Abstract
Autophagy is a highly-conserved cellular degradation and recycling system that is essential for cell survival during nutrient starvation. The loss of viability had been used as an initial screen to identify autophagy-defective (atg) mutants of the yeast Saccharomyces cerevisiae, but the mechanism of cell death in these mutants has remained unclear. When cells grown in a rich medium were transferred to a synthetic nitrogen starvation media, secreted metabolites lowered the extracellular pH below 3.0 and autophagy-defective mutants mostly died. We found that buffering of the starvation medium dramatically restored the viability of atg mutants. In response to starvation, wild-type (WT) cells were able to upregulate components of the respiratory pathway and ROS (reactive oxygen species) scavenging enzymes, but atg mutants lacked this synthetic capacity. Consequently, autophagy-defective mutants accumulated the high level of ROS, leading to deficient respiratory function, resulting in the loss of mitochondria DNA (mtDNA). We also showed that mtDNA deficient cells are subject to cell death under low pH starvation conditions. Taken together, under starvation conditions non-selective autophagy, rather than mitophagy, plays an essential role in preventing ROS accumulation, and thus in maintaining mitochondria function. The failure of response to starvation is the major cause of cell death in atg mutants.
Introduction
Unlike the ubiquitin-proteasome system that requires strict recognition of targets [1], autophagy is a non-selective degradation system induced under starvation conditions that mediates the recycling of cytoplasmic components to supply amino acids [2], [3]. Despite normal gene transcription, autophagy-deficient yeast cells have impaired protein translation during starvation due to inadequate supplies of amino acids [4]. Autophagy is critical for starvation-induced differentiation or development of yeast, nematodes, flies, and mice [5]. Accumulating data indicate that autophagy plays important roles in maintaining cellular homeostasis, and is relevant to several diseases [6], [7].
Mitochondria generate ATP through respiratory chain activity, but reactive oxygen species (ROS) are generated as by-products of cellular respiration. ROS induce damages to membrane, DNA, protein, and organelles, therefore mechanisms regulating the function and quantity of mitochondria are essential for eukaryotic cell function. Autophagy contributes for mitochondria maintenance by their clearance [8], and this process is mediated by selective type of autophagy termed mitophagy [9]–[12].
In S. cerevisiae, autophagy-defective (atg) mutants exhibit reduced viability during nitrogen starvation [13], but the mechanism of cell death has been unclear. Here, we show that non-selective autophagy is necessary to maintain the synthesis of respiratory chain components and ROS scavengers. In autophagy-deficient cells, mitochondrial dysfunction induced during starvation leads to impaired cell viability.
Results
pH buffering allows atg mutants to survive nitrogen starvation
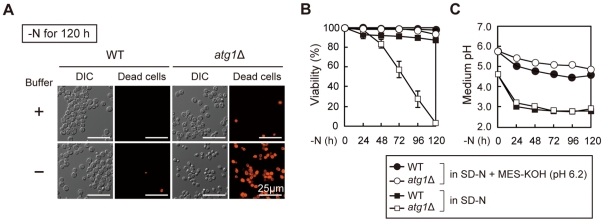
Autophagy is required for cell survival under conditions of nitrogen starvation in yeast [13], and to better understand the cellular events occurring during starvation we examined wild-type (WT) or autophagy-deficient atg1Δ cells during nitrogen starvation. Logarithmically growing atg1Δ cells were transferred to a synthetic medium without nitrogen sources (SD-N, referred to as non-buffered starvation medium), and their viability was monitored for five days using phloxine B, a reagent that stains dead cells. As previously reported, atg1 mutant cells exhibited decreased viability after two days of starvation, and almost completely atg1Δ cells died after five days (3.0% of survival cells). In contrast, 87.2% of WT cells were viable at that time (Figure 1A and 1B). The loss of viability was used as an initial screen to obtain atg mutants [13], but the cause of cell death in autophagy-defective mutants remained unclear. During these cultures, due to secreted metabolites, the pH of both the WT and atg1Δ starvation media decreased and reached a plateau below pH 3.0 (Figure 1C). We hypothesized that maintaining a neural pH could promote cell survival, and when we added MES-KOH (pH 6.2) to the starvation medium (hereafter referred to as buffered starvation medium), the extracellular pH was maintained near pH 5.0 after five days (Figure 1C). Under these conditions, 93.4% of atg1Δ cells remained viable after 120 hours of nitrogen starvation (Figure 1A and 1B), and similar rates of viability were seen for other atg mutants (atg2Δ, atg7Δ, atg11Δ, atg15Δ) (Figure 2C). To confirm that pH buffering was responsible for these observations, we used MES-NaOH (pH 6.2) as buffer, and it also maintained the viability of the autophagy-defective mutants (Figure S1). Additionally, MES-KOH (pH 5.0) slightly restored cell viability, but to a lower extent than MES-KOH (pH 6.2) (Figure S1). Cell viability in the non-buffered starvation medium containing KCl such that concentration of potassium ion is equivalent to that in MES-KOH (pH 6.2) buffering media did not improve (Figure S1). Thus, under conditions of nitrogen starvation, autophagy mutants are very sensitive to the extracellular pH, and more acidic medium leads to greater cell death.
Figure 1. The viability of atg mutants in buffered starvation medium.
(A) WT and atg1Δ cells grown in YEPD medium were transferred to SD-N or SD-N +50 mM MES-KOH (pH 6.2) medium for 120 hours, and dead cells stained by phloxine B were observed by fluorescent microscopy. Scale bar, 25 µm. (B–C) WT and atg1Δ cells grown nitrogen starved as (A) for the indicated times. Cell viability (B) and medium pH (C) were examined by phloxine B staining and pH meter, respectively. These data represent the average of three independent experiments and bars indicate standard deviations.
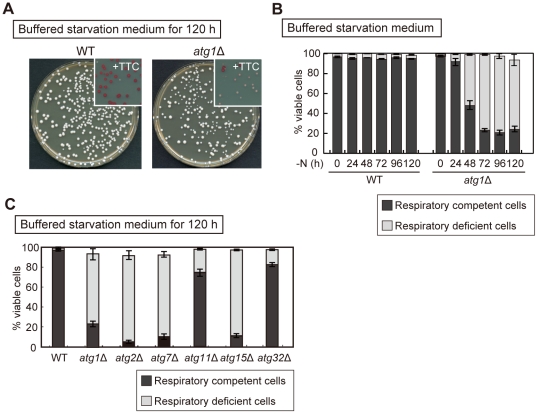
Figure 2. Loss of respiratory function in atg mutants during nitrogen starvation.
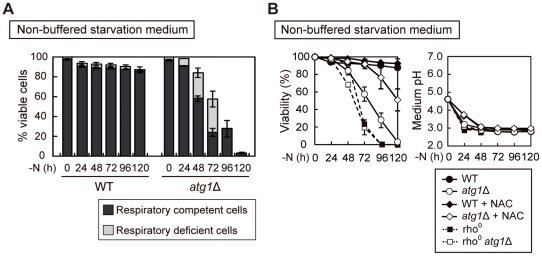
(A) WT and atg1Δ cells grown in YEPD medium were transferred to SD-N +50 mM MES-KOH (pH 6.2). After 120 hours, cultures were diluted 5.0×105 fold and plated onto YEPD agar. The plates were incubated at 30°C for four days. Insets indicate the plate overlaid with TTC agar to examine the respiratory competency of formed colonies. (B) WT, and atg1Δ cells were nitrogen-starved as (A). Cell viability and respiratory competency was determined by phloxine B staining and TTC overlay technique as (A), respectively. The black and gray areas indicate the percentage of viable cells that are respiratory competent or respiratory deficient, respectively. (C) WT, atg1Δ, atg2Δ, atg7Δ, atg11Δ, atg15Δ, and atg32Δ cells were nitrogen-starved as (A). Cell viability and respiratory competency was determined by phloxine B staining and TTC overlay technique as (A), respectively. The black and gray areas indicate the percentage of viable cells that are respiratory competent or respiratory deficient, respectively. These data represent the average of three independent experiments and bars indicate standard deviations.
Autophagy is required for the maintenance of respiratory function during starvation
Buffered starvation medium dramatically improves cell viability of atg mutants under long nitrogen starvation (Figure 1A and 1B). We determined the cell viability also by colony forming ability on YEPD plates and obtained similar results with phloxine B staining (data not shown). In the course of this experiment, we noticed that atg1Δ cells formed small colonies at high rate (Figure 2A). In many cases in yeast, small colonies reflect impaired respiratory function, so we examined the respiratory competency of the cells under starvation conditions using 2,3,5-triphenyltetrazolium chloride (TTC), which stains respiratory competent colonies red [14]. Most of the small colonies derived from atg1 mutants were not colored (Figure 2A), implying that autophagy-deficient cells lost their respiratory activity. To better understand the kinetics of this observation, we monitored the respiratory competency of atg1Δ cells by TTC staining. After 24 hours of nitrogen starvation, atg1Δ cells gradually lost their respiratory activity, and by 72 hours 75.5% of the atg1Δ cells lost their respiratory function, although only 5.0% of WT cells showed respiratory defect (Figure 2B). Other autophagy-defective mutants (atg2Δ, atg7Δ) that are unable to form autophagosomes lost respiratory function with similar kinetics (Figure 2C).
Autophagy plays a role to retain mtDNA during starvation
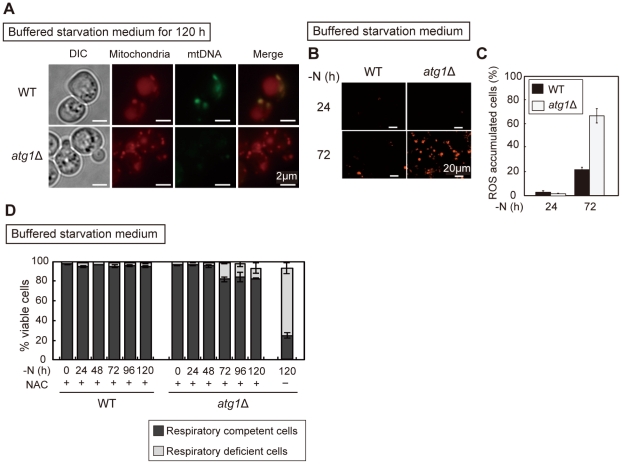
We next analyzed the mechanism by which autophagy-deficient cells lose their respiratory function. We examined if mitochondria retained mtDNA (mitochondria DNA) in atg mutant cells. Mitochondria were visualized by mitochondria targeted mCherry, and mtDNA was labeled with SYBR green. Both WT and atg1Δ cells contained mtDNA until 24 hours of nitrogen starvation (Figure S2). After 120 hours of starvation, the mitochondria in WT cells retained mtDNA, but nearly all of the atg1Δ cell lost their mtDNA (Figure 3A). We confirmed that the small colonies derived from culture grown in buffered starvation medium were comprised of rho0 cells, mitochondria DNA deficient cells, by DAPI staining (Figure S3), which stains both nuclear DNA and mtDNA. These results suggest that autophagy-defective mutants become rho0 cells during nitrogen starvation.
Figure 3. Mitochondrial defect in atg mutants.
(A) WT and atg1Δ cells expressing mitochondria targeted mCherry cultured in SD-N +50 mM MES-KOH (pH 6.2) medium for 120 hours were observed by fluorescent microscopy. Mitochondrial DNA was stained with SYBR green I, and mitochondria were visualized by mitochondria targeted mCherry. Scale bar, 2 µm. (B–C) WT and atg1Δ cells were transferred to SD-N +50 mM MES-KOH (pH 6.2) medium for the indicated time. ROS accumulation was detected by DHE staining (B). Each photo contains about 200 cells. Scale bar, 20 µm. (C) shows quantification of ROS accumulated cells (n>200 cells). (D) WT and atg1Δ cells were transferred to SD-N +50 mM MES-KOH (pH 6.2) with 10 mM NAC for the indicated time. Cell viability was determined by phloxine B staining. Cells from these cultures were plated on YEPD agar and overlaid with TTC agar to examine respiratory competency. The black and gray areas indicate the percentage of viable cells that are respiratory competent or respiratory deficient, respectively. These data represent the average of three independent experiments and bars indicate standard deviations.
ROS are a major factor contributing to the loss of respiratory function
ROS induce mutations in mtDNA, leading to the loss of respiratory function in mice [15]. Additionally, autophagy-deficient yeast cells accumulate ROS to a higher extent than WT cells during stationary phase [16]. We hypothesized that accumulated ROS cause loss of mtDNA in atg mutant cells, and investigated ROS accumulation during nitrogen starvation using dihydroethidium (DHE). When both WT and atg1Δ cells were transferred to buffered starvation medium, ROS levels transiently increased. After 72 hours, the atg1Δ cells (67.0% ROS accumulated cells) accumulated more ROS than WT cells (21.2% ROS accumulated cells) (Figure 3B and 3C). When the antioxidant N-acetylcysteine (NAC) was added to the starvation medium, ROS accumulation was suppressed, and the loss of respiratory function by atg1Δ cells was abrogated (Figure 3D). These results suggest that autophagy is required to prevent excessive ROS accumulation during starvation, and ROS accumulation impairs respiratory function in autophagy-defective mutants.
Inability to increase the expression of respiratory components and ROS scavenger enzyme led ROS accumulation
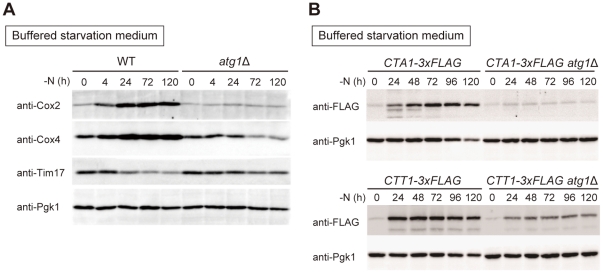
It is known that the electron transport system is the major source of ROS production during respiratory growth [15], [17], [18], so we examined the accumulation of ROS during starvation in rho0 atg1Δ cells. Compared to atg1Δ cells (67.0% ROS accumulated cells), there was little accumulation of ROS in rho0 atg1Δ cells (3.0% of ROS accumulated cells) (Figure S4A and S4B), implying that respiration is the major source of ROS in atg mutants during nitrogen starvation. Alterations in components of the respiratory chain that affect reaction efficiency promote electron transport to oxygen and the generation of ROS [15]. To probe the composition of the respiratory pathway, we examined the expression of Cox4, a subunit of cytochrome C oxidase encoded in the nuclear genome, and Cox2, another cytochrome C oxidase component encoded in the mtDNA [19]. Expression of both Cox2 and Cox4 was increased in WT cells in response to nitrogen starvation, but no significant increase in Cox2 or Cox4 expression occurred in atg1Δ cells (Figure 4A). It is likely that ROS generation is more pronounced in atg mutants because respiratory chain function deviates from optimum conditions. On the other hand, the amount of Tim17, a subunit of the translocase of the mitochondrial inner membrane, slightly decreased in both WT and atg1Δ cells during starvation (Figure 4A), suggesting that mitochondrial protein composition changes during starvation and expression of these newly synthesized proteins depends on intact autophagy.
Figure 4. Altered expression of respiratory components and ROS scavengers by atg mutant cells.
(A) WT and atg1Δ cells were transferred to SD-N +50 mM MES-KOH (pH 6.2) medium for the indicated time. Lysates were prepared using the alkaline-trichloroacetic acid method and subjected to immunoprecipitation with anti-Cox2, anti-Cox4, anti-Tim17 and anti-Pgk1. Pgk1 was used as loading control. (B) WT and atg1Δ cells expressing Cta1-3×FLAG or Ctt1-3×FLAG were nitrogen-starved as in (A) for the indicated time. Cell lysates were prepared as (A) and subjected to immune precipitation with anti-FLAG and anti-Pgk1. Pgk1 was used as loading control.
Several pathways have evolved to scavenge ROS, and we examined the expression of Cta1 and Ctt1, both of which catalyze the decomposition of ROS to water and oxygen [20], [21]. Expression of both Cta1 and Ctt1 was immediately increased upon shifting WT cells to nitrogen starvation medium, but expression of these scavenger proteins was not relatively increased in atg1Δ cells upon starvation (Figure 4B). Thus, the inability to increase the expression of respiratory chain components and ROS scavengers likely leads to the accumulation of ROS in autophagy-defective cells.
Vacuolar degradation by non-selective autophagy, rather than selective autophagy, is important for the maintenance of respiratory function
Autophagy is involved in not only the maintenance of amino acid levels but also cellular homeostasis by sequestration of aberrant organelles, therefore we assessed another possibility that sequestration of dysfunctional organelles by autophagy contributes for the maintenance of respiratory functions. Mitochondria is selectively degraded in the vacuoles via autophagy termed mitophagy [9], [10], and Atg32, a mitochondria-anchored protein, is essential for this process [11], [12]. However, a substantial fraction of atg32Δ cells retained their respiratory function after 120 hours of nitrogen starvation (Figure 2C), indicating that mitophagy is not significantly involved in the maintenance of respiratory function. Other selective types of autophagy such as the Cvt (cytoplasm to vacuole targeting) pathway and pexophagy as well as mitophagy require the Atg11 protein [22], but atg11Δ cells did not show respiratory defect. In contrast, atg15Δ cells, which cannot perform the intravacuolar disintegration of autophagic bodies [23], inner membrane-bound structures of autophagosomes, exhibited significant respiratory defect (Figure 2C). Taken together, vacuolar degradation of cytoplasmic components by non-selective autophagy, rather than selective autophagy, is crucial for the maintenance of respiratory function during nitrogen starvation.
Defective respiration leads to cell death in atg mutants
In atg mutants, the time course of the appearance of the respiratory-deficient cells in buffered starvation medium temporally corresponded to the loss of cell viability seen in non-buffered starvation medium. We monitored the respiratory activity of autophagy-defective mutants in non-buffered starvation medium, and after 96 hours of starvation nearly all of the surviving atg1Δ cells exhibited respiratory activity (Figure 5A), indicating that respiratory-deficient cells were selectively eliminated.
Figure 5. Viability of respiratory deficient cells during nitrogen starvation.
(A) WT and atg1Δ cells were transferred to non-buffered SD-N medium for the indicated time. Cell viability was determined by phloxine B staining. Nitrogen-starved cells were plated onto YEPD agar and overlaid with TTC agar to examine the respiratory competency of formed colonies. The black and gray areas indicate the percentage of viable cells that are respiratory competent or respiratory deficient, respectively. (B) WT, atg1Δ, rho0, and rho0 atg1Δ cells grown in YEPD medium were transferred to SD-N with or without 10 mM NAC for the indicated time. In the presence of NAC, medium pH was adjusted by using KOH. Cell viability and medium pH were examined by phloxine B staining and pH meter, respectively. These data represent the average of three independent experiments and bars indicate standard deviations.
We next determined whether the cell death observed in non-buffered starvation medium was due to the loss of respiratory function. The viability of respiratory-defective rho0 cells, which completely lack mtDNA, was monitored in non-buffered starvation medium (Figure 5B). After 72 hours of nitrogen starvation 57.6% of atg1Δ cells were viable, but only 22.8% of rho0 cells survived, though the pH of the media of both cultures decreased at the same rate (Figure 5B). We confirmed that autophagy is normally induced in rho0 cells (Figure S5A). When starved in the buffered starvation media, both rho0 and rho0 atg1Δ cells showed high viability (Figure S5B). In the presence of NAC, lethality of atg1Δ cells was rescued in non-buffered starvation medium (Figure 5B). NAC raises the medium pH slightly, but the extent of restored viability seems not to be explained by this buffering effect (See Figure S1). Taken together, these data suggest that respiratory deficient cells are exquisitely sensitive to low pH under conditions of nitrogen starvation compared with respiratory competent cells. Our data indicate that accumulated ROS in atg mutants impaired respiratory function, which renders cells sensitive to the low pH environment, leading to greater cell death in non-buffered starvation media.
Discussion
In this study, we showed links between impaired autophagy, mitochondrial injury and dysfunction, and, ultimately, cell death under conditions of nitrogen starvation.
We previously reported that autophagy-defective mutants cannot maintain the amino acid pool needed for protein synthesis, and this impairs the ability of cells to upregulate the expression of starvation-induced genes [4]. In the present study, we found proteins involved in cellular respiration encoded both chromosomally (i.e. Cox4) and by the mtDNA (i.e. Cox2) upregulated under starvation conditions (Figure 4A). The ROS scavenger proteins Cta1 and Ctt1 also augmented under such conditions and autophagy is required for upregulation of these starvation-induced proteins (Figure 4B), consequently atg mutants show the increased production of ROS and reduced ROS scavenging activity (Figure 3B and 3C), likely leading mitochondria dysfunction (Figure 2B). Consistently, treatment of cells with the antioxidant NAC suppressed the loss of respiratory activity and restored cell viability during starvation in atg mutants (Figure 3D and 5B). In this context, autophagy is essential for synthesis of minimal essential proteins to maintain mitochondrial function during nitrogen starvation.
Autophagy could contribute to the elimination of damaged mitochondria and, in this way, prevent the accumulation of ROS [8]. Mitochondria are non-selectively incorporated into autophagosomes during starvation [24], but they are also selectively degraded via mitophagy, in which Atg32 is requisite [11], [12]. However, atg32Δ cells showed nearly normal respiratory activity (Figure 2C), indicating that selective mitochondria elimination is not responsible for the observed respiratory defects caused in atg mutants under starvation conditions. On the other hand, cells lacking ATG15, which can form autophagosomes but not degrade the contents [23], exhibited respiratory deficient cells (Figure 2C). It is likely that sequestration of cytoplasmic components is not sufficient to retain respiratory function, but impaired autophagic degradation of cytoplasmic components is responsible for the maintenance of respiratory function.
In this study, we proposed that supply of amino acids via autophagy is important for the maintenance of respiratory function under starvation conditions. After 120 hours nitrogen starvation atg mutants exhibit respiratory defects dramatically (WT: 3.8%, atg1Δ: 75.5%) (Figure 2B). Recently, Zhang et al. presented that stationary phase atg mutant cells shows higher spontaneous petite frequencies up to 6% than about 3% in WT cells [16]. The total cellular amino acid levels in autophagy deficient cells drastically decrease under starvation conditions compared to those under nutrient rich conditions [4]. It is likely that amino acid pool in nitrogen starved atg mutant is smaller than that in stationary phase atg mutant. It might account for the difference between rates of respiratory deficient cells of Zhang's result and our result.
The mechanisms by which excess ROS can induce the loss of mtDNA in autophagy-defective mutants remain unclear. It is known that the loss of mtDNA is caused by inhibition of mtDNA replication during cell division [25]. Defect in mtDNA replication for several generations decreases inherited mtDNA number, and finally cells become rho0 mutants. However, nitrogen-starved cells are arrested at the first cell cycle [24], therefore a different unknown mechanism independent of cell proliferation must be involved.
We suggested that the maintenance of mitochondria function is important for cell survival during cell starvation. Recently, the relationship between autophagy and chronological life span had been discussed [8], [26], and Fabrizio et al. presented that deletion of genes involved in autophagy or mitochondria functions shorten life span in Saccharomyces cerevisiae [27]. Matecic et al. reported that raising medium pH to 6.0 also induces the extension of chronological life span in atg mutants [28]. Thus, essential factors for long-term survival and adaptation to starvation conditions have common features, and neutralizing medium suppress short life span or starvation induced cell death in atg mutants. Shorten chronological life span in autophagy-defective mutants may be at least partly attributed to mitochondria dysfunction.
We revealed that buffered starvation medium allows autophagy deficient cells to survive five days of nitrogen starvation (Figure 1A and 1B). It allows us to investigate autophagy during long nitrogen starvation. Until now, we observed that lipid droplets are transported to the vacuole in an autophagy dependent manner under prolonged starvation (manuscript in preparation). Thus, buffered starvation conditions will be useful to elucidate novel autophagic processes.
Autophagy has been implicated in a variety of essential cellular functions, and we have established an important role for autophagy in maintaining mitochondrial function by supporting essential protein synthesis, and mitochondria dysfunction is the major cause of starvation-induced cell death in autophagy-defective mutants. Further experiments in yeast will provide mechanistic insights into physiological roles of autophagy.
Materials and Methods
Yeast Strains and Media
The yeast strains used in this study are listed in Table S1. The mutant strains were generated using standard genetic and molecular biology techniques. To investigate nitrogen-starved cells, cells grown in rich medium (YEPD medium: 1% yeast extract, 2% peptone, and 2% glucose) were shifted to nitrogen starvation medium (SD-N medium: 0.17% yeast nitrogen base without amino acids and ammonium sulfate and 2% glucose). Yeast cells were grown in YEPD medium to an OD600 of 1.4 at 30°C. These cells were washed with distilled water, resuspended in SD-N medium and cultured for the indicated times at 30°C. The rho0 derivative was obtained by ethidium bromide treatment as described [25].
Microscopy and live cell imaging
Cells were observed using an inverted microscope (IX71 or IX81; Olympus) equipped with differential interference contrast optics, epifluorescence capabilities, 60× (UPlanApo 60×, NA: 1.40; Olympus), 100× (UPlanApo 100×, NA: 1.35; Olympus) or 150× (UApo N 150×, NA: 1.45; Olympus) objective lenses, a EM-CCD camera (ImagEM Enhanced; HAMAMATSU PHOTONICS) or a monochrome CCD camera (CoolSNAP HQ; Roper) or, and filter sets for mCherry, DHE staining, DAPI staining and SYBR-Green I staining. Images were captured using image acquisition and analysis software (AQUACOSMOS or MetaMorph 7.0r4; Molecular Devices).
Immunoblotting
Samples corresponding to 5.0×10−2 OD600 units of cells were separated by SDS-PAGE followed by the western blotting and immunodecoration. After treatment with enhanced chemiluminescence reagents, proteins were detected using a luminescent image analyzer (LAS-4000 mini; Fujifilm). Anti-Cox2, Anti-Cox4, and Anti-Tim17 were gifts from Dr. Endo, Nagoya University, Japan. To detect GFP, Flag, and Pgk1, anti-GFP antiserum (Roche), Anti-Flag antiserum (Sigma), and Anti-Pgk1 (Invitrogen) were used, respectively.
Determination of cell viability
Cells viability was determined by phloxine B (final concentration 5 µg/ml) staining. Cell cultures were examined by fluorescence microscopy with a blue filter. Brightly fluorescent cells were counted as dead cells. About 200 cells were counted at each time point.
Measurement of medium pH
Medium pH was measured using a pH electrode/meter (Horiba).
Triphenyltetrazolium chloride staining
Respiratory competency was determined by the TTC staining method. TTC agar overlay was carried out as described [14].
Detection of reactive oxygen species (ROS)
Reactive oxygen species were detected by DHE (dihydroethidium; final concentration 5 µg/ml) staining. Cells were incubated at 30°C for 20 min, washed with distilled water, resuspended in distilled water and examined by fluorescence microscopy with a blue filter.
Supporting Information
The viability of atg1 mutants in various starvation medium. atg1Δ cells grown in YEPD medium were transferred to the indicated starvation medium for the indicated time. Cell viability and medium pH were examined by phloxine B staining and pH meter, respectively. (○); atg1Δ cells in SD-N +50 mM MES-KOH (pH 6.2), (⧫); atg1Δ cells in SD-N +50 mM MES-NaOH (pH 6.2), (◊); atg1Δ cells in SD-N +50 mM MES-KOH (pH 5.0), (▪); atg1Δ cells in SD-N added 5 mM KCl to adjust at a same potassium concentration with MES-KOH (pH 6.2). These data represent the average of three independent experiments and bars indicate standard deviations.
(TIF)
Mitochondria DNA in nitrogen-starved atg mutants. WT and atg1Δ cells expressing mitochondria targeted mCherry cultured in SD-N +50 mM MES-KOH (pH 6.2) medium for 24 hours were observed by fluorescent microscopy. Mitochondrial DNA was stained with SYBR green I, and mitochondria were visualized by mitochondrial targeting mCherry. Scale bar, 5 µm.
(TIF)
DNA in cells derived from small size of colony. atg1Δ cells grown in YEPD medium were transferred to SD-N +50 mM MES-KOH (pH 6.2). After 120 hours, cultures were diluted 5.0×105 fold and plated onto YEPD agar. The plates were incubated at 30°C for four days. Cells derived from large or small size of colony culture in YEPD medium. Yeast cells were grown to an OD600 of 0.6 at 30°C and 2.5 µg/ml DAPI was added to the medium to detect both nuclear DNA and mtDNA. Before subjecting to microscopy cells were washed with distilled water, resuspended in distilled water and observed by fluorescence microscopy. Scale bar, 5 µm.
(TIF)
ROS generation in respiratory defective cells. (A–B) WT, atg1Δ, rho0, and rho0 atg1Δ cells were transferred to SD-N +50 mM MES-KOH (pH 6.2) medium for 72 hours. ROS accumulation was detected by DHE staining (A). Each photo contains about 200 cells. Scale bar, 20 µm. (B) Quantification of ROS accumulated cells (n>200 cells). This data represents the average of three independent experiments and bars indicate standard deviations.
(TIF)
Respiratory deficient cells during nitrogen starvation. (A) WT, atg1Δ, rho0, and rho0 atg1Δ cells expressing GFP-Atg8 were transferred to SD-N +50 mM MES-KOH (pH 6.2) medium for the indicated time. During autophagy process, GFP-Atg8 (depicted by arrow) is delivered to the vacuole, and hydrolyzed to generate free GFP (depicted by arrowhead). Generation of free GFP indicates transport of the marker to the vacuole. Lysates were prepared using a Multi-Beads Shocker (model MB601NIHS, Yasui Kikai Co. Osaka, Japan) and subjected to immunoprecipitation with anti-GFP and anti-Pgk1. Pgk1 was used as loading control. (B) WT, atg1Δ, rho0, and rho0 atg1Δ cells were transferred to SD-N or SD-N +50 mM MES-KOH (pH 6.2). Cell viability was examined by phloxine B. This data represents the average of three independent experiments and bars indicate standard deviations.
(TIF)
Yeast strains used in this study.
(PDF)
Acknowledgments
We thank the lab members of Y.O. for helpful discussions and technical support. We are grateful to Toshiya Endo (Nagoya University, Nagoya, Japan) for the gift of antibodies.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kornitzer D, Ciechanover A. Modes of regulation of ubiquitin-mediated protein degradation. J Cell Physiol. 2000;182:1–11. doi: 10.1002/(SICI)1097-4652(200001)182:1<1::AID-JCP1>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 2.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 3.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 4.Onodera J, Ohsumi Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem. 2005;280:31582–31586. doi: 10.1074/jbc.M506736200. [DOI] [PubMed] [Google Scholar]
- 5.Mizushima N. The pleiotropic role of autophagy: from protein metabolism to bactericide. Cell Death Differ. 2005;12:1535–1541. doi: 10.1038/sj.cdd.4401728. [DOI] [PubMed] [Google Scholar]
- 6.Kuma A, Mizushima N. Physiological role of autophagy as an intracellular recycling system: With an emphasis on nutrient metabolism. Semin Cell Dev Biol. 2010;21:683–690. doi: 10.1016/j.semcdb.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takeda K, Yoshida T, Kikuchi S, Nagao K, Kokubu A, et al. Synergistic roles of the proteasome and autophagy for mitochondrial maintenance and chronological lifespan in fission yeast. Proc Natl Acad Sci USA. 2010;107:3540–3545. doi: 10.1073/pnas.0911055107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lemasters JJ. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005;8:3–5. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- 10.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 12.Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 14.Ogur M, St John R, Nagai S. Tetrazolium overlay technique for population studies of respiration deficiency in yeast. Science. 1957;125:928–929. doi: 10.1126/science.125.3254.928. [DOI] [PubMed] [Google Scholar]
- 15.Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, et al. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320:661–664. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Qi H, Taylor R, Xu W, Liu LF, Jin S. The role of autophagy in mitochondria maintenance: characterization of mitochondrial functions in autophagy-deficient S. cerevisiae strains. Autophagy. 2007;3:337–346. doi: 10.4161/auto.4127. [DOI] [PubMed] [Google Scholar]
- 17.Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 18.Semenza GL. Oxygen-dependent regulation of mitochondrial respiration by hypoxia-inducible factor 1. Biochem J. 2007;405:1–9. doi: 10.1042/BJ20070389. [DOI] [PubMed] [Google Scholar]
- 19.Carr HS, Winge DR. Assembly of cytochrome c oxidase within the mitochondrion. Acc Chem Res. 2003;36:309–316. doi: 10.1021/ar0200807. [DOI] [PubMed] [Google Scholar]
- 20.Hartig A, Ruis H. Nucleotide sequence of the Saccharomyces cerevisiae CTT1 gene and deduced amino-acid sequence of yeast catalase T. Eur J Biochem. 1986;160:487–490. doi: 10.1111/j.1432-1033.1986.tb10065.x. [DOI] [PubMed] [Google Scholar]
- 21.Cohen G, Fessl F, Traczyk A, Rytka J, Ruis H. Isolation of the catalase A gene of Saccharomyces cerevisiae by complementation of the cta1 mutation. Mol Gen Genet. 1985;200:74–79. doi: 10.1007/BF00383315. [DOI] [PubMed] [Google Scholar]
- 22.Yorimitsu T, Klionsky DJ. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell. 2005;16:1593–1605. doi: 10.1091/mbc.E04-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Epple UD, Suriapranata I, Eskelinen EL, Thumm M. Aut5/Cvt17p, a putative lipase essential for disintegration of autophagic bodies inside the vacuole. J Bacteriol. 2001;183:5942–5955. doi: 10.1128/JB.183.20.5942-5955.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119:301–311. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall RM, Mattick JS, Nagley P, Cobon GS, Eastwood FW, et al. The action of structural analogues of ethidium bromide on the mitochondrial genome of yeast. Mol Biol Rep. 1977;3:443–449. doi: 10.1007/BF00808386. [DOI] [PubMed] [Google Scholar]
- 26.Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, et al. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–91. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 27.Fabrizio P, Hoon S, Shamalnasab M, Galbani A, Wei A, et al. Genome-wide screen in Saccharomyces cerevisiae identifies vacuolar protein sorting, autophagy, biosynthetic, and tRNA methylation genes involved in life span regulation. PLoS Genet. 2010;6:e1001024. doi: 10.1371/journal.pgen.1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matecic M, Smith DL, Pan X, Maqani N, Bekiranov S, et al. A microarray-based genetic screen for yeast chronological aging factors. PLoS Genet. 2010;6:e1000921. doi: 10.1371/journal.pgen.1000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The viability of atg1 mutants in various starvation medium. atg1Δ cells grown in YEPD medium were transferred to the indicated starvation medium for the indicated time. Cell viability and medium pH were examined by phloxine B staining and pH meter, respectively. (○); atg1Δ cells in SD-N +50 mM MES-KOH (pH 6.2), (⧫); atg1Δ cells in SD-N +50 mM MES-NaOH (pH 6.2), (◊); atg1Δ cells in SD-N +50 mM MES-KOH (pH 5.0), (▪); atg1Δ cells in SD-N added 5 mM KCl to adjust at a same potassium concentration with MES-KOH (pH 6.2). These data represent the average of three independent experiments and bars indicate standard deviations.
(TIF)
Mitochondria DNA in nitrogen-starved atg mutants. WT and atg1Δ cells expressing mitochondria targeted mCherry cultured in SD-N +50 mM MES-KOH (pH 6.2) medium for 24 hours were observed by fluorescent microscopy. Mitochondrial DNA was stained with SYBR green I, and mitochondria were visualized by mitochondrial targeting mCherry. Scale bar, 5 µm.
(TIF)
DNA in cells derived from small size of colony. atg1Δ cells grown in YEPD medium were transferred to SD-N +50 mM MES-KOH (pH 6.2). After 120 hours, cultures were diluted 5.0×105 fold and plated onto YEPD agar. The plates were incubated at 30°C for four days. Cells derived from large or small size of colony culture in YEPD medium. Yeast cells were grown to an OD600 of 0.6 at 30°C and 2.5 µg/ml DAPI was added to the medium to detect both nuclear DNA and mtDNA. Before subjecting to microscopy cells were washed with distilled water, resuspended in distilled water and observed by fluorescence microscopy. Scale bar, 5 µm.
(TIF)
ROS generation in respiratory defective cells. (A–B) WT, atg1Δ, rho0, and rho0 atg1Δ cells were transferred to SD-N +50 mM MES-KOH (pH 6.2) medium for 72 hours. ROS accumulation was detected by DHE staining (A). Each photo contains about 200 cells. Scale bar, 20 µm. (B) Quantification of ROS accumulated cells (n>200 cells). This data represents the average of three independent experiments and bars indicate standard deviations.
(TIF)
Respiratory deficient cells during nitrogen starvation. (A) WT, atg1Δ, rho0, and rho0 atg1Δ cells expressing GFP-Atg8 were transferred to SD-N +50 mM MES-KOH (pH 6.2) medium for the indicated time. During autophagy process, GFP-Atg8 (depicted by arrow) is delivered to the vacuole, and hydrolyzed to generate free GFP (depicted by arrowhead). Generation of free GFP indicates transport of the marker to the vacuole. Lysates were prepared using a Multi-Beads Shocker (model MB601NIHS, Yasui Kikai Co. Osaka, Japan) and subjected to immunoprecipitation with anti-GFP and anti-Pgk1. Pgk1 was used as loading control. (B) WT, atg1Δ, rho0, and rho0 atg1Δ cells were transferred to SD-N or SD-N +50 mM MES-KOH (pH 6.2). Cell viability was examined by phloxine B. This data represents the average of three independent experiments and bars indicate standard deviations.
(TIF)
Yeast strains used in this study.
(PDF)