Abstract
The transcription factors Runx2 and Osx are necessary for osteoblast and odontoblast differentiation, while Dspp is important for odontoblast differentiation. The relationship among Runx2, Osx, and Dspp during tooth and craniofacial bone development remains unknown. In this study, we hypothesized that the roles of Runx2 and Osx in the regulation of osteoblast and odontoblast lineages may be independent of one another. The results showed that Runx2 expression overlapped with Osx in dental and osteogenic mesenchyme from E12 to E16. At the later stages, from E18 to PN14, Runx2 and Osx expressions remained intense in alveolar bone osteoblasts. However, Runx2 expression was down-regulated, whereas Osx expression was clearly seen in odontoblasts. At later stages, Dspp transcription was weakly present in osteoblasts, but strong in odontoblasts where Osx was highly expressed. In mouse odontoblast-like cells, Osx overexpression increased Dspp transcription. Analysis of these data suggests differential biological functions of Runx2, Osx, and Dspp during odontogenesis and osteogenesis. Abbreviations: E, embryonic day; PN, post-natal day; Dspp, dentin sialophosphoprotein; Osx, Osterix.
Keywords: Runx2, Osx, Dspp, odontoblast, osteoblast, tooth development
Introduction
Tooth development involves sequential and reciprocal interactions between dental epithelial and mesenchymal cells and proceeds through a series of cytodifferentiations in specific spatial-temporal patterns. Epithelial and mesenchymal cells differentiate into ameloblasts and odontoblasts, respectively (Linde and Goldberg, 1993). Odontoblasts synthesize and secrete collagenous and non-collagenous proteins (NCPs) to form dentin extracellular matrix (DECM). Among NCPs, dentin sialophosphoprotein (Dspp) is a phosphorylated protein representing a major component of non-collagenous DECM, highly expressed in odontoblasts and essential for dentinogenesis (D’Souza et al., 1997; Butler, 1998). Mutations of Dspp gene are associated with various forms of dentin genetic disorders (Xiao et al., 2001; Rajpar et al., 2002; Kim and Simmer, 2007). Dentinogenesis is a complex process in which multiple signaling pathways converge to induce dentin formation and is controlled by many growth and transcription factors (Thesleff, 2003).
Transcription factor Runx2 is necessary for osteoblast and odontoblast differentiation and regulates many bone- and tooth-related gene expressions. Runx2 determines the lineage of osteoblasts and odontoblasts from mesenchymal cells (Ducy et al., 1997; Lian et al., 2006). The temporal-spatial Runx2 expression pattern during bone and tooth formation has been described (Jiang et al., 1999; Bronckers et al., 2001; Yamashiro et al., 2002). For example, Runx2-deficient mice showed impaired tooth formation, progressing only to the cap/early bell stages (D’Souza et al., 1999). Moreover, persons with Runx2 gene mutations display dental disorders, with supernumerary teeth, abnormal tooth eruption, and tooth hypoplasia (Komori et al., 1997; Lee et al., 1997; Mundlos et al., 1997).
Osterix (Osx or Sp7) is another osteoblast-specific transcription factor and is expressed in tooth germ mesenchymal cells (Nakashima et al., 2002). Genetic studies have shown that cortical bone and bone trabeculae formation is abolished in Osx knock-out mice. Also, in Osx null mice, expression of type I collagen and osteoblast marker genes is reduced in mesenchymal cells. However, Runx2 expression in Osx null mice is unaffected, whereas no Osx transcripts are detected in skeletal elements of Runx2 null mice, indicating that Osx acts as a downstream gene of Runx2 in the osteoblast differentiation signaling pathway (Nakashima et al., 2002). Recent studies have found that the effect of Osx on its target genes is involved in other signaling pathways, independent of Runx2 (Lee et al., 2003; Ulsamer et al., 2008). Although osteoblasts and odontoblasts originate from mesenchymal cells and have several common characteristics, bone and dentin display some different physical and biological functions. Thereby, the molecular mechanisms regulating the expression of tooth-/bone-related genes in odontoblasts, especially in the later stages of tooth development, somehow differ from those of osteoblasts (Chen et al., 2005; James et al., 2006). Previously, we and other laboratories observed differential Runx2 expression patterns between alveolar bone osteoblasts and dental cells during tooth formation (Bronckers et al., 2001; Yamashiro et al., 2002; Chen et al., 2005). However, the Osx expression pattern during tooth development has not been described. Furthermore, the relationship among Runx2, Osx, and Dspp during development of teeth and craniofacial bones remains unclear. In this study, we investigated the spatial-temporal expression patterns of Runx2, Osx, and Dspp at different stages of development of teeth and craniofacial bones, as well as studied the effect of Osx on Dspp transcription in mouse odontoblast-like cells.
Materials & Methods
Animals and Tissue Preparation
All experimental procedures involving the use of animals were approved by the University of Texas Health Science Center at San Antonio (UTHSCSA), TX. ICR mice were purchased from Harlan-Laboratory Animals Inc. (Indianapolis, IN, USA). Mouse tissues were dissected and fixed in 4% paraformaldehyde overnight. After demineralization in 15% EDTA, samples were dehydrated in increasing concentrations of ethanol, embedded in paraffin, sectioned, and prepared for in situ hybridization assay.
In situ Hybridization
32P-rUTP-labeled antisense riboprobes corresponding to Runx2 (Chen et al., 2002), Osx (Nakashima et al., 2002), and Dspp (Chen et al., 2005) were generated. The in situ hybridization was performed as described previously (Gluhak-Heinrich et al., 2003).
Cell Culture
Mouse immortalized odontoblast-like (MO6-G3) cells were grown at 33°C under 5% CO2 in α-MEM with 10% fetal calf serum, 100 units/mL penicillin/streptomycin, 50 µg/mL ascorbic acid, and 10 mM Na β-glycerophosphate (MacDougall et al., 1995). Mouse bone marrow stromal ST2 cells and primary calvarial osteoblasts isolated at post-natal day 3 from C57BL6 mice (Harlan-Laboratory Animals Inc.) were grown in α-MEM supplemented with 10% fetal calf serum, 100 units/mL penicillin/streptomycin, at 37°C under 5% CO2.
Immunohistochemistry
MO6-G3 cells were cultured on glass slides and fixed with methanol/acetone (1:1). Cells were treated with 10% normal donkey or goat serum for 60 min at room temperature. Immunohistochemistry was performed with a 1:100 dilution of primary polyclonal antibodies specific for Dsp and Osx (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA). A negative control of mouse IgG I was purchased from Dakocytomation (Carpinteria, CA, USA). The cells were incubated at 4°C overnight and then washed, followed by incubation with the secondary antibodies (goat-anti-rabbit or donkey-anti-goat, 1:3000) with Alexa Fluo® 488 (Molecular Probes, Eugene, OR, USA). Images of Alexa Fluo® 488 staining were obtained at the Core Optical Imaging Facility at UTHSCSA, under the same parameters, with a Nikon inverted microscope.
Effect of Osx on Dspp and Runx2 Expression
To assess the effect of Osx on Dspp and Runx2 expression, we transfected MO6-G3 and ST2 cells with either Osx or an empty expression vector. After 48 hrs, RNA was isolated with the use of a RNA STAT-60 kit (Tel-Test, Inc. Friendswood, TX, USA), treated with DNase I, and purified with the RNeasy Mini Kit (Qiagen Inc., Valencia, CA, USA). For quantitative real-time PCR (qRT-PCR), amplification reactions were assayed in real-time on an ABI 7500 (Applied Biosystems, Foster City, CA, USA) with SYBR Green chemistry as described previously (Chen et al., 2005). Differential Dspp and Runx2 expressions between Osx and mock expression in MO6-G3 and ST2 cells were calculated and normalized to SYBR activity. We used analysis of variance with a t test to determine significant differences between the control and treated groups. All values were expressed as means ± SD from 3 independent experiments performed in triplicate.
Results
Expression Patterns of Runx2, Osx, and Dspp in Developing Teeth
To understand the relationship among Runx2, Osx, and Dspp during tooth and craniofacial bone development, we studied Runx2, Osx, and Dspp mRNA expression patterns at different stages of tooth and craniofacial bone development, using an in situ hybridization assay. At E12, Runx2 and Osx expression was intense in the mesenchymal condensates of forming bones and teeth in the developing maxillary and mandibular arches (Appendix Figs. 1B, 1C). Overall, Osx expression levels were relatively weaker than those of Runx2, while Dspp transcripts were not detectable at E12 (Appendix Fig. 1D).
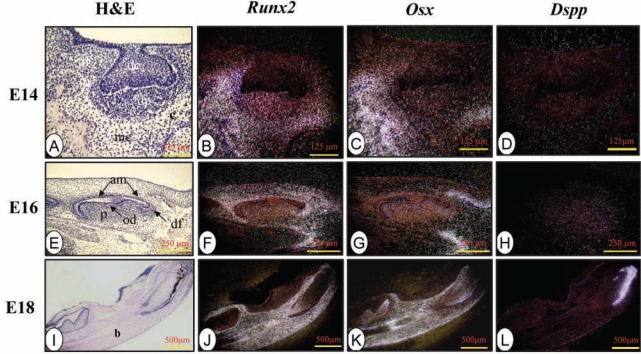
At the cap stage (E14), Runx2 mRNA was highly expressed in mesenchymal cells in alveolar bone, dental papilla, and follicle (Fig. 1B). Osx was almost co-expressed with Runx2 in these same areas (Fig. 1C). However, Runx2 and Osx mRNA expression was barely seen in dental epithelium. In addition, there was no Dspp signal in dental and osteogenic mesenchyme (Fig. 1D).
Figure 1.
Runx2, Osx, and Dspp expression patterns in developing teeth and surrounding tissues from E14 to E18. Runx2 mRNA was clearly detected in dental and osteogenic mesenchyme from E14 to E16, but its expression was down-regulated in odontoblast, ameloblast, and dental pulp cells at E18 of the mandibular incisor and first molar. Osx expression mostly overlapped with Runx2 expression from E14-16. At E18, its expression was intense in ameloblasts, odontoblasts, and dental pulp cells. Dspp transcripts were absent at E14, but its transcript signals were slightly detected in dental and osteogenic mesenchyme at E16. At E18, Dspp transcripts were evident in odontoblasts and pre-ameloblasts in the mandibular incisor and first molar. am, ameloblasts; b, alveolar bone; c, Meckel’s cartilage; de, dental epithelium; df, dental follicle; dp, dental papilla; mc, mesenchymal condensates; od, odontoblasts; p, dental pulp cells.
At the bell stage (E16), both Runx2 and Osx transcripts were expressed in differentiating osteogenic mesenchyme, ameloblasts, odontoblasts, and dental pulp cells (Figs. 1F, 1G). At this stage, there was a weak Dspp signal in odontoblasts, ameloblasts, dental pulp cells, and surrounding tissues (Fig. 1H).
At E18, Runx2 expression was dramatically down-regulated in the odontoblasts, ameloblasts, and dental pulp cells, except for cells near the mesenchyme within alveolar bone in the developing incisor and molar. Its signal was apparent in differentiating alveolar bone osteoblasts (Fig. 1J, Appendix Figs. 1F, 1J). Osx expression in the alveolar bone osteoblasts overlapped with Runx2 expression. However, Osx expression remained intense in odontoblasts and dental pulp cells (Fig. 1K, Appendix Figs. 1G, 1K). At this stage, the Dspp mRNA signal was clear in differentiating and differentiated odontoblasts, and in pre-ameloblasts in the incisor and molar (Fig. 1L, Appendix Figs. 1H, 1L).
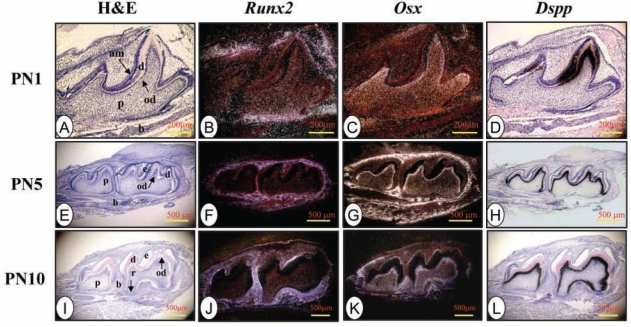
At PN 1, Runx2 mRNA was highly expressed in osteoblasts, but its signal was weakly detected in odontoblasts and dental pulp cells in the developing incisor and molar (Fig. 2B, Appendix Fig. 2B). In contrast, Osx transcripts were intense in odontoblasts and dental pulp cells, in addition to its expression in osteoblasts (Fig. 2C, Appendix Fig. 2C). At this stage, Dspp signal was apparent in differentiating and differentiated odontoblasts and differentiating ameloblasts, but its signal was weakly detected in osteoblasts (Fig. 2D, Appendix Fig. 2D). At PN 5, Runx2, Osx, and Dspp expression patterns were similar to those at PN1. However, the Osx signal was more intense in odontoblasts where Dspp mRNA was highly expressed (Figs. 2G, 2H).
Figure 2.
Runx2, Osx, and Dspp expression patterns in developing teeth and surrounding tissues at post-natal days (PNs) ranged from PN 1 to PN 10. Runx2 mRNA was detected in osteogenic mesenchyme and down-regulated in ameloblasts, odontoblasts, and dental pulp cells. In contrast, Osx expression was seen in odontoblasts and dental pulp cells. Dspp mRNA was strongly expressed in odontoblasts and weakly expressed in ameloblasts at post-natal (PN) days 5 and 10 during tooth development.
Similar to PN5, at PN 8 to 14, Runx2 mRNA was weakly detected in odontoblasts and dental pulp cells in developing molars, whereas its signal was apparently seen in alveolar bone osteoblasts, cells within the cemento-enamel junction, and roots (Fig. 2J, Appendix Figs. 2F, 2J, 2N, 2R). Unlike Runx2, high Osx expression was still present in odontoblasts and dental pulp cells. Osx gene expression was also seen in bone, cemento-enamel junction, and roots overlapped with Runx2 expression (Fig. 2K, Appendix Figs. 2G, 2K, 2O, 2S). Dspp transcripts from PN 8 to PN14 were dominantly expressed in odontoblasts (Fig. 2L, Appendix Figs. 2H, 2L, 2P, 2T). Notably, high Osx and Dspp expression levels overlapped in odontoblasts at the later stages of tooth development. Runx2 expression in mouse osteoblast cell lines was higher than in the mouse odontoblast-like cells (Appendix Fig. 3).
Expression of Osx and Dspp in Mouse Odontoblast-like Cells
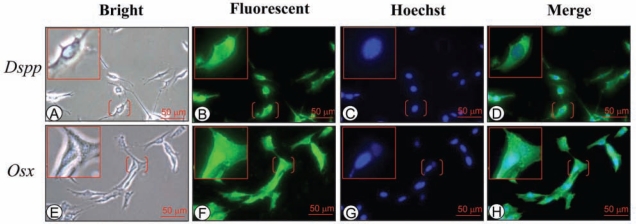
To assess cellular localization of Dspp and Osx expression, we used mouse odontoblast-like cells to detect Osx and Dspp expression by immunohistochemistry. Dspp was expressed in the cytoplasm in MO6-G3 cells, while Osx signal was found in the nucleus and cytoplasm (Fig. 3F).
Figure 3.
Osx and Dspp expression in mouse odontoblast-like cells. Immortalized mouse odontoblast-like cells (MO6-G3) were photographed by light microscopy (A,E). Expression of Osx and Dspp proteins in MO6-G3 cells was analyzed by immunostaining with primary anti-Dsp or anti-Osx antibody (B and F, respectively). Cells were stained with Hoechst for the nucleus (C,G). Panels D and H are composites of B-C and F-G, respectively. Dspp signal was detected in the cytoplasm in the mouse odontoblast-like cells. Osx protein was expressed in both the nucleus and cytoplasm.
Effect of Osx Overexpression on Dspp Expression
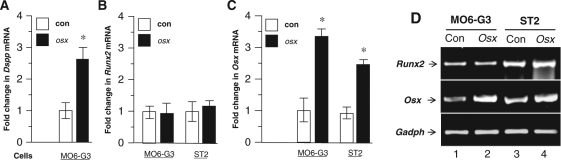
Since expression of Osx and Dspp genes was observed in mouse odontoblasts (Figs. 1, 2), we further examined whether forced expression of Osx is able to regulate Dspp gene expression in mouse odontoblast-like cells. Osx overexpression caused a 2.7-fold increase in Dspp mRNA levels compared with the control group by qRT-PCR analysis (Fig. 4A), indicating that Osx enhances Dspp transcription in mouse odontoblast-like cells. In contrast, there was no change in Runx2 expression levels in Osx-transfected odontoblast-like and osteoblast cells (Figs. 4B, 4D).
Figure 4.
Effect of Osx on Dspp and Runx2 expressions. Mouse odontoblast-like and osteoblast cells were transfected with either Osx or empty expression plasmid as a control. After 48-hour transfection, total RNA was used for qRT-PCR and semi-quantitative PCR. The primer sequences used for qRT-PCR were as follows: Dspp, forward 5′-AACTCTGTGGCTGTGCCTCT-3′ and reverse 5′-TATTGACTCGGAGCCATTCC-3′; Runx2, forward 5′-AGTGCTCTAACCACAGTCCATGCA-3′ and reverse 5′-TACAAACCATACCCAAGTACCTGTTT-3′; Osx, forward 5′-ATGGCGTCCTCTCTGCTTGA-3′ and reverse 5′-TTGAGAAGGGAGCTGGGTAG-3′; and cyclophilin, forward 5′-GGTGACTTCACACGCCATAA-3′ and reverse 5′-CATGGCCTCCACAATATTCA-3′. The mRNA expression without Osx overexpression (empty vector transfectant) in MO6-G3 or ST2 cells is designated as a 1.0-fold increase. Expression levels of Runx2, Osx, and Dspp mRNAs in MO6-G3 or ST2 cells with Osx transfectants are represented as fold-changes in relation to the control. All data are represented by mean ± SD from 3 independent experiments performed in triplicate. *P values lower than 0.05 were considered statistically significant. For semi-quantitative PCR, a pair of primers of the glyceraldehyde 3-phosphate dehydrogenase (GADPH) gene was used as an internal positive control, as follows: forward 5′-CCATGGAGAAGGCCGGG-3′ and reverse 5′-CAAAGTCATGGATGACC-3′. The primers of Runx2 and Osx are described above. Quantitative RT-PCR was used to detect mRNA expression levels of Dspp (A), Runx2 (B), and Osx(C) in MO6-G3 or ST2 cells with or without Osx transfection. D. Semi-quantitative PCR analysis of Runx2, Osx, and GADPH mRNA expressions in MO6-G3 and ST2 cells transfected with or without Osx expression vector.
Discussion
In this present study, we investigated Runx2, Osx, and Dspp gene expression patterns at different stages of mouse tooth development using an in situ hybridization assay. At E12, Runx2 transcripts were seen in dental and osteogenic mesenchyme in developing maxillary and mandibular bones. With tooth development at the cap and bell stages, Runx2 signaling was detected in mesenchymal cells within the alveolar bone, dental papilla, and follicle. At E16, Runx2 transcripts were expressed in ameloblasts, odontoblasts, and dental pulp cells, in addition to the osteogenic mesenchyme, but its signal was remarkably down-regulated in ameloblasts, odontoblasts, and dental pulp cells at E18 and the later stages during tooth development. For the Osx gene, its expression pattern was similar to that of Runx2 from E12 to E16. Unlike Runx2 expression, Osx transcripts remained intense in odontoblasts and dental pulp cells at the late stages, from E18 to PN14. In particular, Osx expression was apparent in odontoblasts in which Dspp was highly expressed. This suggests that the mechanisms regulating osteoblast and odontoblast lineages differentially involve Runx2 and Osx. Runx2 null mice have shown a complete arrest of tooth development in cap/early bell stages prior to odontoblast differentiation (D’Souza et al., 1999). Furthermore, Runx2 up-regulated Dspp transcription in mouse pre-odontoblast-like cells, but down-regulated Dspp activity in mouse odontoblast-like cells (Gaikwad et al., 2001; Chen et al., 2005). Therefore, Runx2 may be involved in tooth development prior to the bell stage, but not the later stages. Differential effects of Runx2 on dentin matrix protein 1 (Dmp1) gene expression during bone and tooth development have been reported (Feng et al., 2002). Like Dspp, Dmp1 is one member of the SIBLING gene family (Fisher and Fedarko, 2003). These investigators found that Runx2 expression overlapped with Dmp1 expression at E14.5-E19.5 in normal developing bones and teeth. Dmp1 expression was absent in developing bones in Runx2 null mice, whereas Dmp1 expression in developing teeth was unaffected. Based on different Runx2 and Osx expression patterns during tooth development, we hypothesized that Runx2 regulates bone and tooth development at the early stages (E12-16). Runx2 may regulate bone and tooth development directly or via a Runx2-related signaling pathway such as Osx. With tooth development, Runx2 expression was down-regulated in ameloblasts, odontoblasts, and dental pulp cells. Thus, Runx2 may not be involved in odontoblast and dental pulp cell differentiation and the regulation of dental gene expression such as Dspp at the later stages of tooth formation. Recently, it was observed that Runx2 inhibits the terminal differentiation of odontoblasts and Dspp expression in Runx2 transgenic mice (Miyazaki et al., 2008). In contrast, Osx continues to be expressed in these cells and may induce cell differentiation and tooth-related gene expression at the later stages of tooth development via a Runx2-independent signaling pathway (Celil and Campbell, 2005; Ulsamer et al., 2008). Furthermore, our study showed that Osx overexpression resulted in a Dspp transcription increase in mouse odontoblast-like cells, but had no effect on Runx2 expression in both the mouse odontoblast-like and osteoblast cells. This suggests that Osx directly affects Dspp expression independent of Runx2.
In addition, our study demonstrated that Runx2 and Osx were highly expressed in osteogenic mesenchyme through all stages of craniofacial bone development. This suggests that both Runx2 and Osx are involved in osteoblast differentiation at the early and later stages of osteogenesis, independently or in concert (Nakashima et al., 2002; Celil and Campbell, 2005; Ulsamer et al., 2008).
Finally, Runx2 and Osx were expressed in tooth roots, suggesting that they may be involved in tooth eruption and movement (Zou et al., 2003).
Supplementary Material
Acknowledgments
We are grateful to Dr. Benoit de Crombrugghe (M.D. Anderson Cancer Center, University of Texas, Houston, USA) for providing the Osx mammalian expression construct. We thank Dr. Howard Dang for critical reading of the manuscript.
Footnotes
This work was supported by NIDCR Grants DE113221 and DE014484.
A supplemental appendix to this article is published electronically only at http://jdr.sagepub.com/supplemental.
References
- Bronckers AL, Engelse MA, Cavender A, Gaikwad J, D’Souza RN. (2001). Cell-specific patterns of Cbfa1 mRNA and protein expression in postnatal murine dental tissues. Mech Dev 101:255-258 [DOI] [PubMed] [Google Scholar]
- Butler WT. (1998). Dentin matrix proteins. Eur J Oral Sci 106:204-210 [DOI] [PubMed] [Google Scholar]
- Celil AB, Campbell PG. (2005). BMP-2 and insulin-like growth factor-I mediate osterix (Osx) expression in human mesenchymal stem cells via the MAPK and protein kinase D signaling pathways. J Biol Chem 280:31353-31359 [DOI] [PubMed] [Google Scholar]
- Chen S, Gu TT, Sreenath T, Kulkarni AB, Karsenty G, MacDougall M. (2002). Spatial expression of Cbfa1/Runx2 isoforms in teeth and characterization of binding sites in the DSPP gene. Connect Tissue Res 43:338-344 [DOI] [PubMed] [Google Scholar]
- Chen S, Rani S, Wu Y, Unterbrink A, Gu TT, Gluhak-Heinrich J, et al. (2005). Differential regulation of dentin sialophosphoprotein expression by Runx2 during odontoblast cytodifferentiation. J Biol Chem 280:29717-29727 [DOI] [PubMed] [Google Scholar]
- D’Souza RN, Cavender A, Sunavala G, Alvarez J, Ohshima T, Kulkarni AB, et al. (1997). Gene expression patterns of murine dentin matrix protein 1 (Dmp1) and dentin sialophosphoprotein (DSPP) suggest distinct developmental functions in vivo. J Bone Miner Res 12:2040-2049 [DOI] [PubMed] [Google Scholar]
- D’Souza RN, Åberg T, Gaikwad J, Cavender A, Owen M, Karsenty G, et al. (1999). Cbfa1 is required for epithelial-mesenchymal interactions regulating tooth development in mice. Development 126:2911-2920 [DOI] [PubMed] [Google Scholar]
- Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. (1997). Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89:747-754 [DOI] [PubMed] [Google Scholar]
- Feng JQ, Zhang J, Dallas SL, Lu Y, Chen S, Tan X, et al. (2002). Dentin matrix protein 1, a target molecule for Cbfa1 in bone, is a unique bone marker gene. J Bone Miner Res 17:1822-1831 [DOI] [PubMed] [Google Scholar]
- Fisher LW, Fedarko NS. (2003). Six genes expressed in bones and teeth encode the current members of the SIBLING family of proteins. Connect Tissue Res 44(Suppl 1):33-40 [PubMed] [Google Scholar]
- Gaikwad JS, Hoffmann M, Cavender A, Bronckers AL, D’Souza RN. (2001). Molecular insights into the lineage-specific determination of odontoblasts: the role of Cbfa1. Adv Dent Res 15:19-24 [DOI] [PubMed] [Google Scholar]
- Gluhak-Heinrich J, Ye L, Bonewald LF, Feng JQ, MacDougall M, Harris SE, et al. (2003). Mechanical loading stimulates dentin matrix protein 1 (DMP1) expression in osteocytes in vivo. J Bone Miner Res 18:807-817 [DOI] [PubMed] [Google Scholar]
- James MJ, Jarvinen E, Wang XP, Thesleff I. (2006). Different roles of Runx2 during early neural crest-derived bone and tooth development. J Bone Miner Res 21:1034-1044 [DOI] [PubMed] [Google Scholar]
- Jiang H, Sodek J, Karsenty G, Thomas H, Ranly D, Chen J. (1999). Expression of core binding factor Osf2/Cbfa-1 and bone sialoprotein in tooth development. Mech Dev 81:169-173 [DOI] [PubMed] [Google Scholar]
- Kim JW, Simmer JP. (2007). Hereditary dentin defects. J Dent Res 86:392-399 [DOI] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, et al. (1997). Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89:755-764 [DOI] [PubMed] [Google Scholar]
- Lee B, Thirunavukkarasu K, Zhou L, Pastore L, Baldini A, Hecht J, et al. (1997). Missense mutations abolishing DNA binding of the osteoblast-specific transcription factor OSF2/CBFA1 in cleidocranial dysplasia.Nat Genet 16:307-310 [DOI] [PubMed] [Google Scholar]
- Lee MH, Kwon TG, Park HS, Wozney JM, Ryoo HM. (2003). BMP-2-induced Osterix expression is mediated by Dlx5 but is independent of Runx2. Biochem Biophys Res Commun 309:689-694 [DOI] [PubMed] [Google Scholar]
- Lian JB, Stein GS, Javed A, van Wijnen AJ, Stein JL, Montecino M, et al. (2006). Networks and hubs for the transcriptional control of osteoblastogenesis. Rev Endo Metabolic Disorders 7:1-16 [DOI] [PubMed] [Google Scholar]
- Linde A, Goldberg M. (1993). Dentinogenesis. Crit Rev Oral Biol Med 4:679-728 [DOI] [PubMed] [Google Scholar]
- MacDougall M, Thiemann F, Ta H, Hsu P, Chen LS, Snead ML. (1995). Temperature sensitive Simian Virus 40 Large T antigen immortalization of murine odontoblast cell cultures: establishment of clonal odontoblast cell line. Connect Tissue Res 33:97-103 [DOI] [PubMed] [Google Scholar]
- Miyazaki T, Kanatani N, Rokutanda S, Yoshida C, Toyosawa S, Nakamura R, et al. (2008). Inhibition of the terminal differentiation of odontoblasts and their transdifferentiation into osteoblasts in Runx2 transgenic mice. Arch Histol Cytol 71:131-146 [DOI] [PubMed] [Google Scholar]
- Mundlos S, Otto F, Mundlos C, Mulliken JB, Aylsworth AS, Albright S, et al. (1997). Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell 89:773-779 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, et al. (2002). The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 108:17-29 [DOI] [PubMed] [Google Scholar]
- Rajpar MH, Koch MJ, Davies RM, Mellody KT, Kielty CM, Dixon MJ. (2002). Mutation of the signal peptide region of the bicistronic gene DSPP affects translocation to the endoplasmic reticulum and results in defective dentine biomineralization. Hum Mol Genet 11:2559-2565 [DOI] [PubMed] [Google Scholar]
- Thesleff I. (2003). Epithelial-mesenchymal signalling regulating tooth morphogenesis. J Cell Sci 116(Part 9):1647-1648 [DOI] [PubMed] [Google Scholar]
- Ulsamer A, Ortuño MJ, Ruiz S, Susperregui AR, Osses N, Rosa JL, et al. (2008). BMP-2 induces Osterix expression through up-regulation of Dlx5 and its phosphorylation by p38. J Biol Chem 283:3816-3826 [DOI] [PubMed] [Google Scholar]
- Xiao S, Yu C, Chou X, Yuan W, Wang Y, Bu L, et al. (2001). Dentinogenesis imperfecta 1 with or without progressive hearing loss is associated with distinct mutations in DSPP. Nat Genet 27:201-204; erratum in Nat Genet 27:345, 2001 [DOI] [PubMed] [Google Scholar]
- Yamashiro T, Åberg T, Levanon D, Groner Y, Thesleff I. (2002). Expression of Runx1, -2 and -3 during tooth, palate and craniofacial bone development. Mech Dev 119(Suppl 1):107-110 [DOI] [PubMed] [Google Scholar]
- Zou SJ, D’Souza RN, Ahlberg T, Bronckers AL. (2003). Tooth eruption and cementum formation in the Runx2/Cbfa1 heterozygous mouse. Arch Oral Biol 48:673-677 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.