Abstract
Template independent polymerases, and terminal deoxynucleotidyl transferase (TdT) in particular, have been widely used in enzymatic labeling of DNA 3′-ends, yielding fluorescently-labeled polymers. The majority of fluorescent nucleotides used as TdT substrates contain tethered fluorophores attached to a natural nucleotide, and can be hindered by undesired fluorescence characteristics such as self-quenching. We previously documented the inherent fluorescence of a set of four benzo-expanded deoxynucleoside analogs (xDNA) that maintain Watson–Crick base pairing and base stacking ability; however, their substrate abilities for standard template-dependent polymerases were hampered by their large size. However, it seemed possible that a template-independent enzyme, due to lowered geometric constraints, might be less restrictive of nucleobase size. Here, we report the synthesis and study of xDNA nucleoside triphosphates, and studies of their substrate abilities with TdT. We find that this polymerase can incorporate each of the four xDNA monomers with kinetic efficiencies that are nearly the same as those of natural nucleotides, as measured by steady-state methods. As many as 30 consecutive monomers could be incorporated. Fluorescence changes over time could be observed in solution during the enzymatic incorporation of expanded adenine (dxATP) and cytosine (dxCTP) analogs, and after incorporation, when attached to a glass solid support. For (dxA)n polymers, monomer emission quenching and long-wavelength excimer emission was observed. For (dxC)n, fluorescence enhancement was observed in the polymer. TdT-mediated synthesis may be a useful approach for creating xDNA labels or tags on DNA, making use of the fluorescence and strong hybridization properties of the xDNA.
INTRODUCTION
Synthetic nucleobase analogs have attracted broad interest for their utility in studying biological processes (1–5), their medicinal value (6–8), and their potential to expand the genetic alphabet (9–12). Among these, fluorescent nucleobase analogs show particular utility as probes of biological structure and activity due to their ease of enzymatic or synthesizer-based incorporation into DNA, and their intimate association with the stacked DNA structure (13–19). Fluorescent nucleobases have been widely employed in DNA microarray gene expression analysis (20), fluorescence in situ hybridization (21), and single-nucleotide polymorphism analysis (22,23).
Fluorescent nucleobases can be close variants of the canonical bases, or can be structurally quite distinct from the natural structures (24–26). Previously we have described a set of size-expanded nucleoside analogs (xDNA) containing an added benzene ring inserted into the nucleobase of all four natural bases (27–29). While maintaining Watson–Crick hydrogen-bonded base pairing, these large-sized nucleobases stack more strongly than their natural counterparts, resulting in double helices that are larger and more thermally stable than natural DNA (30). The expansion of the π-system also results in the four xDNA nucleobases being fluorescent, with high quantum yields and emission in the violet–blue range (31,32). The fluorescence properties of single xDNA monomers in DNA and of short oligomers of xDNA have been studied recently (33). To date, xDNA nucleobases have been incorporated into oligonucleotides only via DNA synthesizer incorporation of phosphoramidite derivatives, and enzymatic incorporation has been largely unexplored. Only one xDNA nucleoside triphosphate has yet been described in a preliminary report (34).
Enzymatic incorporation of fluorescent nucleotides into DNA is useful for the detection of biological processes (35–38) and in DNA sequencing (39–41). In the well-known TUNEL assay, terminal deoxynucleotidyl transferase (TdT) is used to label the 3′-ends of fragmented DNAs in cells undergoing apoptosis with a fluorescently-labeled deoxyuridine triphosphate (FAM-dUTP) (42–45). The enzyme can incorporate dozens of successive nucleotide monomers into a homopolymer, resulting in distinct labeling. As TdT is a template-independent polymerase (46), the resulting labeled oligomer is a single strand of DNA.
Because of its large size, xDNA is a challenging substrate for DNA polymerases. Previous studies on the enzymatic incorporation of natural nucleoside triphosphates opposite an xDNA template base with the Klenow fragment of DNA Pol I have shown that, although pairing selectively with their corresponding xDNA base, the pair synthesis and extension is hindered due to size constraints of the resulting base pair (1,47). A repair enzyme with greater steric flexibility was able to synthesize four consecutive bases of DNA on an xDNA template, but still with lowered efficiency relative to natural DNA pairs (48). However, other classes of polymerases should also be considered, since they display widely varied and specialized functions. TdT in particular is of interest because it does not require a template strand to polymerize DNA; thus we hypothesized that it might be able to incorporate xDNA nucleotides successively, as there would be no large-sized double-stranded structure to consider. The enzymatic synthesis of xDNA homopolymers could potentially be of interest in multiple respects. For example, the homopolymers could be used as affinity tags, akin to polyA tails (49) but with higher affinity from the stronger helix-forming tendencies of xDNA. Second, strings of xDNA bases can display unusual fluorescence characteristics (33), such as excimers in poly(dxA), which are otherwise not available in natural DNA bases or in fluorescein-labeled thymidine (the common TdT substrate). Third, they could potentially be used as improved reporters in assays that employ TdT.
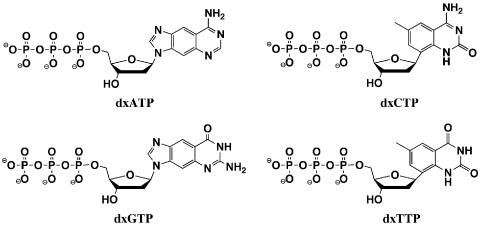
Here we report the synthesis of xDNA nucleoside triphosphates (Figure 1) and their substrate properties with the TdT enzyme. We find that all four are efficient substrates for the enzyme using a natural DNA oligodeoxynucleotide as primer. Experiments show that, in addition to having incorporation efficiencies comparable to those of natural DNA triphosphates, as many as thirty consecutive monomers could be incorporated. Finally, we find that oligonucleotide strands containing multiple dxA and dxC monomers undergo distinct fluorescence changes from their respective triphosphate monomers in aqueous solutions, and when attached to DNA primers on glass support beads. The results suggest the use of these inherently fluorescent nucleotides in applications that involve end-labeling of nucleic acids.
Figure 1.
Expanded DNA nucleoside triphosphates used in this study.
EXPERIMENTAL PROCEDURE
Preparation of xDNA nucleoside triphosphate derivatives
Syntheses of nucleosides dxA, dxC, dxG and dxT were carried out according to previous methods (28,29,31). Details are given in the Supplementary Data. In brief, the free nucleoside compounds were treated with phosphorus oxychloride at 0°C in trimethylphosphate, followed by addition of tributylammonium pyrophosphate and tributylamine. Although the synthesis of the dxA triphosphate has been reported earlier (34), our synthesis followed a different one-pot procedure described by Ludwig (50), which requires no protection of the exocyclic amine or 3′-hydroxyl.
Purification of nucleotides dxATP, dxCTP, dxGTP and dxTTP required a two-step procedure involving first anion exchange fast protein liquid chromatography using a diethylaminoethyl cellulose column followed by reverse-phase HPLC (Supplementary Data). The purified triphosphate derivatives were subsequently converted to the sodium salt, and were characterized by mass spectrometry and phosphorus NMR (Supplementary Data).
TdT-mediated incorporation of multiple nucleoside triphosphates
A DNA primer (5′-ATA CCA AAG T-3′) was 5′-end-labeled with [γ-32P] ATP, purified and desalted using a microspin column. A primer solution consisting of unlabeled primer, labeled primer and buffer consisting of potassium acetate (200 mM), Tris–acetate (800 mM, pH 7.9), magnesium acetate (40 mM), CoCl2 (1 mM) and dithiothreitol (4 mM) were allowed to incubate with an enzyme solution containing TdT (4 U/ µl) for 3 min at 37°C [1 U of TdT is defined by the supplier as the amount of enzyme that will incorporate 1 nmol dATP in 1 h at 37°C using d(A)18 as primer]. After 3 min, dNTP was added to make a final [dNTP] of 10 µM. The reaction was allowed to proceed for 1 h, then was quenched with loading buffer (95% formamide, 20 mM EDTA, 0.05% xylene cyanol and bromophenol blue). The lengths of the reaction products were determined by running quenched reaction samples on 20% denaturing polyacrylamide gels, exposing radiolabeled images to a phosphor screen, and analyzed by phosphorimaging.
Steady-state kinetics of TdT single-nucleotide insertion into DNA primers
The steady-state kinetics of single-nucleotide insertion reactions were followed and evaluated according to previous methods (51). The above procedure was used with the exception of a reduced incubation time from 1 h to 1 min to suppress multiple incorporations and ensure <20% primer extension to meet the requirement for the initial rates method. To initiate reaction, an equal volume of dNTP solution, containing a variable dNTP concentration, was added to the primer/enzyme solution. Further details can be found in the Supplementary Data (p. 6).
The extents of reaction were determined by running quenched reaction samples on 20% denaturing polyacrylamide gels, exposing radiolabeled images to a phosphor screen, and analyzed by phosphorimaging. The relative velocities of dNTP incorporation were calculated as v = ([S] I1/(I0+I1))/t, where Ip = product band intensity (n + 1), I0 = n band intensity (starting primer), [S] = concentration of substrate, and t = reaction time (min). Vmax and Km values were extracted from Hanes–Woolf plots of [S]/v versus [S]. [S] = [dNTP] and were regarded as constant during the course of each reaction. Error estimates were obtained from standard deviations of reactions run in at least triplicate.
Fluorescence methods for study of dxATP and dxCTP incorporation by TdT
TdT enzyme (1 U/µl, cloned from calf thymus, New England Biolabs) was added to a solution containing primer (5′-ATA CCA AAG T-3′) and TdT buffer consisting of potassium acetate (200 mM), Tris–acetate (800 mM, pH 7.9), magnesium acetate (40 mM) and CoCl2 (1 mM). The mixture was allowed to incubate for 3 min at 37°C. A solution of either dxATP or dxCTP was then added and the reaction was allowed to proceed for 1 h at 37°C. The reaction was then quenched and heated to denature the enzyme. Fluorescence measurements were made on the oligonucleotides after purification using size-exclusion chromatography (NAPTM-10 containing Sephadex G-10, GE Healthcare). Further details can be found in the Supplementary Data (p. 8).
For fluorescence time course studies, fluorescence was measured directly from the crude solution by taking 10 µl aliquots from the reaction mixture, and then mixing in 1 ml fluorescence cuvettes containing PIPES buffer solution (100 mM NaCl, 10 mM MgCl2 and 10 mM Na•PIPES, pH 7.0). The fluorescence spectra of the resulting oligonucleotides were measured at varied time points with a Spex-Fluorolog-3 series fluorimeter. Excitation wavelengths were 330 nm for dxC fluorescence analysis and 333 nm for dxA. Fluorescence curves were normalized through the fluorescence measurement of a blank solution containing PIPES buffer solution and subtraction of this data from the resulting solutions containing oligonucleotide strands.
Controlled pore glass bead-primer fluorescence studies
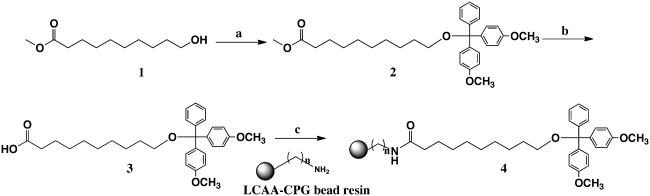
Synthesis of a DMT-protected non-cleavable linker compound 4 (Scheme 1) was followed as reported earlier (52), with the exception that controlled pore glass (CPG) beads were used instead of polystrene beads for the solid support and a different peptide coupling agent (HATU) was used for step c.
Scheme 1.
Synthesis of CPG solid support with primer attached. (a) dimethoxytrityl chloride, N, N-diisopropylethylamine, 4-dimethylaminopyridine, dichloromethane, 2 h, room temparature, 92% (b) LiOH-H2O, THF/MeOH/H2O (3:1:1), 24 h, room temparature, 59% (c) HATU (peptide coupling agent), 4-dimethylaminopyridine, N, N-diisopropylethylamine, diemethylformamide, room temparature, 75%.
Conjugation of a dT25 primer to CPG (4) was accomplished by reverse 3′–5′ automated DNA synthesis (1 µmol scale) on an ABI 392 DNA synthesizer. dT-5′-CE phosphoramidite (Glen Research) was used under standard β-cyanoethyl phosphoramidite chemistry coupling conditions (overall coupling yield, 62%). The CPG-oligomers were deprotected in concentrated ammonium hydroxide (55°C, 16 h), then filtered and washed with water, dichloromethane and ethyl acetate, and dried under vacuum overnight.
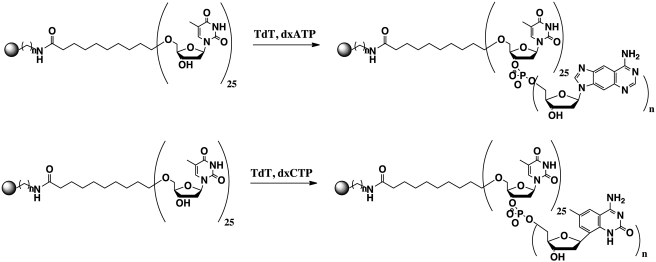
A suspension of CPG beads containing primers (∼5 µM) was mixed with the TdT enzyme (1 U/µl) in TdT buffer [potassium acetate (200 mM), Tris–acetate (800 mM, pH 7.9), magnesium acetate (40 mM) and CoCl2 (1 mM)]. Following incubation of the enzyme/primer mixture, dxCTP or dxATP solutions (final [dxCTP] = 140 µM, final [dxATP] = 68 µM) were added and the reaction was allowed to proceed for 1 h at 37°C (Scheme 2). The total reaction volume was 20 µl. The resulting CPG beads were filtered, washed with deionized water and dried under vacuum, then imaged using an epifluorescence microscope (Nikon Eclipse E800 equipped with ×4 and ×20 objective). An excitation filter (330–380 nm) was used for beads incubated with dxATP and dxCTP. The emission filter for beads incubated with dxATP was a >420 nm long pass, while the emission filter for beads incubated with dxCTP was >400 nm.
Scheme 2.
Addition of dxATP and dxCTP to (dT)25 primer attached to CPG bead.
RESULTS
Incorporation of multiple xDNA and natural nucleoside triphosphates with TdT
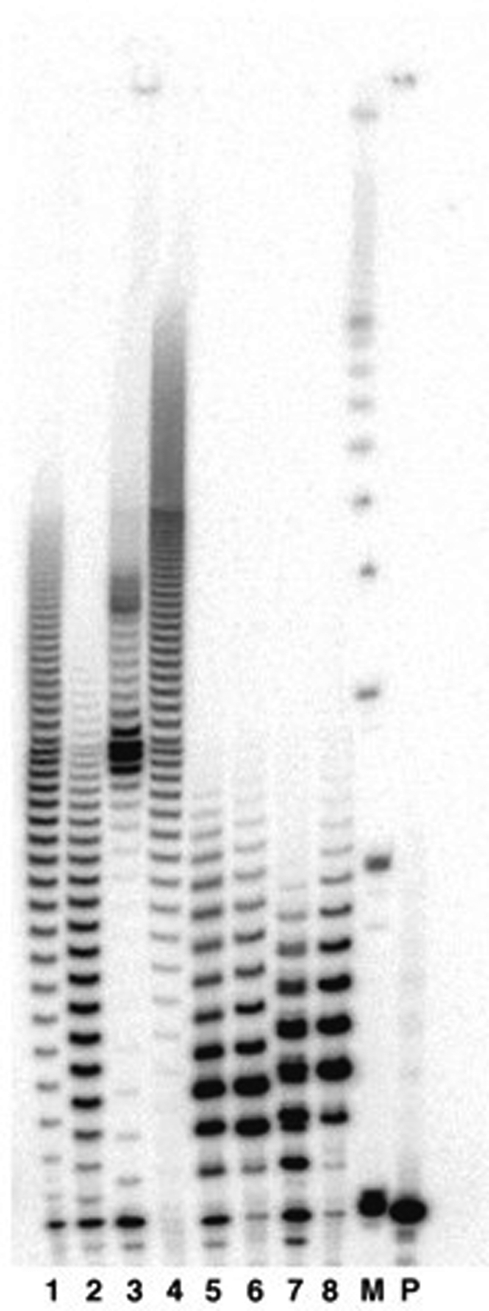
Analysis after TdT labeling reactions using PAGE gels showed extensive incorporation of each of the four natural nucleoside triphosphates (Figure 2, lanes 1–4) and incorporation of 1–20 xDNA nucleoside triphosphates (lanes 5–8) under the same conditions. Among the natural dNTPs, stalling of deoxyguanosine incorporation (lane 2) was observed after ∼30 nt, possibly due to the formation of G-quadruplexes. The longest synthesized polymers were seen for dATP (lane 4, up to nearly 100 nt) consistent with previous studies showing the preference of TdT for this base (53). The xDNA oligomers fell into a narrower range of lengths and showed little differences in length among the four. Under the conditions of the experiment, the most populated length was a trimer, but lengths of at least 15 nt were present in all cases. It should be noted that polymer lengths varied with enzyme activity, concentration and source as well as dNTP concentration; an example is given in the Supplementary Figure S1 showing lengths for xDNA polymers of 30 nt and greater [except (dxT)n, which seemed to be limited to shorter lengths].
Figure 2.
Autoradiogram of a polyacrylamide gel showing TdT insertion of xDNA and DNA nucleoside triphosphates after 1 h. Lane 1: incorporation of dTTP, 10 µM; lane 2: dGTP, 10 µM; lane 3: dCTP, 10 µM; lane 4: dATP, 10 µM; lane 5: dxTTP, 10 µM; lane 6: dxGTP, 10 µM; lane 7: dxCTP, 10 µM; lane 8: dxATP, 10 µM. M: 10 bp size marker; P: unreacted DNA primer (sequence 5′-dATACCAAAGT-3′).
Steady-state kinetics of TdT insertion of single nucleoside triphosphates on a DNA primer
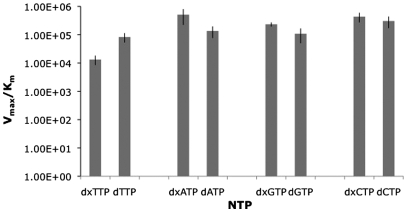
To evaluate the inherent efficiency of enzymatic incorporation of the initial xDNA nucleotides, we carried out steady-state kinetics studies of primer extension, measuring insertion of the first nucleotide. The activities with natural DNA nucleotides were also measured for comparison. Data for efficiencies (as Vmax/Km) are plotted in Figure 3; numerical values are given in the Supplementary Table S1. Interestingly, the single-nucleotide incorporation efficiencies of the expanded nucleoside triphosphates were comparable to those of their natural congeners. The expanded deoxythymidine (dxTTP) had the lowest efficiency of all nucleotides, and was ∼6-fold lower than the natural deoxythymidine (dTTP). However, the expanded deoxyadenosine (dxATP) had the highest incorporation efficiency, which was higher than that of natural deoxyadenosine by a factor of 4. The expanded deoxyguanosine and deoxycytosine (dxGTP and dxCTP) analogs had incorporation efficiencies nearly the same as, or slightly higher than, their natural counterparts.
Figure 3.
Steady-state incorporation efficiencies (as Vmax/Km) for the single nucleotide incorporation of expanded and natural nucleoside triphosphates into a primer by TdT. Data were obtained by gel electrophoretic analysis using radiolabeled primers.
Solution fluorescence studies of dxATP and dxCTP incorporation
Previous studies with synthetic xDNA oligomers have revealed that two of the monomers, dxC and dxA, give distinct changes in their emission spectra in oligomeric form as compared to the monomers (33). To investigate whether this could be observed in TdT reactions with these monomers, we measured the spectra of purified products, and of reactions over varied time points.
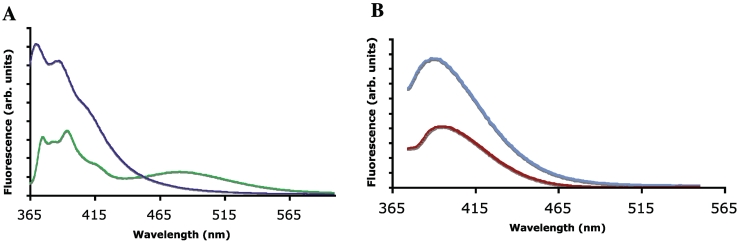
After incubation of an oligodeoxynucleotide primer with dxATP and TdT, size-exclusion chromatography was used to remove excess dNTP. Spectra of the remaining oligonucleotide-enriched fraction showed a distinct change in the spectrum relative to the monomer dxATP (Figure 4A). While the monomer showed an emission maximum at ∼382 nm, the purified product strand showed an emission spectrum with peaks typical of the monomer dxATP as well as a distinct long-wavelength peak at ∼488 nm, characteristic of excimer emission by adjacent dxA bases (33).
Figure 4.
Spectra of xDNTP monomers and of TdT reaction products partially purified by size exclusion. (A) Normalized emission spectra of TdT reaction products with dxATP (green) and monomer dxATP (purple) in buffer (excitation 333 nm). (B) Normalized emission spectrum of the TdT reaction product with dxCTP (blue) and monomer dxCTP (red) in PIPES buffer (excitation 330 nm). Spectra were measured in PIPES buffer (100 mM NaCl, 10 mM MgCl2 and 10 mM Na•PIPES, pH 7.0).
The product of the TdT reaction with dxCTP using the same DNA primer was also purified by size exclusion. While both the monomer and TdT reaction product had an emission maximum at ∼395 nm (Figure 4B), the TdT reaction product showed an ∼2-fold increase in fluorescence, consistent with a previous observation of increased fluorescence in dxC oligomers relative to the monomer components (33).
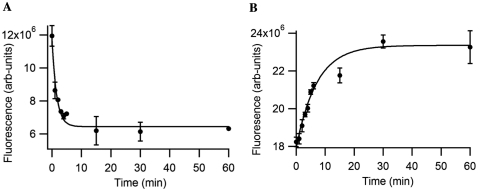
We then monitored the reactions over time by removing aliquots and measuring emission changes during the course of the polymerizations with a 10-mer DNA primer. TdT reactions containing dxATP showed a decrease in fluorescence at the emission maximum of 385 nm over 30 min, as expected with a decrease in dxATP monomer over time (Figure 5A). This is consistent with a previous observation of lowered quantum yield of dxA in synthetic homo-oligomers (33). In contrast, time course studies of the fluorescence emission of the TdT reactions containing dxCTP showed an increase in fluorescence at the emission maximum of 395 nm, again consistent with previous observations in synthesized xDNAs (Figure 5B). Thus the results confirm that TdT-mediated reaction products yield emission changes that can be monitored as they proceed.
Figure 5.
Changes in fluorescence emission during TdT-mediated polymerization of xDNA. (A) dxATP incorporation measured at 385 nm. Reaction aliquots measured in PIPES buffer (100 mM NaCl, 10 mM MgCl2 and 10 mM Na•PIPES, pH 7.0, excitation 333 nm). (B) dxCTP incorporation measured at 395 nm (excitation 330 nm). Curves are exponential fits to the data.
Fluorescence imaging of dxATP and dxCTP incorporation on solid support
TdT-mediated DNA polymerization reactions have been shown to be useful in microscopic imaging on surfaces; for example, in observing DNA fragmentation in apoptosis in fixed cells (44,54) and high-throughput sequencing in microplate wells and microarrays (55). To test whether products of xDNA polymerization could be imaged in a similar way, we carried out reactions on 25-mer DNA primers conjugated to controlled pore glass (CPG), and captured images after incubation with TdT and xdNTPs. For these studies we again chose dxATP and dxCTP, since these were shown previously to give the largest changes in fluorescence in oligomeric form.(33).
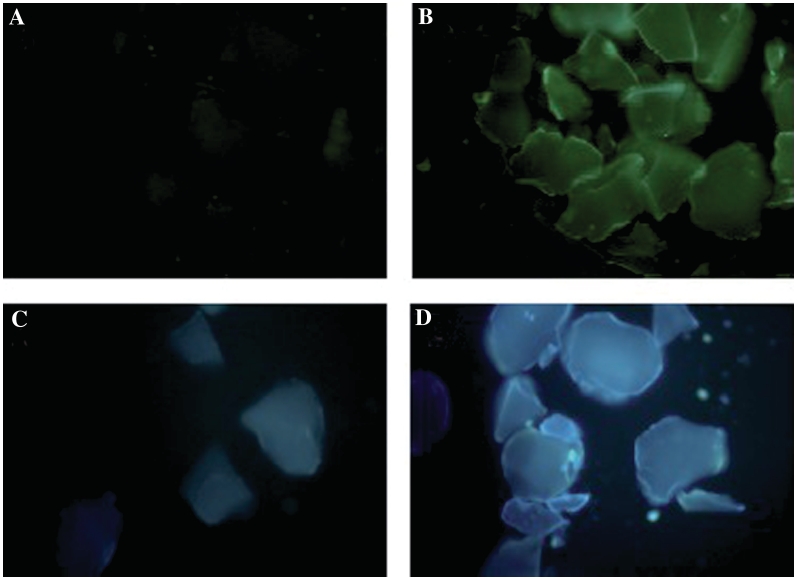
Images of CPG beads after TdT reactions with these unnatural monomers under an epifluorescence microscope revealed visible fluorescence changes for both dxATP and dxCTP incorporation (Figure 6). Blue/green fluorescence (>420 nm) was observed after TdT was added to a reaction mixture containing the primer-substituted CPG-beads and dxATP (Figure 6A and B). The same procedure was used to image glass substrates following reaction of TdT with the CPG-attached primer and dxCTP. Images measured at >400 nm showed a blue fluorescence enhancement of the beads after addition of TdT to a mixture containing the CPG beads and dxCTP (Figure 6C and D).
Figure 6.
Fluorescence microscopy images of TdT-mediated xDNA reaction products on CPG with (dT)25 primer attached. (A) [dxATP] = 68 µM, TdT omitted. (B) [dxATP] = 68 µM, [TdT] = 1 U/µl. (C) [dxCTP] = 140 µM, TdT omitted. (D) [dxCTP] = 140 µM, [TdT] = 1 U/µl. Excitation 330–380 nm for all images; emission was measured with a 420 nm long-pass filter for (A), (B) and 400 nm long-pass filter for (C and D).
DISCUSSION
Our experiments confirm that the template-independent polymerase TdT can successfully synthesize oligomers of size-expanded DNA (xDNA) in lengths ranging from 3 to 30 nt. The initial enzymatic efficiencies are similar to those of natural dNTPs with this enzyme. The data further show that two of the monomers (dxC and dxA) yield changes in fluorescence in the reaction products, which can be observed in the purified product oligomers and imaged on solid supports. Moreover, the reactions produce changes in emission intensity in solution that can be followed over time, allowing reaction progress to be directly monitored spectroscopically.
The majority of previous studies of TdT incorporating nucleotides use either natural nucleotides, or tethered fluorophores attached to natural nucleotides, such as fluorescein-dUTP, used in the TUNEL assay (42–45). Only a few studies have used TdT to incorporate unnatural nucleotides not tethered to fluorophores. In two studies, the non-natural nucleotides incorporated were indole derivatives in which fluorescence was not described (56,57), and in the other study, we documented the incorporation of 3–4 fluorescent pyrene nucleotide analogs (58) which stalled strongly at that point. The current studies are distinct from these precedents not only because of the changes in fluorescence of the incorporated dxA and dxC analogs, but also because of the strong abilities of xDNA to hybridize to natural DNA, which has previously been documented (59). The aforementioned pyrene and indole oligomers have no ability to hybridize.
While our data show that all four xDNA nucleotides were incorporated at the ends of a primer multiple times by TdT, the number of nucleotides incorporated was significantly less than that of the natural deoxynucleoside triphosphates (Figure 2). This suggests that while the initial incorporation is efficient, later nucleotides are added less efficiently, implying that some property of oligomeric xDNAs beyond short sequences acts to inhibit further extension to some degree. Two properties of single-stranded xDNA that are distinct from DNA are stronger stacking and increased hydrophobicity (1). It is possible that such properties might lead to unproductive conformations or associations with the enzyme. Previous experiments with other unnatural nucleotides with this enzyme have also shown truncated products to be common (56,58).
The fluorescence characteristics of TdT-synthesized products observed here are consistent with those seen earlier with synthesized xDNAs. In a previous study of synthetic xDNA oligomers, long-wavelength excimer emission (490–590 nm) from adjacent dxA residues appeared when more than two dxA molecules were attached to a natural oligonucleotide, and the intensity of the band increased proportional to the number of dxA nucleotides on the strand (33). Here, we observed an apparent excimer band at 465–515 nm on a purified olignonucleotide strand containing multiple dxA nucleotides incorporated with TdT (Figure 4A). Fluorescence studies of dxCTP incorporation displayed over an order of magnitude increase in fluorescence following purification of the strand (Figure 4B) and a significant increase observed over time as the monomer was consumed and the fluorescence-enhanced polymer was synthesized.
The current findings suggest possible uses of TdT-mediated xDNA synthesis. Enzymatic incorporation of fluorescent xDNA nucleotides offers some possible advantages over the incorporation of nucleotides containing tethered fluorophores. The incorporation of directly stacked chromophores may be able to more accurately and intimately report on DNA structure and environment since xDNA can hybridize as part of the helix, and thus structural perturbation is minimized upon hybridization. In addition, as stated previously, we have shown that an oligonucleotide strand comprised of all xDNA bases can bind a natural complement strand with much higher affinity than DNA (59). Thus it is possible that homopolymer xDNA strands synthesized by TdT could be used as affinity tags for separation of DNAs to which they are attached. Beyond this, there may be advantages in the fluorescence as well. For example, fluorescein-dU is known to self-quench strongly in homooligomers (33), while dxC oligomers have enhanced emission. Moreover, fluorescein-dU oligomers do not change their emission wavelength, while dxA oligomers yield a strong red-shifted excimer emission.
A practical advantage of using TdT for the incorporation of xDNA nucleotides is that it obviates the use of a DNA synthesizer for creating homopolymers, in applications where mixtures of lengths are acceptable. Further studies will be useful in applying the unusual fluorescence and hybridization properties of these expanded analog oligonucleotide strands in biological applications. It will also be of interest to study the photophysical origins of the fluorescence changes that occur in greater detail. Finally, exploration of other enzymes that may be able to efficiently incorporate expanded nucleotide analogs will also be worthy of study.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: National Institutes of Health grant (GM063587).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We acknowledge the US National Institutes of Health (GM063587, GM067201) for support, and we thank the National Science Foundation (CHE-0639053) for support of fluorescence instrumentation in the Department of Chemistry.
REFERENCES
- 1.Krueger AT, Lu HG, Lee AHF, Kool ET. Synthesis and properties of size-expanded DNAs: toward designed, functional genetic systems. Acc. Chem. Res. 2007;40:141–150. doi: 10.1021/ar068200o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peng XH, Hong IS, Li H, Seiclman MM, Greenberg MM. Interstrand cross-link formation in duplex and triplex DNA by modified pyrimidines. J. Am. Chem. Soc. 2008;130:10299–10306. doi: 10.1021/ja802177u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu J, Li NS, Sengupta RN, Piccirilli JA. Synthesis and biochemical application of 2′-O-methyl-3′-thioguanosine as a probe to explore group I intron catalysis. Biorg. Med. Chem. 2008;16:5754–5760. doi: 10.1016/j.bmc.2008.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hendrickson CL, Devine KG, Benner SA. Probing minor groove recognition contacts by DNA polymerases and reverse transcriptases using 3-deaza-2′-deoxyadenosine. Nucleic Acids Res. 2004;32:2241–2250. doi: 10.1093/nar/gkh542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vineyard D, Zhang X, Donnelly A, Lee I, Berdis AJ. Optimization of non-natural nucleotides for selective incorporation opposite damaged DNA. Org. Biomol. Chem. 2007;5:3623–3630. doi: 10.1039/b712480e. [DOI] [PubMed] [Google Scholar]
- 6.Kantarjian H, Garcia-Manero G, O'Brien S, Faderl S, Ravandi F, Westwood R, Green SR, Chiao JH, Boone PA, Cortes J, et al. Phase I clinical and pharmacokinetic study of oral sapacitabine in patients with acute leukemia and myelodysplastic syndrome. J. Clin. Oncol. 28:285–291. doi: 10.1200/JCO.2009.25.0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biron KK, Noblin JE, Demiranda P, Elion GB. Uptake, distribution, and anabolism of acyclovir in herpes-simplex virus-infected mice. Antimicrob. Agents Chemother. 1982;21:44–50. doi: 10.1128/aac.21.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sehgal PB, Derman E, Molloy GR, Tamm I, Darnell JE. 5,6-Dichloro-1-Beta-D-ribofuranosylbenzimidazole inhibits initiation of nuclear heterogeneous Rna chains in hela-cells. Science. 1976;194:431–433. doi: 10.1126/science.982026. [DOI] [PubMed] [Google Scholar]
- 9.Yang ZY, Chen F, Chamberlin SG, Benner SA. Expanded genetic alphabets in the polymerase chain reaction. Angew. Chem. Inter. Ed. 2010;49:177–180. doi: 10.1002/anie.200905173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kimoto M, Mitsui T, Yokoyama S, Hirao I. A unique fluorescent base analogue for the expansion of the genetic alphabet. J. Am. Chem. Soc. 2010;132:4988–4989. doi: 10.1021/ja100806c. [DOI] [PubMed] [Google Scholar]
- 11.Leconte AM, Hwang GT, Matsuda S, Capek P, Hari Y, Romesberg FE. Discovery, characterization, and optimization of an unnatural base pair for expansion of the genetic alphabet. J. Am. Chem. Soc. 2008;130:2336–2343. doi: 10.1021/ja078223d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirao I, Ohtsuki T, Fujiwara T, Mitsui T, Yokogawa T, Okuni T, Nakayama H, Takio K, Yabuki T, Kigawa T, et al. An unnatural base pair for incorporating amino acid analogs into proteins. Nat. Biotechnol. 2002;20:177–182. doi: 10.1038/nbt0202-177. [DOI] [PubMed] [Google Scholar]
- 13.Allan BW, Reich NO. Targeted base stacking disruption by the EcoRI DNA methyltransferase. Biochemistry. 1996;35:14757–14762. doi: 10.1021/bi9615708. [DOI] [PubMed] [Google Scholar]
- 14.Seela F, Feiling E, Gross J, Hillenkamp F, Ramzaeva N, Rosemeyer H, Zulauf M. Fluorescent DNA: the development of 7-deazapurine nucleoside triphosphates applicable for sequencing at the single molecule level. J. Biotechnol. 2001;86:269–279. doi: 10.1016/s0168-1656(00)00418-1. [DOI] [PubMed] [Google Scholar]
- 15.Okamoto A, Tainaka K, Nishiza K, Saito I. Monitoring DNA structures by dual fluorescence of pyrene derivatives. J. Am. Chem. Soc. 2005;127:13128–13129. doi: 10.1021/ja053609e. [DOI] [PubMed] [Google Scholar]
- 16.Hawkins ME. Synthesis, purification and sample experiment for fluorescent pteridine-containing DNA: tools for studying DNA interactive systems. Nature Protocols. 2007;2:1013–1021. doi: 10.1038/nprot.2007.150. [DOI] [PubMed] [Google Scholar]
- 17.Ikeda S, Kubota T, Kino K, Okamoto A. Sequence dependence of fluorescence emission and quenching of doubly thiazole orange labeled DNA: effective design of a hybridization-sensitive probe. Bioconj. Chem. 2008;19:1719–1725. doi: 10.1021/bc800201m. [DOI] [PubMed] [Google Scholar]
- 18.Jeong HS, Kang S, Lee JY, Kim BH. Probing specific RNA bulge conformations by modified fluorescent nucleosides. Org. Biomol. Chem. 2009;7:921–925. doi: 10.1039/b816768k. [DOI] [PubMed] [Google Scholar]
- 19.Hirose W, Sato K, Matsuda A. Synthesis and properties of new fluorescent nucleosides and oligodeoxynucleotides derived from 5-formyl-2′-deoxyuridine. Nucleic Acids Symp. Ser. 2009;53:135–136. doi: 10.1093/nass/nrp068. [DOI] [PubMed] [Google Scholar]
- 20.Schena M, Shalon D, Heller R, Chai A, Brown PO, Davis RW. Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc. Natl Acad. Sci. USA. 1996;93:10614–10619. doi: 10.1073/pnas.93.20.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lichter P, Tang CJC, Call K, Hermanson G, Evans GA, Housman D, Ward DC. High-resolution mapping of human chromosome-11 by insitu hybridization with cosmid clones. Science. 1990;247:64–69. doi: 10.1126/science.2294592. [DOI] [PubMed] [Google Scholar]
- 22.Chen XN, Kwok PY. Template-directed dye-terminator incorporation (TDI) assay: a homogeneous DNA diagnostic method based on fluorescence resonance energy transfer. Nucleic Acids Res. 1997;25:347–353. doi: 10.1093/nar/25.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okamoto A, Tanaka K, Fukuta T, Saito I. Design of base-discriminating fluorescent nucleoside and its application to T/C SNP typing. J. Am. Chem. Soc. 2003;125:9296–9297. doi: 10.1021/ja035408l. [DOI] [PubMed] [Google Scholar]
- 24.Shafiee M, Gosselin G, Imbach JL, Eriksson S, Maury G. Synthesis of new fluorescent nucleoside analogues and application to the study of human deoxycytidine kinase. Nucleosides Nucleotides. 1999;18:717–719. doi: 10.1080/15257779908041552. [DOI] [PubMed] [Google Scholar]
- 25.Zozulya V, Blagoi Y, Dubey I, Fedoryak D, Makitruk V, Ryazanova O, Shcherbakova A. Anchorage of oligonucleotide hybridization by tethered phenazine nucleoside analogue. Biopolymers. 2003;72:264–273. doi: 10.1002/bip.10403. [DOI] [PubMed] [Google Scholar]
- 26.Li YF, Soni PB, Liu LF, Zhang X, Liotta DC, Lutz S. Synthesis of fluorescent nucleoside analogs as probes for 2′-deoxyribonucleoside kinases. Bioorg. Med. Chem. Lett. 2010;20:841–843. doi: 10.1016/j.bmcl.2009.12.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu HB, Gao JM, Lynch SR, Saito YD, Maynard L, Kool ET. A four-base paired genetic helix with expanded size. Science. 2003;302:868–871. doi: 10.1126/science.1088334. [DOI] [PubMed] [Google Scholar]
- 28.Liu HB, Gao JM, Maynard L, Saito YD, Kool ET. Toward a new genetic system with expanded dimensions: size-expanded analogues of deoxyadenosine and thymidine. J. Am. Chem. Soc. 2004;126:1102–1109. doi: 10.1021/ja038384r. [DOI] [PubMed] [Google Scholar]
- 29.Liu HB, Gao JM, Kool ET. Size-expanded analogues of dG and dC: synthesis and pairing properties in DNA. J. Org. Chem. 2005;70:639–647. doi: 10.1021/jo048357z. [DOI] [PubMed] [Google Scholar]
- 30.Liu HB, Gao JM, Kool ET. Helix-forming properties of size-expanded DNA, an alternative four-base genetic form. J. Am. Chem. Soc. 2005;127:1396–1402. doi: 10.1021/ja046305l. [DOI] [PubMed] [Google Scholar]
- 31.Leonard NJ, Sprecker MA, Morrice AG. Defined dimensional changes in enzyme substrates and cofactors - synthesis of lin-benzoadenosine and enzymatic evaluation of derivatives of benzopurines. J. Am. Chem. Soc. 1976;98:3987–3994. doi: 10.1021/ja00429a040. [DOI] [PubMed] [Google Scholar]
- 32.Gao JM, Liu HB, Kool ET. Expanded-size bases in naturally sized DNA: evaluation of steric effects in Watson-Crick pairing. J. Am. Chem. Soc. 2004;126:11826–11831. doi: 10.1021/ja048499a. [DOI] [PubMed] [Google Scholar]
- 33.Krueger AT, Kool ET. Fluorescence of size-expanded DNA bases: reporting on DNA sequence and structure with an unnatural genetic set. J. Am. Chem. Soc. 2008;130:3989–3999. doi: 10.1021/ja0782347. [DOI] [PubMed] [Google Scholar]
- 34.Leonard NJ, Scopes DIC, Vanderlijn P, Barrio JR. Dimensional probes of enzyme binding-sites of adenine-nucleotides - biological effects of widening adenine ring by 2.4 A. Biochemistry. 1978;17:3677–3685. doi: 10.1021/bi00611a001. [DOI] [PubMed] [Google Scholar]
- 35.Sandin P, Stengel G, Ljungdahl T, Borjesson K, Macao B, Wilhelmsson LM. Highly efficient incorporation of the fluorescent nucleotide analogs tC and tC(O) by Klenow fragment. Nucleic Acids Res. 2009;37:3924–3933. doi: 10.1093/nar/gkp266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Srivatsan SG, Tor Y. Synthesis and enzymatic incorporation of a fluorescent pyrimidine ribonucleotide. Nat. Protoc. 2007;2:1547–1555. doi: 10.1038/nprot.2007.222. [DOI] [PubMed] [Google Scholar]
- 37.Cox WG, Singer VL. Fluorescent DNA hybridization probe preparation using amine modification and reactive dye coupling. BioTechniques. 2004;36:114–122. doi: 10.2144/04361RR02. [DOI] [PubMed] [Google Scholar]
- 38.Mandal SS, da Silva EF, Reha-Krantz LJ. Using 2-aminopurine fluorescence to detect base unstacking in the template strand during nucleotide incorporation by the bacteriophage T4 DNA polymerase. Biochemistry. 2002;41:4399–4406. doi: 10.1021/bi015723p. [DOI] [PubMed] [Google Scholar]
- 39.Smith LM, Sanders JZ, Kaiser RJ, Hughes P, Dodd C, Connell CR, Heiner C, Kent SBH, Hood LE. Fluorescence detection in automated DNA-sequence analysis. Nature. 1986;321:674–679. doi: 10.1038/321674a0. [DOI] [PubMed] [Google Scholar]
- 40.Prober JM, Trainor GL, Dam RJ, Hobbs FW, Robertson CW, Zagursky RJ, Cocuzza AJ, Jensen MA, Baumeister K. A system for rapid DNA sequencing with fluorescent chain-terminating dideoxynucleotides. Science. 1987;238:336–341. doi: 10.1126/science.2443975. [DOI] [PubMed] [Google Scholar]
- 41.Tang C, Shi XL, Li XJ, Lu ZH. DNA sequencing by synthesis with degenerate primers. J. Genet. Genomics. 2008;35:545–551. doi: 10.1016/S1673-8527(08)60074-0. [DOI] [PubMed] [Google Scholar]
- 42.Shchepotin IB, McRae DA, Shabahang M, Buras RR, Evans SRT. Hyperthermia and verapamil inhibit the growth of human colon cancer xenografts in vivo through apoptosis. Anticancer Res. 1997;17:2213–2216. [PubMed] [Google Scholar]
- 43.Ramsey-Ewing A, Moss B. Apoptosis induced by a postbinding step of vaccinia virus entry into Chinese hamster ovary cells. Virology. 1998;242:138–149. doi: 10.1006/viro.1997.8985. [DOI] [PubMed] [Google Scholar]
- 44.Steensma DP, Timm M, Witzig TE. Flow cytometric methods for detection and quantification of apoptosis. Methods Mol. Med. 2003;85:323–332. doi: 10.1385/1-59259-380-1:323. [DOI] [PubMed] [Google Scholar]
- 45.Chinetti G, Griglio S, Antonucci M, Torra IP, Delerive P, Majd Z, Fruchart JC, Chapman J, Najib J, Staels B. Activation of proliferator-activated receptors alpha and gamma induces apoptosis of human monocyte-derived macrophages. J. Biol. Chem. 1998;273:25573–25580. doi: 10.1074/jbc.273.40.25573. [DOI] [PubMed] [Google Scholar]
- 46.Kossel H, Roychoud R. Synthetic polynucleotides - terminal addition of riboadenylic acid to deoxyoligonucleotides by terminal deoxynucleotidyl transferase as a tool for specific labelling of deoxyoligonucleotides at 3′-ends. Eur. J. Biochem. 1971;22:271–276. doi: 10.1111/j.1432-1033.1971.tb01541.x. [DOI] [PubMed] [Google Scholar]
- 47.Krueger AT, Lu H, Hojland T, Liu H, Gao J, Kool ET. Towards the replication of xDNA, a size-expanded unnatural genetic system. Nucleic Acids Symp. Ser. 2008;52:455–456. doi: 10.1093/nass/nrn231. [DOI] [PubMed] [Google Scholar]
- 48.Delaney JC, Gao JM, Liu HB, Shrivastav N, Essigmann JM, Kool ET. Efficient replication bypass of size-expanded DNA base pairs in bacterial cells. Angew. Chem. Int. Ed. 2009;48:4524–4527. doi: 10.1002/anie.200805683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Proudfoot N. Poly(a) signals. Cell. 1991;64:671–674. doi: 10.1016/0092-8674(91)90495-k. [DOI] [PubMed] [Google Scholar]
- 50.Ludwig J. A new route to nucleoside 5′-triphosphates. Acta Biochimica Et Biophysica Hungarica. 1981;16:131–133. [PubMed] [Google Scholar]
- 51.Boosalis MS, Petruska J, Goodman MF. DNA-polymerase insertion fidelity - gel assay for site-specific kinetics. J. Biol. Chem. 1987;262:14689–14696. [PubMed] [Google Scholar]
- 52.Gao JM, Strassler C, Tahmassebi D, Kool ET. Libraries of composite polyfluors built from fluorescent deoxyribosides. J. Am. Chem. Soc. 2002;124:11590–11591. doi: 10.1021/ja027197a. [DOI] [PubMed] [Google Scholar]
- 53.Delarue M, Boule JB, Lescar J, Expert-Bezancon N, Jourdan N, Sukumar N, Rougeon F, Papanicolaou C. Crystal structures of a template-independent DNA polymerase: murine terminal deoxynucleotidyltransferase. EMBO J. 2002;21:427–439. doi: 10.1093/emboj/21.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perry SW, Epstein LG, Gelbard HA. Simultaneous in situ detection of apoptosis and necrosis in monolayer cultures by TUNEL and trypan blue staining. BioTechniques. 1997;22:1102–1106. doi: 10.2144/97226st01. [DOI] [PubMed] [Google Scholar]
- 55.Zhou SY, Kassauei K, Cutler DJ, Kennedy GC, Sidransky D, Maitra A, Califano J. An oligonucleotide microarray for high-throughput sequencing of the mitochondrial genome. J. Mol. Diag. 2006;8:476–482. doi: 10.2353/jmoldx.2006.060008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berdis AJ, McCutcheon D. The use of non-natural nucleotides to probe template-independent DNA synthesis. ChemBioChem. 2007;8:1399–1408. doi: 10.1002/cbic.200700096. [DOI] [PubMed] [Google Scholar]
- 57.Smith CL, Simmonds AC, Felix IR, Hamilton AL, Kumar S, Nampalli S, Loakes D, Hill F, Brown DM. DNA polymerase incorporation of universal base triphosphates. Nucleosides Nucleotides. 1998;17:541–554. [Google Scholar]
- 58.Cho YJ, Kool ET. Enzymatic synthesis of fluorescent oligomers assembled on a DNA backbone. ChemBioChem. 2006;7:669–672. doi: 10.1002/cbic.200500515. [DOI] [PubMed] [Google Scholar]
- 59.Gao JM, Liu HB, Kool ET. Assembly of the complete eight-base artificial genetic helix, xDNA, and its interaction with the natural genetic system. Angew. Chem. Int. Ed. 2005;44:3118–3122. doi: 10.1002/anie.200500069. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.