Abstract
According to current estimations ∼95% of multi-exonic human protein-coding genes undergo alternative splicing (AS). However, for 4000 human proteins in PDB, only 14 human proteins have structures of at least two alternative isoforms. Surveying these structural isoforms revealed that the maximum insertion accommodated by an isoform of a fully ordered protein domain was 5 amino acids, other instances of domain changes involved intrinsic structural disorder. After collecting 505 minor isoforms of human proteins with evidence for their existence we analyzed their length, protein disorder and exposed hydrophobic surface. We found that strict rules govern the selection of alternative splice variants aimed to preserve the integrity of globular domains: alternative splice sites (i) tend to avoid globular domains or (ii) affect them only marginally or (iii) tend to coincide with a location where the exposed hydrophobic surface is minimal or (iv) the protein is disordered. We also observed an inverse correlation between the domain fraction lost and the full length of the minor isoform containing the domain, possibly indicating a buffering effect for the isoform protein counteracting the domain truncation effect. These observations provide the basis for a prediction method (currently under development) to predict the viability of splice variants.
INTRODUCTION
The decade following the publication of the complete human genome has seen dramatic developments in sequencing technologies, which in turn generated a plethora of new sequence data, both genomic and expressed (i.e. cDNAs and ESTs). Alternative splicing (AS) is one of the fields that gained the most from these revolutionary advances. While in the pre-genomic era most complexities in human biology setting apart Homo sapiens from lower organisms were attributed to a high number of human genes, estimated to be at least 100 000 (1), 10 years later much of this proteomic diversity is thought to come from the high number of sequence variants generated by AS, coming from only ∼24 000 protein-coding genes (2). Recent studies found that AS events could be detected in ∼92–97% of multi-exonic human genes (3,4). Advancement of new technologies such as tissue-specific quantitative microarray (5) and next-generation sequencing (4) makes it possible to measure the relative abundance of AS events (3,6).
To this day, most information about AS is generated on the nucleic acid (DNA or RNA) level. While splice junctions observed in a processed RNA have been fruitful in delineating different aspects of tissue-specific AS variants (7) it has also become clear that many splice variants are not translated into functional proteins. One of the mechanisms to control this is non-sense-mediated decay (NMD), which effectively prevents the expression of splice variants with a premature stop codon. NMD has been shown to have a widespread coupling with AS (8). Another surveillance system, the unfolded protein response (UPR) (9) and the related ERAD (endoplasmic reticulum-associated degradation) (10) operates on splice variants that result in misfolded proteins in the ER (11,12).
According to recent estimations (2), there are ∼24 000 protein-coding genes in the human genome, with an average of four isoforms per gene (13), the number of splice variants ranging from 0 to 5–6 per gene (14) for most gene families. However, there is still a lot of uncertainty about the real number of splice variants that appear as functional proteins for each gene, due to the limited information about splice variants detected on the protein level. Only recently, due to technological developments in proteomics has this problem been tackled in a systematic way in a genome-wide fashion (15,16). Tanner et al. (15) combined mass spectrometry with searches in the human genome to validate 39 000 exons on the translation level and this lead to the confirmation of 40 AS events. Tress (16) and Power (17) used a similar technique to discover multiple alternative gene products for over a hundred Drosophila genes and three human genes in platelet, respectively.
AS has been observed to be coupled with intrinsic protein disorder as such proteins are naturally less prone to mis-folding and thus to degradation (18). In a recent study, we have shown in connection with this phenomenon that chimeric proteins generated by chromosomal translocation also tend to survive with much higher frequency if they are disordered, for the same reason (19). It has also become apparent (by one of us, H. Hegyi) that natural proteins contain only intact globular domains (20). We found that with the exception of ∼10% of the protein families in Pfam (in total containing more than 10 000 protein families as of today), globular Pfam domains (21) can be reliably used to eliminate ‘mispredicted’ proteins (generated usually by automatic pipelines, based on genomic DNA and ESTs) where a large fraction of a globular domain is missing from the protein in question (20,22). This approach proved very useful in the automatic annotation process of proteins in the Trembl (23) and Ensembl (24) databases to pinpoint potential errors in the correct delineation of the protein sequences in question. It has also been observed before that AS tends to avoid globular domains (25). However, the authors also noted that 28% of AS variants do have split domains.
There is a large gap between structural and sequence information available of isoforms of proteins generated by AS. While there is a plethora of sequences of alternative splice variants for human proteins, only a handful of protein structures of such variants (26) exist. Various researchers predict various numbers of viable sequence variants (27,28) but only a handful of bioinformatics studies have dealt with the problem. However, the discipline will apparently play a large role in determining the functional splice variants by eliminating those that exist as proteins only in silico.
Here we studied in detail how AS affects the integrity of globular domains. Using human proteins in Swissprot, we find that there is a significant difference with respect to domain truncation size distribution between provisional splice variants and those for the existence of which there is at least some evidence. We also find that in those cases when there is a severe truncation of a globular domain cutting the domain in half the exposed hydrophobic surface is usually small, comparable to that of an intact (sub)domain. Another survival strategy of the incomplete domain is to have a substantial amount of intrinsic disorder around the splice site of the AS event. These observations together will form the foundation of a server currently under construction we shall name Domain Integrity Verification of AS or DIVAS for short.
MATERIALS AND METHODS
Sequence analysis
We used the human SwissProt data set of the UniProt knowledgebase (23,29) as a source of protein sequence, function and AS information. We used the Swissprot nomenclature to nominate the major and minor isoforms (i.e. the alternatively spliced variants), and to categorize the splice events as deletion, substitution and insertion. In total 7101 major and 13 437 minor isoforms were extracted.
Whereas Swissprot is extensively annotated and highly reliable for the major isoforms of the proteins, there is relatively little information supplied for the splice variants, especially regarding their existence as viable proteins. For this reason, we have generated two more data sets, with increasing level of confidence with respect to the existence of protein products.
First, we have selected those splice variants which have a ‘name’ or ‘synonym’ in the Swissprot annotation (as opposed to a single serial number). This group contains 6057 splice variants of 3958 human proteins. Whereas there is no explicit evidence for the existence of these splice variants as proteins, their existence at the level of mRNA has been confirmed by more than one independent study, which increases the likelihood that they represent viable alternatives to the major isoform. Furthermore, their number is sufficiently high for statistically rigorous analyses. We call this group ‘named’ throughout this article.
Next, we created a group of minor isoforms for the existence of which as a protein there is evidence in the literature, usually an expression study incorporating a western blot. By sifting through the literature provided in the annotation part of the relevant proteins in Swissprot, we collected 505 such human isoforms. Because their existence is confirmed at the protein level, they represent the ultimate test case for our concepts, although their low number in certain cases does not allow rigorous statistical analysis. These experimentally verified minor variants, enlisted in Supplementary Table S1, are termed ‘verified’ throughout the article.
To investigate the localisation of splice events in human proteins, we divided the Swissprot splice events into three groups. The most frequently occurring splice type was the deletion of a particular protein region (n = 10 634), which could result from either exon skipping or alternative initiation. All splice events were mapped to the major isoform. We also collected substitution (n = 6635) and insertion splice events (n = 1467). It must be noted that the number of insertions is skewed by the fact that Swissprot designates the longest isoform to be the major one by default (23). For all three splice events, we performed random controls. We randomly selected a protein from the data set of the major isoforms and randomly picked a splice event from the splice-event data set of that type, also randomly choosing the splice site in the main isoform in question. We always matched the size of the random data set to that of the real splice event data set.
Domain analysis
Domain information was obtained from the Pfam-A.full file of the Pfam database (v23.0; 21), by extracting Swissprot and Pfam identification numbers and the locations of the latter. We also reconstructed all minor isoforms using the corresponding annotations in Swissprot. To investigate domain changes in the minor isoforms we recorded all the instances when AS occurs within the boundaries of a Pfam domain, truncating it, inserting extra amino acids or substituting a part of a domain with a different sequence. We paid special attention to the cases when a domain got truncated by an AS event.
To determine the statistical significance of the truncation events we generated a control group where we used the same minor-isoform data set but randomized the location of the AS events. As we worked with the same sample size as in the original data set, we were able to compare directly the occurrence of the domain truncations in the random and original data sets. We also compared the distribution of the relative domain truncation sizes among the original (all human Swissprot) splice variants, the ‘named’ ones and the randomized control group.
Disorder analysis
Disorder patterns were mapped to all major isoform sequences by using the IUPred (30) predictor. The disorder of a region was quantified as the percentage of the disordered amino acids in the region, with a threshold of 0.5 for the predicted scores of the individual residues.
3D analysis
To validate our results of sequence-based domain truncations in 3D, we also carried out a structural analysis of the PDB entries of the affected domains to see how the truncations caused by AS might affect the survival of the domains in question. The approaches that have been applied have been discussed subsequently.
Alternatively spliced variants in PDB
We collected all the human proteins that have the 3D structure of at least two isoforms in PDB. In order to accomplish this, we compared the sequences of both the major and minor isoforms to ‘seqres’, containing the sequences of all chains in PDB, using Blastp (31). Keeping only those entries from both databases that matched each other with a percentage identity of 90% or higher, we selected those pairs of matches where the major and a minor isoform of the same protein could be mapped to different PDB entries (both with a near perfect score). The procedure can be formalized as follows.
One isoform of a human protein has a higher sequence similarity to ‘entry 1’ of seqres than to ‘entry 2’ whereas another isoform is more similar to ‘entry 2’ than to ‘entry 1’.
We also require that the difference between ‘entry 1’ and ‘entry 2’ is in a region where both isoforms exist or it could happen that ‘entry 1’ and ‘entry 2′ simply map to two different domains of the protein in question.
We needed this elaborate procedure in case the isoform and the PDB entry were not 100% identical (e.g. the latter underwent genetic engineering to facilitate the crystallization of the protein) but based on the overall sequence match the PDB entry in question corresponds to that particular isoform and is in an ‘orthologous’ relationship with another isoform corresponding to a different PDB entry.
This approach resulted in 15 pairs of PDB entries that appear to have been derived from different splice variants of the same entry in Swissprot (Table 1). One protein, FGFR_HUMAN has three isoforms with a structure in PDB.
Table 1.
Human alternatively spliced proteins in Swissprot with minimum two isoforms in PDB (FGFR2_HUMAN has three isoforms in PDB)
| Swissprot ID | Isoform numbers | PDB ID1 | PDB ID2 | Alt splic type | Difference | Description |
|---|---|---|---|---|---|---|
| CHKA_HUMAN | Isoforms 1,2 | 3f2r_A | 2cko_A | Insertion* | 18 | Longer isoform 3f2r has an insertion of 18 amino acids disordered in the longer variant. In 2cko, 7 residues around the site of insertion are also disordered |
| CRK_HUMAN | Isoforms 1,2 | 2eyz_A | 2eyy_A | C-term + domain | 96 | Longer isoform 2eyz has three SH3 domains whereas shorter 2eyy has only two. The shorter version is oncogenic |
| CSF3_HUMAN | Isoforms 1,2 | 1gnc_A | 1pgr_A | Insertion* | 3 | Longer isoform 1gnc_A has a 3 amino acid insertion, missing from structure (i.e. disordered) |
| EDA_HUMAN | Isoforms 1,3 | 1rj7_A | 1rj8_A | Insertion | 2 | 1rj7_A has a 2-amino-acid insertion compared to 1rj8_A, structure determined. Distinct receptor specificities |
| FGFR2_HUMAN | Isoforms 19,14 | 1nun_B | 3dar_A | C-term + domain | 120 | 1nun_B consists of two Ig-like domains, 3dar_A has only one |
| FGFR2_HUMAN | Isoforms 1,19 | 1djs_A | 1nun_B | Insertion | 2 | 1djs_A has a 2-amino-acid insertion compared to 1nun_B, both structured |
| GHR_HUMAN | Isoforms 1,4 | 1axi_B | 2aew_A | Insertion* | 27 | 1axi_B is 27-amino-acid longer in N-terminus, disordered |
| GNAS2_HUMAN | Isoforms 1,2 | 1azs_C | 1cul_C | Insertion* | 14 | 1azs_C has a 14-amino-acid insertion compared to 1cul_C, disordered; 1 cul_C has 5-amino-acid disordered around the place of insertion |
| KHK_HUMAN | Isoforms 1,2 | 2hqq_A | 3b3l_A | Alt exon | 44 | Same length substitution (alternative exons), both structures are fully ordered |
| MK08_HUMAN | Isoforms 1,3 | 3elj_A | 1ukh_A | Alt exon | 12 | Same length substitution (alternative exons), both structures are fully ordered |
| NRX1A_HUMAN | Isoforms 1,2 | 2r1b_A | 3bod_A | Insertion* | 30 | 2r1b_A has a 30-amino-acid insertion, half of it is ordered and forms a protruding-long helix |
| PTN13_HUMAN | Isoforms 1,4 | 1q7x_A | 3pdz_A | Insertion | 5 | 1q7x_A has a 5-amino-acid insertion, fully ordered; they have a different affinity to tumor suppressor protein APC |
| RAC1_HUMAN | Isoforms 1,2 | 1ryf_A | 1i4t_D | Insertion* | 19 | 1ryf_A has a 19-amino-acid insertion, disordered, also in the preceeding 15 amino acids; the insertion induces a conformational change, PMID:14625275 |
| ST2B1_HUMAN | Isoforms 1,2 | 1q1z_A | 1q1q_A | Alt N-term | 10 | 1q1q_A and 1q1z_A both form helical structures in their N-termini upon pregnenolone binding, have different specificity |
| UAP1_HUMAN | Isoforms 1,2 | 1jvd_A | 1jv1_A | Insertion* | 17 | 1jvd_A has a 17-amino-acid insertion, fully disordered. 1jv1_A is structured at the place of insertion |
The isoform numbers and the PDB identifiers representing the two isoforms are listed. The type of AS and the number of residues the two PDBs differ from each other (Difference) are indicated. Asterisk next to the number shows if intrinsic disorder was also observed in one or both isoforms.
The occurrence of structural domains in the splice variants in Swissprot
We compared both the major and minor isoforms of human proteins to ‘seqres’ and wherever feasible to SCOP sequences (32), the latter containing the sequences of structural domains of the PDB entries. For both of these large-scale comparisons, we used the Blastp program (31). We recorded sequence matches with a percentage identity of 60% or higher and collected those hits where >10% of the domain match was missing due to a truncation caused by AS.
Hydrophobic surface analysis of PDB entries
For those PDB matches where a structural domain was interrupted by an alternative splice site, we carried out a detailed structural analysis focusing on the hydrophobic surface potentially exposed due to domain interruption by AS. We used the CHASA (33) method to calculate the hydrophobic surface area as follows:
We determined the nonpolar (hydrophobic) surface area of the full PDB chain in question.
We extrapolated the ideal hydrophobic surface values for gradually smaller intact domains of the same type using Chothia’s formula for globular domains (34).
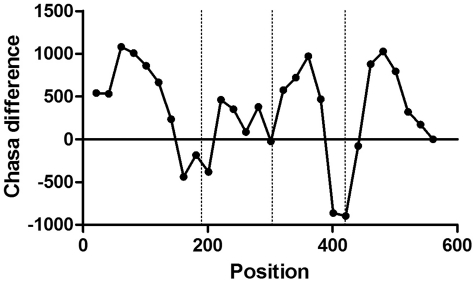
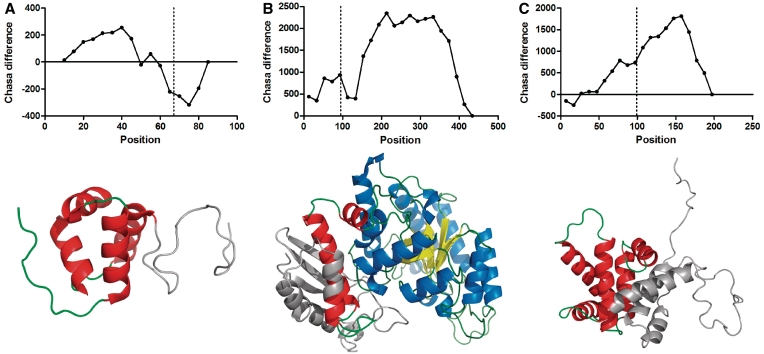
We also determined the hydrophobic surface values of the actual ‘subdomains’ of the PDB chain in question by gradually truncating it (removing the coordinates) in steps of 5, 10 or 20 residues, depending on the original length of the chain (<101 residues, between 101 and 300, or >300 residues, respectively). We calculated the difference between the two values at each truncation point we named CHASA-diff. A typical graph of the differing values is shown for 1c47 chain A (Figure 5).
We determined the local minima for each hydrophobic surface difference curve generated with CHASA-diff and compared them with domain truncation sites in our verified data set, to see if there is a correlation between them, i.e. if truncations preferentially happen in the vicinity of these local minima of hydrophobic surface differences. Rather than using only the exact PDB structures belonging to their corresponding Swissprot entries, we compared their sequences using Blastp, and allowed 60% or better sequence identity between a splice variant and a PDB sequence for a splice variant to be considered for the analysis. We accepted as valid only those local minima that were on average at least 200 kcal deeper than the two nearest local maxima on each side, and only those splice sites that were in the vicinity of such local minima, no farther than 13 residues in the amino acid sequence. To justify the values for these two parameters we carried out a perturbation for both of them, explained in the ‘Results’ section.
Figure 5.
CHASA-diff analysis of 1c47 phosphoglucomutase-1 with SCOP domain boundaries indicated by vertical lines. The domain boundaries separating the four domains in the PDB structure coincide with minima in CHASA-diff, indicating relatively small hydrophobic surface areas.
Perl scripting
Wherever not indicated otherwise calculations were done by self-made Perl scripts.
RESULTS
Sequence analysis of minor isoforms of human proteins in Swissprot
Using the human Swissprot subset of the UniProt Knowledgebase we created a data set containing 20 538 isoforms (7101 major and 13 437 minor), to investigate the effects of the splice events on the protein structure and viability. In delineating the major and minor isoforms, we followed the nomenclature of Swissprot, which usually nominates the longest splice variant to be the major isoform, unless there is reason to believe otherwise.
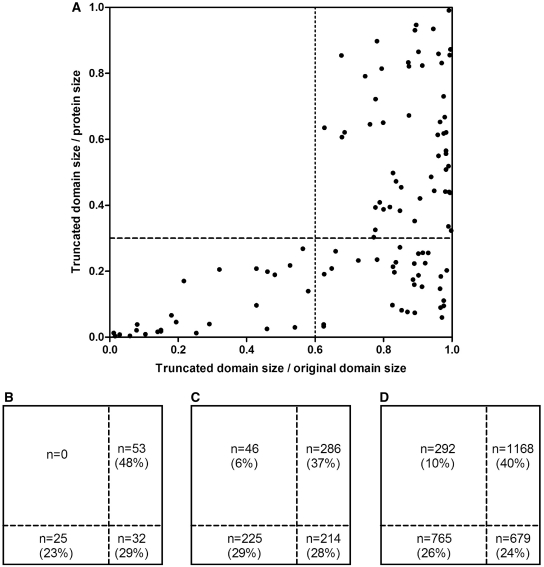
At first, we determined the relative length of the truncated domains (i.e. the remaining part divided by the full length of the domain) and related these values to the full length of the containing proteins. It must be noted that all sequence analysis was carried out using the Pfam domain annotations whereas all structural domain analysis was done with SCOP domains. The results are shown in Figure 1. In Figure 1A, the results for the ‘verified’ group are shown, each truncated domain indicated with a dot, whereas in Figure 1B–D data are shown in terms of actual numbers, for the ‘verified’, ‘named’ and the total number of alternative splice variants (in Swissprot), respectively. (For the definition of ‘verified’, ‘named’ and ‘random’ groups of splice variants see ‘Materials and Methods’ section.) For the ‘verified’ group, all the truncated domains satisfy at least one of the following two criteria: truncated domain size/original domain size >0.6 OR truncated ‘domain size/protein length’ <0.3, i.e. the upper left quadrant of the rectangle is empty (Figure 1A and B). However, for the named group and the total of Swissprot, this area is increasingly populated (6 and 10%, respectively). According to a χ2-test, the difference is significant both between the ‘verified’ and the ‘named’ group (P = 0.011) and between ‘named’ and Swissprot (P = 0.0002).
Figure 1.
‘Retained portion’ of the truncated domains (i.e. the remaining part divided by the full length of the domain) versus their ‘relative length’ related to the full length of the containing minor isoform (i.e. the remaining part of domain divided by the full length of the protein). (A) Each truncated domain indicated by a dot, shown for the ‘verified’ group. (B) Same data as in (A) but the population of the four quadrangles indicated with percentage numbers. (C) Data for the named group, same representation as in (B). (D) Data for the total number of alternative splice variants in Swissprot.
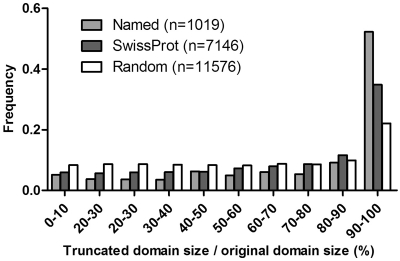
After summarizing all splice events in the ‘named’/all Swissprot/randomized Swissprot sets, it became apparent that splice sites preferably avoid globular domains: while 11 576 out of the total of 33 223 (∼35%) randomized splice sites in the Swissprot splice variants fall into a domain, this value for the actual splice variants without randomization is 7146 (out of 33 223, ∼22%) and further decreases to ∼9% for the ‘named’ set (1019 out of 10 743 domains). However, even when the splice site falls into a globular domain, the relative length of the remaining domain is not evenly distributed between 0 and 1 but strongly biased towards values close to 1, as shown in Figure 2 (also apparent from Figure 1). This bias is the most apparent again in the ‘named’ group; i.e. there is a strong selection against severe domain truncation in globular domains to preserve the structural integrity of such domains. In contrast, in the randomly generated data set the frequencies of the various truncations were almost uniform (except for the minor truncations caused by the overrepresented small sized splice events). The difference between Swissprot and the ‘named’ group was again highly significant (P < 0.0001).
Figure 2.
Percentage distribution of relative domain size of domains truncated by AS in the ‘named’/all Swissprot/randomized Swissprot sets. Bin size increment is 0.1.
In an earlier study, Dunker and co-workers (18) found that AS is associated with protein disorder, a plausible association considering how much less the intrinsically disordered regions are affected structurally by major changes in sequence when compared to ordered regions. However in their study, no experimental evidence for the actual existence of the alternative protein products was taken into consideration, and no attempt was made to address the three different types of AS events (deletion, substitution and insertion) separately either. As we have considerably more data, both at the mRNA (the ‘named’ group) and protein (‘verified’ group) level, we could establish the significance of differences between observed splicing events and chance occurrence in most cases.
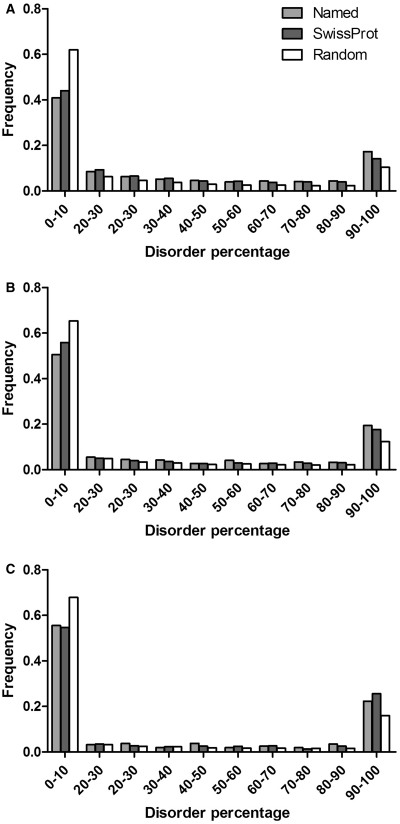
The most frequently occurring splice events are the deletions. Comparing the frequency distribution of percentage disorder in the deleted protein region (Figure 3) with the control groups using the χ2-probe, we found statistically significant differences between all groups (difference between ‘named’ and Swissprot, P = 0.024; between Swissprot and ‘random’, P < 0.0001). Significant differences could also be observed for substitutions (for the full region replaced, significance of difference between ‘named’ and Swissprot, P = 0.01, whereas between Swissprot and random, P < 0.0001, data not shown). Due to the relatively small sample size of insertions (1467, compared to 6635 substitutions and 10 634 deletions, as described in ‘Materials and Methods’ section), significance could not be established between the ‘named’ group and Swissprot, however Swissprot was significantly more disordered than the ‘random’ group (P < 0.0001).
Figure 3.
Frequency distribution of percentage disorder in protein regions deleted (A) substituted (B) or disrupted by an insertion (C) by AS in the ‘named’ group/all of Swissprot/randomized Swissprot. The three groups were significantly different from one another, established by χ2-tests (P < 0.05), regarding deletions (A) and substitutions (B) but not for insertions (C), due to the small sample number of the named group. For further details see ‘Results’ section.
Alternative splice variants in PDB
We collected all the human proteins that have the 3D structure of at least two isoforms in PDB. The results of this exhaustive search procedure (described in ‘Materials and Methods’ section) are shown in Table 1. Despite the large number of minor isoforms for human proteins in Swissprot we found only 14 proteins for which we could find the structure of at least two isoforms in PDB. For one protein, FGFR_HUMAN, there are three different PDB structures corresponding to three different isoforms. We indicated in the table, the type of AS that occurred for the isoforms of each protein. The most frequently occurring AS type is the insertion (here we did not distinguish between insertions and deletions), which occurs nine times out of the 15 cases altogether. The length of the insert varies between 2 (EDA_HUMAN) and 30 (NRX1A_HUMAN) amino acids.
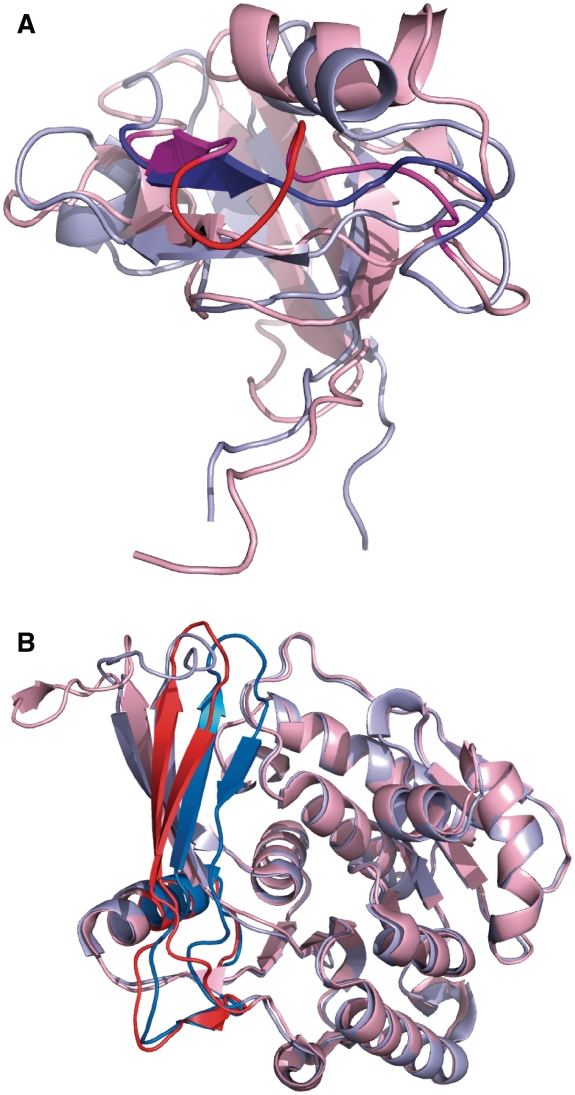
The longest insert that has an ordered structure at the site of the insertion in both isoforms is 5 amino acids in the PDZ2 domain of the protein tyrosine phosphatase PTN13_HUMAN where the two splice variants have a different affinity to the tumor suppressor protein APC (35). The structural alignment of the two isoforms is shown in Figure 4A.
Figure 4.
Structural isoforms of alternatively spliced human proteins. (A) Structural alignment of the major and minor isoform in the PDZ2 domain of the protein tyrosine phosphatase PTN13_HUMAN. The two splice variants have a different affinity to the tumor suppressor protein APC (35). Isoform 4 has a 5-residue-long insertion (colored bright red) compared to the main isoform. This was the longest insertion in a splice variant in PDB where both variants are ordered at the alternative splice site. (B) The structural alignment of the two isoforms of the hexokinase KHK_HUMAN (PDB codes: 2hqqA, 3b3lA). The minor isoform contains a 44-residue substitution, which represents a paralogous local exon duplication (indicated with solid blue and red colors in 2 hqqA and 3b3lA, respectively). AS of the KHK gene selects either one or the other of two adjacent, homologous 135-bp exons.
In those proteins where the insertion is longer than 10 amino acids usually both isoforms are disordered at the site of the insertion. This is the case for CHKA_HUMAN, GNAS2_HUMAN, NRX1A_HUMAN and RAC1_HUMAN. In the latter, 15 residues preceding the insertion site are also disordered, besides the insertion itself, which, however, induces a conformational change in the partner protein it interacts with (36).
NRX1A_HUMAN is an interesting case study where half of the 30-residue-long insertion is ordered and forms a long protruding helix in the longer isoform.
In two proteins, CRK_HUMAN and FGFR2_HUMAN one isoform had an extra domain on the C-terminus, in both cases a full-sized domain, without being interrupted by an alternative splice site. In one isoform of GHR_HUMAN, there was an alternative N-terminus; however the extra amino acids in the longer isoform are all disordered in the relevant pdb structure, ‘1axi’.
Aside from the latter alternatives when one isoform has an extra domain, the largest difference between the structures of two isoforms was a 44-amino-acid-long substitution in the hexokinase KHK_HUMAN. AS of the KHK gene selects either one or the other of two adjacent 135-bp exons, which represent the evolutionary descendants of a paralogous local exon duplication (37). The structural alignment of the two isoforms is shown in Figure 4B. The two structures are fairly similar even though their sequences share only 35% identity. So far no physiological function has been found for the minor isoform (37).
Accessible non-polar surface analysis of the human splice variants
Hydrophobic surface analysis of PDB entries
As described in ‘Materials and Methods’ section, we used a procedure we called CHASA-diff to calculate the hydrophobic-surface difference of the truncated domains at the position of truncation compared to an intact domain of the same type and of the size that is left of the domain after truncation. The procedure is illustrated in Figure 5 for PDB chain 1c47A. It consists of four domains as determined by SCOP (32), also indicated in Figure 5 (throughout this section, we used domain definitions and boundaries as delineated by SCOP). As it is apparent from the figure the domain boundaries coincide with hydrophobic surface minima, in accordance with the notion that globular domains fold in a way that buries most of the hydrophobicity inside of the domain, building up the hydrophobic core.
Extending the analysis to splice variants with sufficient similarity to a PDB entry
We analyzed all the truncated domains in the ‘verified’ and ‘named’ sets, with at least 60% sequence identity to a PDB chain. After running CHASA-diff for the PDB chain, it was checked if the truncation site was sufficiently close to a local minimum in the CHASA-diff curve and if the local minimum was sufficiently deep. As explained in ‘Materials and Methods’ section, a local minimum was accepted if it was at least 200 kcal deeper than the average of its two neighboring maxima. For the vicinity of a truncation site from a local minimum to be accepted a threshold of 13 residues was chosen.
We perturbed the latter values by changing the energy threshold at first to 100, then to 300 kcal whereas for the vicinity threshold, we chose at first 14 then 12 residues. For each perturbed value and also for the unperturbed ones, we calculated the ratio of the error rate (i.e. the number of ‘bad’ versus all splice variants in that category) for the unverified and the verified group. Taken the unperturbed values of 200 kcal and 13 residues, we found this value to be (65/364)/(1/16) = 3.03. Changing the energy threshold to 300 kcal this ratio changed to (79/364)/(2/16) = 1.74; whereas for the threshold of 100 kcal this ratio resulted in (59/364)/(1/16) = 2.59. In other words, threshold = 200 kcal resulted in the maximum discriminating power between the unverified and the verified group. Perturbing the vicinity threshold produced similar values.
We paid special attention to those splice variants where a domain lost 10% or more of its original length due to AS. Using Blastp, requiring at least 60% sequence identity for matches between these splice variants in the ‘named’ set and chains in PDB we got 380 distinct matches for 331 isoforms of 187 human proteins in Swissprot matching the sequences of 154 different chains in PDB. After evaluating the truncated domains for their newly exposed hydrophobic surface, we found that 310 of the 380 matches (81.6%) satisfied the above two criteria about the truncation site being sufficiently close to a local minimum in the CHASA-diff curve of the matching PDB chain.
We also looked at the truncated domain length/isoform length ratio for these structural matches in the named set, to see if there is any difference between the 310 energetically favorable and the 70 unfavorable matches. In accordance with the results summarized in Figure 1, we found that while the average for this ratio for the favorable set was 0.199 ± 0.011, for the unfavorable set it was 0.423 ± 0.037, i.e. more than twice as big than for the favorable ones.
The results for the ‘verified’ set are shown in Table 2. We found that 11 out of the 12 (91.7%) isoform matches with a truncated domain with experimental evidence for the existence of the isoform as a functioning (or malfunctioning, if its existence was implicated in cancer) protein met the two criteria described above, the only exception being isoform 2 of ENOA_HUMAN. The 3D structure and the CHASA-diff values for 1pdy, the most matching PDB chain are shown in Figure 6A, with the truncated portion indicated. The truncation site (at 94 of the 433 residue pdb chain) is at a local maximum, presumably cutting the smaller first domain into half. However, it should be also noted that the remaining portion of the domain, i.e. residues 94–137 is only a small fraction (0.12) of the total length of the protein, ENOA_HUMAN-2 as also shown in Table 2. As seen in Figure 1 and confirmed for the structural matches for the ‘named’ set, this is another indicator of the survival of an isoform. We show the truncated structures for two more splice variants, EDF1_HUMAN-2 and BID_HUMAN-4, represented by PDB chains 1x57A (Figure 6B) and 2bidA (Figure 6C), respectively. The corresponding CHASA-diff profiles are also indicated. All the rest of the 12 truncated domains listed in Table 2 were acceptable by CHASA-diff, using the threshold values for the depth of, and distance from, the nearest hydrophobic energy minimum (200 kcal and 13 residues, respectively).
Table 2.
Truncated PDB chain/SCOP domain matches with a human Swissprot alternative splice variant and acceptable truncation points based on CHASA-diff analysis
| Swissprot splice variant | PDB chain | PDB |
Sw |
Percentage ID | len PDB | len Sw | Potential breakpoints | Chasa | TrDom/ Swlen | SCOP domain | SCOP domain |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beg | End | beg | end | beg | end | len | |||||||||
| O15392-3|BIRC5_HUMAN | 1xoxA | 1 | 76 | 1 | 76 | 96 | 117 | 137 | 107,117,87,7 | + | 0.51 | d1xoxa1 | 1 | 70 | 111 |
| O60869-2|EDF1_HUMAN | 1x57A | 8 | 66 | 71 | 129 | 100 | 91 | 139 | 75,80,85,10,1 | + | 0.42 | d1x57a1 | 1 | 59 | 78 |
| P06733-2|ENOA_HUMAN | 1pdyA | 94 | 433 | 1 | 338 | 75 | 434 | 341 | 433,233,133,33,13 | – | 0.12 | d1pdya2 | 93 | 137 | 139 |
| P37231-2|PPARG_HUMAN | 1hraA | 8 | 79 | 109 | 179 | 63 | 80 | 475 | 80,30,5 | + | 0.15 | d1hraa_ | 8 | 79 | 80 |
| P51991-2|ROA3_HUMAN | 1x4bA | 22 | 109 | 7 | 94 | 76 | 116 | 356 | 110,100,10 | + | 0.25 | d1x4ba1 | 15 | 102 | 103 |
| P55957-4|BID_HUMAN | 2bidA | 99 | 197 | 1 | 99 | 100 | 197 | 99 | 197,87,47,17,7 | + | 1.00 | d2bida_ | 99 | 197 | 197 |
| P62993-2|GRB2_HUMAN | 1gbrA | 10 | 68 | 1 | 59 | 100 | 74 | 176 | 69,74,19 | + | 0.34 | d1gbra_ | 10 | 68 | 74 |
| P62993-2|GRB2_HUMAN | 1fhsA | 50 | 112 | 60 | 122 | 100 | 112 | 176 | 102,112,62,12 | + | 0.36 | d1fhsa_ | 50 | 112 | 112 |
| Q01167-2|FOXK2_HUMAN | 1d5vA | 4 | 84 | 258 | 338 | 61 | 94 | 614 | 94,89,84,59,4 | + | 0.13 | d1d5va_ | 4 | 84 | 94 |
| Q07820-2|MCL1_HUMAN | 1wsxA | 6 | 65 | 171 | 230 | 85 | 162 | 271 | 152,162,52,12 | + | 0.22 | d1wsxa_ | 6 | 65 | 162 |
| Q9NR12-2|PDLI7_HUMAN | 1wf7A | 14 | 100 | 11 | 91 | 62 | 103 | 423 | 93,98,103,18 | + | 0.21 | d1wf7a_ | 14 | 100 | 103 |
| Q9NR12-3|PDLI7_HUMAN | 1wf7A | 14 | 100 | 11 | 91 | 62 | 103 | 153 | 93,98,103,18 | + | 0.57 | d1wf7a_ | 14 | 100 | 103 |
The table contains the following columns: Swissprot splice variant, accession number and identifier of splice variant; PDB chain, name and chain of matched PDB entry; PDB beg, PDB end, Sw beg, Sw end: beginning and end of a Blastp match between the sequences of the PDB chain and splice variant, respectively; Percentage ID, proportion of matching residues; len PDB, length of PDB chain in residues; len Sw, length of Swissprot splice variant; potential breakpoints, positions in the PDB chain where a truncation would be permitted by CHASA-diff, based on the exposed hydrophobicity value differences from an ideal value for an intact globular domain of that size (see text for more details), bold residue number is the closest to the actual truncation in the splice variant in question; Chasa, + if there is an acceptable minimum in the vicinity of the breakpoint (within 13 residues in the sequence) as determined by Chasa-diff, – otherwise; TrDom/Swlen, truncated domain length divided by the total length of Swissprot; SCOP domain, matching SCOP domain identifier; SCOP domain beg, end and len, beginning, end of match and length of SCOP domain, respectively.
Figure 6.
Selected truncated domains from Table 2, PDB structures and CHASA-diff hydrophobic values. The structural portions shown in gray are removed by AS. In the CHASA-diff graphs the position of truncation is indicated by a vertical line. (A) 1x57A. The AS removes residues 67–91, which are actually disordered, thus no hydrophobicity issues arise in this region. CHASA-diff usually predicts negative values for disordered segments. (B) 1pdyA. This was the only match between a verified splice variant and a PDB structure (with 75% sequence identity between isoform 2 of ENOA_HUMAN and 1pdyA) where the calculated exposed hydrophobic surface (CHASA difference between the actual and theoretical values) at the position of truncation at residue 94 was ‘prohibitively’ high. Interestingly, Pfam predicts a coiled-coil structure for this region over a 20-residue stretch. (C) 2bidA. Our method allows a truncation at residue 87, the actual splice is at residue 99, which is within the accepted vicinity, set at 13 residues.
DISCUSSION
The number of new alternative splice variants incorporated in various databases has been steadily increasing in recent years [Uniprot (38), Refseq (39), Ensembl (24), ASTD (40), AS-ALPS (41)], reaching the point when practically all multi-exonic genes have been found to generate alternatively spliced variants. However, the specific instances when a minor isoform is produced are still largely undetermined. As only ∼10–20% of all AS events are conserved across two or more species (42,43), it has been suggested that AS is also used as a tool to regulate the expression of functional isoforms via NMD (44) or in protein degradation pathways (45). According to a recent paper by Melamud and Moult (46) a considerable amount of AS is the result of a stochastic process dependent only on the number of introns and expression level of a gene and has no function at all (46).
The major cause of the uncertainty regarding the functional isoforms is the small number of splice variants that have been seen as expressed proteins. By painstakingly sifting through the available annotation for any minor variants for human proteins in Swissprot, we identified only 505 such variants, corresponding to <5% of all minor variants of human proteins recorded in the 2008 release of Swissprot.
Even less is known about the 3D structure of the splice variants. While it is becoming increasingly clear, also supported by our results, that intrinsic protein disorder plays a significant role in AS (18) we also found several globular domains that are able to survive severe truncations imposed by AS. Only a few previous works paid attention to this important issue, i.e. structural aspects of AS, with widely varying conclusions. While Stetefeld and Ruegg concluded that AS may result in small changes in protein structure (26), Birzele found evidence in the literature that splicing events may represent transitions between different folds in the protein sequence-structure space (28). Wang, on the other hand, found that splicing tends to preserve the fold and it typically takes place on the protein surface (47). Melamud and Moult found that most splice variants would result in proteins whose stability would be severely compromised (13), in line with their previous paper about most AS being the result of a stochastic process, without any biological meaning (46).
One way to settle this issue in a conclusive way is to look at the experimental evidence, i.e. the 3D structure of proteins that have undergone AS. An extensive search identified only 14 human proteins in Swissprot (Table 1) that have the structure of at least one minor variant in PDB and this, although not enough to make sweeping conclusions about the structural aspects of AS tends to support the conservative view, i.e. relatively small changes allowed for globular folds to survive AS. Aside from insertions/deletions/substitutions in the minor isoforms, in two cases an entire domain gets deleted on the C-terminus, and in one case, ST2B1_HUMAN, the alternative isoform starts with an alternative N-terminus. However, in none of the minor isoforms does a globular domain get substantially truncated: the longest insertion/deletion that is accommodated by a fold without the help of intrinsic disorder is 5 amino acids, missing from 3pdzA.
While all of the above approaches captured some truths about the structural aspects of AS, they are mostly observational studies, not using any rigorous measurements to make assumptions about the existence of a splice variant that has not been seen as a mature protein. In this article, we analyzed several different parameters for different sets of isoforms of human proteins, namely: (i) the length distribution of truncated domains; (ii) the ratio of the lengths of the truncated domains and the containing isoforms; (iii) the intrinsic protein disorder of the isoforms at the splice site; and (iv) the newly exposed hydrophobic surface created by the truncation and the difference between an intact and the truncated domain (CHASA-diff).
We found that all of the above measurements differ significantly for a set of alternatively spliced variants from random controls and the extent to which a difference can be observed usually depends on the evidence we have for the physical existence of the isoforms as proteins. We paid the most attention to those 505 minor isoforms for the existence of which there is experimental evidence in the literature. In this set, we found 12 incidences (listed in Table 2) when a domain sufficiently similar (>60% sequence identity) to a structural domain in SCOP was truncated by AS, 11 of which were predicted to survive the truncation using CHASA-diff with the chosen parameters.
Regarding CHASA-diff, we also found that the local minima often coincide with domain boundaries as explicitly shown for PDB chain 1c47A in Figure 5. This is a plausible finding as the primary driving force in the formation of globular structure is hydrophobicity (48) and it is also the bottleneck in the survival or degradation of a globular domain truncated by an alternative splice site. It is further proof of the validity of our approach to predict the survival of a truncated domain: if we see a minimum in CHASA-diff within a domain similar in depth to that of a boundary between two domains, we have good reason to assume that this is a valid truncation site, especially if the other parameters regarding the length and disorder of the protein are favorable, too.
It must be noted that the validity of such prediction is largely independent of the genetic mechanism that produced the domain truncation. We used this approach before to analyze fusion proteins generated by chromosomal translocations (19), a genetic process that also can produce truncated globular domains. We found that the truncation site of a protein kinase is remarkably close to the boundary between the two sub-domains, also associated with a minimum in hydrophobic surface energy (19).
An expanded data set to include all the splice variants from other organisms is under construction and it will apparently help to form a more nuanced view of the circumstances that determine the survival of AS variants. The expanded data set together with the observed values in hydrophobic surface area, intrinsic protein disorder and domain truncation distribution will be benchmarked to achieve the most discriminating power between the negative and positive data sets. The method is currently under development and will be made available to the public as a server we shall name Domain Integrity Verification of AS or DIVAS for short.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Hungarian Scientific Research Fund (grant number PD76286); Marie Curie International Reintegration Grant from the European Commission (grant number 046572 to H.H.); Hungarian Scientific Research Fund (grant numbers K60694, NK71582); Hungarian Ministry of Health (grant number ETT 245/2006 to P.T.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Liang F, Holt I, Pertea G, Karamycheva S, Salzberg SL, Quackenbush J. Gene index analysis of the human genome estimates approximately 120,000 genes. Nat. Genet. 2000;25:239–240. doi: 10.1038/76126. [DOI] [PubMed] [Google Scholar]
- 2.Flicek P, Aken BL, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Coates G, Fairley S, et al. Ensembl's 10th year. Nucleic Acids Res. 2010;38:D557–D562. doi: 10.1093/nar/gkp972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 5.Pan Q, Shai O, Misquitta C, Zhang W, Saltzman AL, Mohammad N, Babak T, Siu H, Hughes TR, Morris QD, et al. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol. Cell. 2004;16:929–941. doi: 10.1016/j.molcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 6.Hashimoto S, Qu W, Ahsan B, Ogoshi K, Sasaki A, Nakatani Y, Lee Y, Ogawa M, Ametani A, Suzuki Y, et al. High-resolution analysis of the 5′-end transcriptome using a next generation DNA sequencer. PLoS ONE. 2009;4:e4108. doi: 10.1371/journal.pone.0004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 8.Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl Acad. Sci. USA. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kincaid MM, Cooper AA. ERADicate ER stress or die trying. Antioxid. Redox. Signal. 2007;9:2373–2387. doi: 10.1089/ars.2007.1817. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y, Chang A. Heat shock response relieves ER stress. Embo J. 2008;27:1049–1059. doi: 10.1038/emboj.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vashist S, Kim W, Belden WJ, Spear ED, Barlowe C, Ng DT. Distinct retrieval and retention mechanisms are required for the quality control of endoplasmic reticulum protein folding. J. Cell Biol. 2001;155:355–368. doi: 10.1083/jcb.200106123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vashist S, Ng DT. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J. Cell Biol. 2004;165:41–52. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Melamud E, Moult J. Structural implication of splicing stochastics. Nucleic Acids Res. 2009;37:4862–4872. doi: 10.1093/nar/gkp444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin P, Fu GK, Wilson AD, Yang J, Chien D, Hawkins PR, Au-Young J, Stuve LL. PCR isolation and cloning of novel splice variant mRNAs from known drug target genes. Genomics. 2004;83:566–571. doi: 10.1016/j.ygeno.2003.09.023. [DOI] [PubMed] [Google Scholar]
- 15.Tanner S, Shen Z, Ng J, Florea L, Guigo R, Briggs SP, Bafna V. Improving gene annotation using peptide mass spectrometry. Genome Res. 2007;17:231–239. doi: 10.1101/gr.5646507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tress ML, Bodenmiller B, Aebersold R, Valencia A. Proteomics studies confirm the presence of alternative protein isoforms on a large scale. Genome Biol. 2008;9:R162. doi: 10.1186/gb-2008-9-11-r162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Power KA, McRedmond JP, de Stefani A, Gallagher WM, Gaora PO. High-throughput proteomics detection of novel splice isoforms in human platelets. PLoS One. 2009;4:e5001. doi: 10.1371/journal.pone.0005001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Romero PR, Zaidi S, Fang YY, Uversky VN, Radivojac P, Oldfield CJ, Cortese MS, Sickmeier M, LeGall T, Obradovic Z, et al. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc. Natl Acad. Sci. USA. 2006;103:8390–8395. doi: 10.1073/pnas.0507916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hegyi H, Buday L, Tompa P. Intrinsic structural disorder confers cellular viability on oncogenic fusion proteins. PLoS Comput. Biol. 2009;5:e1000552. doi: 10.1371/journal.pcbi.1000552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagy A, Hegyi H, Farkas K, Tordai H, Kozma E, Banyai L, Patthy L. Identification and correction of abnormal, incomplete and mispredicted proteins in public databases. BMC Bioinformatics. 2008;9:353. doi: 10.1186/1471-2105-9-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tress ML, Martelli PL, Frankish A, Reeves GA, Wesselink JJ, Yeats C, Olason PI, Albrecht M, Hegyi H, Giorgetti A, et al. The implications of alternative splicing in the ENCODE protein complement. Proc. Natl Acad. Sci. USA. 2007;104:5495–5500. doi: 10.1073/pnas.0700800104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boutet E, Lieberherr D, Tognolli M, Schneider M, Bairoch A. UniProtKB/Swiss-Prot. Methods Mol. Biol. 2007;406:89–112. doi: 10.1007/978-1-59745-535-0_4. [DOI] [PubMed] [Google Scholar]
- 24.Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kriventseva EV, Koch I, Apweiler R, Vingron M, Bork P, Gelfand MS, Sunyaev S. Increase of functional diversity by alternative splicing. Trends Genet. 2003;19:124–128. doi: 10.1016/S0168-9525(03)00023-4. [DOI] [PubMed] [Google Scholar]
- 26.Stetefeld J, Ruegg MA. Structural and functional diversity generated by alternative mRNA splicing. Trends Biochem. Sci. 2005;30:515–521. doi: 10.1016/j.tibs.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 27.Yura K, Shionyu M, Hagino K, Hijikata A, Hirashima Y, Nakahara T, Eguchi T, Shinoda K, Yamaguchi A, Takahashi K, et al. Alternative splicing in human transcriptome: functional and structural influence on proteins. Gene. 2006;380:63–71. doi: 10.1016/j.gene.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 28.Birzele F, Csaba G, Zimmer R. Alternative splicing and protein structure evolution. Nucleic Acids Res. 2008;36:550–558. doi: 10.1093/nar/gkm1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Consortium UniProt. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2008;36:D190–D195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dosztanyi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 31.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fleming PJ, Fitzkee NC, Mezei M, Srinivasan R, Rose GD. A novel method reveals that solvent water favors polyproline II over beta-strand conformation in peptides and unfolded proteins: conditional hydrophobic accessible surface area (CHASA) Protein Sci. 2005;14:111–118. doi: 10.1110/ps.041047005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chothia C. The nature of the accessible and buried surfaces in proteins. J. Mol. Biol. 1976;105:1–12. doi: 10.1016/0022-2836(76)90191-1. [DOI] [PubMed] [Google Scholar]
- 35.Kachel N, Erdmann KS, Kremer W, Wolff P, Gronwald W, Heumann R, Kalbitzer HR. Structure determination and ligand interactions of the PDZ2b domain of PTP-Bas (hPTP1E): splicing-induced modulation of ligand specificity. J. Mol. Biol. 2003;334:143–155. doi: 10.1016/j.jmb.2003.09.026. [DOI] [PubMed] [Google Scholar]
- 36.Fiegen D, Haeusler LC, Blumenstein L, Herbrand U, Dvorsky R, Vetter IR, Ahmadian MR. Alternative splicing of Rac1 generates Rac1b, a self-activating GTPase. J. Biol. Chem. 2004;279:4743–4749. doi: 10.1074/jbc.M310281200. [DOI] [PubMed] [Google Scholar]
- 37.Trinh CH, Asipu A, Bonthron DT, Phillips SE. Structures of alternatively spliced isoforms of human ketohexokinase. Acta. Crystallogr. D Biol. Crystallogr. 2009;65:201–211. doi: 10.1107/S0907444908041115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.(2009) The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 37:D169–D174. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koscielny G, Le Texier V, Gopalakrishnan C, Kumanduri V, Riethoven JJ, Nardone F, Stanley E, Fallsehr C, Hofmann O, Kull M, et al. ASTD: The Alternative Splicing and Transcript Diversity database. Genomics. 2009;93:213–220. doi: 10.1016/j.ygeno.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 41.Shionyu M, Yamaguchi A, Shinoda K, Takahashi K, Go M. AS-ALPS: a database for analyzing the effects of alternative splicing on protein structure, interaction and network in human and mouse. Nucleic Acids Res. 2009;37:D305–D309. doi: 10.1093/nar/gkn869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan Q, Bakowski MA, Morris Q, Zhang W, Frey BJ, Hughes TR, Blencowe BJ. Alternative splicing of conserved exons is frequently species-specific in human and mouse. Trends Genet. 2005;21:73–77. doi: 10.1016/j.tig.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 43.Thanaraj TA, Clark F, Muilu J. Conservation of human alternative splice events in mouse. Nucleic Acids Res. 2003;31:2544–2552. doi: 10.1093/nar/gkg355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saltzman AL, Kim YK, Pan Q, Fagnani MM, Maquat LE, Blencowe BJ. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol. Cell. Biol. 2008;28:4320–4330. doi: 10.1128/MCB.00361-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katzenberger RJ, Marengo MS, Wassarman DA. Control of alternative splicing by signal-dependent degradation of splicing-regulatory proteins. J. Biol. Chem. 2009;284:10737–10746. doi: 10.1074/jbc.M809506200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Melamud E, Moult J. Stochastic noise in splicing machinery. Nucleic Acids Res. 2009;37:4873–4886. doi: 10.1093/nar/gkp471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang P, Yan B, Guo JT, Hicks C, Xu Y. Structural genomics analysis of alternative splicing and application to isoform structure modeling. Proc. Natl Acad. Sci. USA. 2005;102:18920–18925. doi: 10.1073/pnas.0506770102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dyson HJ, Wright PE, Scheraga HA. The role of hydrophobic interactions in initiation and propagation of protein folding. Proc. Natl Acad. Sci. USA. 2006;103:13057–13061. doi: 10.1073/pnas.0605504103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.