Abstract
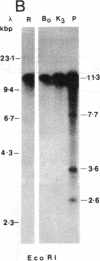
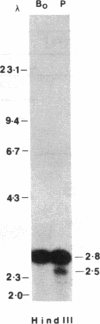
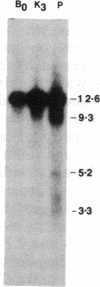
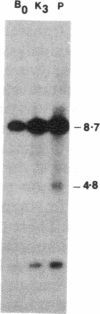
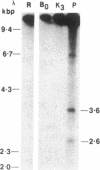
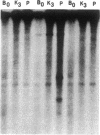
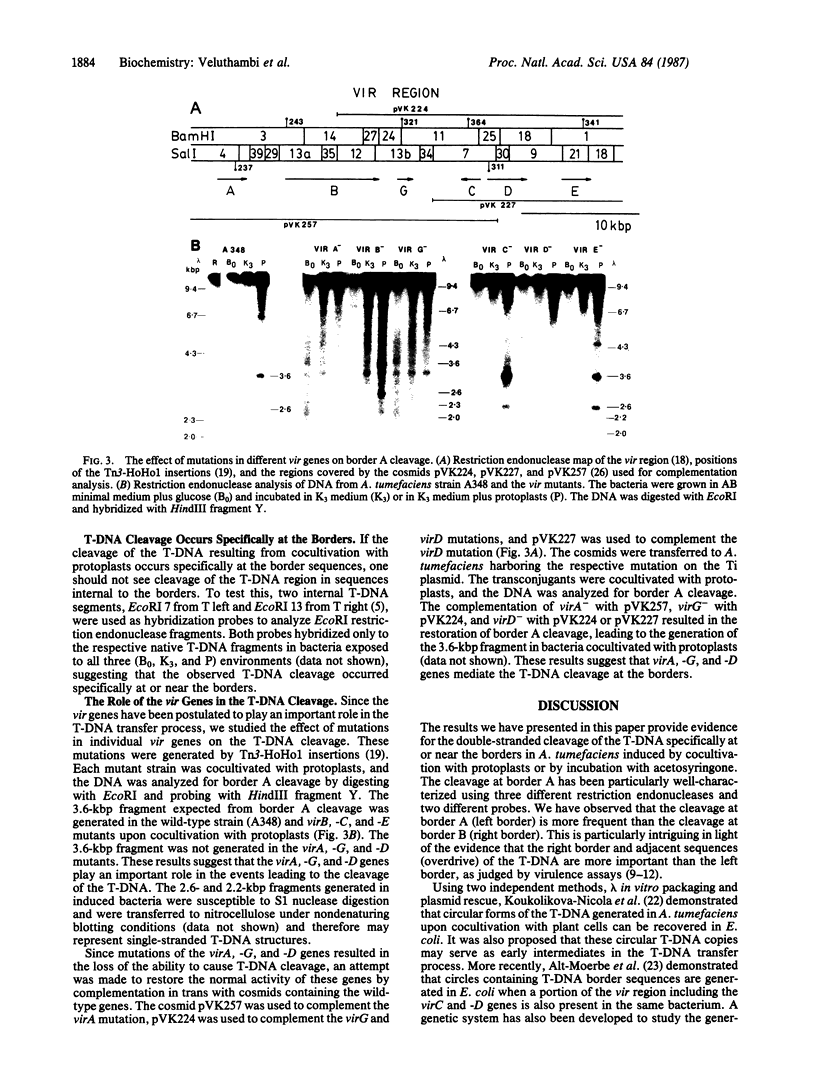
Agrobacterium tumefaciens transfers the T-DNA portion of its Ti plasmid to the nuclear genome of plant cells. Upon cocultivation of A. tumefaciens strain A348 with regenerating tobacco leaf protoplasts, restriction endonuclease fragments of the T-DNA were generated that are consistent with double-stranded cleavage of the T-DNA at the border sequences. The T-DNA border cleavage was also induced by acetosyringone, a compound that induces many of the virulence genes. T-DNA cleavage did not occur in Agrobacterium strains harboring Tn3-HoHo1 insertions in the virA, -D, or -G genes. Insertion mutations in virB, -C, or -E did not have any effect on the T-DNA cleavage. Complementation of the mutations in virA, -D, or -G with cosmids containing the respective wild-type genes restored the T-DNA cleavage. Since virA and -G are essential in regulating the expression of other vir genes in response to plant signal molecules, the virD gene product(s) appear to mediate double-stranded T-DNA border cleavage.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alt-Moerbe J., Rak B., Schröder J. A 3.6-kbp segment from the vir region of Ti plasmids contains genes responsible for border-sequence-directed production of T region circles in E. coli. EMBO J. 1986 Jun;5(6):1129–1135. doi: 10.1002/j.1460-2075.1986.tb04337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan M. W., Chilton M. D. T-DNA of the Agrobacterium Ti and Ri plasmids. Annu Rev Genet. 1982;16:357–384. doi: 10.1146/annurev.ge.16.120182.002041. [DOI] [PubMed] [Google Scholar]
- Bolton G. W., Nester E. W., Gordon M. P. Plant phenolic compounds induce expression of the Agrobacterium tumefaciens loci needed for virulence. Science. 1986 May 23;232(4753):983–985. doi: 10.1126/science.3085219. [DOI] [PubMed] [Google Scholar]
- Caplan A., Herrera-Estrella L., Inzé D., Van Haute E., Van Montagu M., Schell J., Zambryski P. Introduction of genetic material into plant cells. Science. 1983 Nov 18;222(4625):815–821. doi: 10.1126/science.222.4625.815. [DOI] [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas C. J., Staneloni R. J., Rubin R. A., Nester E. W. Identification and genetic analysis of an Agrobacterium tumefaciens chromosomal virulence region. J Bacteriol. 1985 Mar;161(3):850–860. doi: 10.1128/jb.161.3.850-860.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner R. C., Knauf V. C. Transfer of Agrobacterium DNA to Plants Requires a T-DNA Border But Not the virE Locus. Science. 1986 Feb 14;231(4739):725–727. doi: 10.1126/science.231.4739.725. [DOI] [PubMed] [Google Scholar]
- Garfinkel D. J., Nester E. W. Agrobacterium tumefaciens mutants affected in crown gall tumorigenesis and octopine catabolism. J Bacteriol. 1980 Nov;144(2):732–743. doi: 10.1128/jb.144.2.732-743.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelvin S. B., Karcher S. J., DiRita V. J. Methylation of the T-DNA in Agrobacterium tumefaciens and in several crown gall tumors. Nucleic Acids Res. 1983 Jan 11;11(1):159–174. doi: 10.1093/nar/11.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille J., van Kan J., Schilperoort R. trans-Acting virulence functions of the octopine Ti plasmid from Agrobacterium tumefaciens. J Bacteriol. 1984 May;158(2):754–756. doi: 10.1128/jb.158.2.754-756.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinegardner R. T. An improved fluorometric assay for DNA. Anal Biochem. 1971 Jan;39(1):197–201. doi: 10.1016/0003-2697(71)90476-3. [DOI] [PubMed] [Google Scholar]
- Iyer V. N., Klee H. J., Nester E. W. Units of genetic expression in the virulence region of a plant tumor-inducing plasmid of Agrobacterium tumefaciens. Mol Gen Genet. 1982;188(3):418–424. doi: 10.1007/BF00330043. [DOI] [PubMed] [Google Scholar]
- Jen G. C., Chilton M. D. The right border region of pTiT37 T-DNA is intrinsically more active than the left border region in promoting T-DNA transformation. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3895–3899. doi: 10.1073/pnas.83.11.3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauf V. C., Nester E. W. Wide host range cloning vectors: a cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid. 1982 Jul;8(1):45–54. doi: 10.1016/0147-619x(82)90040-3. [DOI] [PubMed] [Google Scholar]
- Peralta E. G., Hellmiss R., Ream W. Overdrive, a T-DNA transmission enhancer on the A. tumefaciens tumour-inducing plasmid. EMBO J. 1986 Jun;5(6):1137–1142. doi: 10.1002/j.1460-2075.1986.tb04338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peralta E. G., Ream L. W. T-DNA border sequences required for crown gall tumorigenesis. Proc Natl Acad Sci U S A. 1985 Aug;82(15):5112–5116. doi: 10.1073/pnas.82.15.5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. B., O'Hara P. J., Kwok W., Montoya A. L., Lichtenstein C., Gordon M. P., Nester E. W. DNA from the A6S/2 crown gall tumor contains scrambled Ti-plasmid sequences near its junctions with plant DNA. Cell. 1982 Jul;29(3):1005–1014. doi: 10.1016/0092-8674(82)90464-0. [DOI] [PubMed] [Google Scholar]
- Stachel S. E., An G., Flores C., Nester E. W. A Tn3 lacZ transposon for the random generation of beta-galactosidase gene fusions: application to the analysis of gene expression in Agrobacterium. EMBO J. 1985 Apr;4(4):891–898. doi: 10.1002/j.1460-2075.1985.tb03715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachel S. E., Nester E. W. The genetic and transcriptional organization of the vir region of the A6 Ti plasmid of Agrobacterium tumefaciens. EMBO J. 1986 Jul;5(7):1445–1454. doi: 10.1002/j.1460-2075.1986.tb04381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachel S. E., Zambryski P. C. virA and virG control the plant-induced activation of the T-DNA transfer process of A. tumefaciens. Cell. 1986 Aug 1;46(3):325–333. doi: 10.1016/0092-8674(86)90653-7. [DOI] [PubMed] [Google Scholar]
- Thomashow M. F., Nutter R., Montoya A. L., Gordon M. P., Nester E. W. Integration and organization of Ti plasmid sequences in crown gall tumors. Cell. 1980 Mar;19(3):729–739. doi: 10.1016/s0092-8674(80)80049-3. [DOI] [PubMed] [Google Scholar]
- Wang K., Herrera-Estrella L., Van Montagu M., Zambryski P. Right 25 bp terminus sequence of the nopaline T-DNA is essential for and determines direction of DNA transfer from agrobacterium to the plant genome. Cell. 1984 Sep;38(2):455–462. doi: 10.1016/0092-8674(84)90500-2. [DOI] [PubMed] [Google Scholar]
- Yadav N. S., Vanderleyden J., Bennett D. R., Barnes W. M., Chilton M. D. Short direct repeats flank the T-DNA on a nopaline Ti plasmid. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6322–6326. doi: 10.1073/pnas.79.20.6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., Iwahashi M., Yanofsky M. F., Nester E. W., Takebe I., Machida Y. The promoter proximal region in the virD locus of Agrobacterium tumefaciens is necessary for the plant-inducible circularization of T-DNA. Mol Gen Genet. 1987 Jan;206(1):174–177. doi: 10.1007/BF00326554. [DOI] [PubMed] [Google Scholar]
- Zambryski P., Depicker A., Kruger K., Goodman H. M. Tumor induction by Agrobacterium tumefaciens: analysis of the boundaries of T-DNA. J Mol Appl Genet. 1982;1(4):361–370. [PubMed] [Google Scholar]