Abstract
Protein phosphatases, in coordination with protein kinases, play crucial roles in regulation of signaling pathways. To identify protein tyrosine phosphatases (PTPs) and serine–threonine (ser–thr) phosphatases in the Strongylocentrotus purpuratus genome, 179 annotated sequences were studied (122 PTPs, 57 ser–thr phosphatases). Sequence analysis identified 91 phosphatases (33 conventional PTPs, 31 dual specificity phosphatases, 1 Class III Cysteine-based PTP, 1 Asp-based PTP, and 25 ser–thr phosphatases). Using catalytic sites, levels of conservation and constraint in amino acid sequence were examined. Nine of 25 receptor PTPs (RPTPs) corresponded to human, nematode, or fly homologues. Domain structure revealed that sea urchin-specific RPTPs including two, PTPRLec and PTPRscav, may act in immune defense. Embryonic transcription of each phosphatase was recorded from a high-density oligonucleotide tiling microarray experiment. Most RPTPs are expressed at very low levels, whereas nonreceptor PTPs (NRPTPs) are generally expressed at moderate levels. High expression was detected in MAP kinase phosphatases (MKPs) and numerous ser–thr phosphatases. For several expressed NRPTPs, MKPs, and ser–thr phosphatases, morpholino antisense-mediated knockdowns were performed and phenotypes obtained. Finally, to assess roles of annotated phosphatases in endomesoderm formation, a literature review of phosphatase functions in model organisms was superimposed on sea urchin developmental pathways to predict areas of functional activity.
Keywords: Phosphatase, PTP, DSP, MKP, Serine-threonine phosphatase, PPP, PPM, Genome, Strongylocentrotus, Urchin
Introduction
Although often overlooked, the protein tyrosine and serine–threonine (ser–thr) phosphatases play critical roles in regulation of numerous cellular activities. They control cell receptors, signaling cascades, cytoplasmic regulatory proteins, transcription factors, posttranslational processes, and, in some instances, even act as switches between signaling pathways, activating one while inhibiting another. Most phosphatases regulate these processes by dephosphorylating specific substrate proteins (enzymes, regulatory proteins, receptors, transcription factors, etc.), generally counteracting the effects of kinases, proteins that phosphorylate substrates. Often, dephosphorylation inactivates the substrate, although, in other cases, the substrate is activated. Some phosphatases perform functions in addition to dephosphorylation. For example, there are phosphatases, such as DUSP11 (SHP2), that also act as adaptor proteins, linking components in a signal transduction pathway (Gomperts et al., 2003). Other phosphatases have completely lost the ability to dephosphorylate proteins and, instead, perform novel functions. In humans, about half of the myotubularin related protein tyrosine phosphatases (MTMRs) are catalytically inactive. Instead of dephosphorylating proteins, they are thought to modulate activities of catalytically active MTMRs by dimerizing with them (Laporte et al., 2003).
Protein phosphorylation and dephosphorylation generally occurs at tyrosine, serine, or threonine residues. Proteins that dephosphorylate tyrosine residues are called protein tyrosine phosphatases (PTPs) whereas those that act on serine or threonine sites are called serine–threonine (ser–thr) phosphatases.
This activity is dependent on the type of catalytic domain present. PTPs are categorized together because they all contain an invariant motif, CxxxxxR (CX5R), which forms the active site of their catalytic domains. These are monomeric proteins and well-known members include the receptor protein tyrosine phosphatases (RPTPs), nonreceptor protein tyrosine phosphatases (NRPTPs), and dual specificity phosphatases (DSPs). DSPs differ notably from other PTPs in that they dephosphorylate both serine/threonine and tyrosine sites.
There are three families of ser–thr phosphatases: the PPPs (Phosphoprotein phosphatases), the PPMs (Phosphoprotein phosphatases activated by magnesium), and the FCPs (TFIIF stimulated CTD phosphatases). PPPs are categorized together, in part, because each has a single conserved PP2A catalytic domain (PP2Ac domain). These polymeric proteins are activated when a single catalytic subunit interacts with one or more regulatory subunits. Identity of the regulatory subunit influences when or where the catalytic subunit acts. By utilizing different regulatory partners, one catalytic subunit can perform numerous cellular activities.
The other ser–thr phosphatases are monomeric proteins. PPMs are a fairly large family that contain a PP2C catalytic domain (PP2Cc) and depend on the presence of magnesium or manganese for catalytic activity. The FCPs are a minor group. They contain the signature motif of phosphotransferases and phosphohydrolases, DXDX(T/V), but are otherwise highly divergent from the PPPs and PPMs.
Since most protein phosphatases were discovered within the last 20 years, much remains to be learned about roles they play and how they are regulated. Initially, identifying phosphatase functions was quite difficult. Many early experiments relied on chemical inhibitors such as okadaic acid (a ser–thr inhibitor), calyculin (a ser–thr inhibitor), and sodium orthovanadate (a PTP inhibitor). These inhibitor experiments gave investigators a general idea about which types of phosphatases were acting, but not which specific phosphatase or pathway was affected. Other attempts to block single phosphatases were thwarted by the fact that more than one phosphatase acted on the substrate. In these instances, it was necessary to block several phosphatases to see an effect. Also, if catalytic activity was controlled by regulatory subunits, knocking out the catalytic subunit affected several cell processes simultaneously, again making it difficult to identify the substrate or cell signaling pathway. As genomic data become available and, as more is learned about phosphatase functions in different model systems, it has become much easier to study specific roles of phosphatases in developmental processes.
This paper describes protein tyrosine and ser–thr phosphatases that were identified in silico using the Strongylocentrotus purpuratus genome. Expression of these proteins in the sea urchin embryo is also evaluated using high density oligonucleotide tiling microarrays and EST databases. Finally, several of the identified phosphatases were blocked in developing sea urchins using morpholino antisense oligonucleotide injection and their phenotypes are described.
One process that has been thoroughly investigated by the sea urchin community is endomesoderm specification. Although much is now known about this process, little has been reported about roles of phosphatases in regulation of sea urchin endomesodermal pathways. An additional goal of this paper is to identify potential functions of S. purpuratus protein phosphatases in endomesoderm formation. This should be a valuable resource to those wishing to study phosphatase function in development.
Materials and methods
Searching for phosphatase domains in the sea urchin genome
To find putative phosphatases in the sea urchin genome, sequences of known chordate phosphatases were obtained from NCBI Entrez Protein site (http://www.ncbi.nlm.nih.gov/) and blasted against Baylor’s Human Genome Sequencing Center Strongylocentrotus purpuratus BLAST site (http://www.hgsc.bcm.tmc.edu/blast/blast.cgi?organism=Spurpuratus). The Baylor site retrieved matching sequences from the genome of a single urchin (compiled by the Baylor gene assembly team). Information concerning assembly of the genome appears in Weinstock and The Sea Urchin Genome Sequencing Consortium (2006). If retrieved sequence E values were 10−30 or less, identity of the urchin sequence was confirmed by reciprocal comparison against the non-redundant NCBI BLASTP site (http://www.ncbi.nlm.nih.gov/BLAST/). Throughout this paper, this process will be referred to as “back blasting.” Putative urchin phosphatase sequences were then archived in the Baylor College of Medicine Sea Urchin Genome Project site (http://annotation.hgsc.bcm.tmc.edu/Urchin). To identify the seven genes reported in Wessel et al. (1995), their deduced amino acid sequences were blasted against Baylor’s Strongylocentrotus purpuratus BLAST site.
The domain detection sites Pfam (http://www.sanger.ac.uk/Sortware/Pfam/), NCBI (http://www.ncbi.nlm.nih.gov/BLAST/), and SMART (http://smart.embl-heidelberg.de/smart/set_mode.cgi?NORMAL=1) were used to learn about structure of putative phosphatases. To find missing portions of sequences: A) Tiling data were examined to determine whether the gene was expressed in areas of the scaffold missing in the annotated sequence, B) Genboree (http://www.genboree.org/java-bin/login.jsp) was used to find alternate sequence predictions for the gene based on contig data from sites such as the NIDCR S. purpuratus Genome Search page (http://urchin.nidcr.nih.gov/blast/index.html) or NCBI, C) Sequences were checked for overlapping identical regions, D) PTPR D1 and D2 trees were compared to identify the second catalytic domain, and E) back blasting results not used in phylogenetic analyses were reexamined. In all cases, orientation of genes in scaffolds was considered and data were checked to ascertain that both parts of the gene were on the same DNA strand (positive or negative). Transmembrane domains were detected at TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) and the Mouse Genome Informatics site (http://www.informatics.jex.org/) was useful for determining alternate phosphatase names.
Evaluating protein expression
Tiling array data generated for the S. purpuratus genome project (Samanta et al., 2006) were examined. In each case, the raw data were evaluated and peak heights were measured for each predicted exon. Quantitative values were calculated for each gene by averaging all exon peaks when there were 5 peaks or less. When 6 or more peaks were present, low and high values were eliminated and remaining peaks were averaged. Averages were graded as no expression (<3), very low expression (3–4.99), low expression (5–9.99), moderate expression (10–29.99), or high expression (>30).
EST data were used to find expressed sequences undetected by the tiling array. If EST data for an annotated gene appeared in Geneboree, the gene was considered expressed even if tiling data were inconclusive. Also, annotated sequences were blasted against all sea urchin ESTs at the NCBI BLAST Sea Urchin Sequences site (http://www.ncbi.nlm.nih.gov/genome/seq/BlastGen/BlastGen.cgi?taxid=7668). Annotated sequences that matched an EST sequence were back blasted to the Baylor S. purpuratus Blast site. Sequences with E values of zero were accepted as expressed.
Finding human and/or invertebrate homologues
Representative phosphatase sequences for homologue analysis were collected from several sources. Vertebrate, Drosophila melanogaster, and Cae-norhabditis elegans RPTPs and NRPTPs were downloaded from the Novo Nordisk Science protein tyrosine phosphatase database (http://ptp.cshl.edu or http://science.novonordisk.com/ptp) while sequences for other phosphatases were found in GenBank (http://www.ncbi.nih.gov/entrez/query.fcgi?db=Protein), UniProt (http://www.pir.uniprot.org/), or Ensembl (http://www.ensembl.org/index.html). Potential cnidarian (sea anemone) homologues were obtained by domain searches of the Nemtostella vectensis genome at http://www.stellabase.org. (Accession numbers for non-sea urchin phosphatases used in these analyses appear in Suppl. 9). Sequences were aligned using ClustalX. Conserved regions were selected in McClade, and classification of the phosphatases by phylogenetic analysis was performed in PAUP. Rooted or unrooted neighbor-joining trees were generated, followed by bootstrap analysis of the data. Two thousand repetitions were used for bootstrap analysis.
For the RPTP analysis, most sequences were partial. Therefore phylogenetic comparison was limited to PTPc domain 1 (D1) or PTPc domain 2 (D2). Since PTPcD1 and D2 sequences segregate in neighbor joining trees, it was easy to distinguish S. purpuratus D1 sequences from D2 sequences by running them with known HsPTPc D1 and HsPTPc D2 sequences (not shown). Using this information, separate PTPc D1 and D2 trees were produced. The RPTP D2 neighbor-joining tree was used to confirm phylogenetic relationships observed in the D1 tree. Similarity of sea urchin RPTP D1 domains to those of other species was evaluated by neighbor-joining, maximum likelihood, maximum parsimony (not shown), and Bayesian analysis.
When duplications appeared in a tree, pairwise comparison of the genes was performed (http://www.ncbi.nlm.nih.gov/blast/bl2seq/wblast2.cgi) to ensure that they were not identical. Genboree was also used to detect null sequences (artifacts of genome sequencing), to compare locations of overlapping sequences in different scaffolds (to determine whether two sequences were complementary ends of the same protein), and to compare exon/intron boundaries (and neighboring sequences) when searching for haplotype pairs.
Determining potential functions of identified sea urchin phosphatases
To identify potential functions of annotated S. purpuratus phosphatases in endomesoderm formation, a list of sea urchin endomesodermal signaling pathways (and components that comprise these pathways) was generated. In addition, a list of genes known to affect sea urchin endomesoderm formation was obtained from the Endomesoderm Network site (http://sugp.caltech.edu/endomes/). Using Pubmed (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi) and Endnote, activities of phosphatases in endomesodermal signaling pathways were collected and tabulated.
Design and microinjection of anti-phosphatase MASOs
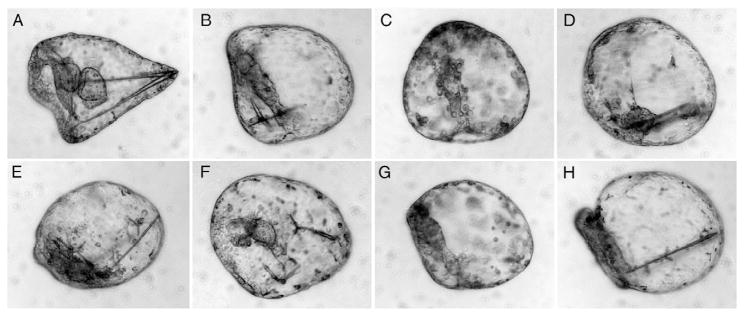
A list of phosphatase sequences from mouse was compiled and used to perform BLAST searches of S. purpuratus expressed sequence tags (ESTs) to identify phosphatases expressed in the embryo. ESTs encoding N-terminal protein sequences (based on similarity to the N-termini of mouse phosphatases) were then used to obtain morpholino antisense oligonucleotides (MASOs) from Genetools, LLC (Corvallis, OR). In all cases except one, the MASO sequences were targeted to the translation initiation site of respective mRNAs; the one exception is DUSP1/2/4/5, for which the GLEAN3 model was used to design a splice-blocking MASO that targets the first exon-intron junction (Table 1). The anti-phosphatase MASOs as well as a standard non-specific control MASO (Genetools) were initially injected by standard methods into fertilized eggs, at three different concentrations (injection solutions containing 200, 400, or 800 μM MASO and 120 mM KCl). Injected embryos developed at 15 °C, with periodic observation and notation of their phenotypes. Additional microinjections of several MASOs were subsequently carried out at the most effective dose for each MASO, and digital light micrographs were obtained on a Zeiss Axiovert 20 microscope equipped with an Axiocam MRm. Images were processed in Photoshop.
Table 1.
A survey of the Class I Cysteine-based conventional PTPs in the S. purpuratus genome and their predicted expression based on tiling and EST data
| Category | PTP gene | Alternative names | Urchin gene | Annotated ID | Tiling data | EST data |
|---|---|---|---|---|---|---|
| PTPs | ||||||
| A. Class I Cys-Based PTPs | ||||||
| A. 1. Conventional PTPs | ||||||
| A. 1.1. Receptor PTPs | ||||||
| R1/6 | PTPRC | CD45, LCA | None | |||
| R2A | PTPRD | RPTP-delta | PTPR D/F/S | SPU_020050 | Moderate (11.6) | No |
| PTPRF | LAR | |||||
| PTPRS | RPTP-sigma, PTPNU3, CRYPalpha | |||||
| R2B | PTPRK | RPTP-kappa | None | |||
| PTPRM | RPTP-mu | |||||
| PTPRT | RPTP-rho | |||||
| PTPRU | PTP-psi, PTP-lambda, PTP-omicron | |||||
| R3 | PTPRB | PTP-beta | PTPRH-like1 | SPU_016937 | Low (6.6) | No |
| PTPRH | SAP 1, PTPBEM2 | PTPRH-like2 | SPU_028717 | Very Low (4.8) | No | |
| PTPRJ | CD148; DEP1; RPTP-eta; PTPBYP | PTPRscav | SPU_004193/005860 | Very Low (4.5) | No | |
| PTPRO | PTP-phi, PTP-BK, PTPBEM1, PTPoc, PTPcryp2 | PTPRO-like | SPU_001461 | Very Low (3.8) | Yes | |
| PTPRQ | PTPS31, PTPGMC | |||||
| PTPRV | OST-PTP, Esp, OST | |||||
| R4 | PTPRA | RPTP-alpha | None | |||
| PTPRE | RPTP-epsilon | |||||
| R5 | PTPRG | RPTP-gamma | PTPR G/Z1 | SPU_011335 | No | No |
| PTPRZ | RPTP-zeta | PTPR G/Z2 | SPU_021599 | Moderate (12.4) | Yes | |
| DmPTP99A | ||||||
| R7 | PTPRR | PC12-PTP1, PCPTP, PCPTP1, PTPBR7, PTP-SL | PTPRR | SPU_008670 | Very Low (4.4) | No |
| PTPN5 | STEP, STEP61 | |||||
| PTPN7 | HePTP, LCPTP, BPTP-4, C920001D21Rik | |||||
| R8 | PTPRN | IA-2, Islet cell antigen 512 | None | |||
| PTPRN2 | IA-2β, Phogrin, PTPRP, RPTP-pi, PTPNP, acPTPIA2beta | |||||
| R9 | DmPTP69D-like | PTP69D-likc | SPU_027101 | Low (5.7) | No | |
| Urchin Specific Group 1 | PTPRFn1 | SPU_016143/016144 | Very Low (4.3) | Yes | ||
| PTPRFn2 | SPU_027014/003581* | No | Yes | |||
| PTPRLec1 | SPU_019852 | No | No | |||
| PTPRLec2 | SPU_022839 | Very Low (3.4) | Yes | |||
| PTPRLec3 | SPU_020604 | Very Low (3.9) | Yes | |||
| PTPRLec4 | SPU_016053 | No | No | |||
| PTPRLec5 | SPU_023162 | No | No | |||
| PTPRLec6 | SPU_024820 | No | No | |||
| Urchin Specific Group 2 | PTPRY1 | SPU_015923 | Very low (4) | No | ||
| PTPRY2 | SPU_020542 | Low (6.12) | No | |||
| Urchin Specific Group 3 | PTPRW1 | SPU_027290/025766* | No | No | ||
| PTPRW2 | SPU_024074 | No | No | |||
| Miscellaneous Urchin RPTPs | PTPRiz | SPU_023115 | Very Low (3.3) | Yes | ||
| PTPRorph1 | SPU_016411 | Low (6.5) | Yes | |||
| PTPRorph2 | SPU_008466 | Moderate (12.1) | No | |||
| PTPRY3 | SPU_028575 | Very Low (3.2) | Yes | |||
| A. 1.2. Non-receptor PTPs | ||||||
| NT1 | PTPN1 | PTPIB | PTPN1/2 | SPU_020281 | Moderate (23) | Yes |
| PTPN2 | MPTP, PTP-S, TCPTP | |||||
| NT2 | PTPN6 | HCP, PTP1C, SHP1, SH-PTP1 | PTPN6/11 | SPU_013810 | Moderate (14.6) | Yes |
| PTPN11 | PTP1D, PTP2C, SH-PTP3, SHP2, SH-PTP2, Syp | |||||
| NT3 | PTPN9 | PTP-MEG2 | PTPN9 | SPU_019984/000776 | Moderate (21) | Yes |
| NT4 | PTPN12 | PTPG1, PTP-P19, PTP-PEST, RKPTP | PTPN 12/18/22 | SPU_019920 | Moderate (16) | No |
| PTPN18 | BDP, PTP20, PTP-HSCF, PTPK1 | |||||
| PTPN22 | LYP, LyPTP, PEP | |||||
| NT5 | PTPN3 | PTPH1 | PTPN3/4 | SPU_005885 | Low (7) | No |
| PTPN4 | PTP-MEG1, TEP, PTPtep | |||||
| NT6 | PTPN14 | PEZ, PTP36, PTPD2 | PTPN 14/21 | SPU_028592 | Moderate (15.9) | Yes |
| PTPN21 | PTP2E, PTPD1, PTP-RL10 | |||||
| NT7 | PTPN13 | FAP-1, PTP1E, PTP-BAS, PTP-BL, PTPL 1, RIP, PTPBA14 | PTPN13 | SPU_022501 | Moderate (15) | No |
| NT8 | PTPN20 | TypPTP; PTPTyp | None | |||
| NT9 | PTPN23 | DKFZP564F0923, HD-PTP, HDPTP, KIAA 1471, PTP-TD14 | PTPN23 | SPU_007282 | Moderate (10.8) | No |
Human PTPs and a few Drosophila PTPs (indicated by Dm) are shown under “PTP Gene” and S. purpuratus homologues or novel genes appear under “Urchin Gene”. Expression data are presented as well. Yellow highlighted genes are novel. Blue areas indicated genes that are homologous or very similar to human genes. MASOs shown in red block development. Note. Comparison of exon/intron boundaries suggests that none of the duplicate forms are haplotype pairs.
Second domain is hypothesized based on A) clade formed in D2 tree, B) found on the same DNA strand, C) orientation of sequences at ends of scaffolds.
Results and discussion
Most human PTPs and ser–thr phosphatases have S. purpuratus homologues
The protein tyrosine phosphatases
There are four PTP superfamily classes: Class I Cysteine-based PTPs, Class II Cysteine-based PTPs, Class III Cysteine-based PTPs, and Asp-based PTPs (Alonso et al., 2004b). These proteins are categorized as PTPs because they all contain an invariant motif, CxxxxxR (CX5R), which forms the active site of the catalytic domain.
Almost all PTPs are Class I Cysteine-Based PTPs, a group that can be subdivided into two categories: conventional PTPs (which dephosphorylate at tyrosine residues) and dual specificity protein phosphatases or VH1-like phosphatases (which primarily dephosphorylate at tyrosine and/or nearby serine/threonine residues).
Class II and III Cysteine-based PTPs are classified separately from Class I Cysteine-based PTPs because they evolved from distinct lineages. Class II Cysteine-based PTPs are closely related to bacterial arsenate reductases while Class III Cysteine-based PTPs are derived from bacterial rhodanese-like enzymes. Both of these classes are small. Humans, for example, have only one Class II Cysteine-based PTP and 3 Class III Cysteine-based PTPs.
Asp-based PTPs utilize a distinct catalytic mechanism from other PTPs. Class I, II, and III Cysteine-based PTPs, utilize cysteine as the nucleophile in the catalytic domain and form thiol–phosphate intermediates during substrate dephosphorylation (Alonso et al., 2004b). Asp-based PTPs differ in that their catalytic reactions are metal-dependent and rely on a nucleophilic aspartic acid to form a phospho-aspartate intermediate (Rebay et al., 2005). Like Class II and III Cysteine-based PTPs, Asp-based PTPs only have a few members.
Class I cysteine-based PTPs
Conventional PTPs
The conventional PTPs include two groups: receptor protein tyrosine phosphatases (RPTPs), which are membrane-associated proteins (with the exception of a few splice variants), and nonreceptor protein tyrosine phosphatases (NRPTPs), which are cytoplasmic. Most data about these phosphatases come from studies of human proteins.
Structurally, RPTPs contain an intracellular and extra-cellular region. Intracellular portions of the protein tend to be highly conserved due to the fact that the PTP catalytic (PTPc) domain is intracellular. All RPTPs have either one or two PTPc domains. In cases where two PTPc domains are present, the domain closest to the cell membrane, PTPc D1 (D1), dephosphorylates substrate proteins while the distal domain, PTPc D2 (D2), is thought to aid in aligning the RPTP to substrate proteins. Of these, D1 is more highly conserved and is retained in RPTPs that have only one PTPc domain.
Extracellular portions of RPTPs vary quite a bit. In humans, there are at least 23 different RPTPs and most of this diversity is due to variation in extracellular domains. Functions of these domains are less well understood. In some cases, they may influence which extracellular signals the RPTP responds to. In other cases, they allow the protein to perform functions independent of substrate dephosphorylation. Extracellular domains found in human RPTPs include fibronectin 3 repeats (FN3), immunoglobulin (Ig) sequences, glycosylation sites, and MAM domains (Fig. 1).
Fig. 1.
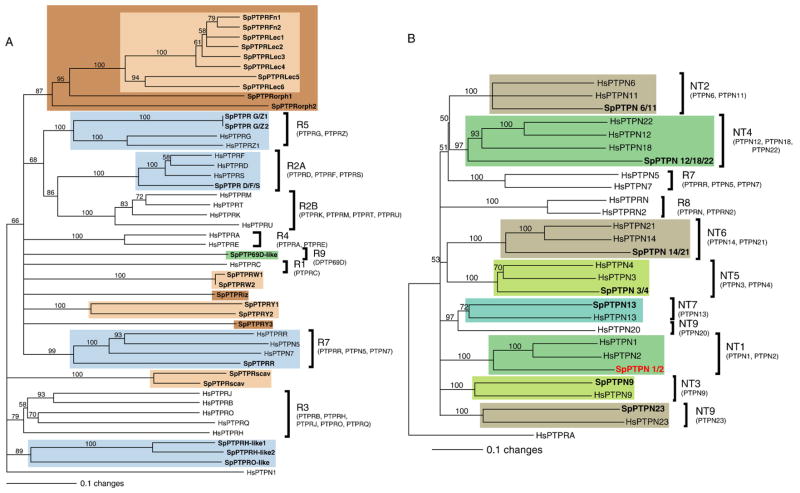
A) Phylogenetic relationships of S. purpuratus RPTPs to human forms. In this neighbor-joining tree (generated using sequences that align with residues 226–503 of HsPTPRA; outgroup=HsPTPN1), the light blue clades contain S. purpuratus homologues of human RPTPs or human-like RPTPs, the green clade contains a S. purpuratus RPTP homologous to invertebrate RPTPs, and the tan or brown clades are orphan S. purpuratus RPTPs. RPTP subtypes are indicated at right edge. In all trees, SpPTPRG/Z1 and SpPTPRG/Z2 are in the R5 RPTP clade, and SpPTPR D/F/S clusters with the R2A RPTPs. SpPTPRH-like1, SpPTPRH-like2, SpPTPRO-like, and SpPTPRscav group near the R3 subgroup, but are only weakly similar to this subgroup (see Suppl. 1 and 2). Three back blast to PTPRO or PTPRH, but, if they are R3 PTPRs, they are probably divergent. In all 3 trees, SpPTPRR is a member of the R7 clade (PTPRR, PTPN5, and PTPN7). One other phosphatase, SpPTP69D-like, forms a clade with the invertebrate specific Type R9. This is weakly supported and probably divergent. A tree generated using RPTP D2 sequences appears in Suppl. 3. B) Phylogenetic relationships of S. purpuratus NRPTPs to human forms. In this neighbor-joining tree (generated using sequences that align with residues 31–279 of HsPTPN1), highlighted clades contain S. purpuratus homologues of human NRPTPs. NRPTP and RPTP subtypes are indicated at right edge. In both trees bootstrap values are shown at the dendogram node (maximum=100) and the horizontal distance indicates percent divergence (0.1 scale=10 substitution events per 100 amino acids). MASOs were produced and tested for SpPTPN 1/2, shown in red. Sp=Strongylocentrotus purpuratus, Hs=Homo sapiens.
RPTPs have been implicated in numerous processes including the regulation of density dependent growth (PTPRJ and possibly PTPRB and PTPRO; Schaapveld et al., 1997), regulation of adherens junction complexes (PTPRF, PTPRK, PTPRM; Wadham et al., 2003), regulation of axon guidance in motor neurons (DLAR, DPTP10D, and DPTP99A; Desai et al., 1997), and neuronal differentiation (PTPRA; Hertog et al., 1999).
Phylogenetic studies have clearly shown that NRPTPs share a common ancestor with the RPTPs. These phosphatases only have one PTPc domain (similar to PTPc D1) and, like the RPTPs, they often have additional domains. These other domains can aid in cellular localization of the NRPTP (Ex: PEST domains target activity to the cell membrane/cytoskeletal interface and SEC14 domains target activity to the perinuclear region), regulation of substrate access (Ex: SH2 domains), or other protein–protein interactions. Domain types that have been found in human NRPTPs include PEST, FERM, SH2, SH3, BRO1, KIND, PDZ, B41, and SEC14 domains (Andersen et al., 2001; Bhaduri and Sowdhamini, 2003; Alonso et al., 2004b; Gandhi et al., 2005). Some of the known functions of NRPTPs include regulation of Ha–Ras dependent cell growth (PTPN23; Cao et al., 1998), coupling of cell protrusion and retraction during cell migration (PTPN12; Sastry et al., 2006), blocking apoptosis (PTPN13; Foehr et al., 2005; Ivanov et al., 2006), promoting vesicle fusion (PTPN9; Huynh et al., 2004), inhibiting signal transduction by Src family kinases (PTPN18; Wang et al., 2001), and promoting cell migration while reducing cell–cell adhesion by dephosphorylating β-catenin (PTPN14 and PTPN1; Wadham et al., 2003).
Based on domain structure, protein function, and homologue analysis of conventional PTPs, Andersen et al. (2001, 2005) concluded that there are 10 types of RPTPs (R1/6, R2A, R2B, R3, R4, R5, R6, R7, R8, and R9) and 9 types of NRPTPs (NT1–NT9). With the exception of Type R9 RPTPs (found in Drosophila), these are all present in humans (See representatives of the RPTP categories in Fig. 4.) (For reviews see Brady-Kalnay and Tonks, 1995; Schaapveld et al., 1997; Neel and Tonks, 1997; Barford et al., 1998; Stoker and Dutta, 1998; Hertog et al., 1999; Andersen et al., 2001; Bhaduri and Sowdhamini, 2003; Alonso et al., 2004b; Gandhi et al., 2005). Although this classification was initially based on structural similarities, phylogenetic analysis using PTPc D1 sequences has shown that extracellular domain structure may be a poor predictor of relatedness (Andersen et al., 2001, 2005). In fact, two of the RPTP clades, R7 and R8, are probably more closely related to the NRPTPs than to the RPTPs.
Fig. 4.
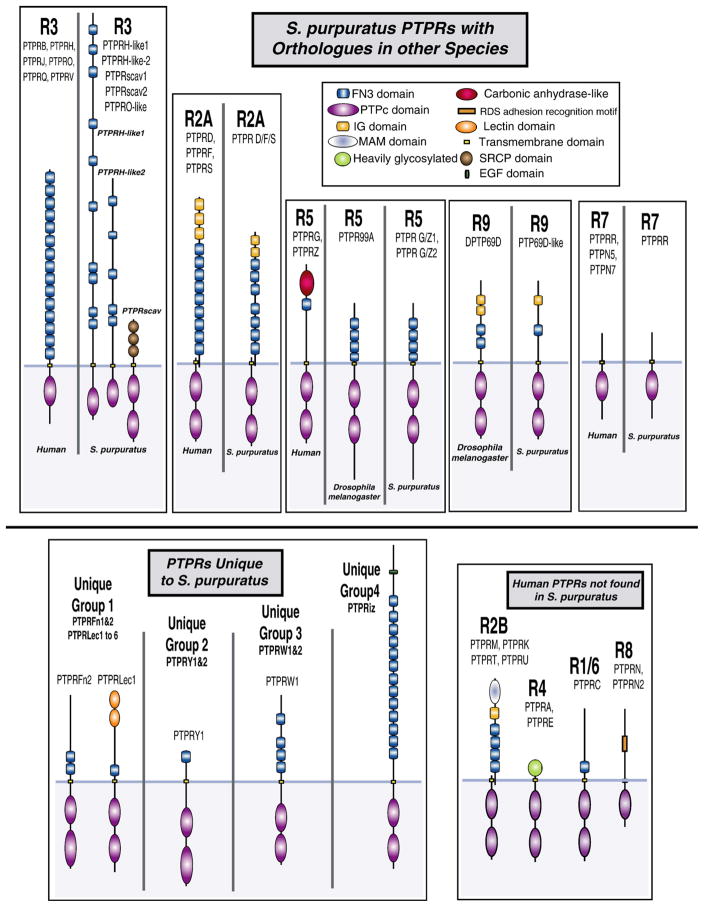
Hypothesized domain structures of S. purpuratus RPTP homologues compared to known human and Drosophila RPTPs. Predictions were based on: (A) results of Pfam, NCBI, and SMART domain searches, (B) known domain structures of human, Drosophila, and C. elegans RPTPs, (C) comparison of partial S. purpuratus sequences from the same clade, and (D) use of the modified S. purpuratus sequences described in methods. PTPRH-like2 contains 5 to 11 FN3 repeats, while PTPH-like 1 has 10–28 FN3 repeats (5 and 10 domains are shown based on the assumption that e-values less than 10−3 are insignificant). The only portion of SpPTPRR that was found was the PTPc domain. Structure of this protein was postulated based on PTPRR proteins in other species. Transmembrane domains were identified in 15 of 25 S. purpuratus RPTPs (all except SpPTPR D/F/S, G/Z1, H-like2, Lec3, Lec6, orph1, R, W1, W2, and Y2). Several sequences lacking transmembrane domains were partial. Since software sometimes misses sequences, transmembrane domains have been drawn here in known or expected positions. Human and Drosophila structures were redrawn from figures on the Novo Nordisk Science protein tyrosine phosphatase database (http://ptp.cshl.edu or http://science.novonordisk.com/ptp). RPTP domain structure raw data are presented in Suppl. 5 and 6.
One difficulty in studying conventional PTPs is the nomenclature. When these were first being discovered and characterized, the same protein was often given different names. In this paper, PTP nomenclature is simplified as in Alonso et al. (2004b). RPTPs are identified using letters of the alphabet (Ex: PTPRA, PTPRS) and NRPTPs are indicated using numbers (Ex: PTPN1, PTPN11). For alternate names see Tables 1–3.
Table 3.
A survey of the serine–threonine phosphatases in the S. purpuratus genome and predicted expression based on tiling and EST data
| Category | Gene | Alternative names | Urchin gene | Annotated ID | Tiling data | EST data |
|---|---|---|---|---|---|---|
| PPPs | ||||||
| A. PP1 Subfamily | ||||||
| A. 1. PP1 | PP1c alpha | Ser/thr protein phosphatase I, catalytic subunit alpha;PPP1-A;PP-1a | PP1 alpha/beta/gamma | SPU_000642/011110 | Very Low (3.4) | No |
| PP1c beta | Ser/thr protein phosphatase I, catalytic subunit beta;PPP1-B;PP-1b | PP1 beta | SPU_001788 | Moderate (15.5) | Yes | |
| PP1c gamma | Ser/thr protein phosphatase I, catalytic subunit gamma | |||||
| Urchin Specific PP1 | PP1-like | SPU_008700 | Moderate (20.5) | Yes | ||
| Morpholino PP1 | PP1 alpha-Partial seq* | SPU_006956 | Moderate (23) | Yes | ||
| B. PP2 Subfamily | ||||||
| B.1. PP2A | PP2A alpha | Ser/thr protein phosphatase 2A, catalytic subunit alpha | PP2A alpha/beta | SPU_025182 | High (68.6) | Yes |
| PP2A beta | Ser/thr protein phosphatase 2A, catalytic subunit beta | |||||
| B.2. PP4 | PP4c | Serine/threonine protein phosphatase 4 catalytic subunit; Pp4; PP-X | PP4c | SPU_027953 | High (100) | Yes |
| B.3. PP6 | PP6c | Serine/threonine protein phosphatase 6, catalytic subunit; Ppp6c | PP6c | SPU_011983 | Low (7.2) | Yes |
| C. PP5 Subfamily | ||||||
| C.1. PP5 (PPP5) | PP5c | Serine/threonine protein phosphatase 5, catalytic subunit | PP5c | SPU_028312 | Low (6.7) | Yes |
| D. PP7 Subfamily | ||||||
| D.1. PP7 | PPEF-1 | Ser/thr protein phosphatase with EF-hand motifs 1, PP7, PPP7 | None | |||
| PPEF-2 | Ser/thr protein phosphatase with EF-hand motifs 2 | PPEF-2 | SPU_008844/011860/028868 | Moderate (10.8) | No | |
| E. PP3 Subfamily | ||||||
| E.1. PP3 (PP2B)(PPP3) | CalcineurinA α | PPP3A alpha, PP2B A alpha | Calcineurin A alpha/beta/gamma | SPU_018404/016657 | Moderate (22.2) | Yes |
| CalcineurinA β | PPP3A beta, PP2B A beta | |||||
| CalcineurinA γ | PPP3A gamma, PP2B A gamma | |||||
| PPMs | ||||||
| A. PP2C Subfamily | ||||||
| Ppm1a | Protein phosphatase 1A, PP1A, PP2C alpha | Ppm 1a/b | SPU_009810 | Moderate (15) | Yes | |
| Ppm1b | Protein phosphatase 1B, PP1B, PP2C beta | |||||
| Ppm1d | Protein phosphatase 1D, PP1D | Ppm1d | SPU_020266 | High (68.8) | Yes | |
| Ppm1e | Protein phosphatase 1E, PP1E | Ppm1e | SPU_006378 | Very Low (3.9) | Yes | |
| Ppm1f | Protein phosphatase 1F, PP1F | |||||
| Ppm1g | Protein phosphatase 1G, PP1G | Ppm1g | SPU_011351 | Moderate (15) | Yes | |
| Ppm1k | Protein phosphatase 1K, PP1K | Ppm1k | SPU_011705/017161 | No | No | |
| Ppm1l | Protein phosphatase 1L, PP1L | Ppm1l | SPU_006132 | Low (7.6) | Yes | |
| ILKAP | Integrin-linked kinase-associated protein phosphatase 2C | ILKAPa | SPU_011070 | No | No | |
| ILKAPb | SPU_011294 | No | No | |||
| Ppm1h | Protein phosphatase 1H | Ppm1h/j/m | SPU_007363 | No | No | |
| Ppm1j | Protein phosphatase 1J, PP1J | |||||
| Ppm1m | Protein phosphatase 1M, PP1M | |||||
| PDP1 | Pyruvate dehydrogenase phosphatase 1 | PDP1/2 | SPU_025951 | Moderate (21.5) | Yes | |
| PDP2 | Pyruvate dehydrogenase phosphatase 2 | |||||
| TA-PP2C | T-cell activation protein phosphatase 2C | TA-PP2C | SPU_017346 | Moderate (28.1) | No | |
| Tab1 | Map3k7ip1;MAP kinase kinase kinase 7 interacting protein 1 | Tab 1 | SPU_005254 | Very Low (4.0) | Yes | |
| Novel Group 1 | Ppm1g-like A | SPU_014625 | Moderate (22.4) | No | ||
| Ppm1g-like B | SPU_004300 | Moderate (17.3) | Yes | |||
| Novel Protein 2 | Ppm1h-like | SPU_026428 | Very Low (3.9) | No | ||
| FCPs | ||||||
| A. FCP | ||||||
| Ctdp1 | Carboxy Terminal Domain, RNA Pol II, polypeptide A, phosphatase | Ctdp1* | SPU_021493 | Low (5.7) | Yes |
Human PTPs are shown under “PTP Gene” and S. purpuratus homologues or novel genes appear under “Urchin Gene”. Yellow highlighted genes are novel. Blue areas indicated genes that are homologous or very similar to human genes. MASOs produced for genes shown in red block development. Note. Comparison of the exon/intron boundaries of Ppm1g-like A and B suggests that these are not haplotype pairs.
Sequence unconfirmed by phylogenetic analysis.
To identify homologues of human conventional PTPs, human sequences were back blasted against the S. purpuratus genome. Using this technique, 49 putative SpRPTP and 12 putative SpNRPTP sequences were detected (Table 1, Suppl. 10). Seventeen other sequences were also weakly similar to conventional PTPs (most were partial sequences).
Because many RPTP sequences are long and because extracellular regions are poorly conserved, this group was difficult to analyze. Many of the sequences crossed scaffolds and back blasting to find extracellular sequences was frequently useless. Because of these complications, full-length S. purpuratus sequences were not used to identify human RPTP and NRPTP homologues. Instead, PTPc domain analysis was performed (the standard procedure to identify conventional PTPs).
Since all RPTPs contain a D1 domain, more detailed analysis was performed on these data. Homologues to the S. purpuratus RPTP D1 sequences were detected using three methods: neighbor-joining (Fig. 1), maximum likelihood (Suppl. 1), and Bayesian analysis (Suppl. 2). Results showed that only 9 of 25 S. purpuratus D1 sequences grouped with RPTP subgroups and that only 3 of the 8 RPTP subgroups have sea urchin homologues strongly supported by bootstrap analysis (R2A, R5, and R7). Type R3-like and R9-like S. purpuratus RPTPs were also found, but these clades (when observed) were weakly supported. In contrast, the other 16 sea urchin sequences appear to be orphans, defining at least 2–3 new clades (these results are supported by high bootstrap values and by neighbor-joining, maximum likelihood, and Bayesian analysis). Intriguingly, one novel group of SpPTPRs always forms a single clade (SpPTPRLec1–6 and SpPTPRFn1–2), suggesting that it arose by lineage specific expansion (the production of paralogues by duplication of an orthologue after speciation; for more about this, see Beane et al., 2006). Further, none of these sequences are identical or located on the same scaffold, indicating that they are likely to be real and distinct genes. Based on Bayesian and maximum likelihood analyses these genes and the other orphans may be distantly related to Drosophila melanogaster CG7180PA (a PTP with no clear human orthologue) and Nematostella vectensis 39268×. Based on tiling microarrays and EST data, 72% of the SpRPTPs are expressed during development (Table 1). With the exception of SpPTPR G/Z2, PTPR D/F/S and PTPRorph2 (present at moderate levels), expression of these genes is either undetectable, very low, or low (see Table 1).
The RPTP D2 neighbor-joining tree (Suppl. 3) corroborates findings in the D1 trees. Again, sea urchin homologues to the R2A and R5 RPTPs were detected, as was the SpPTPRLec/Fn clade and the SpPTPRY1/2 clade. Partial sequences containing the missing D2 domains of SpPTPRW1 and SpPTPRFn2 may have also been identified. This is supported by three observations: A) The detected D2 sequence is on the same strand as the known D1 sequence, B) these genes occupy complementary locations in each scaffold, and C) the D1 and D2 genes occupy the same clade in both trees.
As with most of the phosphatases studied, fewer NRPTPs were found in the sea urchin than in humans. While humans have 15 NRPTPs (Alonso et al., 2004b), only 8 are present in S. purpuratus (Fig. 1B). Since Type R8 RPTPs (PTPRN and PTPRN2) are thought to be more closely related to NRPTPs than to RPTPs, they were included in the S. purpuratus NRPTP tree. Eight of 10 NRPTP types were represented in the S. purpuratus genome. Only the Type NT8 (PTPN20) NRPTP and the Type R8 RPTP were missing. No novel NRPTPs were detected. All S. purpuratus NRPTPs were expressed at moderate levels except PTPN3/4 (expressed at low levels) (see Table 1).
Dual specificity phosphatases
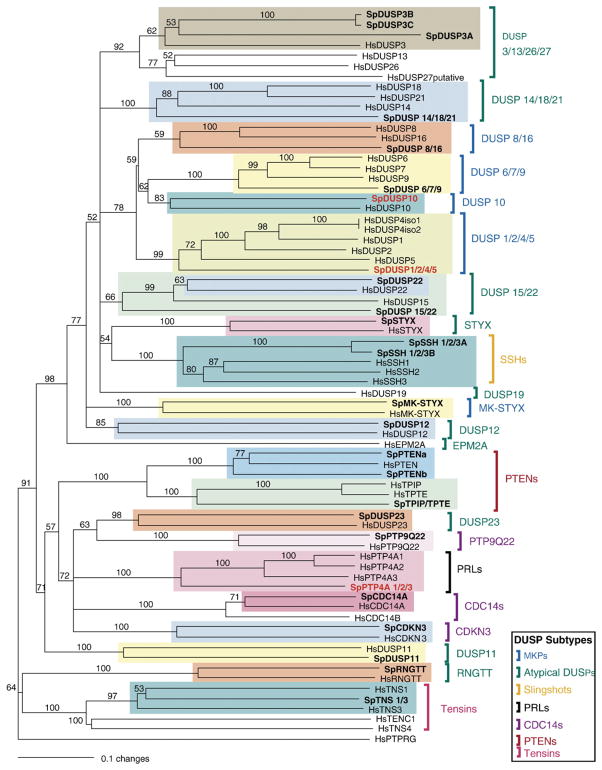
The dual specificity protein phosphatases (DSPs) include 8 subfamilies: MAP kinase phosphatases (MKPs), atypical DSPs, Slingshots, PRLs, CDC14s, PTENs, tensins, and myotubularins. In S. purpuratus, 32 DSPs were found, far fewer than the 61 found in humans. Of these, at least one member of each major subfamily was present: 5 MKPs, 12 atypical DSPs, 2 Slingshots, 1 PRL, 3 CDC14s, 4 PTENs, and 5 myotubularins (Fig. 2, Table 2).
Fig. 2.
Phylogenetic relationships of S. purpuratus DSPs to human forms. In this neighbor-joining tree (generated using sequences that align with residues 306–531 of HsDUSP1; outgroup=HsPTPRG), highlighted clades contain S. purpuratus homologues of human DSPs. Bootstrap values are shown at the dendogram node (maximum=100) and the horizontal distance indicates percent divergence (0.1 scale=10 substitution events per 100 amino acids). MASOs were produced and tested for sequences shown in red. For relationships of myotubularins, CDC25s, or EYAs, see Suppl. 4A–C. Sp=Strongylocentrotus purpuratus, Hs=Homo sapiens.
Table 2.
A survey of the Class I Cysteine-based DSPs, Class II Cysteine-based PTPs, Class III Cysteine-based PTPs, and Asp-based PTPs in the S. purpuratus genome and their predicted expression based on tiling and EST data
| Category | PTP gene | Alternative names | Urchin gene | Annotated ID | Tiling data | EST data |
|---|---|---|---|---|---|---|
| PTPs | ||||||
| A. Class I Cys-Based PTPs | ||||||
| A. 2. DSPs or VH1-like | ||||||
| A.2. 1.MKPs | ||||||
| DUSP1 | 3CH134, CL100/HVH1, Erp, MKP-1, PTPN10 | DUSP 1/2/4/5 | SPU_021143 | Moderate (21.5) | Yes | |
| DUSP2 | PAC-1 | |||||
| DUSP4 | hVH2/TYP, MKP-2 | |||||
| DUSP5 | hVH3/B23 | |||||
| DUSP6 | MKP-3/rVH6, PYST1 | DUSP 6/7/9 | SPU_028124 | High (37.7) | Yes | |
| DUSP7 | B59; MKP-X, PYST2 | |||||
| DUSP9 | MKP-4, PYST3 | |||||
| DUSP8 | HB5, hVH5, M3/6 | DUSP 8/16 | SPU_003348 | Moderate (17.3) | Yes | |
| DUSP16 | MKP-7, MKP-M | |||||
| DUSP10 | MKP-5 | DUSP10 | SPU_006513 | High (49.7) | Yes | |
| MK-STYX | MK-STYX/DUSP24 | SPU_000663 | Low (5.4) | No | ||
| A.2. 2. Atypical DSPs | ||||||
| DUSP3 | VHR; T-DSP11 | DUSP3A | SPU_016669 | No | No | |
| DUSP3B (Tandem Dupl) | SPU_016406 | No | No | |||
| DUSP3C (Tandem Dupl) | SPU_016409 | No | No | |||
| DUSP11 | PIR1 | DUSP11 | SPU_001665 | No | Yes | |
| DUSP12 | G KAP; HYVH1; LMV-DSP4; YVH1 | DUSP12 | SPU_003149 | No | Yes | |
| DUSP13A | BEDP | None | ||||
| DUSP13B | TMDP; TS-DSP6 | |||||
| DUSP26 | VHP; similar to RIKEN cDNA 0710001B24 | |||||
| DUSP27 | DUPD1; FMDSP; similar to cyclophilin | |||||
| DUSP14 | MKP6; MKP-L | DUSP 14/18/21 | SPU_000652 | Very Low (3.1) | Yes | |
| DUSP18 | DUSP20; LMW-DSP20 | |||||
| DUSP21 | BJ-HCC-26; LMW-DSP21; tumor antigen | |||||
| DUSP15 | Q9H1R2; VHY | DUSP 15/22A | SPU_013773 | No | No | |
| DUSP22 | JKAP; JSPI; LMW-DSP2: MKPX; TS-DSP2; VHX | DUSP 15/22B | SPU_022023 | Low (8.1) | No | |
| DUSP19 | DUSP17; LDP-2; SKRP1; TS-DSP1 | None | ||||
| DUSP23 | MOSP; similar to RIKEN cDNA 2810004N20 | DUSP23 | SPU_004533 | No | No | |
| DUSP24 | MGC1136 | MK-STYX/DUSP24 | SPU_000663/018623 | Low (5.4) | No | |
| DUSP25* | FLJ20442; LMW-DSP3; VHZ | |||||
| EPM2A | Laforin | None | ||||
| RNGTT | mRNA capping enzyme | RNGTT | SPU_008892 | No | Yes | |
| STYX | STYX | SPU_011259 | No | No | ||
| A. 2. 3. Slingshots | ||||||
| SSH1 | Slingshot 1 | SSH 1/2/3A | SPU_002366 | Very Low (3.1) | Yes | |
| SSH2 | Slingshot 2 | SSH 1/2/3B | SPU_003158 | Low (8.2) | Yes | |
| SSH3 | Slingshot 3 | None | ||||
| A. 2. 4. PRLs | ||||||
| PTP4A1 | PRL-1 | PTP4A 1/2/3 | SPU_008895 | High (31.4) | Yes | |
| PTP4A2 | OV-1; PRL-2 | |||||
| PTP4A3 | PRL-3 | |||||
| A. 2. 5. CDC14s | ||||||
| CDC14A | CDC14A | SPU_015765 | High (63.4) | Yes | ||
| CDC14B | ||||||
| CDKN3 | KAP | CDKN3 | SPU_003479 | Low (5.5) | Yes | |
| PTP9Q22 | PTP9Q22 | SPU_012073 | Low (7.0) | No | ||
| A. 2. 6. PTENs | ||||||
| PTEN | MMAC1; TEP1 | PTENa | SPU_009522 | High (33.4) | Yes | |
| PTENb | SPU_023882/003142 | Low (7) | No | |||
| TPIP | TPIPα, TPTE and PTEN homologous | TPTE/TPIP | SPU_002928 | No | No | |
| TPTE | PTEN-like; PTEN2 | |||||
| A. 2. 7. Tensins | ||||||
| TNS1 | Tensin 1 | TNS 1/3 | SPU_005602 | Low (7.4) | Yes | |
| TNS3 | Tensin 3 | |||||
| TNS4 | Tensin 4 | |||||
| TENCI | Tensin like C1 domain containing phoshatase, Tensin 2 | |||||
| A. 2. 8. Myotubularins | ||||||
| MTM1 | Myotubularin | None | ||||
| MTMR1 | MTMR 1/2 | SPU_008287 | Very Low (3.3) | No | ||
| MTMR2 | ||||||
| MTMR3 | FYVE-DSPI | MTMR 3/4 | SPU_024752 | Moderate (11) | Yes | |
| MTMR4 | FYVE-DSP2 | |||||
| MTMR5 | SBF1 | MTMR 5/13 | SPU_005099 | Low (7.7) | No | |
| MTMR13 | CMT4B2; SBF2 | |||||
| MTMR6 | MTMR 6/7/8 | SPU_022669 | No | Yes | ||
| MTMR7 | ||||||
| MTMR8 | ||||||
| MTMR9 | LIP-STYX | MTMR9 | SPU_021537/018586 | Low (5.5) | No | |
| MTMR10 | None | |||||
| MTMR11 | CRAα/β | |||||
| MTMR12 | 3-PAP | |||||
| MTMR14* | hEDTP; hJumpy; FLJ22075 | None | ||||
| MTMR15* | KIAA1018 | None | ||||
| B. Class 11 Cys-Based PTPs | ||||||
| ACP1 | BHPTP; LMPTP; low Mr PTP; LMWPTP | None | ||||
| C. Class III Cys-Based PTPs | ||||||
| CDC25A | CDC25 A/B/C | SPU_019568 | Low (5.4) | Yes | ||
| CDC25B | ||||||
| CDC25C | ||||||
| D. Asp-Based PTPs | ||||||
| EYA1 | EYA 1/2/4 | SPU_013869 | Moderate (14.4) | No | ||
| EYA2 | ||||||
| EYA4 |
Human PTPs are shown under “PTP Gene” and S. purpuratus homologues or novel genes appear under “Urchin Gene”. MASOs were produced for genes shown in red. No novel genes were detected. Morpholino blocks development. Note: Comparison of exon/intron boundaries in duplicate forms suggests that none of these are haplotype pairs.
Sequence not available so not in orthologue analysis.
The MKPs are one of the most studied DSP groups. By dephosphorylating MAP kinases (MAPKs) at dual sites, both the threonine and tyrosine residues in MAPK motif -pTXpY-, MKPs halt MAPK activity. In vertebrates, there are three MAPKs: Erk, JNK/SAPK, and p38. While some MKPs only interact with one MAPK (for example, DUSP6/MKP-3 blocks Erk activity), others can dephosphorylate two or all three MAPKs (for example, DUSP4/MKP-2 blocks both Erk and JNK) (Ducruet et al., 2005). With the exception of DUSP10 (MKP-5), DUSP8 (VH5), and DUSP16 (MKP-7), MKPs contain two domains, a DUSP catalytic domain and an N-terminal MAPK binding domain (MKB domain). Other MKPs possess a third domain. The function of this domain is unknown in DUSP10/MKP-5, but, in DUSP8/VH5 and DUSP16/MKP-7, the third domain (a PEST domain) is thought to facilitate ubiquitination and proteolysis when phosphorylated (Farooq and Zhou, 2004). Five homologues of human DSPs were found in the sea urchin: SpDUSP 1/2/4/5 (nuclear DSPs that act on all MAPK types), SpDUSP 6/7/9 (cytosolic DSPs that act on Erks), SpDUSP 8/16 (cytosolic DSPs that dephosphorylate Jnk and p38), SpDUSP10 (also cytosolic, dephosphorylating Jnk and p38) and SpMK-STYX (MKP functions from Alonso et al., 2004a). With the exception of SpMK-STYX (present at low levels), all were expressed at moderate to high levels during development.
Much less is known about atypical DSPs. These tend to be smaller proteins (less than 250 amino acids) that lack the MKB domain (Alonso et al., 2004a) and three dephosphorylate MAPKs (DUSP3, DUSP22, and DUSP14/MKP-6) (Farooq and Zhou, 2004). S. purpuratus homologues of human atypical DSPs identified include SpDUSP11, SpDUSP12, SpDUSP14/18/21, SpDUSP15/22a and b, SpDUSP23, SpRNGTT, SpSTYX, and three copies of SpDUSP3: SpDUSP3A, SpDUSP3B, and SpDUSP3C (Fig. 2). Two of these, DUSP3B and 3C, were formed by tandem duplication. Also, the identification of SpDUSP15/22b is suspicious. This is definitely a DSP, but there appears to be a genome assembly error. The DUSP15/22b sequence is incomplete and lies between exons of SpJAK. Based on the tiling microarrays and EST data, only 5 atypical DSPs are expressed in developing urchins: SpDUSP11, SpDUSP12, SpDUSP14/18/21, SpMK-STYX/DUSP24, and SpRNGTT (Table 2). None are expressed above low levels.
Other DSP subfamilies also have DUSP domains, but are more distantly related to the MKPs and atypical DSPs. These include the Slingshot, PRL, Cdc14, PTEN, and myotubularin subfamilies. Members of the Slingshot (SSH) subfamily (SSH1, SSH2, and SSH3 in humans) regulate the actin cytoskeleton (Niwa et al., 2002; Baum, 2002; Huang et al., 2006). By dephosphorylating cofilin, an actin-binding protein that severs F actin, SSHs indirectly activate disassembly at the slow-growing ends of actin filaments. In the sea urchin, two homologues of human SSH were found, SpSSH1/2/3A, and SpSSH1/2/3B (Fig. 2). During development, SpSSH1/2/3A and B are expressed at very low and low levels respectively (Table 2).
Another subfamily, the PRLs (phosphatase for regenerating liver), is involved in regulation of growth, cell proliferation, cell migration, and differentiation of specific tissues (such as digestive epithelial cells). These phosphatases are among the smallest known (usually 140–180 aa in length) and are fairly simple in structure, with only one catalytic domain and no regulatory or docking domains (Kozlov et al., 2004). Localization and activity of these phosphatases may be regulated through a prenylation site at the C terminus. Humans have 3 PRLs: PTP4A1, 2, and 3. Only one, expressed at high levels, was found in S. purpuratus, SpPTP4A1/2/3 (Fig. 2, Table 2).
Members of the Cdc14 subfamily (including Cdc14A, Cdc14B, CDKN3, and PTP9Q22) interact with cyclin dependent kinases (CDKs/cyclins), inactivating them by removing phosphates (Torres-Rosell et al., 2005; Hannon et al., 1994). Effects of this differ depending on the cyclins involved. For example, Cdc14 promotes exit from mitosis when one group of cyclins associates with it, but when bound to a different cyclin, Cdc14 regulates mitotic entry (Trautmann and McCollum, 2002). Cdc14s have also been linked to nucleolar rDNA segregation during anaphase and to rDNA condensation. Three S. purpuratus homologues of human Cdc14s were found: SpCdc14A, SpCDKN3, and SpPTP9Q22. No Cdc14B homologue was detected (Fig. 2). During development, SpCDKN3 and PTP9Q22 are expressed at low levels whereas SpCdc14A is expressed at high levels (Table 2).
Because strong sequence similarity exists between members of the PTEN subfamily (PTEN, PTPE, PTIP) and other PTPs, and because members of this subfamily have DUSP domains, it was originally thought that PTENs dephosphorylated proteins at serine/threonine or tyrosine sites. With the exception of one study (Raftopoulou et al., 2004), all work in this area has shown that the primary substrates of PTENs are D3-phosphorylated inositol phospholipids (Wishart and Dixon, 2002). Raftopoulou et al. recently suggested that, in some cases, PTENs also dephosphorylate focal adhesion kinase (FAK), a protein. Three members of this subfamily were found in S. purpuratus: SpPTENa, SpPTENb, SpTPTE/TPIP (Fig. 2). Although BLAST searches classify SpPTENb as a homologue of TPIP, neighbor-joining analysis does not support this, but categorizes it as a PTEN homologue. All subfamily members except SpTPTE/TPIP were expressed during embryonic development (Table 2).
Another group of proteins that shares sequence homology with the PTENs (Haynie and Ponting, 1996; Lo, 2004) is the tensins (Tensin1, Tensin3, Tensin4, and Tenc1). These proteins have previously been classified with the PTPs (Alonso et al., 2004a,b) although, with the exception of Tenc1 (Hafizi et al., 2005), they lack phosphatase activity. Instead, Tensin1, Tensin3, and Tensin4 are generally categorized as phosphoproteins or focal adhesion associated proteins (Chen et al., 2002; Lo, 2004; Cui et al., 2004). Structurally, the tensins have domains that bind integrin β (PTB domain), actin (ABD domains), and tyrosine-phosphorylated proteins (SH2 domain) (Lo, 2004). They affect cell motility, cell shape, and cell proliferation (Chen et al., 2002; Lo, 2004; Cui et al., 2004; Hafizi et al., 2005), link receptor tyrosine kinase signaling to control of cell adhesion (Cui et al., 2004; Lo, 2004), and regulate Akt/PKB signaling (Hafizi et al., 2005). Low levels of SpTensin 1/3 were expressed in S. purpuratus.
PTENs are not the only PTPs that dephosphorylate lipids. Members of the myotubularin subfamily, myotubularin (MTM1) and the myotubularin-related proteins (MTMRs), are also lipid phosphatases. Humans have 14 myotubularins: MTM1 and MTMR 1 to 13. Phylogenetic analysis has shown that metazoans have six types of myotubularins (Wishart et al., 2001). Three clades (HsMTMR5/13, HsMTMR9, and HsMTMR10/11/12) contain catalytically inactive forms that lack the ability to dephosphorylate while the other three clades (HsMTM1/MTMR1/2, HsMTMR3/4, and HsMTMR6/7/8) actively dephosphorylate phosphatidylinositol 3-monophosphate (PtdIns3P) and phosphatidylinositol 3,5-biphosphate (PtdIns3, 5P2). Lipid intermediates produced by dephosphorylation act in vacuolar transport and membrane turnover (Laporte et al., 2003). Because the catalytically active and inactive forms interact, it is thought that inactive forms may regulate active myotubularins by heterodimerizing with them (Laporte et al., 2003). Five of the six myotubularin types are present in S. purpuratus (Suppl. 4A). Three catalytically active forms were found: SpMTMR1/2, SpMTMR3/4, and SpMTMR6/7/8. But only two non-catalytic myotubularins were annotated: SpMTMR9 and SpMTMR5/13. Homologues of HsMTM1, HsMTMR10, HsMTMR11, or HsMTMR12 were not observed. With the exception of SpMTMR6/7/8 (not expressed), S. purpuratus MTMRs were expressed at levels ranging from very low to moderate during development (Table 2).
Class II and III cysteine-based PTPs
Only one Class II Cysteine-based PTP is found in humans, ACP (aka the low molecular weight PTPs) (Alonso et al., 2004b). This is a 180 kDa protein that can be alternatively spliced to form 4 different mRNA isoforms. The catalytic domain contains a typical PTP motif, but other portions of the phosphatase have limited similarity to the PTPs. This protein regulates growth factor induced mitotic growth by deactivating tyrosine kinase receptors that interact with growth factors (i.e. insulin, ephrin, and PDGF receptors) (Raugei et al., 2002). It also influences cytoskeletal activities by dephosphorylating p190RhoGAP, a regulator of Rho. No HsACP homologue was found in the S. purpuratus genome.
Class III Cysteine-based PTPs are critical in cell cycle regulation where they dephosphorylate cyclin dependent kinases (Cdk/cyclin). Humans produce three Class III Cysteine-based PTPs: Cdc25A, Cdc25B, and Cdc25C. Two of these, Cdc25B and Cdc25C, regulate progression of cells from G2 to M phase, and Cdc25A, in addition to modulating the G2 to M transition, also regulates the G1 to S transition and contributes to S phase and G2 phase progression (Kristjánsdóttir and Rudolph, 2004; Ducruet et al., 2005). Cdc25 phosphatases are also targets of cell checkpoint proteins. If cells are exposed to ultraviolet light, ionizing irradiation, replication inhibition, or other components that damage DNA, cell checkpoint proteins (such as Chk1-, Chk2-, and p38) phosphorylate Cdc25, resulting in cytoplasmic sequestration or ubiquitin/proteosome-mediated destruction of the phosphatase. Since Cdc25 is necessary for progression of cell cycle, phosphorylation by checkpoint proteins causes mitotic arrest, preventing replication of damaged DNA. These are short proteins (423–566 aas in humans) with a regulatory domain at the N-terminus and a catalytic domain closer to the C-terminus. In S. purpuratus, only one Cdc25 is found, SpCdc25A/B/C (Suppl. 4B). This protein is expressed at low levels during development (Table 2).
Asp-based PTPs
Eyes absent (EYA) is the only member of the Asp-based PTP Class and is notable because, in addition to dephosphorylating proteins, it acts in coordination with Sine oculis (SO) as a transcriptional coactivator (Rebay et al., 2005). It has been implicated in several developmental processes including formation of the eyes, kidneys, ears, and muscle. Recent in vitro data suggest that substrates dephosphorylated by EYA include the CTD domain of RNA polymerase II and EYA itself. In humans, there are 4 forms of EYA: HsEYA1, HsEYA2, HsEYA3, and HsEYA4. In S. purpuratus, only one form was found, SpEYA1/2/4 (Suppl. 4C). This gene is expressed at moderate levels in developing embryos (Table 2).
The serine–threonine phosphatases
As mentioned in the introduction, many ser–thr phosphatases are multimeric proteins, with a catalytic subunit whose activity and localization are controlled by regulatory subunits. It was not within the scope of this study to identify both catalytic and regulatory subunits; instead efforts were focused on identification of the catalytic subunits. In S. purpuratus, 57 sequences were initially identified that blasted to ser–thr phosphatase sequences (catalytic and regulatory). Of these, 25 contained sequences of ser–thr phosphatase catalytic domains complete enough to be used in homologue analysis (2 others were discarded due to redundancy) (Table 3, Suppl. 10). Eighty-eight percent of the annotated sequences containing a catalytic domain were expressed (Table 3).
PPP phosphatases
The PPP family consists of five subfamilies: PP1 (Protein phosphatase 1), PP2A/PP4/PP6, PP5, PP7, and PP2B. Studies of PPP evolutionary relationships found that all 5 subfamilies are present in both humans (deuterostomes) and Drosophila (ecdysozoan protostomes) and that most, with the exception of PP7, are conserved throughout the Eukaryota. Subfamilies are classified in a manner consistent with the findings of Cohen (2004). These investigators reported that the PP1 and PP2A/PP4/PP6 subfamilies share a common ancestor. They also found that PP5 and PP7 phosphatases might be related to the PP2B phosphatases, sharing a distant common ancestor. Two members of the PP2B/PP5/PP7 clade, PP2B and PP7, are only activated in the presence of calcium while other members of the PPP family do not depend on ions for activation. All members of the PPP family contain a catalytic domain that is approximately 280 amino acids in length called the PP2A domain. Representatives of each PPP subfamily are found in the sea urchin (Fig. 3A and B, Table 3).
Fig. 3.
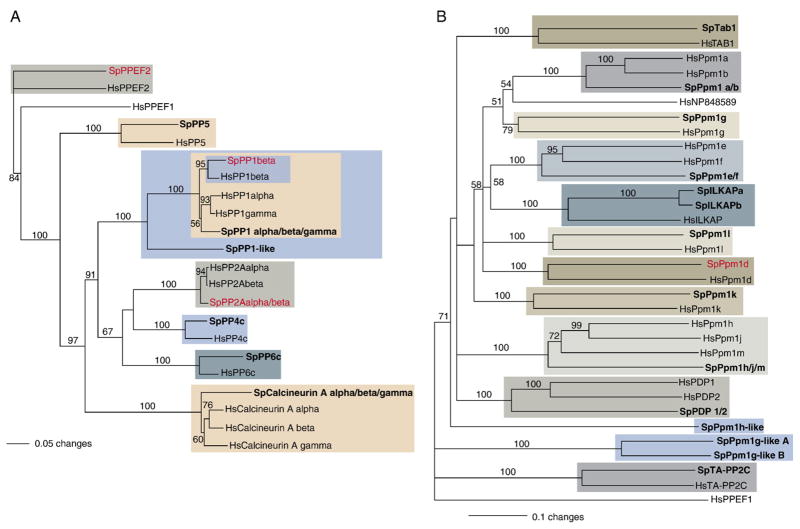
(A) Phylogenetic relationships of S. purpuratus PPPs to human forms (unrooted neighbor-joining tree generated using sequences that align with residues 59–322 of HsPP1alpha). S. purpuratus homologues to human PPPs were found for all subfamilies. Clades containing these homologues are highlighted. (B) Phylogenetic relationships of S. purpuratus PPMs to human forms (rooted neighbor-joining tree generated using sequences that align with residues 60–343 of HsPpm1d; outgroup=HsPPEF1). Sequences highlighted in blue are novel PPMs. Other highlighted sequences are S. purpuratus proteins with human homologues. In both trees, bootstrap values are shown at the dendogram node (maximum=100) and the horizontal distance indicates percent divergence (0.1 scale=10 substitution events per 100 amino acids). MASOs were produced and tested for sequences shown in red. Sp=Strongylocentrotus purpuratus, Hs=Homo sapiens.
The PP1 subfamily
PP1 phosphatases (also known as the PPP1 subfamily) are among the most highly conserved eukaryotic proteins. In fact, an isoform present in Giardia lamblia is 72% identical to human PP1 (Ceulemans et al., 2002)! Humans have 3 catalytic subunits: PP1 alpha, PP1 beta, and PP1 gamma. These catalytic subunits cannot function independently, but must heterodimerize with variable regulatory proteins (over 50 in humans) that influence when and where they act (Ceulemans and Bollen, 2004). PP1 activity is also regulated by kinases. By phosphorylating threonine residues at the C-terminus of the catalytic subunit, kinases can reduce PP1 activity.
PP1 phosphatases function in various cell activities. They dephosphorylate transcription factors so they can be reused, process mRNA, downregulate ion pumps and channels, maintain G1 and G2 phases, allow exit from mitosis, control gene transcription by mRNA Polymerase II, and induce relaxation of actomyosin fibers (Ceulemans and Bollen, 2004). Three PP1 homologues were identified in S. purpuratus: SpPP1 alpha/beta/gamma, SpPP1 beta, and SpPP1-like (Fig. 3A). Two of these, SpPP1-like and SpPP1 beta, are expressed at moderate levels in developing S. purpuratus and SpPP1 alpha/beta/gamma is expressed at very low levels (Table 3). SpPP1-like groups close to the PP1 clade, but does not cluster with any specific metazoan PP1 group (Drosophila, Nematostella, and human sequences were tested, not shown). It may be a highly divergent form or perhaps a pseudogene.
PP2A/PP4/PP6 subfamily
Like PP1, members of the PP2A/PP4/PP6 subfamily also interact with regulatory domains. In humans, catalytic subunits of two PP2A phosphatases have been identified, PP2A alpha and PP2A beta. Normally, the catalytic subunit (C subunit) associates with a second 65 kDa subunit (the A subunit or P65) to form the core enzyme. A second subunit then associates with the core enzyme. This subunit, the B subunit, can vary quite a bit. There are 4 families of B subunits: B (PR55), B′ (PR61), B″ (PR 72, PR130, PR59, and PR48), and B‴ (PR93, PR110). The PP2A (or PPP2) phosphatases are one of the most commonly utilized phosphatases in the cell. These phosphatases regulate canonical and non-canonical Wnt signaling, regulate G2/M transition, initiate and terminate translation, respond to DNA damage, and act in apoptosis. Kinases dephosphorylated by PP2A phosphatases include Erks, calmodulin-dependent kinases, PKA, PKB, PKC, and Cdks (Janssens and Goris, 2001). In S. purpuratus, one PP2A protein (SpPP2A alpha/beta) was identified (Fig. 3A), and, as expected, it is expressed at high levels during development (Table 3).
The other two members of this subfamily, PP4 and PP6, are also highly conserved and are found in both deuterostomes and ecdysozoan protostomes. Like the PP2A phosphatases, PP4 and PP6 each bind a regulatory subunit to form a core protein. The core protein then interacts with a third variable subunit that influences phosphatase activity (Cohen et al., 2005). PP4 is involved in numerous cell activities including chromatin regulation, centrosome maturation, spliceosomal assembly, and regulation of signaling pathways (NF-kB and target of rapamycin) (Cohen et al., 2005). Less is known about PP6. Studies of yeast homologues have shown that this phosphatase is important for the G1/S transition, initiation of translation, and cell shape regulation (Bastians and Ponstingl, 1996; Cohen, 2004). S. purpuratus has one PP4 catalytic subunit (SpPP4c) and one PP6 catalytic subunit (SpPP6c) (Fig. 3A). PP4c is expressed at high levels and PP6c is present at low levels during sea urchin development (Table 3).
The PP5 and PP7 subfamilies
Like all members of the PPP family, PP5 and PP7 phosphatases both have a catalytic domain similar to PP2A. In PP7/PPEF (Protein phosphatase 7/Protein phosphatase with EF hand domains) phosphatases, however, this catalytic domain contains inserts that are absent in PP2A phosphatases (Luan, 2003). These inserts vary in length and there is evidence that they may be autoinhibitory regions (Andreeva and Kutuzov, 1999). It is also known that, like PP2B phosphatases, PPEF activity is calcium dependent. Protein activity is controlled in two ways, by calcium binding to C-terminal EF hand sequences and by interaction of calmodulin with the N-terminal domain (Kutuzov et al., 2002). PP7 phosphatases are important for vision. In mammals, PP7 expression is primarily limited to the retina and the brain (Cohen, 2004), and, if this gene is disrupted (either in mammals or Drosophila), photoreceptor function is impaired (Andreeva and Kutuzov, 1999; Ramulu et al., 2001). Although PP7 performs a different roles in mammals, Drosophila Rdg5, a PP7 homologue, is known to be necessary in dephosphorylation of rhodopsin.
PP5 probably originated from the same ancestral protein as PP7/PPEF, but it differs from PP7/PPEF in both structure and function. PP5 activity is not calcium dependent, but depends on interactions with regulatory proteins or lipids. The regulatory domain of the catalytic subunit is approximately 200 aa long, lies near the N-terminal, and contains 3 tetratricopeptide (TTP) tandem repeats. By interacting with regulatory molecules, PP5 influences atrial natriuretic peptide signaling, ion channel activity, steroid signaling, and RNA polymerase I and II function (Andreeva and Kutuzov, 1999).
In S. purpuratus, homologues were identified for both HsPP5 and HsPP7 (Fig. 3A). SpPP7 is expressed at moderate levels during development and SpPP5 is expressed at low levels (Table 3).
The PP2B Subfamily
The PP2B phosphatases (also known as PPP3 phosphatases) are activated in response to intracellular increases in calcium. When calcium levels rise in the cell, calcium binds both of the regulatory subunits (calmodulin and calcineurin B) needed to activate the catalytic subunit (calcineurin A, also known as PP2B alpha or PPP3A) (Gomperts et al., 2003; Luan, 2003). In humans, there are 3 catalytic subunits, calcineurin A alpha, calcineurin A beta, and calcineurin A gamma. This phosphatase acts in numerous calcium dependent processes and regulates nuclear factor of activated T-cells (NF-AT) mediated transcription. It plays important roles in apoptosis, cardiac physiology, embryonic development, and regulation of metabolic processes (Groenendyk et al., 2004). A single homologue of HsCalcineurin A, SpCalcineurin A alpha/beta/gamma, is present in S. purpuratus (Fig. 3A). A second HsCalcineurin A homologue may also be present. SPU_018404 contains a PP2A domain and back blasts to calcineurin A, however the sequence was too short to analyze using neighbor-joining. Both SPU_018404 and SpCalcineurin A alpha/beta/gamma are expressed at moderate levels in developing embryos (Table 3).
PPM phosphatases
The PPM phosphatases only contain one subfamily, the PP2C subfamily. Unlike the PPP phosphatases, these ser–thr phosphatases are a structurally diverse group of monomeric proteins that are activated in response to magnesium or manganese. Although, like the PPP phosphatases, these proteins act on serine–threonine residues, they probably evolved independently of other ser–thr phosphatases (Schweighofer et al., 2004). A few of the cell activities regulated by members of this subfamily include spliceosome assembly during pre-mRNA splicing (Ppm1g; Murray et al., 1999), cell cycle (Ppm1a and b; Cheng et al., 2000), glycolysis to tricarboxylic acid cycle transition (PDP; Maj et al., 2006), p38 and JNK signaling (Ppm1a; Takekawa et al., 1998), and TAK1 signaling (Ppm1b; Hanada et al., 2001). Eleven homologues of human PP2Cs were identified: SpPpm1a/b, SpPpm1d, SpPpm1e/f, SpPpm1g, SpPpm1h/j/m, SpPpm1k, SpPpm1l, SpILKAP (a and b), SpILKAPb, SpTAB1, SpTA-PP2C, and SpPDP1/2. No human homologues were found for the PP2C phosphatases SPU_026428 (SpPpm1h-like), SPU_014625 (SpPpm1g-likeA), or SPU_004300 (SpPpm1g-likeB) (Fig. 3B). All of these proteins are expressed during development except SpPpm1h, SpPpm1k and SpILKAP (Table 3).
FCPs
The FCP (TFIIF associated CTD phosphatase) family is a small group of phosphatases that are highly divergent from other ser–thr phosphatases except that they have the signature motif of phosphotransferases and phosphohydrolases, DXDX (T/V) (Kamenski et al., 2004). Also, they typically contain 2 conserved regions, the N-terminal FCP1 homology region, which includes the DXDX(T/V) sequence, and the C-terminal BRCT (breast cancer protein-related) domain which binds the CTD domain of RNA polymerase II. By dephosphorylating RNAPII at serine residues, FCP1/CTDP regulates mRNA transcription. One FCP, SpCTDP1, was found in S. purpuratus. Based on EST and tiling data, this gene is expressed at low levels during sea urchin development (Table 3).
Overview of the phosphatases found in S. purpuratus
A total of ninety-one phosphatases were identified in this study. Sixty-six are PTPs and 25 are ser–thr phosphatases. Based on tiling and EST data, it is estimated that 79% of the PTPs and 88% of the ser–thr phosphatases are expressed during S. purpuratus development. Highest levels of expression were observed in the MKPs, PRLs, Cdc14s, PTENs, PPPs, and PPMs. Genes expressed at moderate levels include the RPTPs (a few), NRPTPs, MKPs, myotubularins (one), EYA, and most of the PPPs and PPMs.
Why are some phosphatases highly expressed during development while others are only weakly expressed? The phosphatases transcribed at moderate to high levels regulate MAPK pathways, cell motility, cell cycle control, growth, transcription, organelle assembly, apoptosis, spliceosomal assembly, PI3K-PBP/ATK signaling, and ion channel activities. (For a detailed summary of the activities of phosphatases expressed at moderate to high levels, see Supplemental Table 8.) Many of these are multimeric proteins. By associating with different regulatory subunits they perform a wide array of cellular functions (Examples: PP1, PP2A, and PP4). Since they are frequently utilized, it may be advantageous for the cell to express high levels of these proteins. Some of the highly expressed monomeric forms act in commonly utilized signaling cascades or signal transduction pathways (for example MKPs in MAPK signaling cascades or PTPN6/11 in PTK/Ras signal transduction). Others perform critical functions associated with morphogenesis such as regulating the actin cytoskeleton, cell division, and gene transcription.
Many phosphatases, such as the RPTPs and the myotubularins, are weakly expressed or not present at all. Why are they present at such low levels? In some cases, these proteins may perform specialized developmental tasks that are limited to a small group of cells or only occur for a short period of time. Alternately, these proteins may be exceptionally efficient in their roles or they may be more important in the adult than the embryo. Those that are never expressed may be pseudogenes.
It was common to find that phosphatases in the human genome were absent in S. purpuratus. There may be a number of reasons for this. First, gene duplication was common in chordate evolution. Since any common ancestor shared between S. purpuratus and humans would be basal to the phylum Chordata, it is not surprising that a single S. purpuratus homologue is often found for multiple vertebrate genes (Example: SpMTMR6 is homologous to HsMTMRs 6, 7, and 8).
Other genes missing in the S. purpuratus genome may be present, but were not identified in this study. Although every effort was made to be thorough and comprehensive, we have been conservative in the identification of genes. If two sequences were highly similar (95–100% identical) and possibly overlapped, they were identified as a single sequence. Also, several sequences had to be omitted because they were too incomplete for analysis, improperly assembled in the genome, improperly identified by software, or had other problems associated with this sort of in silico analysis.
Homologues may also be missing because, in some cases, two genes with similar functions could have evolved in the urchin. In this situation, loss of the gene homologous to that in humans would not have been detrimental. Other genes may be absent in the sea urchin because they were unnecessary. Potential candidates include PTPRA, PTPRE, PTPRC, PTPN20, DUSP13, CDC14B, or MTM1. Even though these genes are expressed in members of both the protostomes and other deuterostomes, they are absent in S. purpuratus (or they just were not detected).
Sea urchin specific RPTPs were found using domain analysis
Most of the S. purpuratus RPTPs identified in this study have no obvious homologues. One way to better characterize these novel proteins is by domain analysis. As mentioned before, extracellular domains commonly found in RPTPs include the fibronectin type III (FN3), immunoglobulin-like (Ig), carbonic anhydrase-like (CA), and Meprin/A5/μ (MAM) domains as well as RDGS adhesion recognition motifs and heavily glycosylated regions (Andersen et al., 2005). Domain prediction programs detected six extracellular domain types in S. purpuratus: FN3, Ig, EGF, MAM, Lectin C (CLECT), and scavenger receptor cysteine-rich (SRCR) domains. Sequences corresponding to MBOAT, RVT, and xanthine uracil permease (Xan_ur_permease) were also predicted, but these are probably from neighboring genes or, in the case of RVT, an error in contig assembly (Raw data presented in Suppl 5 and 6). As in other invertebrates, extracellular domain structure did not necessarily correlate to similarity in catalytic domain structure (probably because evolution of extracellular regions is less constrained than that of PTPc domains).
Although S. purpuratus RPTPs were seldom identical to their homologues, they do share some structural similarities (Fig. 4). Like the Type R2A human homologues, SpPTPR D/F/S has two PTPc domains, a series of FN3 repeats and Ig domains (although only 2, not 3 were found). Also, SpPTP69D-like (a Type 9 RPTP) is structurally similar to DmPTP69D. Both have Ig and FN3 domains as well as two catalytic domains. They differ in that SpPTP69D only has one Ig and Fn3 domain while DmPTP69D has two of each. The RPTP most similar to its homologue is SpPTPR G/Z. This Type R5 RPTP is nearly identical to PTP99A, a Drosophila R5 RPTP.
One gene, SpPTPR D/F/S, was previously identified in S. purpuratus. Using PCR primers designed to conserved portions of the PTPc domain, Wessel et al. (1995) identified seven S. purpuratus RPTPs and deduced partial amino acid sequences (83–85 aa in length) for these (6 of these sequences correspond to those in this study). SpPTPR D/F/S appears to be SpPTPSp108. Using Rnase protection assays, they also found that this gene is expressed at moderately high levels in ovary, egg, blastula, and gastrula stages and that expression drops slightly at the pluteus stage. Thus, SpPTPR D/F/S may be a good candidate for developmental studies, where, like HsPTPRF it may help regulate junctional complexes.
Domain structure of another identified homologue, SpPTPRR (a Type R7 RPTP), remains a mystery. The SpPTPRR sequence is incomplete, containing only the PTPc domain. Since this sequence is at the end of the scaffold and no extracellular sequence has been found, the suggested SpPTPRR structure (Fig. 4) was based solely on known conformations of other PTPRR genes.
In the phylogenetic analysis, four S. purpuratus RPTP Type R3-like genes were identified: SpPTPRH-like 1, SpPTPRH-like2, SpPTPRscav, and SpPTPRO-like (Fig. 4, Suppl. 5 and 6). These probably correspond to four of the RPTPs identified by Wessel et al. (1995): PTPSp11 (PTPRH-like1), PTPSp135 (PTPRH-like2), PTPSp111 (PTPRscav), and PTPSp107 (PTPRO-like). Two of these proteins, PTPRH-like1 and PTPRH-like2, are structurally similar to human Type R3 PTPRs, with only one PTPc domain and a series of extracellular FN3 repeats. Structure of PTPRO-like was unclear because the sequence was incomplete. Wessel et al. determined that PTPRH-like2 is expressed at higher levels than PTPRH-like1 and that, in both, highest expression occurs during gastrulation (20× that in the egg or blastula). Based on this, PTPRH-like1 and 2 may act during gastrulation. Highest levels of PTPRO-like mRNA were observed in the ovary and egg, and expression decreased to half this level in the blastula, gastrula, and pluteus stages.
The other S. purpuratus Type R3-like gene is PTPRscav (SpPTPSp111 in Wessel et al., 1995). Two SpPTPRscav sequences are present in the neighbor-joining tree, but these are actually overlapping pieces of the same protein. The fully assembled sequence contains 3 extracellular SRCR repeats and 2 intracellular PTPc domains (most human Type 3 RPTPs have only one) (Fig. 4). Wessel et al. (1995) reported that this gene is expressed throughout development, and that highest mRNA levels are present in the egg (1.5×–2× more than in subsequent developmental stages).
To determine whether potential homologues exist for SpPTPRscav, SMART was queried for proteins containing both PTPc and SRCR domains. No matches were found, suggesting that PTPRscav may be novel. Interestingly, SRCR domains are conserved throughout the animal kingdom, both in soluble and membrane-bound proteins (Sarrias et al., 2004). In mammalian myeloid cells, SRCR domain-containing proteins are thought to facilitate immune responses and, in sea urchins, SRCR domain-containing proteins found in coelomocytes (SpSRCR1 and SpSRCR5) are also presumed to function in the immune response (Pancer et al., 1999). Other sea urchin SRCR domain-containing proteins, present in sperm, bind an egg jelly peptide (Dangott et al., 1989).
At least 4 unique RPTP groups were found in S. purpuratus: Unique group 1, Unique group 2, Unique group 3, and PTPRiz (Fig. 4, Suppl. 5 and 6). Unique group 1, the PTPRLec clade, contains 8 members (SpPTPRLec1–6 and SpPTPRFn1–2). Four of these proteins (SpPTPRLec1, 3, 4 and 5) have 1–2 CLECT domains, 1 FN3 domain (weakly conserved in PTPRLec3 and 4), and 2 PTPc domains (Fig. 4, Supp. 5 and 6). Two proteins in this clade differ in structure (SpPTPRFn1 and SpPTPRFn2), containing 2 FN3 repeats and 2 PTPc domains. Sequences of the remaining members may be incomplete.
To identify potential homologues of the PTPRLec proteins, SMART was queried for proteins that contain both PTPc and CLECT domains. Only one protein was found, Gallus gallus PTPRQ isoform 1 precursor (UPI0000448F75, glomerular mesangial cell receptor PTP). This protein, whose function is unknown, contains 1 inactive PTPc domain, 3 VWD domains, 1 CLECT domain, and 23 FN3 repeats. Although it may be distantly related to PTPRLec phosphatases, it bears little resemblance. CLECT domains are found throughout the Metazoa (and in some non-metazoan groups). They generally bind carbohydrate moieties and are typically found in proteins involved in humoral/immune response, cell adhesion, or regulation of endocytosis (Zelensky and Gready, 2005).
The other two sea urchin specific groups, unique groups 2 (SpPTPRY1, SpPTPRY2) and 3 (SpPTPRW1 and SpPTPRW2) are predicted to contain PTPc D1 and D2 domains and FN3 domains. Unique group 2 has at least 1 FN3 domain and Unique group 3 has 4 FN3 domains, 3 spaced close together near the PTPc D1 domain and 1 that is closer to the carboxy terminal. Wessel et al. (1995) identified SpPTPRW1 as SpPTPSp12. This gene was not detectable in the Rnase protection assays, the tiling microarrays, or the EST datasets. If it is expressed, it is probably present at very low levels.
Finally, there is a fourth unique group with only one representative, SpPTPRiz (Fig. 4, Suppl. 5 and 6). This RPTP has a single EGF binding domain at the carboxy-terminal followed by 13 FN3 repeats and 2 PTPc domains.
Further research will be necessary to determine functions of these novel phosphatases, however the domain types observed (FN3, Ig, Clec, EGF, MAM, SRCR) suggest that several of these RPTPs could be involved in cell–cell interactions, cell adhesion, or physiological receptor activity. For more about known and hypothesized roles of RPTP extracellular domains, see Brady-Kalnay and Tonks (1995); Bixby (2001).
Blocking expression of phosphatases interferes with endomesoderm formation
Previous studies of phosphatases in the sea urchin have mainly focused on their roles in fertilization (Kumano et al., 2001; Whalley et al., 1991; Tash et al., 1988; Brokaw, 1987; Murofushi et al., 1986), regulation of the egg cytoskeleton (Tosuji et al., 2000; Asano and Mabuchi, 2001; Henson et al., 2003), and cell cycle control (Hiriyanna et al., 1995; Johnston et al., 1994; Patel and Whitaker, 1991; Arion and Meijer, 1989; Marc et al., 2004; Philipova et al., 2005a,b; Tosuji et al., 1992, 2003; Gliksman et al., 1992; Wright and Schatten, 1995). Only a few types of phosphatases have been examined, primarily PP1, PP2A, DUSP1/2, or calcineurin, although, as mentioned above, Wessel et al. (1995) cloned and examined expression of several urchin-specific RPTPs. One reason so little is known is that most studies relied on chemical inhibitors that affect specific phosphatase groups. For example, sodium orthovanadate and other vanadate inhibitors were used to block PTP activity whereas okadaic acid or calyculin were used to block type1 and type 2A ser–thr phosphatases.
Inhibitor experiments must be cautiously evaluated. Sometimes these inhibitors affect proteins other than those of interest. For example, when okadaic acid and calyculin were first used, investigators thought that these would allow one to distinguish PP1 from PP2A. Okadaic acid is ~100 times more efficient at blocking PP2A activity than PP1 activity, but calyculin blocks both equally. When calyculin inhibited a reaction, but okadaic acid did not, investigators originally concluded that PP1 activity was responsible. Alternately, when calyculin and okadaic acid both blocked a reaction, they concluded that PP2A was acting. As new PPP phosphatases, also sensitive to okadaic acid and calyculin, were discovered, it became clear that the “PP2A” effects might instead be PP4, PP5, and/or PP6 effects. The same thing has happened with sodium orthovanadate. It blocks PTPs, but can also inhibit alkaline phosphatases, acid phosphatases, and aryl sulfatases.
With a few exceptions (Cervello et al., 1999; Wessel et al., 1995), studies of phosphatase activity in the sea urchin have not looked at developmental events following first cleavage. These events are of particular interest to those studying sea urchin development. In recent years, a primary objective of the sea urchin research community has been to understand interactions between genes regulating endomesoderm specification. To better understand these relationships, the sea urchin gene regulatory network was developed (http://sugp.caltech.edu/endomes/). Although much has been learned about signal transduction pathways and transcription factors acting in endomesoderm formation, very little is known about the phosphatases and kinases that regulate these pathways.
To assess roles of the identified urchin phosphatases in endomesoderm formation, a literature review of phosphatase functions in model organisms was superimposed on sea urchin signal transduction pathways (Table 5). PTP and ser–thr phosphatases regulate or are influenced by a number of endomesoderm network genes including Ets, Alx1, Gcm, Snail, Brachyury, and Hox11/13b. Signaling pathways necessary for endomesoderm formation that are regulated by phosphatases include the canonical Wnt, non-canonical Wnt, Delta/Notch, EGF, FGF, PDGF, and VEGF pathways. Table 5 gives examples of phosphatases that may influence endomesoderm formation in the urchin. Based on this compilation, the following sea urchin phosphatases are hypothesized to affect endomesoderm formation: PP1 alpha/beta/gamma, PP1 beta, PP1-like, PP2A alpha/beta, PP4c, PP2B (calcineurin), PTPR D/F/S, PTPRR, PTPN 1/2, PTPN 13, PTPN 14/21, DUSP 6/7/9, PTENa, PTENb, and/or CDC25 A/B/C. Undoubtedly, there are many additional phosphatases critical for endomesoderm specification. For example, phosphatases that influence cell motility (such as PTP4A1) are surely critical for ingression of primary mesenchyme cells and invagination of the archenteron, and those that regulate gene transcription (such as EYA or Ctdp) are likely to be utilized in the production of proteins needed for endomesoderm cell specification and differentiation.
Table 5.
Examples of phosphatases acting in endomeosderm formation
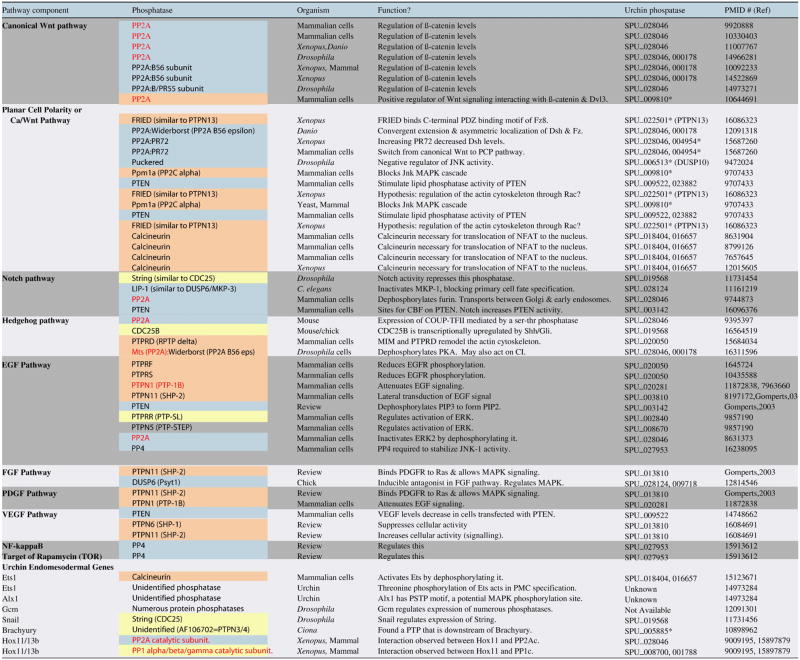 |
Based on the predictions of oligonucleotide tiling arrays, the genes in blue are highly expressed, those in peach are expressed at moderate levels, and those in yellow are present at low or very low levels. Genes that are not highlighted are predicted to be absent in developing sea urchins (or present at extremely low levels). MASOs were produced for genes shown in red. *Potentially similar sea urchin sequence.
What functional data are available concerning the roles of phosphatases in later embryonic development? In one of the only studies looking at roles of phosphatases after first cleavage, Cervello et al. (1999) used chemical inhibitors to block PTPs and ser–thr phosphatases in Paracentrotus lividus. They used sodium orthovanadate to block PTPs, and okadaic acid to block ser–thr phosphatases. In these experiments, embryos exposed to 0.5–1 mM sodium orthovanadate between the hatching blastula and early gastrula stages formed smaller spicules or completely failed to undergo skeletogenesis. They expressed lower levels of type I fibrillar collagen. If, instead, embryos were exposed to 20–40 nM okadaic acid, development proceeded as it normally would, leading the authors to conclude that PTPs regulate micromere differentiation and skeletogenesis in the sea urchin, but that ser–thr phosphatases are not involved.
As we mentioned earlier, much more is known about the effects of okadaic acid now than was known 7 years ago. This inhibitor blocks PP1, PP2A, PP4, PP5, and PP6, but not PP2B, PP7, or PPMs. Also, it is ~100× less efficient at blocking PP1 than the type 2A ser–thr phosphatases. Thus, it is entirely possible that ser–thr phosphatases unaffected by okadaic acid could act in skeletogenesis.
The recent availability of S. purpuratus EST and genomic data have made it much easier to clone individual genes and look at the effects of overexpressing phosphatases or selectively blocking activity by injecting MASOs (a much more specific means of blocking phosphatase activity). To screen for phosphatases that have specific developmental functions in the embryo, a panel of morpholino antisense oligonucleotides (MASOs) targeted to fourteen different developmentally expressed phosphatases was developed (Suppl 7). The MASOs were each injected at three different concentrations into zygotes, and the morphology of the injected embryos was scored at 24, 48, and 96 h post-fertilization (when control embryos were mesenchyme blastula, late gastrula, and pluteus stages, respectively). Five of the fourteen MASOs (PP4c, PP6c, Ppm1a, Ppm1g, and Ptpn9) did not produce consistently obvious abnormalities at any of the doses used. Two MASOs (PP1 alpha and DUSP10) caused cleavage-stage arrest and/or lethality at all doses (data not shown). The remaining seven (PP1 beta, PP2A alpha/beta, PPEF2, Ppm1d, PTP4A 1/2/3, PTPN 1/2, and DUSP 1/2/4/5) are necessary for skeletogenesis, either for patterning or for formation of spicules (Table 4 and Fig. 5). Based on this it is likely that ser–thr phosphatases as well as PTPs affect spiculogenesis. Several phosphatases may also influence formation of the gut. In five cases, PP2A alpha/beta, PPEF2, Ppm1d, PTP4A 1/2/3, and DUSP 1/2/4/5, formation of the endoderm was abnormal or absent. While further controls are necessary both to determine the efficacy of each MASO in depleting its target phosphatase and whether the observed morphological defects are specifically caused by depletion of that phosphatase, results of this preliminary study refute the conclusions of Cervello et al. (1999) that ser–thr phosphatases are unimportant in skeletogenesis and suggest that these phosphatases may indeed play critical roles in endomesoderm formation, warranting further investigation.
Table 4.
Larval phenotypes obtained with anti-phosphatase MASOs
| MASO target | Morphological defects |
|---|---|
| PP1 alpha (SPU_006956) | Late cleavage/early blastula stage arrest at all doses; cell division defects? |
| PP1 beta (SPU_001788) | Skeletal patterning defects (short skeletal rods, sometimes crossing or parallel; oral ectoderm defect?) at medium and high doses |
| PP2A alpha/beta (SPU_025182) | Gastrulation and skeletogenic defects; poorly differentiated/radialized ectoderm at medium and high doses |
| PPEF2 (SPU_028868) | Endoderm and oral ectoderm defects at low and medium dose; some loss of skeletons at high dose |
| Ppm1d (SPU_020266) | Nothing obvious at low and medium doses; global differentiation defects at high dose, with small guts and short spicules, rounded ectoderm |
| PTP4A 1/2/3 (SPU_008895) | Skeletal defects and small gut at medium dose; blastula stage arrest at high dose |
| PTPN 1/2 (SPU_020281) | Failure of skeletogenesis at low dose (ranging from no spicules, to short spicules or in some cases a single spicule); more general differentiation defects at medium and high doses |
| DUSP 1/2/4/5 (SPU_021143) | Endoderm and oral ectoderm malformed or diminished at medium dose |
| DUSP10 (SPU_006513) | Cleavage stage arrest/death at all doses |
Fig. 5.
Examples of 4- to 5-day larval phenotypes obtained by injecting zygotes with the following morpholino antisense oligonucleotide (MASOs): (A) non-specific control MASO (normal pluteus); (B) anti-PP1 beta; (C) anti-PP2A alpha/beta; (D) anti-Ppm1d; (E) anti-PPEF2; (F) anti-PTP4A 1/2/3; (G) anti-PTPN 1/2; and (H) anti-DUSP 1/2/4/5. All embryos are oriented more or less the same, with the oral side toward the left (except in panel C, which is radialized), and the anal side down.
One interesting result of the MASO study was that PP2A alpha/beta, a phosphatase blocked by okadaic acid, lacked skeletogenic or gastrulation defects in the Cervello study, but exhibited these defects in response to MASO injection. A possible explanation for this is that the okadaic acid concentration may have been too low to see a noticeable effect. Many phosphatases overlap in function with other phosphatases. This redundancy is an important means of protecting the embryo during development. If one phosphatase is lost, development proceeds as it should because “backup” phosphatases allow the animal to regulate. In this case, PP2A alpha/beta may dephosphorylate the same protein as a second phosphatase. If levels of PP2A alpha/beta are decreased, the “backup” phosphatase may dephosphorylate well enough for development to proceed “normally.” If PP2A alpha/beta expression is completely abolished, however, the “backup” phosphatase may not be able to compensate and development is grossly disrupted. These sorts of functional redundancies are commonly encountered in phosphatase studies (Table 5).
It is also notable that the phenotypes observed for PP2A alpha/beta, PP1 alpha, PP1 beta, and some of the other multifunctional phosphatases are probably not the result of blocking a single cellular process, but are caused by blocking several. Very careful studies blocking these multifunctional phosphatases in limited regions of the embryo or for limited durations combined with knowledge of their functions in other animals will be useful to learn more about specific functions of these phosphatases in endomesoderm formation.
Conclusions
Numerous PTP and ser–thr phosphatases were detected in the S. purpuratus genome. In general, most phosphatases have been conserved although, as expected, a single phosphatase in S. purpuratus is often homologous to several phosphatases in chordates. One group, the RPTPs, is an exception. Only a few sea urchin RPTPs were homologous to chordate RPTPs and several sea urchin-specific clades were identified. At least one of these (the PTPRLec clade, Unique group 1) probably arose by lineage specific expansion.
Learning more about roles of phosphatases and kinases (see Bradham et al., 2006) during sea urchin development will be critical to understanding how endomesoderm formation and other developmental processes are regulated. Also, much remains to be learned about general roles of phosphatases. Little information is available for the atypical DSPs, and many of the RPTPs. Among the ser–thr phosphatases, detailed studies of PP5, PP6, PP7, and the PPMs would be useful.
Availability of the sea urchin genome will greatly facilitate investigation of phosphatases and proteins in echinoderm development. With genomic data it will be much easier to clone individual phosphatases, predict groups that may overlap in function, and block phosphatase activity in vivo. The S. purpuratus genome also provides an invaluable resource to those studying in deuterostome or metazoan evolution.
Acknowledgments
We would like to acknowledge Arcady Mushegian, Manisha Goel, Kathy Foltz, Richard Hynes, Cyndi Bradham, Mariano Coll, Jenifer Croce, Cynthia Messier, Esther Miranda, Alex Primus, Jonathan Rast, and Gary Wessel for annotating some of the phosphatases discussed. Thanks to Jim Balhoff, Wendy Beane, Simon Wu, and Greg Wray for assistance with the trees. Thanks to Cyndi Bradham for suggesting this project, to Jenifer Croce for reviewing the manuscript and to all of the McClay lab members for their support. This work was financed by several organizations including AHA grant 0420074Z to CAB, NIH grants HD14483, HD039948-03, and GM61464 to DRM, and NIH grant GM070840 to JC. Morpholinos were purchased using funds from the Stowers Institute for Medical Research. K. D.W. was supported by the U.S. Army Medical Research and Materiel Command under W81XWH-04-1-0324.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ydbio.2006.08.050.
References
- Alonso A, Rojas A, Godzik A, Mustelin T. The dual-specific protein tyrosine phosphatase family. Top Curr Genet. 2004a;5:333–358. [Google Scholar]
- Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004b;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Andersen JN, Mortensen OH, Peters GH, Drake PG, Iversen LF, Olsen OH, Jansen PG, Andersen HS, Tonks NK, Moller NPH. Structural and evolutionary relationships among protein tyrosine phosphatase domains. Mol Cell Biol. 2001;21 (21):7117–7136. doi: 10.1128/MCB.21.21.7117-7136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JN, Del Vecchio RL, Kannan N, Gergel J, Neuwald AF, Tonks NK. Computational analysis of protein tyrosine phosphatases: practical guide to bioinformatics and data resources. Methods. 2005;35:90–114. doi: 10.1016/j.ymeth.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Andreeva AV, Kutuzov MA. RdgC/PP5-related phosphatases: novel components in signal transduction. Cell Signal. 1999;11 (8):555–562. doi: 10.1016/s0898-6568(99)00032-7. [DOI] [PubMed] [Google Scholar]
- Arion D, Meijer L. M-phase-specific protein kinase from mitotic sea urchin eggs: cyclic activation depends on protein synthesis and phosphorylation but does not require DNA or RNA synthesis. Exp Cell Res. 1989;183 (2):361–375. doi: 10.1016/0014-4827(89)90397-2. [DOI] [PubMed] [Google Scholar]
- Asano Y, Mabuchi I. Calyculin-A, an inhibitor for protein phosphatases, induces cortical contraction in unfertilized sea urchin eggs. Cell Motil Cytoskelet. 2001;48 (4):245–261. doi: 10.1002/cm.1013. [DOI] [PubMed] [Google Scholar]
- Barford D, Das AK, Egloff MP. The structure and mechanism of protein phosphatases: insights into catalysis and regulation. Annu Rev Biophys Biomol Struct. 1998;27:133–164. doi: 10.1146/annurev.biophys.27.1.133. [DOI] [PubMed] [Google Scholar]
- Bastians H, Ponstingl H. The novel human protein serine/threonine phosphatase 6 is a functional homologue of budding yeast Sit4p and fission yeast ppe1, which are involved in cell cycle regulation. J Cell Sci. 1996;109:2865–2874. doi: 10.1242/jcs.109.12.2865. [DOI] [PubMed] [Google Scholar]
- Baum B. Winging it—Actin on the fly. Dev Cell. 2002;2:125–133. doi: 10.1016/s1534-5807(02)00119-3. [DOI] [PubMed] [Google Scholar]
- Beane WS, Voronina E, Wessel GM, McClay DR. GTP-binding proteins and their regulators in S. purpuratus: GTPases and the prediction of “Reduced Complexity” in early deuterostomes. Dev Biol 2006 [Google Scholar]
- Bhaduri A, Sowdhamini R. A genome-wide survey of human tyrosine phosphatases. Protein Eng. 2003;16 (12):881–888. doi: 10.1093/protein/gzg144. [DOI] [PubMed] [Google Scholar]
- Bixby JL. Ligands and signaling through receptor-type tyrosine phosphatases. Life. 2001;51:157–163. doi: 10.1080/152165401753544223. [DOI] [PubMed] [Google Scholar]
- Bradham CA, Foltz KR, Beane WS, Arnone MI, Rizzo F, Coffman JA, Mushegian A, Goel M, Morales J, Geneviere AM, Lapraz F, Robertson AJ, Kelkar H, Loza-Coll M, Townley IK, Raisch M, Roux MM, Lepage T, Gache C, McClay DR, Manning G. The sea urchin kinome: a first look. Dev Biol. 2006;300 (1):180–193. doi: 10.1016/j.ydbio.2006.08.074. [DOI] [PubMed] [Google Scholar]
- Brady-Kalnay SM, Tonks NK. Protein tyrosine phosphatases as adhesion receptors. Curr Opin Cell Biol. 1995;7:650–657. doi: 10.1016/0955-0674(95)80106-5. [DOI] [PubMed] [Google Scholar]
- Brokaw CJ. A lithium-sensitive regulator of sperm flagellar oscillation is activated by cAMP-dependent phosphorylation. J Cell Biol. 1987;105 (4):1789–1798. doi: 10.1083/jcb.105.4.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Zhang L, Ruiz-Lozano P, Yang Q, Chien KR, Graham RM, Zhou M. A novel putative protein–tyrosine phosphatase contains a BRO1-like domain and suppresses Ha-ras-mediated transformation. J Biol Chem. 1998;273 (33):21077–21083. doi: 10.1074/jbc.273.33.21077. [DOI] [PubMed] [Google Scholar]
- Cervello M, Sanfilippo R, Isola G, Virruso L, Scalia G, Cammarata G, Gambino R. Phosphorylation-dependent regulation of skeletogenesis in sea urchin micromere-derived cells and embryos. Dev Growth Differ. 1999;41 (6):769–775. doi: 10.1046/j.1440-169x.1999.00479.x. [DOI] [PubMed] [Google Scholar]
- Ceulemans H, Bollen M. Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol Rev. 2004;84:1–39. doi: 10.1152/physrev.00013.2003. [DOI] [PubMed] [Google Scholar]
- Ceulemans H, Stalmans W, Bollen M. Regulator-driven functional diversification of protein phosphatase-1 in eukaryotic evolution. BioEssays. 2002;24:371–381. doi: 10.1002/bies.10069. [DOI] [PubMed] [Google Scholar]
- Chen H, Duncan IC, Bozorgchami H, Lo SH. Tensin1 and a previously undocumented family member, tensin2, positively regulate cell migration. Proc Natl Acad Sci U S A. 2002;99 (2):733–738. doi: 10.1073/pnas.022518699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A, Kaldis P, Solomon MJ. Dephosphorylation of human cyclin-dependent kinases by protein phosphatase type 2C alpha and beta 2 isoforms. J Biol Chem. 2000;275 (44):34744–34749. doi: 10.1074/jbc.M006210200. [DOI] [PubMed] [Google Scholar]
- Cohen PTW. Overview of protein serine/threonine phosphatases. Top Curr Genet. 2004;5:1–20. [Google Scholar]
- Cohen PTW, Philp A, Vázquez-Martin C. Protein phosphatase 4—From obscurity to vital functions. FEBS Lett. 2005;579:3278–3286. doi: 10.1016/j.febslet.2005.04.070. [DOI] [PubMed] [Google Scholar]
- Cui Y, Liao YC, Lo SH. Epidermal growth factor modulates tyrosine phosphorylation of a novel tensin family member, tensin3. Mol Cancer Res. 2004;2 (4):225–232. [PubMed] [Google Scholar]
- Dangott LJ, Jordan JE, Bellet RA, Garbers DL. Cloning of the mRNA for the protein that crosslinks to the egg peptide speract. Proc Natl Acad Sci U S A. 1989;86:2128–2132. doi: 10.1073/pnas.86.7.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai CJ, Krueger NX, Saito H, Zinn K. Competition and cooperation among receptor tyrosine phosphatases control motoneuron growth cone guidance in Drosophila. Development. 1997;124 (10):1941–1952. doi: 10.1242/dev.124.10.1941. [DOI] [PubMed] [Google Scholar]
- Ducruet AP, Vogt A, Wipf P, Lazo JS. Dual specificity protein phosphatases: therapeutic targets for cancer and Alzheimer’s disease. Annu Rev Med. 2005;45:725–750. doi: 10.1146/annurev.pharmtox.45.120403.100040. [DOI] [PubMed] [Google Scholar]
- Farooq A, Zhou MM. Structure and regulation of MAPK phosphatases. Cell Signalling. 2004;16:769–779. doi: 10.1016/j.cellsig.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Foehr ED, Lorente G, Vincent V, Nikolich K, Urfer R. FAS associated phosphatase (FAP-1) blocks apoptosis of astrocytomas through dephosphorylation of FAS. J Neuro-Oncol. 2005;74 (3):241–248. doi: 10.1007/s11060-004-7202-x. [DOI] [PubMed] [Google Scholar]
- Gandhi TKB, Chandran S, Peri S, Saravana R, Amanchy R, Prasad TSK, Pandey A. A bioinformatics analysis of protein tyrosine phosphatases in humans. DNA Res. 2005;12:79–89. doi: 10.1093/dnares/12.2.79. [DOI] [PubMed] [Google Scholar]
- Gliksman NR, Parsons SF, Salmon ED. Okadaic acid induces interphase to mitotic-like microtubule dynamic instability by inactivating rescue. J Cell Biol. 1992;119 (5):1271–1276. doi: 10.1083/jcb.119.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomperts BD, Kramer IM, Tatham PER. Signal Transduction. Elsevier Academic Press; San Diego: 2003. [Google Scholar]
- Groenendyk J, Lynch J, Michalak M. Calreticulin, Ca2+, and calcineurin-signaling from the endoplasmic reticulum. Mol Cells. 2004;17 (3):383–389. [PubMed] [Google Scholar]
- Hafizi S, Ibraimi F, Dahlback B. C1-TEN is a negative regulator of the Akt/PKB signal transduction pathway and inhibits cell survival, proliferation, and migration. FASEB J. 2005;19 (8):971–973. doi: 10.1096/fj.04-2532fje. [DOI] [PubMed] [Google Scholar]
- Hanada M, Ninomiya-Tsuji J, Komaki K, Ohnishi M, Katsura K, Kanamaru R, Matsumoto K, Tamura S. Regulation of the TAK1 signaling pathway by protein phosphatase 2C. J Biol Chem. 2001;276 (8):5753–5759. doi: 10.1074/jbc.M007773200. [DOI] [PubMed] [Google Scholar]
- Hannon GJ, Casso D, Beach D. KAP: a dual specificity phosphatase that interacts with cyclin-dependent kinases. Proc Natl Acad Sci. 1994;91:1731–1735. doi: 10.1073/pnas.91.5.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynie DT, Ponting CP. The N-terminal domains of tensin and auxilin are phosphatase homologues. Protein Sci. 1996;5:2643–2646. doi: 10.1002/pro.5560051227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson JH, Kolnik SE, Fried CA, Nazarian R, McGreevy J, Schulberg KL, Detweiler M, Trabosh VA. Actin-based centripetal flow: phosphatase inhibition by calyculin-A alters flow pattern, actin organization, and actomyosin distribution. Cell Motil Cytoskeleton. 2003;56 (4):252–266. doi: 10.1002/cm.10149. [DOI] [PubMed] [Google Scholar]
- Hertog JD, Blanchetot C, Buist A, Overvoorde J, Van Der Sar A, Tertoolen LGJ. Receptor protein–tyrosine phosphatase signalling in development. Int J Dev Biol. 1999;43:723–733. [PubMed] [Google Scholar]
- Hiriyanna KT, Buck WR, Shen SS, Ingebritsen TS. Thiophosphorylated RCM-lysozyme, an active site-directed protein tyrosine phosphatase inhibitor, inhibits G2/M transition during mitotic cell cycle and uncouples MPF activation from G2/M transition. Exp Cell Res. 1995;216 (1):21–29. doi: 10.1006/excr.1995.1003. [DOI] [PubMed] [Google Scholar]
- Huang TY, DerMardirossian C, Bokoch GM. Cofilin phosphatases and regulation of actin dynamics. Curr Opin Cell Biol. 2006;18 (1):26–31. doi: 10.1016/j.ceb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Huynh H, Bottini N, Williams S, Cherepanov V, Musumeci L, Saito K, Bruckner S, Vachon E, Wang X, Kruger J, Chow CW, Pellecchia M, Monosov E, Greer PA, Trimble W, Downey GP, Mustelin T. Control of vesicle fusion by a tyrosine phosphatase. Nat Cell Biol. 2004;6 (9):831–839. doi: 10.1038/ncb1164. [DOI] [PubMed] [Google Scholar]
- Ivanov VN, Ronai Z, Hei TK. Opposite roles of FAP-1 and dynamin in the regulation of Fas (CD95) translocation to the cell surface and susceptibility to Fas ligand-mediated apoptosis. J Biol Chem. 2006;281 (3):1840–1852. doi: 10.1074/jbc.M509866200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens V, Goris J. Protein phosphatase 2A: a highly regulated family of serine/threonine phsophatases implicated in cell growth and signaling. Biochem J. 2001;353:417–439. doi: 10.1042/0264-6021:3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JA, Sloboda RD, Silver RB. Phosphoprotein phosphatase 1 (PP1) is a component of the isolated sea urchin mitotic apparatus. Cell Motil Cytoskeleton. 1994;29 (3):280–290. doi: 10.1002/cm.970290311. [DOI] [PubMed] [Google Scholar]
- Kamenski T, Heilmeier S, Meinhart A, Cramer P. Structure and mechanism of RNA polymerase II CTD phosphatase. Mol Cell. 2004;15 (3):399–407. doi: 10.1016/j.molcel.2004.06.035. [DOI] [PubMed] [Google Scholar]
- Kozlov G, Cheng J, Ziomek E, Banville D, Gehring K, Ekiel I. Structural insights into molecular function of the metastasis-associated phosphatase PRL-3. J Biol Chem. 2004;279 (12):11882–11889. doi: 10.1074/jbc.M312905200. [DOI] [PubMed] [Google Scholar]
- Kristjánsdóttir K, Rudolph J. Cdc25 phosphatases and cancer. Chem Biol. 2004;11:1043–1051. doi: 10.1016/j.chembiol.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Kumano M, Carroll DJ, Denu JM, Foltz KR. Calcium-mediated inactivation of the MAP kinase pathway in sea urchin eggs at fertilization. Dev Biol. 2001;236 (1):244–257. doi: 10.1006/dbio.2001.0328. [DOI] [PubMed] [Google Scholar]
- Kutuzov MA, Solov’eva OV, Andreeva AV, Bennett N. Protein Ser/Thr phosphatases PPEF interact with calmodulin. Biochem Biophys Res Commun. 2002;293 (3):1047–1052. doi: 10.1016/S0006-291X(02)00338-8. [DOI] [PubMed] [Google Scholar]
- Laporte J, Bedez F, Bolino A, Mandel JL. Myotubularins, a large disease-associated family of cooperating catalytically active and inactive phosphoinositide phosphatases. Hum Mol Genet. 2003;12 (2):R285–R292. doi: 10.1093/hmg/ddg273. [DOI] [PubMed] [Google Scholar]
- Lo SH. Tensin. Int J Biochem Cell Biol. 2004;36 (1):31–34. doi: 10.1016/s1357-2725(03)00171-7. [DOI] [PubMed] [Google Scholar]
- Luan S. Protein phosphatases in plants. Annu Rev Plant Biol. 2003;54:63–92. doi: 10.1146/annurev.arplant.54.031902.134743. [DOI] [PubMed] [Google Scholar]
- Maj MC, Cameron JM, Robinson BH. Pyruvate dehydrogenase phosphatase deficiency: orphan disease or an under-diagnosed condition? Mol Cell Endocrinol. 2006;249(1–2):1–9. doi: 10.1016/j.mce.2006.02.003. (Electronic publication) [DOI] [PubMed] [Google Scholar]
- Marc J, Belle R, Morales J, Cormier P, Mulner-Lorillon O. Formulated glyphosate activates the DNA-response checkpoint of the cell cycle leading to the prevention of G2/M transition. Toxicol Sci. 2004;82 (2):436–442. doi: 10.1093/toxsci/kfh281. [DOI] [PubMed] [Google Scholar]
- Murofushi H, Ishiguro K, Takahashi D, Ikeda J, Sakai H. Regulation of sperm flagellar movement by protein phosphorylation and dephophorylation. Cell Motil Cytoskeleton. 1986;6 (2):83–88. doi: 10.1002/cm.970060203. [DOI] [PubMed] [Google Scholar]
- Murray MV, Kobayashi R, Krainer AR. The type 2C Ser/Thr phosphatase PP2Cg is a pre-mRNA splicing factor. Genes Dev. 1999;13:87–97. doi: 10.1101/gad.13.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neel BG, Tonks NK. Protein tyrosine phosphatases in signal transduction. Curr Opin Cell Biol. 1997;9:193–204. doi: 10.1016/s0955-0674(97)80063-4. [DOI] [PubMed] [Google Scholar]
- Niwa R, Nagata-Ohashi K, Takeichi M, Mizuno K, Uemura T. Control of actin reorganization by Slingshot, a family of phosphatases that dephosphorylate ADF/Cofilin. Cell. 2002;108:233–246. doi: 10.1016/s0092-8674(01)00638-9. [DOI] [PubMed] [Google Scholar]
- Pancer Z, Rast JP, Davidson EH. Origins of immunity: transcription factors and homologues of effector genes of the vertebrate immune system expressed in sea urchin coelomocytes. Immunogenetics. 1999;49:773–786. doi: 10.1007/s002510050551. [DOI] [PubMed] [Google Scholar]
- Patel R, Whitaker M. Okadaic acid suppresses calcium regulation of mitosis onset in sea urchin embryos. Cell Regul. 1991;2 (5):391–402. doi: 10.1091/mbc.2.5.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipova R, Kisielewska J, Lu P, Larman M, Huang JY, Whitaker M. ERK1 activation is required for S-phase onset and cell cycle progression after fertilization in sea urchin embryos. Development. 2005a;132 (3):579–589. doi: 10.1242/dev.01607. [DOI] [PubMed] [Google Scholar]
- Philipova R, Larman MG, Leckie CP, Harrison PK, Groigno L, Whitaker M. Inhibiting MAP kinase activity prevents calcium transients and mitosis entry in early sea urchin embryos. J Biol Chem. 2005b;280 (26):24957–24967. doi: 10.1074/jbc.M414437200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raugei G, Ramponi G, Chiarugi P. Low molecular weight protein tyrosine phosphatases: small, but smart. Cell Mol Life Sci. 2002;59:941–949. doi: 10.1007/s00018-002-8481-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raftopoulou M, Etienne-Manneville S, Self A, Nicholls S, Hall A. Regulation of cell migration by the C2 domain of the tumor suppressor PTEN. Science. 2004;303:1179–1181. doi: 10.1126/science.1092089. [DOI] [PubMed] [Google Scholar]
- Ramulu P, Kennedy M, Xiong WH, Williams J, Cowan M, Blesh D, Yau KW, Hurley JB, Nathans J. Normal light response, photoreceptor integrity and rhodopsin dephosphorylation in mice lacking both protein phosphatases with EF hands (PPEF-1 and PPEF-2) Mol Cell Biol. 2001;21:8605–8614. doi: 10.1128/MCB.21.24.8605-8614.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebay I, Silver SJ, Tootle TL. New vision from Eyes absent: transcription factors as enzymes. Trends Genet. 2005;21 (3):163–171. doi: 10.1016/j.tig.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Samanta MP, Tongprasit W, Istrail S, Cameron A, Tu Q, Davidson EH. High resolution transcriptome map of the sea urchin embryo. Dev Biol. 2006 doi: 10.1126/science.1131898. [DOI] [PubMed] [Google Scholar]
- Sarrias MR, Gronlund J, Padilla O, Madsen J, Holmskov U, Lozano F. The scavenger receptor Cysteine-rich (SRCR) domain: an ancient and highly conserved protein module of the innate immune system. Crit Rev Immunol. 2004;24 (1):1–37. doi: 10.1615/critrevimmunol.v24.i1.10. [DOI] [PubMed] [Google Scholar]
- Sastry SK, Rajfur Z, Liu BP, Cote JF, Tremblay M, Burridge K. PTP–PEST couples membrane protrusion and tail retraction via VAV2 and p190RhoGAP. J Biol Chem. 2006 doi: 10.1074/jbc.M600897200. (Electronic publication ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaapveld R, Wieringa B, Hendriks W. Receptor-like protein tyrosine phosphatases: Alike and yet so different. Mol Biol Rep. 1997;24:247–262. doi: 10.1023/a:1006870016238. [DOI] [PubMed] [Google Scholar]
- Schweighofer A, Hirt H, Meskiene I. Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci. 2004;9 (5):1360–1385. doi: 10.1016/j.tplants.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Stoker A, Dutta R. Protein tyrosine phosphatases and neural development. BioEssays. 1998;20:463–472. doi: 10.1002/(SICI)1521-1878(199806)20:6<463::AID-BIES4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Takekawa M, Maeda T, Saito H. Protein phosphatase 2Calpha inhibits the human stress-responsive p38 and JNK MAPK pathways. EMBO J. 1998;17 (16):4744–4752. doi: 10.1093/emboj/17.16.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tash JS, Krinks M, Patel J, Means RL, Klee CB, Means AR. Identification, characterization, and functional correlation of calmodulin-dependent protein phosphatase in sperm. J Cell Biol. 1988;106 (5):1625–1633. doi: 10.1083/jcb.106.5.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Rosell J, Machin F, Aragón L. Cdc14 and the temporal coordination between mitotic exit and chromosome segregation. Cell Cycle. 2005;4 (1):109–112. doi: 10.4161/cc.4.1.1356. [DOI] [PubMed] [Google Scholar]
- Tosuji H, Mabuchi I, Fusetani N, Nakazawa T. Calyculin A induces contractile ring-like apparatus formation and condensation of chromosomes in unfertilized sea urchin eggs. Proc Natl Acad Sci U S A. 1992;89 (22):10613–10617. doi: 10.1073/pnas.89.22.10613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosuji H, Miyaji K, Fusetani N, Nakazawa T. Effect of calyculin A on the surface structure of unfertilized sea urchin eggs. Cell Motil Cytoskeleton. 2000;46 (2):129–136. doi: 10.1002/1097-0169(200006)46:2<129::AID-CM5>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Tosuji H, Fusetani N, Seki Y. Calyculin A causes the activation of histone H1 kinase and condensation of chromosomes in unfertilized sea urchin eggs independently of the maturation-promoting factor. Comp Biochem Physiol, C Toxicol Pharmacol. 2003;135 (4):415–424. doi: 10.1016/s1532-0456(03)00143-1. [DOI] [PubMed] [Google Scholar]
- Trautmann S, McCollum D. Cell cycle: New functions for Cdc14 family phosphatases. Curr Biol. 2002;12:R733–R735. doi: 10.1016/s0960-9822(02)01250-2. [DOI] [PubMed] [Google Scholar]
- Wadham C, Gamble JR, Vadas MA, Khew-Goodall Y. The protein tyrosine phosphatase Pez is a major phosphatase of adherens junctions and dephosphorylates β-catenin. Mol Biol Cell. 2003;14:2520–2529. doi: 10.1091/mbc.E02-09-0577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Lemay S, Tsai S, Veillette A. SH2 domain-mediated interaction of inhibitory protein tyrosine kinase Csk with protein tyrosine phosphatase-HSCF. Mol Cell Biol. 2001;21 (4):1077–1088. doi: 10.1128/MCB.21.4.1077-1088.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock G. The Sea Urchin Genome Sequencing Consortium, 2006. The genome of the sea urchin Strongylocentrotus purpuratus. Science. doi: 10.1126/science.1133609. (in review) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel GM, Clark F, Berg L. A diversity of enzymes involved in the regulation of reversible tyrosine phosphorylation in sea urchin egg and embryos. Comp Biochem Physiol. 1995;110B (3):493–502. doi: 10.1016/0305-0491(94)00212-d. [DOI] [PubMed] [Google Scholar]
- Whalley T, Crossley I, Whitaker M. Phosphoprotein inhibition of calcium-stimulated exocytosis in sea urchin eggs. J Cell Biol. 1991;113 (4):769–778. doi: 10.1083/jcb.113.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart MJ, Dixon JE. PTEN and myotubularin phosphatases: from 3-phophoinositide dephosphorylation to disease. Trends Cell Biol. 2002;12 (12):579–585. doi: 10.1016/s0962-8924(02)02412-1. [DOI] [PubMed] [Google Scholar]
- Wishart MJ, Taylor GS, Slama JT, Dixon JE. PTEN and myotubularin phosphoinositide phosphatases: bring bioinformatics to the lab bench. Curr Opin Cell Biol. 2001;13:172–181. doi: 10.1016/s0955-0674(00)00195-2. [DOI] [PubMed] [Google Scholar]
- Wright SJ, Schatten G. Protein tyrosine phosphorylation during sea urchin fertilization: microtubule dynamics require tyrosine kinase activity. Cell Motil Cytoskeleton. 1995;30 (2):122–135. doi: 10.1002/cm.970300204. [DOI] [PubMed] [Google Scholar]
- Zelensky AN, Gready JE. The C-type lectin-like domain superfamily. FEBS J. 2005:272. doi: 10.1111/j.1742-4658.2005.05031.x. [DOI] [PubMed] [Google Scholar]