with Swing, see http://www.j3d.org/tutorials/quick_fix/swing.html.
The DTI Data Set
Before discussing how individual fibers are generated and displayed, it is necessary to describe the composition of a DTI data set. Typically, diffusion-weighted MRI images are acquired slice-by-slice using echo-planar imaging (EPI) MRI sequences, resulting in an image volume that is organized on a 256 × 256 × N grid, where N is the number of slices acquired (usually between 20 and 40). From seven or more diffusion-weighted image volumes, a DTI data set is generated, such that for each grid point (or “voxel”), we have a 3 × 3 diffusion tensor that describes the speed and direction of water diffusion averaged over the voxel volume. However, most DTI tractography algorithms consider only the direction of maximum diffusion (which is obtained from the diffusion tensor), and it therefore suffices for our purposes to consider a DTI data set as a collection of three-dimensional vectors organized on a regular 3D grid. For more details on DTI, see the review article by Basser and Jones.1
The in vivo DTI data sets used in this study were acquired as part of a routine clinical MRI imaging protocol approved by the Institutional Review Board at our institution.
Modular Fiber Generation
Based on the 3D vector field in a DTI data set, it is possible to construct continuous 3D lines (or “fibers”) that may extend across the entire brain volume. Figure 2 shows examples of “bundles” of many such 3D fibers generated from a DTI data set. Generating such fibers from discrete vector fields requires the development of the so-called fiber “tractography” algorithms, many of which have been proposed in the literature.3 The most elementary of such algorithm simply propagates the fiber along the direction determined by the local diffusion vector. This requires calculating the diffusion vector at arbitrary points within the image volume (not just on the grid points), which will be done here by interpolating between the nearest-neighbor values of the 3D diffusion vectors.
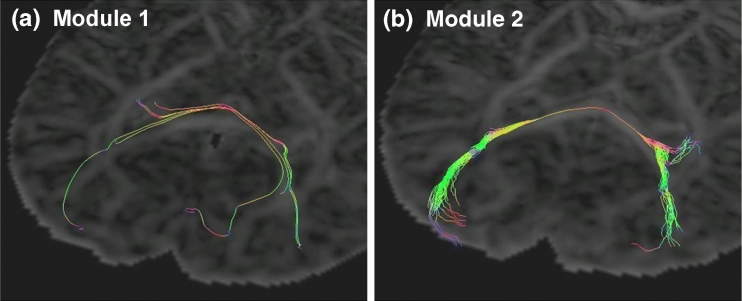
Fig 2.
Comparison of two different tractography algorithms applied to the same DTI data set. (a) Tractography results using Module 1 (see text). Tractography was initiated from five different seed points in close proximity to each other. Fibers are propagated along the direction of maximum diffusion [principal diffusion direction (PDD)], and terminated if the angle between fiber segments in neighboring pixels exceeds 45°. (b) Tractography results after loading Module 2, which generates several fibers from any given seed point. All fibers in (b) originate from the same seed point. In regions of high fractional anisotropy (FA), the fiber propagates along the PDD, whereas in low-FA regions, a random perturbation is added to the PDD. The images in (a) and (b) were acquired as “screenshots” of the interactive GUI described in A Graphical User Interface for Interactive Tractography.
The modular framework presented here contains two key ideas. First, each fiber (or a group of fibers) is encapsulated within a single object that inherits from the javax.media.j3d.BranchGroup class. This “tractography module” contains all code implementing the tractography algorithm, and describes fully the coordinates and appearance (such as line thickness and color coding) of the fiber(s). In this way, fiber tracts can be generated and displayed by simply creating an instance of the tractography module and adding it directly to the SimpleUniverse object. Furthermore, the internal algorithms of the fiber object are completely “invisible” to the SimpleUniverse object, and future modifications or replacements of the tractography module are possible without recompiling the GUI application that contains the SimpleUniverse object. The following shows how such a tractography module may be structured:
import javax.vecmath.Point3f;
import javax.media.j3d.BranchGroup;
import javax.media.j3d.Shape3D;
import org.dti.development.DtiData;
-
public class TractographyModule extends BranchGroup {
- DtiData data;
- Point3f pBegin;
- // constructor
- public TractographyModule(DtiData data, Point3f pBegin) {
- this.data = data;
- this.pBegin = pBegin;
- addChild(new Fiber());
- // add additional Fiber objects, if desired
- }
- // inner class for creating a single fiber as a Shape3D object
- class Fiber extends Shape3D {
- // create single fiber here
- }
- }
The constructor for the tractography module takes two arguments: (1) a reference to a DtiData object containing tensor data, and (2) the coordinates from which to “grow” the fiber(s). The second key component of the proposed modular framework is the formulation of a common interface for accessing DTI data from within the tractography module. In other words, communication between the tractography module and the DTI data happens exclusively through a clearly defined list of methods (the DtiData interface). The proposed interface, which is created as part of a new library org.dti.development, is defined as follows:- package org.dti.development;
- public interface DtiData {
- // Get 3×3 diffusion tensor matrix evaluated at point (x,y,z)
- double[][] getDT(double x, double y, double z);
- // Get eigenvalue. Numbering is in decreasing order, such that
- //ind = 1 (3) corresponds to the principal (minimum) eigenvalue.
- double getLambda(double x, double y, double z, int ind);
- // Get eigenvector (unit length) corresponding to eigenvalue lambda#,
- // where # = 1, 2, or 3
- double[] getEigenVector(double x, double y, double z, int ind);
- //Get Fractional Anisotropy (FA) and Apparent Diffusion Coefficient (ADC)
- double getFA(double x, double y, double z);
- double getADC(double x, double y, double z);
- // Get volume dimensions (unit: pixels) and voxel size (unit: mm)
- int[] getVolumeDimensions();
- double[] getVoxelDimensions();
- } // end of interface DtiData
To comply with the proposed framework, any “client” GUI application must provide a data class that implements this interface. Finally, the client application must provide a way to load new tractography modules, which may be done by providing the name of the tractography module during start-up of the GUI application, e.g., as a command-line argument. Alternatively, tractography modules can be loaded dynamically during runtime. An example of the latter scenario will be presented in the next section.
To summarize, the requirements for adhering to the proposed framework are: (1) to create tractography modules that inherit from BranchGroup, and that access data through the methods of the DtiData object reference passed to its constructor; (2) to ensure that any custom client GUI application or software tool creates a data object that implements the DtiData interface; and (3) to ensure that the client software tool allows the user of the software to select new tractography modules without the need to recompile the application.
Results
A Graphical User Interface for Interactive Tractography
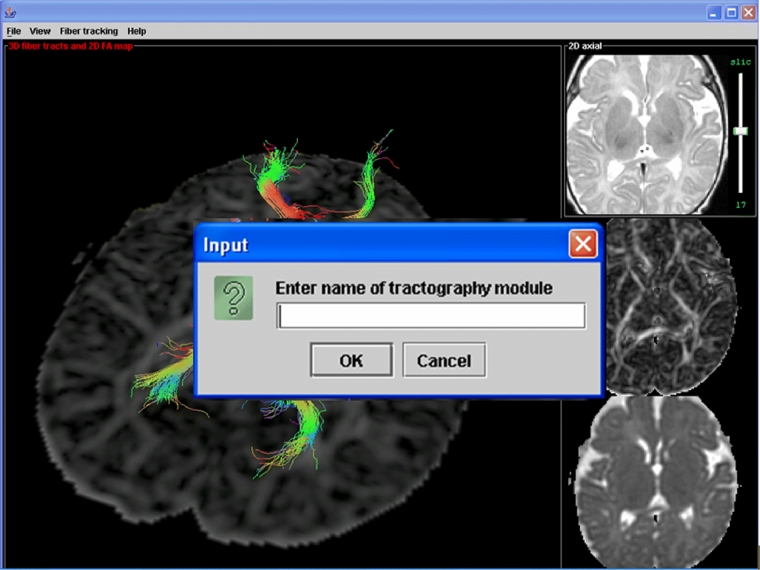
To demonstrate the benefits of the proposed programming framework for rapid algorithm development and testing, a complete graphical user interface (GUI) for interactive tractography was developed using the Java 3D API. This GUI consists of a SimpleUniverse object that is added to a Swing JPanel object, which in turn is added to the content pane of the application main window (a JFrame object). The GUI allows selection of a starting point for fiber tractography, and the resulting fiber(s) are calculated and displayed. Preprocessed diffusion tensor data is stored on hard disk in a platform-independent scientific data format. Data retrieval is handled by a data object that implements the DtiData interface. Tractography modules are loaded by entering the full module name (e.g., “edu.mymrilab.dtimodules.Module1”) in a text input box, which is shown in Figure 1 along with a “screenshot” of the GUI application main window in the background.
Fig 1.
Screenshot of a GUI application for interactive DTI tractography developed to test the proposed modular framework. The GUI allows the user to enter the name of the desired tractography module, which is then loaded and displayed in the 3D window shown in the background in the figure. The three image panels on the right are used for point selection and volume navigation.
After obtaining the module name from the user, the program loads and displays the tractography module in the following way:
import javax.swing.JOptionPane;
import java.lang.ClassLoader;
import java.lang.Class;
import java.lang.reflect.Constructor;
JOptionPane op = new JOptionPane();
String moduleName = (String) op.showInputDialog(“Enter name of tractography module”);
- try {
- ClassLoader cl = this.getClass().getClassLoader();
- Class moduleClass = cl.loadClass(moduleName);
- Class[] classarglist = new Class[]{DtiData.class, Point3f.class};
- Constructor ctor = moduleClass.getConstructor(classarglist);
- BranchGroup tractographyModule = (BranchGroup) ctor.newInstance(new Object[] {data, pStart});
- // add tractographyModule to this application's SimpleUniverse object here
- } catch (ClassNotFoundException mce) {
- System.out.println(“failed to load class”);
}
Tractography Module 1
“Module 1” implements a simple tractography algorithm that propagates the fiber along the direction determined by the local direction of maximum diffusion, and terminates the fiber if the angle between fiber segments in neighboring voxels exceeds 45°. Module 1 belongs to a package with the name edu.univA.mri.dtitractography.modules, and the .class (Java bytecode) files for this module are placed in a folder $root\edu\univA\mri\dtitractography\modules on the local hard disk (alternatively, the module may be packaged in a jar archive), where $root is any folder that is in the Java classpath. Figure 2(a) shows the results of loading this module into the GUI described in the section “A Graphical User Interface for Interactive Tractography” and generating five fibers starting from points in close proximity to each other. Figure 2 uses DTI data obtained in a newborn patient. Details of the image acquisition and preprocessing procedure for this data set have been reported elsewhere.4
Tractography Module 2
Figure 2(b) shows the results of loading “Module 2” into the GUI (without closing the GUI) and initiating fiber growth from a single point. Module 2 generates 50 fibers, all originating from the same starting point. Each fiber propagates along the direction of maximum diffusion [principal diffusion direction (PDD)] in regions of high fractional anisotropy (FA), whereas in low-FA regions, a random perturbation is added to the PDD.5 As in the section “Tractography Module 1,” the angle between fiber segments in neighboring voxels is limited to 45°. Although a detailed clinical comparison of Figure 2(a) and (b) is beyond the scope of this work, it is interesting to note thatModule 2 appears to “capture” the branching (or splitting) of the fiber bundle (right-hand side ofFig 2b), whereas this branching is not apparent in Figure 2(a). This is attributable to the random perturbations added to the fiber trajectory in Module 2, which allows many possible directions to be “sampled.” Module 1, on the other hand, produces exactly the same results every time tractography is initiated from any given seed point (i.e., the algorithmis deterministic), and is therefore very sensitive to the placement of the seed point(s).
Discussion
Although both examples shown in Figure 2 generate collections of individual “streamline” fibers, it is important to note that the proposed framework allows any visual representation of a DTI set, such as 3D surface renderings based on “Fast Marching” tractography6 methods or 3D ellipsoids representing the diffusion tensor at each grid point. The only requirement is that any such tractography module must inherit from javax.media.j3d. BranchGroup, and that it has a constructor that takes an org.dti.development.DtiData object reference followed by a javax.vecmath.Point3f object reference as arguments. As with any Java class, additional constructors are also possible, which enable the creator of the tractography module to implement custom module behavior that is designed for a particular GUI program or other software/processing tools.
It is also important to note that the proposed framework places no restrictions on the appearance and general functionality of the client GUI applications, as long as the GUI program provides (1) access to local data files through an object that implements the org.dti.development.DtiData interface, and (2) a program structure—which need not be based entirely on Java—that can display a javax.media.j3d.BranchGroup object.
Conclusion
We have designed and demonstrated a modular framework for development and interlaboratory dissemination of fiber tractography algorithms based on DTI data. The proposed framework allows new or experimental tractography algorithms and visual representations of white matter pathways to be integrated with existing 3D GUI applications. This framework allows each individual clinical MRI research site to develop its own custom GUI application, while allowing seamless integration and testing of new fiber modules developed either in-house or at other participating laboratories.
Acknowledgments
The author thanks Dr. Ashok Panigrahy for useful discussions and assistance with clinical interpretation and evaluation, and Dr. Stephan Erberich for assistance with data collection and preprocessing.
References
- 1.Basser PJ, Jones DK. Diffusion-tensor MRI: theory, experimental design and data analysis—a technical review. NMR Biomed. 2002;15:456–467. doi: 10.1002/nbm.783. [DOI] [PubMed] [Google Scholar]
- 2.Sotak CH. The role of diffusion tensor imaging in the evaluation of ischemic brain injury—a review. NMR Biomed. 2002;15:561–569. doi: 10.1002/nbm.786. [DOI] [PubMed] [Google Scholar]
- 3.Mori SS, Zijl PC. Fiber tracking: principles and strategies—a technical review. NMR Biomed. 2002;15:468–480. doi: 10.1002/nbm.781. [DOI] [PubMed] [Google Scholar]
- 4.Nielsen JF, Ghugre NR, Panigrahy A: Affine and polynomial mutual information co-registration for artifact elimination in Diffusion Tensor Imaging of newborns. Magn Reson Imaging 22:1319–1323, 2005 [DOI] [PubMed]
- 5.Lazar M, Alexander AL: White matter tractography using random vector (RAVE) perturbation. In: Proceedings of the 10th Annual Meeting of ISMRM, Honolulu, 2002
- 6.Parker GJ, Stephan KE, Barker GJ. Initial demonstration of in vivo tracing of axonal projections in the macaque brain and comparison with the human brain using diffusion tensor imaging and fast marching tractography. NeuroImage. 2002;15:797–809. doi: 10.1006/nimg.2001.0994. [DOI] [PubMed] [Google Scholar]