Abstract
RNA and DNA guanine-rich sequences can adopt unusual structures called Guanine quadruplexes (G4). A quadruplex-prone RNA sequence is present at the 5′-end of the 451-nt-long RNA component of telomerase, hTERC. As this quadruplex may interfere with P1 helix formation, a key structural element for this RNA, we are seeking molecules that would alter this RNA duplex–quadruplex equilibrium. In this work, we present a fluorescence-based test designed to identify G4 ligands specific for the hTERC G-rich motif and that can prevent P1 helix formation. From an initial panel of 169 different molecules, 11 were found to be excellent P1 duplex inhibitors. Interestingly, some of the compounds not only exhibit a strong selectivity for quadruplexes over duplexes, but also demonstrated a preference for G4–RNA over all other quadruplexes. This test may easily be adapted to almost any quadruplex-forming sequence and converted into HTS format.
INTRODUCTION
Guanine-rich sequences can adopt unusual structures called Guanine quadruplexes (G4) based on stacked guanine quartets (1). The human genome possesses number of sequences prone to adopt such structures, as for example in telomeric repeats, in several oncogenic promoters, in ribosomal DNA and in the immunoglobulin switch region. Key biological processes could be affected by the formation of these structures (2,3). Furthermore, formation of G4 has been evidenced in vivo in ciliate telomeres (4), during G-rich sequence transcription (G-loop) (5) and for pilin antigenic variation in Neisseria gonorrhoeae (6).
Quadruplex formation is not limited to DNA sequences and recent studies illustrated the importance of quadruplexes at the RNA level such as hTERT splicing (7), N-RAS 5′UTR (8), prokaryotic translation start (9) and TERRA (10).
A number of pathways potentially link quadruplex formation (at the DNA or RNA level) and telomere maintenance. Indeed, G4 formation has been evidenced for the G-rich strand of telomeric repeats and could therefore affect telomere elongation by telomerase (11–13), the binding of other telomeric factors like hPOT1 (14). G4 formation by the telomeric repeats could also affect general telomere replication (15,16). TERRA, the RNA transcript corresponding to the G-rich strand (17) has been proposed to be involved in telomere function regulation (18) and can form some very stable G4 (10,19). There are G4 prone sequences in the mRNA for the catalytic subunit of telomerase (hTERT) which have been proposed to affect hTERT splicing upon G4 ligand exposure (7). Finally, G4 formation has been reported in c-MYC (20) and hTERT (21,22) promoters and thus G4 ligands could affect hTERT expression and thus telomerase activity.
An evolutionary conserved quadruplex-prone RNA sequence is also present at the 5′-end of the RNA component of telomerase, called hTERC (or hTR) in humans (23). In a previous work, we demonstrated that oligonucleotides mimicking the 5′ of hTERC can form a G4 and that this could interfere with secondary structure, inhibiting the formation of a local RNA double helix called P1 (23). In principle, the ability of hTERC to form a quadruplex could therefore represent a new mechanism of action to account for the telomeric effects of G4 ligands. This prompted us to study in more details the implication of G4 formation in hTERC.
In order to evidence a G4-related effect on hTERC and on telomerase activity or biogenesis, we need to identify G4 ligands that are specific for the G4 formed in hTERC. In this work, we present a fluorescence-based test designed to identify G4 ligands specific for the G4 of hTERC and that can prevent P1 helix formation. As a starting pool of potential ligands, we used a library of compounds already tested in the laboratory, which are mainly known telomeric G4 DNA ligands. We choose to start with a library of known G4 ligands as:
we anticipated positive hits despite a small library side;
we had some experience with these compounds;
affinity for this quadruplex could be compared with other targets;
these molecules have different scaffold and charges and somewhat represent our current level of knowledge concerning the recognition of G-quadruplex; and
some of the compounds are commercially available or may be easily obtained, and implementation of a pilot assay in another lab is relatively straigthforward.
This test relies on a duplex–quadruplex competition: compounds that stabilize G4 RNA should inhibit hybridization of the guanine-rich RNA to its complementary sequence. The system chosen here closely matches the biological situation, as the RNA–RNA duplex matches the P1 helix of hTERC. Compounds that inhibit P1 helix formation are then tested for bona fide RNA G4 binding.
MATERIALS AND METHODS
Oligonucleotides
Fluorescently labelled oligonucleotides (F22, F22m, 37Q and 37Qm32) were purchased from IBA (Göttingen, Germany, RNA HPLC grade, 200 nmol scale).
F22: 6FAM-5′-UGGCCCGUUCGCCCCUCCCGGG-3′
F22m: 6FAM-5′-UGGCCCGUUCGCUUCUCUCGGG-3′
37Q: 5′-GGGUUGCGGAGGGUGGGCCUGGGAGGGGUGGUGGCCA-3′-BHQ1
37Qm32: 5′-GAGUUGCGAAGAGUGAGCCUGAGAGAAGUGAUGGCCA-3′-BHQ1
Non-fluorescent oligonucleotides were purchased on the 200 nmol scale from IBA (HPLC grade) or Eurogentec (Seraing, Belgium, Oligold grade). Unmodified sequences used for competition experiment are reported in Supplementary Table S1.
Chemicals
The ligands were taken from the in house ligand library that we previously used mainly to identify telomeric G4 ligands. These ligands were gifts or collaboration products from many chemistry laboratories: P. Mailliet (Sanofi-Aventis, Vitry sur Seine, France), M.P. Teulade-Fichou (Institut Curie, Orsay, France), S. Neidle (School of Pharmacy, London, UK), K. Shin-ya (University of Tokyo, Tokyo, Japan), G. Pratviel (Laboratoire de Chimie de Coordination, Toulouse, France), F. Guéritte (ICSN, Gif sur Yvette, France), T.C. Chang (Institute of Atomic and Molecular Sciences, Taiwan, ROC) and M. Stevens (School of Pharmaceutical Sciences, University of Nottingam, UK). We also included a family of crown-shape ligands developed by C. Ferroud (CNAM, Paris, France). Porphyrin derivatives were purchased from Frontier Scientific (Logan, Utah, USA) and stock solutions were prepared at 10 mM in water. MST-312 and PIPER were purchased from Calbiochem and resuspended at 10 and 2 mM, respectively, in DMSO. For all other ligands, 1–4 mM DMSO stock solutions were used. Cacodylic acid, KCl, LiOH and LiCl were from Sigma.
Fluorescent test
For the screening test, the fluorescent oligonucleotide (F22) alone or in the presence of the complementary strand (37Q) was incubated at 50 nM in 20 mM Lithium cacodylate (pH 7.2) supplemented with 1 mM KCl and 99 mM LiCl. When present, 37Q was added in slight molar excess (i.e. 75 nM). After a simple annealing procedure (95°C, 15′ then cool down to 40°C at 1°C/min), 22.5 µl of the fluorescent mix were distributed into 96-well qPCR plates (Stratagene) containing 2.5 µl of either a 10 µM solution of ligand (giving a final ligand concentration of 1 µM) or 1% DMSO (giving a final concentration of 0.1%). After centrifugation, the plate was transferred to a real-time thermal cycler (Mx3000p, Stratagene). Data acquisition proceeds using the FAM channel and in the following order (Supplementary Figure S1):
Step 1: 30 steps of 1 min at 37°C with a reading at the end of each step.
Step 2: 15 steps of 1 min at 95°C with a reading at the end of each step.
Step 3: 31 steps of 1 min from 95°C to 37°C with a temperature decrease of 2°C and a reading at the end of each step.
After each ‘qPCR’ experiment, plates were read using a fluorescent imaging system (Typhoon 9410) using a 488-nm excitation wavelength and a 520BP40-emission filter, with a 600-V PMT gain and a +3-mm focus setting.
Data analysis
Data from the qPCR apparatus were exported as text file and processed with Microsoft Excel. No ligand effect was observed during isothermal data acquisition at 37°C (Step 1) and thus only data from Step 3 were analyzed. For each well, fluorescent intensity reading at 37°C after the ‘in machine’ annealing (IF37) was normalized with the fluorescent intensity reading corresponding dissociated state at 82°C (IF82) of the same well to compensate for well-to-well fluorescence variations. In order to evaluate the ability of a given ligand (X) to prevent duplex formation, we defined a ligand efficacy (Effi) corresponding to the fraction of unformed duplex, using normalized fluorescence (NF=IF37/IF82) reading from wells containing the fluorescent oligonucleotide alone (F22) and wells containing the quenched duplex (F22 + 37Q) in the absence of ligand.
Using this parameter, a good ligand that totally prevents duplex formation will have an Effi of 1 and a poor ligand an Effi of 0 (Supplementary Figure S3). Slightly negative Effi value are within the noise limits of the system while more negative Effi value might indicate a conformational change of the duplex induced by the ligand or a strong fluorescence quenching of the FAM. In both cases such molecules are unlikely to be false negative compounds.
Quality of each plate was also assessed by computing a Z′ factor (24), an estimator that take into account the distribution of the minima and maxima value of the plate and also the difference between these maxima and minima. In our case, minima were associated with duplex state controls (F22 + 37Q) and maxima with single-stranded state controls (F22). Most plates contained quadruplicates of the F22 alone and the F22 + 37Q wells used for Effi and Z′ calculation. Dissociation step data (Step 2) and temperature profile data from Step 3 were used to assess quality of starting material and duplex formation, respectively.
For experiments in the presence of competitors, a competitor efficacy (C_Effi) of the competitor Y for the ligand X was define using the following formula:
Using this estimator, a good competitor will have a C_Effi value close to 1 and for a poor competitor, the value will be 0.
Gel electrophoresis
Gel electrophoresis to validate hit compounds was performed as previously described (23).
RESULTS
The test principles
Several fluorescence based tests for G4 ligands have been previously reported (25–28). These tests can be classified into two categories: (i) stabilization of a G4 by a ligand followed by FRET melting and (ii) fluorescent ligand displacement by a G4 ligand. Both tests are designed to identify G4 ligand in the context of a simple equilibrium between a single-strand and a quadruplex. In our system, we wanted to identify G4 ligands which could also work in a duplex–quadruplex equilibrium context, which may be seen as more physiologically relevant, as it represents a common situation for DNA or RNA G-rich sequences. This test is inspired by previous report using fluorescence to follow DNA duplex–quadruplex competition (29,30). A similar test using scintillation proximity signal has been used for high-throughput identification of G4 ligands (31), but this scintillation proximity assay is not easy to set up in a laboratory and this was a ‘turn off’ test, meaning that the presence of a quadruplex ligand would decrease the signal detected. The method combines two short oligos mimicking the sequence around the P1 helix. Using a full length (451 nt) RNA with dual labelling is impossible here: the test is intermolecular, and a conformational change cannot be evidenced in a routine assay with hTERC.
For convenience, we choose to favour fluorescence detection and in order to built a ‘turn on’ system, we choose to combine a fluorescent strand with a complementary strand bearing a quencher. In this system (Figure 1A), the duplex form is associated with a low-fluorescence signal, and the quadruplex with a high-fluorescence signal. Furthermore, because of the well-known quenching properties of guanines and the possible quenching of the fluorescence by the ligand bound to the G4, we decided to insert the fluorescent reporter (FAM) on the non G-rich strand (F22).
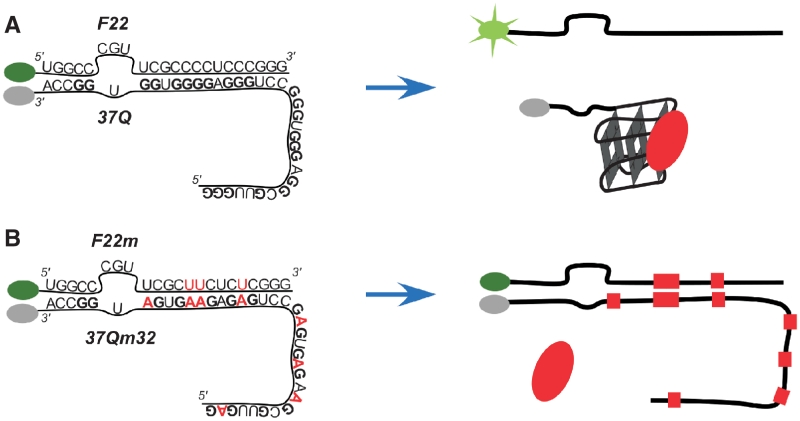
Figure 1.
Principle of the assay. In the presence of a bona fide quadruplex ligand, duplex formation is inhibited in the F22 + 37Q system (quadruplex prone, A) and thus fluorescence of the F22 is high (not quenched) whereas duplex formation is unperturbed on the mutated system (F22m + 37Qm32, non-quadruplex prone, B) and thus F22m fluorescence is low (quenched).
In order to demonstrate that P1-helix formation inhibition was related to G4 formation by the G-rich strand (37Q), we built a control system bearing G to A substitutions in eight positions within the first 32 nt of the sequence (37Qm32). A bone fide G4 ligand should not be able to prevent duplex annealing for this system (Figure 1B).
Setting up the test conditions and parameters
Salt conditions
We chose to work at a medium ionic strength with a total monocation concentration around 100 mM. Playing with lithium and potassium concentrations allowed us to modulate G4 stability while keeping duplex formation relatively unaffected. Following our previous observation regarding duplex–quadruplex competition on the same system (23), we first needed to define KCl–LiCl concentrations that allow full duplex formation in the absence of ligand, but still allow quadruplex formation of the G-rich strand in absence of its complementary sequence as follows.
Working in the absence of KCl allows nearly complete duplex formation but prevent any quadruplex formation on this system at 37°C even in the presence of the ligand 360A that we identified in our previous work (23).
Using 10 mM KCl (with 90 mM LiCl) reduces duplex formation with the fluorescent system by 30% compared to the result obtained with gel electrophoresis (Supplementary Figure S2).
On the other hand, using 1 mM KCl (supplemented with 99 mM LiCl) leads to a duplex formation comparable to the one obtained in 100 mM LiCl while maintaining the ability to inhibit duplex formation with 360A.
For these reasons, we chose the later set of conditions for all further experiments.
Oligonucleotide concentrations
Fluorescence titrations were performed to determine the practical concentrations of the fluorescent and quencher oligonucleotides with the following constraints:
fluorescence signal from the duplex state and the single-stranded state of the fluorescent oligonucleotide should be above background for fluorescence detection with either the qPCR machine and the fluorescent scanner;
concentrations should be well above duplex dissociation constant (Kd) to allow complete duplex formation and
quencher oligonucleotide concentration (37Q) should be as low as possible to allow easy ligand titration; and
quencher oligonucleotide concentration (37Q) should be equal or higher than the concentration of the fluorescent oligonucleotide F22.
Titrations were performed with three concentrations of the fluorescent oligonucleotide (F22) (20, 50 and 100 nM) and concentrations of the quencher oligonucleotides (37Q) ranking 0–40, 0–100 and 0–200 nM, respectively. These titrations showed that the fluorescent signal with 20 nM of F22 with an excess of 37Q was too close to background level while titrations with 50 and 100 nM were qualitatively acceptable. In the three cases, concentration ranges were well above the apparent Kd for duplex formation that we determined to be in the range of 1–2 nM by fitting the titration data (data not shown). Thus, to limit G-rich oligonucleotide concentration, we choose to set the F22 and 37Q concentration to 50 and 75 nM, respectively.
Ligand concentration
In our previous work based on gel electrophoresis (23), we determined that 360A was active at 1 and 2 µM. Preliminary results with 360A on the fluorescent system confirmed that this ligand was still active under conditions of this new test at 1 and 2 µM, but partial quenching of the single-stranded fluorescence was observed at 2 µM of 360A. This led us to choose 1 µM as the reference concentration for screening purposes.
Screen results
In parallel to setting up and validation experiments with 360A, a few reference compounds have been tested: 12 459 (32), PhenDC3 (33) and BRACO19 (34) (Figure 2). These preliminary experiments demonstrated that PhenDC3 was a better G4hTERC ligand than 360A and did not quench the fluorescence of the single strand up to 2 µM. We therefore decide to use this ligand as our ‘standard’ with the aim to identify ligand that would be at least as potent as PhenDC3.
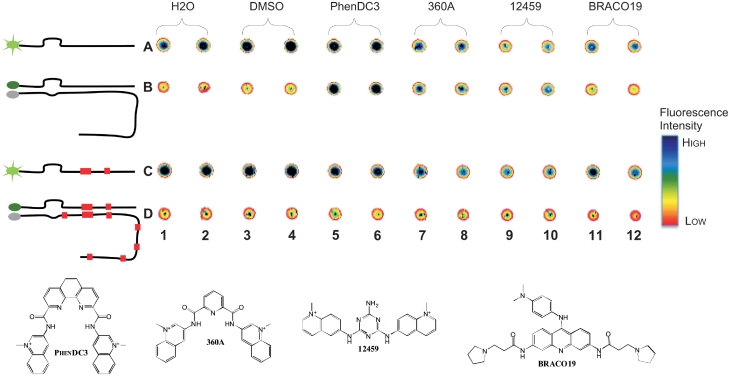
Figure 2.
Proof of principle. Top: post-annealing fluorescence reading of a set-up plate with duplicate of control (H2O, DMSO, 0.05%) and test molecules (PhenDC3, 360A, 12459 and BRACO19 at 1 µM). Lane A, F22 alone; B, F22 + 37Q; C, F22m; D, F22m + 37Qm32. Fluorescence quenching by the ligand is visible for 12459 and 360A to a lesser extend (A9–10, A7–8, C9–10, C7–8). High fluorescence in lane B correspond to molecule that prevent annealing of F22 on 37Q (B5–10) and the low fluorescence in the corresponding row in lane D confirms a quadruplex-related mechanism. Annealing has been perfomed in a real-time thermal cycler and fluorescence annealing profile analysis results in comparable conclusion. Bottom: formulae of the molecules used in this experiment.
Using a 96-wells plate format, we performed >790 independent readings corresponding to 173 independent chemical references and 169 different molecules. Test quality was assessed using Z′ factor calculation for each plate. This estimator was routinely in the 0.7–0.9 range, meaning that the data extracted from these plate are of very good quality (24). In some experiments, this indicator fell in the 0.4–0.5 range indicating medium quality data for this plate. This quality factor was taken into account to define the ‘Hit threshold’ (see below).
Every plate used for the screening contains the standard PhenDC3 to evaluate the test robustness. Under these conditions, our reference compound PhenDC3 presented an Effi of 0.77 ± 0.14. For a positive hit, we decided to fix an arbitrary threshold value of 0.5 for Effi. For a typical high-quality plate (Z′ > 0.7), this corresponds to a signal of >12 SD above the signal of a negative control. Even for a medium quality plate (0.4 < Z′ ≤ 0.7), this threshold corresponds to a hit signal of >5 SD above the signal of a negative control. For good quality data set (Z′ > 0.7), we also define a secondary threshold for medium quality hit with an threshold Effi value down to 0.4, allowing us to identify more hits, including positive controls from the setting up experiment (12 459 and 360A). However, this low-threshold value cannot be used for medium quality data set as it leads to non-reproducible results. For the rest of the manuscript, good hits will refer to molecules with Effi > 0.5 and medium hits to molecules with 0.5 ≥ Effi > 0.4. Figure 3 and Supplementary Figure S3 illustrate a typical result. This experiment was of good quality on the basis of the Z′ factor (0.92) and thus good (red background) and medium (orange background) hits were taken into account. All good hit molecules as defined by their Effi values are associated with a high-fluorescence intensity (dark blue, see well 2E, 3D, 4G, 8B, 9G, 10C and 10F for examples) whereas non hit molecules present a low-fluorescence intensity (red–yellow, see well 2C, 6H, 10H and 11H). For some hits, a clear discrepancy is observed between Effi values and fluorescence images (see well 2A, 9D and 11A for extreme examples). With the aim of an unbiaised hit determination, all hit judged by their Effi values were kept but full renaturation profiles (Supplementary Figure S3A) are usefull to identify potential artefacts.
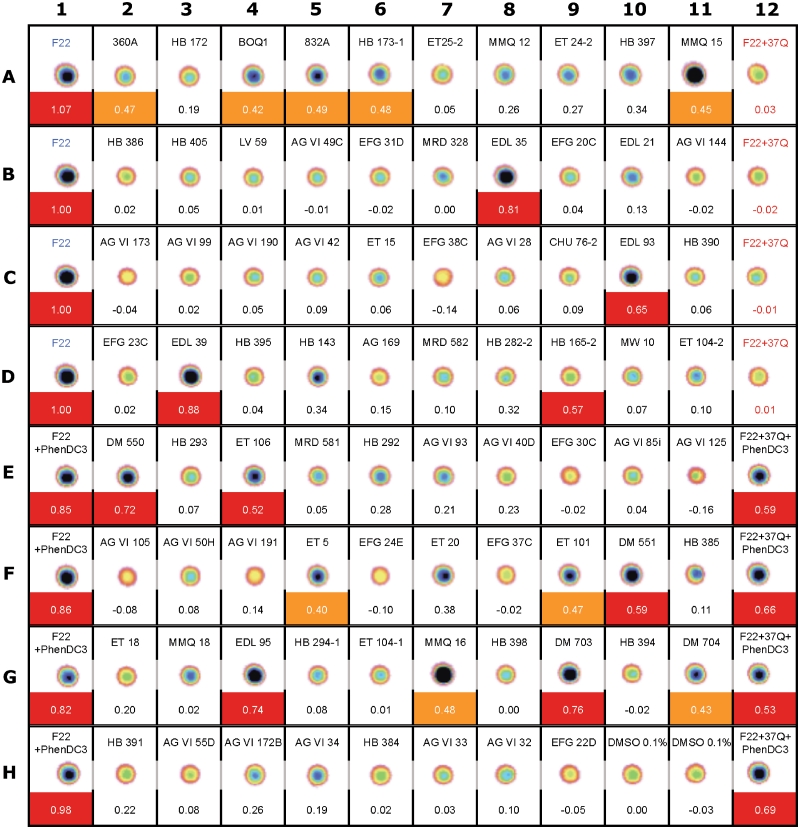
Figure 3.
Effi and post-annealing fluorescence imaging of a typical plate. Each cell correspond to a well of a 96-wells plate used and fluorescence image of each well has been inserted in each cell between the content of the well and the corresponding Effi value. In this experiment, column 1 contains the fluorescent oligonucleotide F22 alone (line A–D) or in the presence of the control compound PhenDC3 (line E–H). Similarely, column 12 contains the duplex system F22 + 37Q alone (line A–D) or in the presence of PhenDC3 (line E–H). Lines A–D of these two columns were used for Effi and Z′ factor (0.92 in this experiment) calculation. Effi values of good or medium hits are highlighted in red and orange, respectively. In this plate, several chemical references correspond to different batches of PhenDC3 (EDL35, DM550 and DM703) and PhenDC6 (EDL39, DM551 and DM704). Also note that BipyDC3 and BipyDC6 correspond to the chemical reference EDL93 and EDL95 respectively.
Using these threshold values, we identify 10 molecules as good hits (307A, 832A, BipyDC3, BipyDC6, HB167, HB173, L2G2, L2H2, PhenDC3, PhenDC6), and 12 as medium hits (12 459, 115 405, 360A, BMVC, BOQ1, HB165, HB282, ET101, ET106, MMQ15, MMQ16, S1T1-7) in this first instance screen. These molecules were then subject to validation tests.
G4 mechanism validation
In order to validate potential hit, experiments were reproduce using in parallel the wild-type system (F22 + 37Q) and a mutated system unable to form any G4 (F22m + 37Qm32). We also included in this test setup the free fluorescent oligonucleotides F22 and F22m to assess for any direct effect of the ligand on these oligonucleotides. At this validation step, we decide to include ‘borderline’ and negative molecules as a mean to validate data from the first instance screen, and thus 30 molecules were tested. Three classes of results are expected:
Class A molecules that are efficient on F22 + 37Q and not on F22m + 37Qm32 (‘true hits’).
Class B molecules efficient on both systems (‘non-specific hits’) and
Class C molecules not efficient on the F22 + 37Q system (‘false positives’) (Table 1).
From this classification, eight molecules come out as ‘good hits’ using a G4 mechanism to prevent P1 helix formation (307A, 832A, BipyDC3, BipyDC6, L2G2, L2H2, PhenDC3 and PhenDC6) and three molecules as G4-related ‘medium hits’ (115405, 12459, 360A). The existence of a non-specific-related mechanism could not be evidenced with any of the selected molecules. Other compounds have been excluded on the basis of reproducibility issues (BMVC, BOQ1, ET101, ET106, HB165, HB167, HB173, HB282, MMQ15, MMQ16, S1T1-7) and further studies would be required to validate or invalidate these hits and decipher experimental noise from molecules stability/solubility issues.
Table 1.
Results summary
| Reproductibility | ss binding | mutant system binding | Effi Li100 | Effi K1 | Effi K10 | |
|---|---|---|---|---|---|---|
| Medium hits | ||||||
| 12 459 | + | + | – | – | + | ++ |
| 115 405 | + | + | – | – | + | ++ |
| 360A | + | – | – | – | + | ++ |
| Good hits | ||||||
| 307A | + | – | – | – | ++ | nd |
| 832A | + | – | – | – | ++ | nd |
| BiPyDC3 | + | – | – | – | ++ | ++ |
| BiPyDC6 | + | – | – | – | ++ | ++ |
| L2G2 | + | – | – | – | ++ | ++ |
| L2H2 | + | – | – | – | ++ | ++ |
| PhenDC3 | + | – | – | – | ++ | ++ |
| PhenDC6 | + | + | – | – | ++ | ++ |
| Negative | ||||||
| AGVI191 | + | nd | – | – | – | – |
| BRACO19 | + | nd | – | – | – | + |
| DOCB | + | * | – | – | nd | nd |
| RHPS4 | + | * | – | – | nd | nd |
| Telomestatin | – | – | – | – | – | ++ |
Reproducibility: +, good or medium hit in all experiments. Ss binding: +, low-fluorescence intensity of F22 alone in the presence of the compound, *, fluorescent molecule (see main text). Mutant system binding: –, the compound was not able to prevent the annealing with the mutated system F22m + 37Qm32. Effi (Li100, K1, K10): –, below 0.4, + ; between 0.4 and 0.5; ++, above 0.5. nd, not determined.
Data obtained with the single-stranded oligonucleotides also illustrate two possible artefacts: fluorescence quenching of the single-stranded oligonucleotides (115405, BMVC, HB167, HB282, PhenDC6) and fluorescence interference in the case of fluorescent ligands (DOCB and RHPS4). The first type of artefact was not detrimental as the extent of fluorescent quenching was low. Concerning the second type, the Effi parameter calculation was still valid for RHPS4 as the fluorescence from the single stranded and the duplex state are both shifted in the same extend to higher values (Supplementary Figure S4). On the other hand, the fluorescence properties of DOCB prevent any reliable Effi calculation, but using the full annealing profile for the wild type and the mutated system reveals no P1 helix formation inhibition (Supplementary Figure S4) allowing us to exclude this compound.
We also tested the effect of K+ concentration in the annealing buffer as mentioned in the test setting up section. Fourteen molecules were tested under these conditions. No molecule was able to prevent P1 helix formation in the absence of K+ (LiCl 100 mM). On the other hand, results with higher K+ content (KCl 10 mM/LiCl 90 mM) confirm a G4-related mechanism for all the class A molecules tested (all the medium or good hits become good hits) and also revealed that some non-hits molecule in low KCl become medium or good hits (BRACO19 and telomestatin). In other words, increasing potassium concentration decreases the strigency of the test but confirms that the process studied is K+-dependent.
In gel test validation
In order to confirm the results obtained by this fluorescent assay, seven hit molecules (360A, PhenDC3, PhenDC6, BipyDC3, BipyDC6, L2G2, L2H2) were tested using the gel-based system reported in our previous publication (23). We also included in this test three control molecules: a true negative control (EDL21) and two molecules that are efficient only with higher K+ content (telomestatin and BRACO19). With a higher KCl content (KCl 10 mM/LiCl 90 mM), we were able to reproduce P1 helix formation inhibition with all the hit molecules. According to this test (Supplementary Figure S5A), the molecules can be split into the following categories regarding P1 helix formation inhibition:
medium: 360A, BipyDC3, BipyDC6;
good: PhenDC3, PhenDC6, L2G2, L2H2; and
inactive or weak: results with the control molecules (EDL21, BRACO19, Telomestatin) are in agreement with the fluorescent test with 10 mM KCl: EDL21 and BRACO19 are not active and the telomestatin seems slightly less efficient than the medium hits.
Using the mutated system, that we also published previously, we confirm that none of these molecules could prevent the formation of the mutated (non-G4-prone) P1 helix (Supplementary Figure S5B). Under more stringent conditions (lower KCl content), which are closer to the fluorescent test conditions, only the PhenDC derivatives are able to prevent P1 helix formation on the G4-prone system and still no ligand appears efficient on the non G4-prone system (Supplementary Figure S5C and D). This result confirms the PhenDC derivatives as very efficient and specific molecules to prevent P1 helix formation on a G4-prone system.
Thus, the results obtained with this gel test with 10 mM KCl are qualitatively comparable to the results from the fluorescence test with 1 mM KCl. Other good hits from the fluorescent test (L2H2, L2G2, BipyDC3 and BipyDC6) were not as efficient with this in gel test with KCl 1 mM. This difference could be related to the non-equilibrium state of the sample during the electrophoresis (partial displacement of the molecule with a lower residency time upon loading in the gel followed by a duplex renaturation in the presence of sucrose in the well), arguing in favor of the fluorescence test.
Specificity profiles
Using the fluorescent test, we also managed to obtain specificity profiles for some ligands. Similarly to what has been done with FRET-melting experiments (35), we evaluated the efficacy of some ligands in the presence of an excess of various non-fluorescently labelled competitor. If the ligand has some affinity for the competitor, this later should trap the ligand and hinder its action of preventing P1 helix formation (Figure 4A). Two set of competition experiments were performed, using either a short list of 7 competitors on 11 ligands (good or medium hits) or a longer list of 21 competitors on PhenDC3, PhenDC6 and L2H2 (Supplementary Table S1). Competition experiments were performed at 1 and 10 µM of competitor, keeping the ligand concentration at 1 µM and KCl at 1 mM. Control experiments performed in the absence of ligand confirmed that the competitors used here did not interfere with the reporter duplex formation (data not shown).
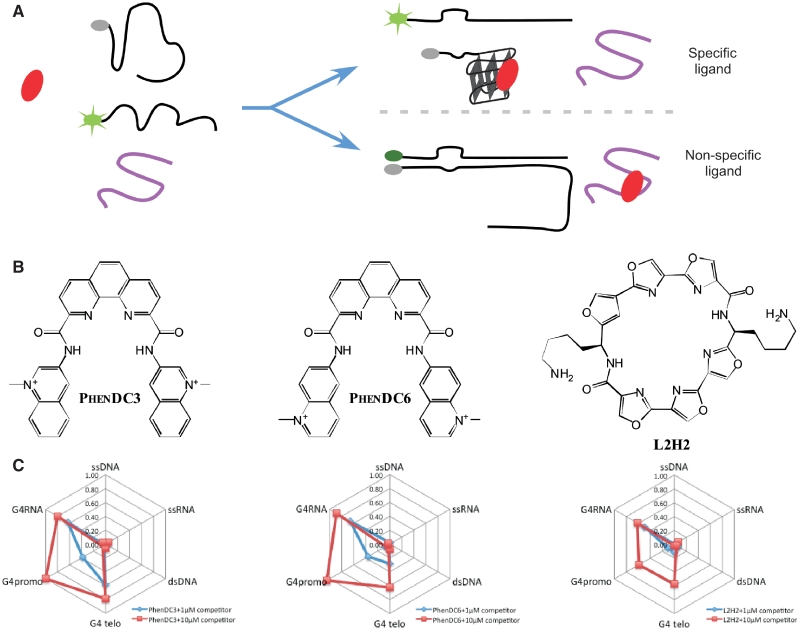
Figure 4.
Selectivity profile. (A) Principle of the selectivity test. (B) Chemical formulae of the best hits. C_Effi values for the three best hits (1 µM) challenged with 21 differents competitors of six types (ssDNA, dsDNA, ssRNA, telomeric G4DNA, promoter type G4DNA, G4RNA) at 1 and 10 µM.
Competition results can be analyzed using five classes of competitors: single-stranded (ss) DNA or RNA, double-stranded (ds) DNA, G4DNA and G4RNA. SsDNA, ssRNA or dsDNA competitors had very little effect on most of the ligands (with the exception of 115405). Ligand efficacies were generaly reduced in presence of 10 µM of G4 competitor, which confirms the preference of these ligands for G4 structures. 12459, L2G2 and L2H2 were the least affected by G4 competitor, suggesting that these ligands exhibit the highest specificity for the hTERC RNA quadruplex. Results obtained at 1 µM of ligand allow a more detailed profiling, and indicate some preferences for G4 RNA versus G4DNA for PhenDC6, 832A, 307A, BipyDC6, L2G2 and L2H2, and no marked preference for PhenDC3 and BipyDC3 (Supplementary Figure S6).
Using a larger panel of quadruplex competitors allowed us to separate the G4 DNA class into telomeric G4DNA, promoter G4DNA and other G4. Using these subclasses for PhenDC3, PhenDC6 and L2H2 enable us to propose that L2H2 and PhenDC6 have a preference for G4RNA (clearly visible at 1 µM of competitor) whereas PhenDC3 has no marked preference (Figure 4C) but a larger number of G4RNA sequences should be used in order to draw a more detailled conclusion.
We have also tested the unusual G4DNA formed by the (sub)telomeric sequence variant AGGG(CTAGGG)3 (36) as competitor for L2H2, PhenDC3 and PhenDC6. This G4 was a very inefficient competitor (Supplementary Figure S6). In this particular quadruplex, only two G quartets are formed and a GCGC quartet at one end and a GC pair at the other end cap them. In this context, terminal stacking by the ligand is expected to be disfavoured on both sides and thus this further argue for a terminal stacking type of interaction of these ligands with the other more classical G4.
DISCUSSION
Critical parameters
This fluorescent test allowed us to identify 11 ligands that prevent the hybridization of a couple of oligonucleotides mimicking the P1 helix of hTERC.
The concentration of cations-especially K+-is a critical parameter in this test as it directly affects the amount of duplex formed in the absence of ligand. When comparing ligand efficacy under different cation conditions, erroneous conclusion can be drawn if care is not taken for the calculation. One should always compare the amount of duplex formed in the presence of the ligand under a given cation condition to the amount of duplex formed in the absence of ligand and under cation conditions that allow quasi-complete duplex formation (LiCl 100 mM or KCl 1 mM/LiCl 99 mM). If KCl 10 mM/LiCl 90 mM conditions are used to evaluate ligand efficacy, Effi would be overestimated as less duplex is formed even in the absence of ligand. Similar care should be taken for C_Effi calculation as this calculation is derived from Effi calculation. These parameters are thus not appropriate for external parameters evaluation even if they remain robust to compare ligands or competitors under the same cation condition. If a wide range of external parameters are to be tested, NF would be more appropriate, but with NF, comparison between the G4-forming system and the non-G4-forming system are not possible as the temperature dependency of fluorescence properties of the duplexes and the ‘open’ form are not comparable for the two systems.
Screening test?
Using the low threshold to identify what we called medium and good hits leads to a high-success rate for the first intention screen (22/169). This 13% success rate reflects the fact that the original library was strongly biased towards G4 ligands and in particular telomeric G4 DNA ligands. The validation tests indicate that the majority (9/12) of the medium hits were false positives whereas the majority of the good hits (8/10) were validated. Therefore, setting the threshold to 0.5 lead to more robust results with an acceptable success rate (6% for the first intention screen and 5% after validation). Using a non-biased library should allow a lower success rate compatible with high throughput approaches.
In the actual configuration of the test, incubation of the ligand in the presence of the preformed duplex would appear unnecessary as we failed to identify any ligand able to restore fluorescence on this substrate. Indeed for competition experiments, this first step of the protocol is not used but with the objective to identify ligand that could affect a preassembled telomerase, we propose to keep this step for genuine library screening.
In order to identify bone fide G4hTERC ligands and rule out general G4 ligands, non-labelled competitors could be added even in the primary screen. Using either a mixture of competitors or a competitor that could mimic both promoter and telomeric type of quadruplexes [like GTERT060 (22)] could be interesting.
Comparision with FRET-melting assays for G4 ligands
There are important differences between the ‘traditional’ FRET-melting assay we developped a decade ago (25,37) and this assays:
instead of looking at a single-strand—quadruplex transition, we are studying a duplex—quadruplex competition. This may be seen as more physiologically relevant, as it represents a common situation for DNA or RNA G-rich sequences, in which local unwiding of the double-helix is required to allow G4 formation.
One can naturally adapt this assay to ‘almost any DNA or RNA quadruplex’ forming sequence (except perhaps for the most stable ones) provided that the Li+/K+ balance is adjusted to allow a duplex formation that can be ‘easily challenged’ by a quadruplex ligand. The hTERC system presented here was chosen as RNA is less often studied and competition with P1 helix formation is biologically relevant.
The test is based on a ‘bi-molecular’ interaction, rather than on a conformational change of an intramolecular quadruplex. This is an important point, as it will allow further developments with methods adapted for true HTS format, such as such as alphascreen or TR-FRET.
The test allows the design of a ‘control duplex’ system, with mutated C- and G-rich strand that retain equivalent duplex potential but lose quadruplex-forming ability. This control is essential to avoid false positive interfering with the assay. A similar control cannot be implemented in the previous assays.
A ‘variety of competitors’ may be tested in this assay. We acknowledge that different unlabelled sequences could be tested in the FRET-melting assay, but a major limitation was that the stability of the competitor had to be significantly larger than the one the labelled quadruplex (otherwise, if the competitor melts at a lower temperature, it cannot act as a duplex or quadruplex competitor when the labelled sequence starts to unfold). This is not a problem here: one does not need to worry about comparing stabilities of the different structures; all that is needed is that the competitor folds at the temperature of the assay. The only limitation is that the competitor should not be capable of hybridization with the complementary C-rich strand. But this is a moderate limitation as compared to the ones we face with FRET melting. We indeed tested an unprecedented number of competitors in this MS (Figure 4 and Supplementary Figure S6).
In principle, one may design an ‘isothermal’ version of this approach (but this requires adding the different components of the reaction in a precise order: the C-rich complementary strand being added last). This is a fundamental difference with a FRET-melting assay, which does provide semi-quantitative information on the stabilization efficiency, but requires scanning a wide temperature range. In this assay, one could restrict the analysis to a single-physiological temperature (37°C): this is an important condition to design an assay with a high-throughput potential.
The two assays are actually complementary, as the assay presented here can be adapted to high-throughput (either to test many ligands or many competitors). On the other hand, FRET melting may provide semi-quantitative information which is more difficult to derive from this method. A similar approach should be easily adapted to test the activity of G4 nucleases. In this case, thermal denaturation is likely to affect the protein activity and thus protein activity should occur in the first step of the annealing procedure, meaning that the complementary strand should be added after the denaturation step.
hTERC targeting?
With this test, we identified ligands that may prevent P1 helix formation in hTERC through a G4 mechanism. The ultimate goal is to find compounds that bind to this full-length RNA and induce a conformational change, but the screening assay developped here cannot be directly transposed to a much larger RNA. Interference with P1-helix formation remains to be demonstrated on the fully folded hTERC embedded in an active telomerase complex. The telomerase substrate is also able to form G4 structures, and thus in the absence of fully specific ligand targeting the G4 of hTERC, evaluation of the activity of these ligands on telomerase activity would require to deconvoluate G4-related effect on hTERC from the effect on the telomeric substrate. This will require the design and characterization of mutant of hTERC with mutations either in the G4-prone sequence and/or in the template of hTERC.
Furthermore the biological consequences of P1 disruption in hTERC remain to be tested. P1 disruption could either alter the template boundary definition (38) and thus prevent processive addition of the telomeric repeat or lead the incorporation of mutated repeat (39) but it could also affect the global folding of hTERC in the telomerase complex by altering long distance interaction (40,41) or finally it could also affect proper telomerase assembly of localization (42).
Up to now, none of articles analyzing the effects of G4 ligands on telomerase activity took into account the possibility that these ligands could also act by binding on the RNA component of telomerase: their effects on telomerase was generally attributed to a binding on the telomeric DNA substrate. In our previous publication (23), we showed that a novel quadruplex-prone target may be involved in these effects. In this article, we also showed that many G4 ligands used to target telomeric G4 can also bind to hTERC. In order to deconvoluate these mechanisms and validate the intended P1 helix-related mechanism, one will need to find a ligand truly specific for hTERC quadruplex, with no binding to the human telomeric motif. The assay developped here allows for such screening. So far, even if some ligands exhibit a preference for hTERC over telomeric DNA, the differential affinity is not sufficient. However, as discussed above, our method will allow the screening of much larger libraries to find promising compounds. These questions remain to be tested and the quest for a G4 ligand specific for a given G4 folding, sequence and chemistry is the next challenge for chemists in the field.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
INSERM (to J.-L.M.); ARC (to J.-L.M.); CNRS-MNHN (to J.-L.M.); ‘Fondation pour la Recherche Médicale, Région Aquitaine’ (to J.-L.M.); CNRS-PIR (to J.L.M.); ANR-09-Blanc-0355 (to J.-L.M.). Funding for open access charge: INSERM U565, CNRS-MNHN UMR7196; Museum National d’Histoire Naturelle, 43 rue Cuvier, 75005 Paris, France.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We wish to thank all the chemists for providing ligands (‘Materials and Methods’ section), D. Morgan, J. Gros, A. De Cian and A. Guédin for technical advices and discussions, A. Ceccaldi for the Z′ factor, C. Guetta and E. Largy for preparing the compounds plate.
REFERENCES
- 1.Gellert M, Lipsett MN, Davies DR. Helix formation by guanylic acid. Proc. Natl Acad. Sci. USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lipps HJ, Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 2009;19:414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat. Struct. Mol. Biol. 2006;13:1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- 4.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps H. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 5.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cahoon LA, Seifert HS. An alternative DNA structure is necessary for pilin antigenic variation in Neisseria gonorrhoeae. Science. 2009;325:764–767. doi: 10.1126/science.1175653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gomez D, Lemarteleur T, Lacroix L, Mailliet P, Mergny JL, Riou JF. Telomerase downregulation induced by the G-quadruplex ligand 12459 in A549 cells is mediated by hTERT RNA alternative splicing. Nucleic Acids Res. 2004;32:371–379. doi: 10.1093/nar/gkh181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumari S, Bugaut A, Huppert J, Balasubramanian S. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wieland M, Hartig JS. RNA quadruplex-based modulation of gene expression. Chem. Biol. 2007;14:757–763. doi: 10.1016/j.chembiol.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 10.Randall A, Griffith JD. Structure of long telomeric RNA transcripts: the G-rich RNA forms a compact repeating structure containing G-quartets. J. Biol. Chem. 2009;284:13980–13986. doi: 10.1074/jbc.M900631200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 12.Sun D, Thompson B, Cathers BE, Salazar M, Kerwin SM, Trent JO, Jenkins TC, Neidle S, Hurley LH. Inhibition of human telomerase by a G-quadruplex-interactive compound. J. Med. Chem. 1997;40:2113–2116. doi: 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- 13.Zhang ML, Tong XJ, Fu XH, Zhou BO, Wang J, Liao XH, Li QJ, Shen N, Ding J, Zhou JQ. Yeast telomerase subunit Est1p has guanine quadruplex-promoting activity that is required for telomere elongation. Nat. Struct. Mol. Biol. 2010;17:202–209. doi: 10.1038/nsmb.1760. [DOI] [PubMed] [Google Scholar]
- 14.Gomez D, O’Donohue MF, Wenner T, Douarre C, Macadré J, Koebel P, Giraud-Panis MJ, Kaplan H, Kolkes A, Shin-ya K, et al. The G-quadruplex ligand telomestatin inhibits POT1 binding to telomeric sequences in vitro and induces GFP-POT1 dissociation from telomeres in human cells. Cancer Res. 2006;66:6908–6912. doi: 10.1158/0008-5472.CAN-06-1581. [DOI] [PubMed] [Google Scholar]
- 15.De Cian A, Cristofari G, Reichenbach P, De Lemos E, Monchaud D, Teulade-Fichou MP, Shin-ya K, Lacroix L, Lingner J, Mergny L. Reevaluation of telomerase inhibition by quadruplex ligands and their mechanisms of action. Proc. Natl Acad. Sci. USA. 2007;104:17347–17352. doi: 10.1073/pnas.0707365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 18.Luke B, Lingner J. TERRA: telomeric repeat-containing RNA. EMBO J. 2009;28:2503–2510. doi: 10.1038/emboj.2009.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saccà B, Lacroix L, Mergny JL. The effect of chemical modifications on the thermal stability of different G-quadruplex-forming oligonucleotides. Nucleic Acids Res. 2005;33:1182–1192. doi: 10.1093/nar/gki257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palumbo SL, Ebbinghaus SW, Hurley LH. Formation of a unique end-to-end stacked pair of G-quadruplexes in the hTERT core promoter with implications for inhibition of telomerase by G-quadruplex-interactive ligands. J. Am. Chem. Soc. 2009;131:10878–10891. doi: 10.1021/ja902281d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim KW, Lacroix L, Yue DJ, Lim JK, Lim JM, Phan AT. Coexistence of two distinct G-quadruplex conformations in the hTERT promoter. J. Am. Chem. Soc. 2010;132:12331–12342. doi: 10.1021/ja101252n. [DOI] [PubMed] [Google Scholar]
- 23.Gros J, Guédin A, Mergny JL, Lacroix L. G-quadruplex formation interferes with P1 helix formation in the RNA component of telomerase hTERC. Chembiochem. 2008;9:2075–2079. doi: 10.1002/cbic.200800300. [DOI] [PubMed] [Google Scholar]
- 24.Zhang JL, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 25.Mergny JL, Lacroix L, Teulade-Fichou MP, Hounsou C, Guittat L, Hoarau M, Arimondo PB, Vigneron JP, Lehn JM, Riou JF, et al. Telomerase inhibitors based on quadruplex ligands selected by a fluorescence assay. Proc. Natl Acad. Sci. USA. 2001;98:3062–3067. doi: 10.1073/pnas.051620698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guyen B, Schultes CM, Hazel P, Mann J, Neidle S. Synthesis and evaluation of analogues of 10H-indolo[3,2-b]quinoline as G-quadruplex stabilising ligands and potential inhibitors of the enzyme telomerase. Org. Biomol. Chem. 2004;2:981–988. doi: 10.1039/b316055f. [DOI] [PubMed] [Google Scholar]
- 27.Paramasivan S, Bolton PH. Mix and measure fluorescence screening for selective quadruplex binders. Nucleic Acids Res. 2008;36:e106. doi: 10.1093/nar/gkn487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monchaud D, Allain C, Teulade-Fichou M. Development of a fluorescent intercalator displacement assay (G4-FID) for establishing quadruplex-DNA affinity and selectivity of putative ligands. Bioorg. Med. Chem. Lett. 2006;16:4842–4845. doi: 10.1016/j.bmcl.2006.06.067. [DOI] [PubMed] [Google Scholar]
- 29.Li JWJ, Tan WH. A single DNA molecule nanomotor. Nano Lett. 2002;2:315–318. doi: 10.1021/nl9011694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alberti P, Mergny JL. DNA duplex-quadruplex exchange as the basis for a nanomolecular machine. Proc. Natl Acad. Sci. USA. 2003;100:1569–1573. doi: 10.1073/pnas.0335459100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guittat L, Lacroix L, Gomez D, Arimondo PB, De Cian A, Pennarun G, Amrane S, Alberti P, Lemarteleur T, Aouali N, et al. In: Dynamical Genetics. Parisi V, Valeria De Fonzo V, Aluffi-Pentini F, editors. Kerala, India: Research Signpost; 2004. pp. 199–236. [Google Scholar]
- 32.Gomez D, Aouali N, Londoño-Vallejo A, Lacroix L, Mégnin-Chanet F, Lemarteleur T, Douarre C, Shin-ya K, Mailliet P, Trentesaux C, et al. Resistance to the short term antiproliferative activity of the G-quadruplex ligand 12459 is associated with telomerase overexpression and telomere capping alteration. J. Biol. Chem. 2003;278:50554–50562. doi: 10.1074/jbc.M308440200. [DOI] [PubMed] [Google Scholar]
- 33.De Cian A, Delemos E, Mergny JL, Teulade-Fichou MP, Monchaud D. Highly efficient G-quadruplex recognition by bisquinolinium compounds. J. Am. Chem. Soc. 2007;129:1856–1857. doi: 10.1021/ja067352b. [DOI] [PubMed] [Google Scholar]
- 34.Gowan SM, Harrison JR, Patterson L, Valenti M, Read MA, Neidle S, Kelland LR. A G-quadruplex-interactive potent small-molecule inhibitor of telomerase exhibiting in vitro and in vivo antitumor activity. Mol. Pharmacol. 2002;61:1154–1162. doi: 10.1124/mol.61.5.1154. [DOI] [PubMed] [Google Scholar]
- 35.De Cian A, Guittat L, Kaiser M, Saccà B, Amrane S, Bourdoncle A, Alberti P, Teulade-Fichou M, Lacroix L, Mergny JL. Fluorescence-based melting assays for studying quadruplex ligands. Methods. 2007;42:183–195. doi: 10.1016/j.ymeth.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 36.Lim KW, Alberti P, Guedin A, Lacroix L, Riou JF, Royle NJ, Mergny JL, Phan AT. Sequence variant (CTAGGG)n in the human telomere favors a G-quadruplex structure containing a G.C.G.C tetrad. Nucleic Acids Res. 2009;37:6239–6248. doi: 10.1093/nar/gkp630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mergny JL, Maurizot JC. Fluorescence resonance energy transfer as a probe for G-quartet formation by a telomeric repeat. Chem. Bio. Chem. 2001;2:124–132. doi: 10.1002/1439-7633(20010202)2:2<124::AID-CBIC124>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 38.Chen JL, Greider C. Template boundary definition in mammalian telomerase. Genes Dev. 2003;17:2747–2752. doi: 10.1101/gad.1140303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim MM, Rivera MA, Botchkina IL, Shalaby R, Thor AD, Blackburn EH. A low threshold level of expression of mutant-template telomerase RNA inhibits human tumor cell proliferation. Proc. Natl Acad. Sci. USA. 2001;98:7982–7987. doi: 10.1073/pnas.131211098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ueda CT, Roberts RW. Analysis of a long-range interaction between conserved domains of human telomerase RNA. RNA. 2004;10:139–147. doi: 10.1261/rna.5118104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X, Nishizuka H, Tsutsumi K, Imai Y, Kurihara Y, Uesugi S. Structure, interactions and effects on activity of the 5′-terminal region of human telomerase RNA. J. Biochem. 2007;141:755–765. doi: 10.1093/jb/mvm081. [DOI] [PubMed] [Google Scholar]
- 42.Cristofari G, Adolf E, Reichenbach P, Sikora K, Terns RM, Terns MP, Lingner J. Human telomerase RNA accumulation in Cajal bodies facilitates telomerase recruitment to telomeres and telomere elongation. Mol. Cell. 2007;27:882–889. doi: 10.1016/j.molcel.2007.07.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.