Abstract
The androgen receptor (AR) is a member of the nuclear hormone receptor family of transcription factors that plays a critical role in regulating expression of genes involved in prostate development and transformation. Upon hormone binding, the AR associates with numerous co-regulator proteins that regulate the activation status of target genes via flux to the post-translational modification status of histones and the receptor. Here we show that the AR interacts with and is directly methylated by the histone methyltransferase enzyme SET9. Methylation of the AR on lysine 632 is necessary for enhancing transcriptional activity of the receptor by facilitating both inter-domain communication between the N- and C-termini and recruitment to androgen-target genes. We also show that SET9 is pro-proliferative and anti-apoptotic in prostate cancer cells and demonstrates up-regulated nuclear expression in prostate cancer tissue. In all, our date indicate a new mechanism of AR regulation that may be therapeutically exploitable for prostate cancer treatment.
INTRODUCTION
The androgen receptor (AR) is a member of the nuclear hormone receptor superfamily of transcription factors that transmits androgenic signals derived from the testes, in the form of testosterone/dihydrotestosterone, to the regulation of genes involved in prostate growth and development (1). It is well accepted that the AR plays a pivotal role in prostate cancer (CaP) development and remains the primary target for therapeutic intervention (2). Unfortunately, these treatments are ineffective and activation of the AR signalling cascade via alternative pathways results in an incurable castrate-resistant form of the disease (3). Akin to other members of the nuclear receptor family, activation of the AR is a multi-step process initiated by binding to its cognate hormone androgen in the cytoplasm that promotes AR dimerization, intra-receptor interaction via the N- and C-termini and nuclear translocation (4). Once in the nucleus, the receptor binds DNA via a central DNA-binding domain at specific androgen response elements (AREs) within target genes, such as prostate specific antigen (PSA), and activates transcription via the concerted action of two separate domains of the receptor; the activation function-1 (AF-1) domain, contained within the N-terminal transactivation domain (TD), and the activation function 2 (AF-2) domain present in the C-terminal ligand-binding domain (LBD) (4).
Given its pleiotrophic effect and importance in prostate cell fate, transcriptional activity of the AR is under extremely tight control and is subject to regulation at multiple levels. The remodelling of the AR upon hormone-binding creates an interface for the recruitment of a host of nuclear co-regulator proteins that are requisites for controlling the transcriptional activation of androgen-dependent genes (5). Of the many co-regulators identified to date, several enzymatic activities have been characterized that can either enhance or attenuate the activity of the AR (6,7). For example, histone acetyltransferase (HAT) and histone deacetylase (HDAC) enzymes function antagonistically within the AR signalling cascade to regulate the deposition of the activating acetylation mark on specific histone proteins at receptor target genes and hence provide a layer of receptor transcriptional regulation (8,9). Moreover, the finding that the HAT proteins TIP60, p300 and PCAF directly acetylate lysines K630, K632 and K633 of the KLKK motif located in the hinge region of the receptor to enhance inherent activity demonstrated a role for direct post-translational modification of the AR in controlling transcriptional output (9–11).
More recently, histone methylation and the enzymes that catalyse this reversible modification have become a major focus of epigenetic research (10,11). Our present view is of a dynamic and intricate mode of transcriptional control that, via discriminate and reversible methylation of target lysine residues in histones H3 and H4 by histone methyltransferase (HMT) and histone demethylase (HDM) enzymes, works to acutely regulate gene expression (12–14). Although not fully understood, flux to acetylation and methylation status of specific lysine residues of histones H3 and H4 within promoter regions confers both structural changes to the local chromatin template and provides distinct binding interfaces that control the interaction of positive and negative components of the transcriptional machinery; culminating in ordered regulation of gene expression (15,16). Three methylation states exist namely, mono-, di and tri-methylation (me1,2,3) that can be either activating or repressive to transcription depending on the site of modification (17). Specifically, methylation of lysine residues 4 and 36 in histone H3 (H3K4, H3K36) are associated with up-regulating gene expression and in the maintenance of euchromatic domains (18,19), while modification of histone H3 lysine 9 and 27 (H3K9, H3K27) (20–22), together with histone H4 lysine 20 (H4K20) (23,24), is generally linked with gene repression and in the formation of heterochromatic regions. In keeping with this notion, activation of the AR signalling cascade results in hypermethylation of histone H3-K4 and demethylation of H3K9 at targeted promoters, implicating an important role for both HMT and HDM enzymes in receptor regulation (8,25). Although the HDMs LSD1 (25), JMJD2C (26) and JHDM2A (27) have been shown to be important for activation of AR function via concerted removal of the repressive H3K9me marks, the HMTs responsible for up-regulating H3K4 methylation and AR activity remain unknown.
The HMT enzyme SET9 was originally isolated from a HeLa cell nuclear extract and was shown to up-regulate transcription by catalysing histone H3-K4 mono-methylation in a SET domain-dependent manner and antagonizing both histone H3-K9 methylation and promoter association of the NURD deacetylase complex (28–30). Subsequent studies, however, have suggested that SET9 is primarily a non-histone protein methyltransferase catalysing modification of several transcriptional regulatory proteins, including p53 (31), TAF10 (32), estrogen receptor (ER) (33) and the RelA subunit of NFκB (34,35).
Similarity between the KLKK motif of the AR hinge domain and the lysine-rich sequences targeted by SET9 in several SET9 substrates suggested a potential involvement of the methylase in AR methylation and regulation. Here we show by immunoprecipitation (IP) and chromatin immunoprecipitation (ChIP) experiments in LNCaP prostate cancer cells that SET9 interacts with the AR and associates with the androgen-responsive PSA promoter in a ligand-dependent manner. Importantly, using both in vitro and in vivo analyses, we demonstrate for the first time that the AR is directly methylated by SET9 at lysine K632 within the KLKK motif of the hinge domain and this modification is required for optimal AR activity in prostate cancer cells. Moreover, we find aberrant SET9 expression in prostate cancer by immunohistochemical analysis implicating a potential role of SET9 in cancer progression. In all, our data highlight a new mode of AR regulation that may be therapeutically exploitable in the future.
MATERIALS AND METHODS
Plasmids and peptides
The following plasmids have been described previously: pPSALuc, pMMTVLuc, pCMV-β-gal, pCR3.1-SRC-1, pFlag-AR (36), pVP16-AR-TD, pM-AR-DBD/LBD (37), Flag-SET9 and Flag-SET9H297A (gifts from Danny Reinberg, Howard Hughes Medical Institute) (31). pFlag-ARK632R was generated using site-directed mutagenesis (Quickchange™, Stratagene) using pFlag-AR as template (primer sequences available on request).
The following AR623–640 peptides encompassing the KLKK motif were generated: ARWT, ARK630R, ARK632R and ARK632R/K633R (SynBioSci, CA).
Immunohistochemical analysis
A tissue micro-array (TMA) containing 76 cancer biopsies and 24 benign biopsies was stained [as described in ref. (38)] using a monoclonal anti-SET9 antibody (clone 5F2.3; Millipore) at a dilution of 1:500. SET9 expression was scored blindly by intensity of staining in each biopsy core as being absent (0), weak (+), moderate (++), or strong (+++). Slides were scanned using a Scanscope GL scanner (Aperio) and analysed using Spectrum™ software (Aperio).
3H Methylation assay
Approximately 0.5 µg of bacterially purified His-tagged AR transactivation domain (AR-TD) (39), DNA-binding domain/Hinge/ligand-binding domain (AR-DBD/H/LBD) (40) proteins or the AR623–640 peptides were incorporated into an in vitro methylation reaction containing 1 µg SET9 (Millipore), 2 µCi 3H S-adenosyl methionine (GE Healthcare) and HMT reaction buffer [250 mM Tris–HCl (pH. 9), 2.5 mM DTT, 1 mM PMSF] and incubated at 30°C for 30 min. Proteins were separated by PAGE and exposed to X-ray film as described (36).
Detailed descriptions of all other materials and methods can be found in the Supplementary Data.
RESULTS
SET9 Interacts with the AR
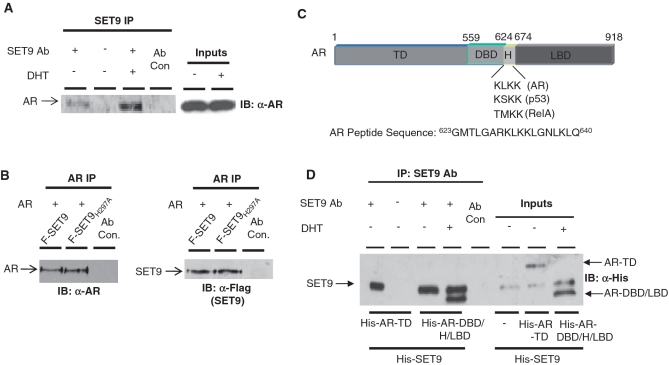
To address a role for SET9 in the AR signalling cascade we first sought to identify an interaction between SET9 and the receptor. Androgen-dependent LNCaP prostate cancer cells were cultured in steroid-depleted media for 40-h prior to treatment with or without 10 nM dihydrotestosterone (DHT) for 8-h and subject to IP using an anti-SET9 antibody. Western analysis of immunoprecipitated material using an anti-AR antibody demonstrated an interaction between endogenous SET9 and AR that is enhanced in the presence of androgen (Figure 1A). To confirm the interaction, the reciprocal IP was performed in AR-null HEK293T cells, grown in serum-containing media, ectopically expressing AR together with wild-type SET9 or the methyltransferase-inactive mutant SETH297A. As shown in Figure 1B, immunoprecipitated AR interacted equally well with both wild-type and mutant forms of SET9.
Figure 1.
AR and SET9 interact in vitro and in vivo. (A) LNCaP cells grown in steroid-depleted media and treated with and without 10 nM DHT for 6 h were subject to IP using an anti-SET9 antibody followed by western analysis using an anti-AR antibody. (B) HEK293T cells were transiently transfected with pFlag-AR and either pFlag-SET9 or pFlag-SET9H297A for 48 h prior to IP using an anti-AR antibody followed by immunoblotting with anti-AR and -Flag antibodies. (C) Diagrammatic representation of the domains of the androgen receptor showing target lysine sequences of SET9 and sequence of AR peptide used in 3H methylation assays. (D) In vitro IP between His-tagged SET9 and N- and C-terminal fragments of the AR using anti-SET9 antibody followed by western analysis using an anti-His antibody.
To address if the interaction between AR and SET9 was direct, we immunoprecipitated purified His-tagged SET9 using an anti-SET9 antibody from a mixture containing either His-tagged N-terminal transactivation domain (AR-TD) (40) or His-tagged DNA-binding domain (DBD)/hinge domain (H)/ligand-binding domain (LBD) (AR-DBD/H/LBD) (39) fragments of the AR (Figure 1C) and 10 nM DHT. Western analysis using an anti-His antibody indicated that SET9 specifically interacts with the DBD/H/LBD domains of the receptor (Figure 1D, lane 4), but not the AR-TD (Figure 1D, lane 1).
The AR is methylated by SET9 in vitro and in vivo
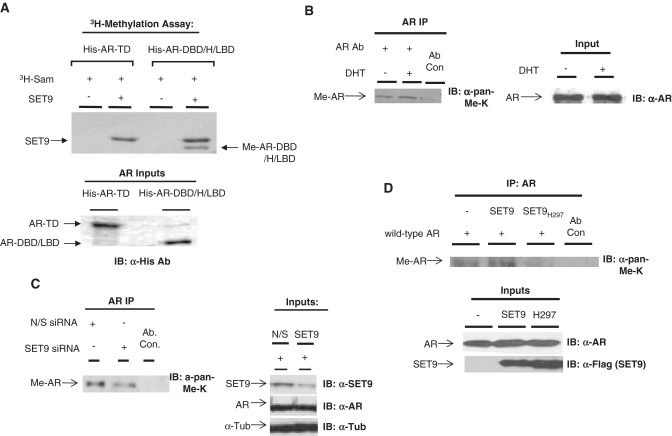
The binding of SET9 to the AR-DBD/H/LBD fragment that contains the 630KLKK633 motif, a sequence that resembles the SET9-methylation sites of p53 [KSK(me)K] (31) and ER [RSK(me)K] (33), suggested a potential role for SET9 in directly methylating the AR. To test this, purified AR-TD or AR-DBD/H/LBD proteins were incorporated into an in vitro methylation assay containing SET9, 3H-labelled S-adenosyl methionine (3H-SAM) and 10 nM DHT. As shown in Figure 2A, the AR-DBD/H/LBD fragment was directly and specifically methylated in the presence of SET9 (Figure 2A, lane 4), but not the AR-TD that remained unmodified (Figure 2A, lane 2), indicating a positive correlation between SET9-AR interaction and receptor methylation. We also detected SET9 auto-methylation as described in ref. (41).
Figure 2.
The AR is methylated by SET9 in vitro and in vivo. (A) An in vitro 3H-SAM methylation assay containing purified SET9 and N- and C-terminal fragments of the AR was incubated for 30 min and samples subject to PAGE, dried and exposed to X-ray film. (B) LNCaP cells grown in steroid-depleted media were treated for 6 h with 10 nM DHT prior to IP using an anti-AR antibody and western analysis using an anti-pan-methyl-lysine antibody (α-pan-methyl-K). (C) LNCaP cells grown in serum-containing media were transiently transfected with either non-silencing (N/S) or SET9 siRNAs for 72 h prior to IP and western analysis as in (B). (D) HEK293T cells were transiently transfected with AR and either SET9 or SET9H297A for 48 h prior to IP and western analysis as in (B).
To assess if the AR is directly methylated in vivo, we immunoprecipitated endogenous AR from LNCaP cells treated with or without 10 nM DHT for 6-h followed by western analysis using an anti-pan-methyl-lysine antibody (Abcam), as previously used to detect both TAF10 (32) and RelA methylation (34). In the absence of androgen, we detected a methylated species at 110 kDa, corresponding to the AR, that became more pronounced upon androgen treatment (Figure 2B, compare lanes 1 and 2), indicating that the AR is methylated in LNCaP cells that is enhanced upon activation of the receptor.
To address the question of whether SET9 is involved in AR methylation in LNCaP cells, we employed short interfering RNA (siRNA) to reduce cellular SET9 levels and compared receptor methylation to a non-silencing siRNA control by IP as described above. As shown in Figure 2C, we achieved ∼70% knockdown of SET9 72-h post-siRNA transfection using SET9 siRNA that resulted in a marked reduction to endogenous receptor methylation (compare lanes 1 and 2) without affecting AR protein levels. This experiment was repeated using an additional anti-methyl-lysine antibody (Abcam) for western analysis and although detection of the methylated AR species was less robust than the anti-pan-methyl-lysine antibody used above, SET9 knockdown attenuated receptor methylation (Supplementary Figure S1) confirming a role for SET9 in AR methylation in vivo.
In an effort to establish a role for the methyltransferase activity of SET9 in AR modification, we over-expressed the AR together with either wild-type SET9 or the methylase-dead SET9H297A mutant, in HEK293T cells and assessed receptor methylation by IP. As shown in Figure 2D, AR methylation is enhanced in the presence of wild-type SET9, but not the methylase-inactive SET9H297A mutant, suggesting that the HMT activity of SET9 is necessary for receptor methylation. The fact that AR methylation levels are reduced below basal levels in the presence of SET9H297A suggest a dominant-negative effect of the mutant over endogenous SET9 (Figure 2D, compare lanes 1 and 3). In all, these in vitro and in vivo findings indicate that SET9 methylates the AR.
Lysine 632 of the AR is methylated by SET9
To delineate the site of SET9-mediated AR methylation we hypothesised that the 630KLKK633 motif would be the preferred site of modification given the similarity with other known SET9 target sequences. To this end, peptides encompassing 623–640 amino acids of the AR containing individual lysine to arginine substitutions at lysine 630, 632 and a double substitution of 632/633, were subject to in vitro methylation and analysed by autoradiography. SET9-mediated methylation of wild-type and K630R peptides was equivalent to modification of histone H3 indicating lysine 630 is not methylated in vitro (Figure 3A, left panel). Importantly, substitution of K632 almost completely abolished methylation and was accomplished by the double lysine 632/633 substitution, indicating that although there is trace methylation of lysine 633 in this assay system, lysine 632 is the preferred site for SET9-mediated methylation of the peptide in vitro (Figure 3A, left panel). To confirm this finding, methylated and non-methylated wild-type AR623–640 peptides were subjected to mass spectrometry analysis. Specifically, one major site of methylation, in the form of mono-methylation, was detected within the peptide at lysine 632 with no modification of lysines 630 and 638, and only very weak methylation at lysine 633 that may an artefact of the methylation reaction (Figure 3A, right panel and Supplementary Figure S2). This finding is consistent with the catalytic mechanism of SET9 for p53 (31), ER (33) and RelA (34).
Figure 3.
Lysine 632 of the AR is methylated by SET9. (A) Left panel: 3H-SAM methylation assay incorporating SET9 and wild-type or point mutants of AR623–640 peptides. Right panel: mass spectrometry analysis of AR peptide (623–640) with (+SET9) and without (−SET9) SET9-mediated methylation. Numbers in italics represent detected fragments in the spectra. A shift of 14 Da at lysine 632 represents methylation and is apparent in both γ and β ion analysis. (B) HEK293T cells transiently transfected with Flag-tagged SET9 and AR or ARK632R were subject to IP using an anti-AR antibody followed by western analysis using anti-Flag and anti-pan-methyl-lysine antibodies.
To investigate whether lysine 632 of the AR is methylated specifically in vivo, HEK293T cells grown in serum-containing media ectopically expressing Flag-SET9 (F-SET9) and either full-length wild-type Flag-AR or Flag-ARK632R, in which lysine 632 was replaced with arginine, were subject to IP using an anti-AR antibody followed by western analysis using both anti-Flag and anti-pan-methyl-lysine antibodies. As shown in Figure 3B (Left Panel), ARK632R is expressed and interacts with SET9 at similar levels to wild-type AR. Methylation of ARK632R, however, is completely abolished compared to wild-type receptor (Figure 3B, right panel) indicating that SET9 methylates AR at lysine K632 in vivo.
SET9 enhances transcriptional activity of the AR
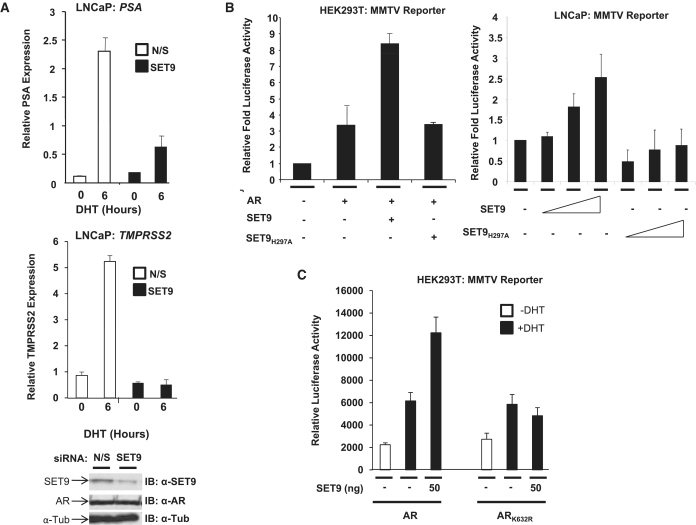
Given that SET9-mediated methylation up-regulates p53 and ER transcriptional activity (31,33), we hypothesised that SET9 would also enhance transcription of androgen-regulated genes. To this end, we depleted endogenous SET9 levels in LNCaP cells by siRNA and assessed hormone-dependent expression of the AR-target genes PSA and TMPRSS2 by quantitative PCR. LNCaP cells grown in steroid-depleted media supplemented with or without 10 nM DHT showed robust reductions in both androgen-dependent PSA (Figure 4A, upper panel) and TMPRSS2 (Figure 4A, lower panel) expression compared to the non-silencing control indicating a role for SET9 in co-activation of the AR.
Figure 4.
SET9 is an AR co-activator. (A) LNCaP cells grown in steroid-depleted media were transiently transfected with non-silencing (N/S) or SET9 siRNAs for 66 h prior to treatment with and without 10 nM DHT for 6 h followed by RNA extraction and real-time PCR analysis of PSA and TMPRSS2 mRNA expression. (B) HEK293T and LNCaP cells were transiently transfected for 48 h with AR (HEK293T cells only) and either SET9 or SET9H297A together with both MMTV-luciferase and β-galactosidase reporters prior to luciferase and β-galactosidase expression analyses. (C) HEK293T cells were transiently transfected and analysed as in (B) with the inclusion of ARK632R. Data represent average of three independent repeats ± standard error (A–C).
To provide evidence of a role for the histone methylase activity of SET9 in AR activation, wild-type SET9 and SET9H297A were ectopically expressed in both LNCaP and HEK293T cells, grown in serum-containing media, together with the AR (HEK293T cells only) and an androgen-responsive MMTV luciferase reporter. As shown in Figure 4B, wild-type SET9, but not the methylase-inactive mutant, enhanced AR activity by ∼2.5-fold in both cell lines indicating that the catalytic activity of SET9 is required for receptor co-activation. Similarly, the HMT activity of SET9 up-regulated AR activity upon the PSA luciferase reporter in HEK293T cells (Supplementary Figure S3A) as well as upon the androgen-responsive AREIII reporter in the osteosarcoma cell line U2OS (Supplementary Figure S3B). In all, this data is consistent with a role for SET9 as an AR co-activator.
To test the requirement of AR methylation at lysine 632 for SET9-mediated receptor co-activation, we examined the effect of SET9 on the activity of wild-type AR and the ARK632R mutant in MMTV reporter assays in HEK293T cells grown in steroid-depleted media. As expected, 10 nM DHT treatment enhanced AR activity by 3-fold (Figure 4C, compare lanes 1 and 2) that was further up-regulated by co-expression of SET9 (Figure 4C, compare lanes 2 and 3). In contrast, although ARK632R responded to androgen stimulation similar to the wild-type receptor, ectopic expression of SET9 failed to stimulate the lysine mutant AR (Figure 4C, compare lanes 5 and 6) indicating that methylation of AR at lysine 632 is necessary and sufficient for co-activation by SET9.
AR methylation enhances inter-domain interaction without affecting receptor stability or cytoplasmic-nuclear shuttling
Having shown a role for receptor methylation in transcriptional co-activation, we next assessed the impact of methylation on the stability of the receptor. Previous reports have shown differing effects of SET9-mediated methylation on protein stability; both p53 and ER are stabilized by SET9 (31,33) while the RelA subunit of NFκB (34) and DNMT1 (41) are both destabilized. To test our hypothesis that SET9 affects AR stability and hence up-regulates transcriptional activity of the receptor, cycloheximide (CHX) time-course experiments were undertaken in HEK293 cells over-expressing AR with and without wild-type or methylase-inactive mutant SET9. As shown in Figure 5A (left panel), in the absence of ectopic SET9, AR levels are reduced by ∼50% after 8 h CHX treatment in serum-containing media which is in line with our previous findings (42). Importantly, expression of SET9 (Figure 5A, middle panel) or SET9H297A (Figure 5A, right panel), had no additional effect on AR protein levels indicating that SET9 does not impact on receptor stability. To test this further, LNCaP cells transiently transfected with SET9 siRNA or control were subject to CHX time-course experiments as above. Endogenous AR destabilization rates in the SET9-depleted cells were equivalent to control cells (Figure 5B). Moreover, overexpression of SET9 or SET9H297A did not affect AR or ARK632R protein levels (Figures 2D and 3C) confirming that SET9 does not regulate AR turnover.
Figure 5.
SET9 enhances AR N- and C-terminal interaction without affecting receptor stability or nuclear shuttling. (A) HEK293T cells transiently transfected with empty vector, SET9 or SET9H297A were treated with 1 µM cycloheximide (CHX) for up to 8 h and then subject to western analysis using anti-AR and anti-α-tubulin antibodies. (B) LNCaP cells transiently transfected with non-silencing (N/S) or SET9 siRNAs were treated with 1 µM CHX and subject to western analysis as in (A). (C) LNCaP cells transiently transfected with N/S or SET9 siRNAs in steroid-depleted media were treated with and without 10 nM DHT for 6 h prior to nuclear-cytoplasmic extraction and western analysis using anti-AR, -PARP1 and -GAPDH antibodies. (D) HEK293T cells transiently transfected with AR or ARK632R were subject to the same experimental procedure as in (C) with the inclusion of an additional 10 nM DHT time-point at 2 h. (E) Mammalian two-hybrid analysis of AR-TD and AR-DBD/H/LBD fragment interaction in HEK293T cells transiently transfected with either SET9, SET9H297A or SRC-1 and Gal4-luciferase and β-galactosidase reporters. Cells were treated with and without 10 nM DHT for 24-h prior to harvesting, and luciferase and β-galactosidase analyses. Data represents the average of three independent experiments performed in quadruplicate ± standard error (asterisk represents statistical significance <0.05).
Another potential mechanism of SET9 co-activation of the AR would be to enhance nuclear import of the hormone-activated receptor. To test this, androgen-dependent nuclear shuttling of the AR was examined in LNCaP cells with and without SET9 knockdown. As shown in Figure 5C, SET9-depletion had no apparent effect on AR translocation to the nucleus after 6 h DHT stimulation compared to control cells (N/S). In addition, we found no difference in kinetics of androgen-induced nuclear movement of wild-type AR versus ARK632R indicating that the presence of SET9 or receptor methylation has no effect on cytoplasmic-nuclear shuttling of the receptor in response to hormone (Figure 5D).
To further decipher the mechanism of SET9-mediated co-activation of the AR, we next investigated the effect of methylation on the N–C terminal interaction of the receptor. Upon hormone-binding, the AR undergoes a major allosteric re-organization that promotes the association of a region of the N-terminal TD, containing FXXLF and WXXLF motifs, with the C-terminal AF-2 domain that is vital for transcriptional activity of the receptor (43,44). Importantly, several components of the AR signalling cascade have been shown to regulate AR inter-domain interaction that correlates with their role as either co-activators or co-repressors. For example, steroid receptor co-activator 1 (SRC-1) facilitates N–C terminal interaction to enhance receptor activity (45), while Rad9 attenuates domain association and hence transcriptional output of the AR (45). Using luciferase reporter-based mammalian two-hybrid assays in HEK293T cells (37), incorporating ectopically expressed Gal4DBD-AR-DBD/LBD and VP16AD-AR-TD proteins, the effect of wild-type SET9 or SET9H297A on AR inter-domain interaction was assessed. As expected, treatment of cells with 10 nM DHT increased interaction between the N- and C-terminal domains of the AR (Figure 5E, compare lanes 1 and 2) which was further stimulated by co-expression of increasing amounts of SRC-1. In the presence of wild-type SET9, inter-domain interaction of the receptor was enhanced to levels equivalent to SRC-1 (Figure 5E, lanes 9–11) without affecting inherent activity of Gal4DBD-AR-DBD/LBD or VP16AD-AR-TD independently (data not shown). Importantly, no enhancement of interaction was seen in the presence of the enzymatically-inactive SET9H297A mutant indicating that the histone methylase activity of SET9 is important for facilitating the N–C terminal association of the receptor. Together, our data indicate that the mechanism of AR co-activation by SET9 is in part dependent upon regulation of inter-domain communication of the receptor without affecting cytoplasmic-nuclear shuttling or stability of the AR.
SET9 associates at the AR-regulated PSA gene and is important for AR recruitment and mono-methylation of histone H3K4
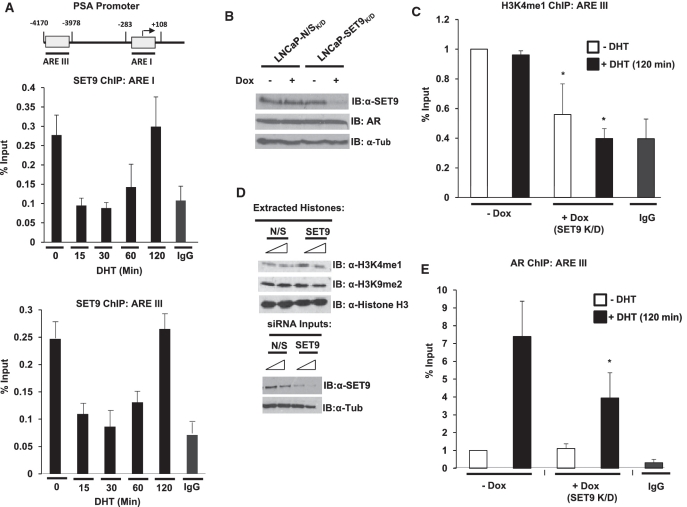
Having established a co-activator role for SET9 in AR-mediated transcription, we next sought to identify if SET9 was recruited to androgen-regulated genes by ChIP analysis. LNCaP cells grown in steroid-depleted media were treated with 10 nM DHT over a time-course of 0–120 min prior to ChIP analysis using an anti-SET9 antibody or an anti-AR antibody control. As expected, AR recruitment to the proximal (ARE I) and distal enhancer (ARE III) regions of the PSA promoter (illustrated in Figure 6A) was androgen-dependent and showed a more robust recruitment to the ARE III region compared to ARE I as described previously (46) (Supplementary Figure S4). In contrast, we found SET9 associated with similar magnitude to both ARE I and III regions of the PSA promoter in the absence of androgen (Figure 6A). After 15 min of hormone treatment, SET9 levels at the PSA promoter rapidly decreased, remained low at 30 min and then increased gradually between 60 and 120 min of androgen stimulation to levels equivalent to the inactive promoter. This dynamic association–disassociation profile of SET9 is very similar to that seen for the binding of SET9 to the IL-8 promoter in response to TNF-α treatment (34), suggesting a common recruitment mechanism for the HMT. Importantly, SET9 was not present on the same promoter elements in the AR null PC3 cell line indicating a dependency on the AR signalling cascade for promoter binding (Supplementary Figure S5).
Figure 6.
SET9 is recruited to the AR-regulated PSA promoter to methylate histone H3K4 and facilitate AR recruitment. (A) LNCaP cells grown in steroid-depleted media for 72-h were treated with 10 nM DHT over a time-course of 0–120 min and then subject to ChIP analysis using an anti-SET9 antibody, or non-specific isotype control, followed by quantitative PCR analysis using primers specific to the proximal (ARE I) and distal (ARE III) regions of the PSA promoter. (B) Representative SET9 knockdown and AR levels from the stable doxycycline-inducible SET9 knockdown LNCaP cell line (LNCaP-SET9K/D) compared to the equivalent non-silencing (N/S) cell line (LNCaP-N/SK/D). (C) ChIP analysis of histone H3K4 mono-methylation (H3K4me1) at ARE III in LNCaP-SET9K/D cells treated with and without doxycycline for 48 h and 10 nM DHT for 0 or 120 min. (D) Representative western analysis of global histone H3K4me1 and H3K9me2 modifications in LNCaP cells transiently transfected with 25 and 50 nM N/S or SET9 siRNAs. (E) An anti-AR antibody was used in ChIP analysis as in (B). Data represents three independent repeats ± standard error (A and E).
Using a stable doxycycline-inducible SET9 knockdown LNCaP cell line, in which SET9 levels were routinely reduced by ∼80% in response to doxycycline treatment, without affecting AR protein levels (Figure 6B), we next addressed the role of SET9 at the PSA promoter. Given that SET9 is a histone H3K4 mono-methylase (28) and has been shown to mono-methylate H3K4 at several NFκB genes (56), we hypothesised that reducing SET9 levels would down-regulate the H3K4me1 mark at the PSA promoter. As shown in Figure 6C, ChIP analysis using an anti-mono-methyl histone H3K4 antibody demonstrated the presence of H3K4me1 marks in the absence of hormone and after 120 min androgen treatment at both ARE I and ARE III (Figure 6C and Supplementary Figure S6A) suggesting a correlation between association of SET9 at the PSA promoter and mono-methylation of histone H3K4 marks. Importantly, in the presence and absence of 120 min androgen stimulation, knockdown of SET9 significantly reduced H3K4me1 at ARE III (Figure 6C, compare lanes 1–3 and 2–4) and at ARE I, albeit not significantly (Supplementary Figure S6A), suggesting a role for SET9 in methylation of H3K4 at the proximal and enhancer regions of the PSA promoter. To demonstrate that the reduction in H3K4me1 in response to SET9 knockdown was not global, histones extracted from LNCaP cells with and without SET9 depletion were subject to western analysis using an anti-H3K4me1 antibody. Reducing cellular SET9 levels had no effect on global methylation of histone H3K4me1 (Figure 6D and Supplementary Figure S7) or methylation of H3K9 (Figure 6D and Supplementary Figure S7), indicating a role for SET9 in regulating specific foci of histone methylation within chromatin.
We next assessed the effect of depleting SET9 levels on hormone-dependent AR recruitment to the PSA promoter by ChIP analysis using an anti-AR antibody. In line with our findings in Supplementary Figure S4, AR was recruited to ARE I and III after 120 min 10 nM DHT treatment in the absence of doxycycline treatment (Figure 6E and Supplementary Figure S6B, compare bars 1 and 2). Upon doxycycline-induced SET9 knockdown, androgen-dependent recruitment of the receptor to ARE III was significantly decreased compared to non-doxycycline-treated cells indicating a role for SET9 in facilitating activated AR binding to the target promoter (Figure 6E). AR recruitment to the proximal ARE I region was also reduced, but not significantly (Supplementary Figure S6B) which may be due to low-level AR binding to this region compared to the ARE III. Importantly, repeating the above experiments with a control cell line that expresses a non-silencing shRNA in response to doxycycline treatment demonstrated no impact on H3K4 methylation or AR recruitment indicating that the effects seen with SET9 depletion are not off-target effects of the shRNA (data not shown).
The above findings indicate multiple roles for SET9 during AR-mediated transcription, including promoter and receptor methylation that are important for AR folding and recruitment to target genes (as illustrated in Figure 8, see Discussion section). To confirm the importance of direct AR methylation in the transcriptional process, we over-expressed wild-type AR or AR632 in AR negative PC3 cells and assessed endogenous PSA expression by quantitative PCR. As expected, wild-type AR markedly up-regulated PSA levels [as described in ref. (47)], while expression of the AR methylation site mutant failed to enhance gene expression indicating that AR methylation is necessary for receptor function in the context of chromatinized genes (Supplementary Figure S8).
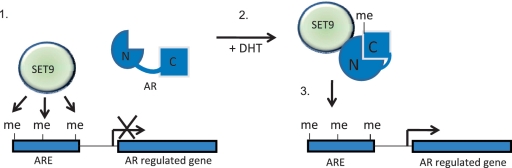
Figure 8.
Postulated model of SET9-mediated regulation of the AR. Our working model is that SET9 enhances AR activity by methylating the receptor and histone H3-K4. Step 1: SET9 is present at androgen response elements (AREs) in the absence of active AR to mono-methylate H3-K4. Step 2: Activation of the AR via dihydrotestosterone (DHT) binding enables interaction of SET9 and the AR followed by methylation of the receptor on lysine 632 that facilitates inter-domain interaction of the N- and C-termini of the AR. Step 3: Methylated AR associates with target genes and enhances transcription.
SET9 is important for androgen-dependent cell proliferation and demonstrates deregulated expression in prostate cancer
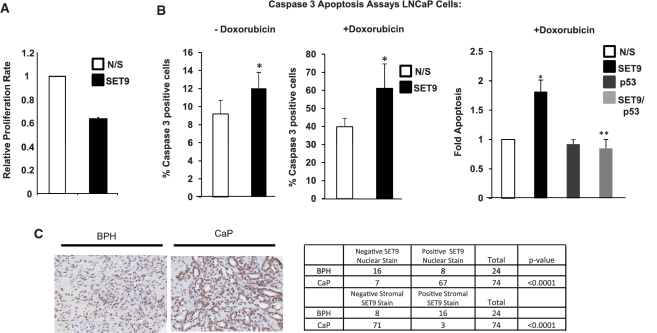
Given SET9 is important for AR-mediated PSA expression in LNCaP prostate cancer cells (Figure 4A), we next sought to investigate the role of SET9 in androgen-dependent cell proliferation. Transient transfection of an siRNA against SET9 resulted in a 40% reduction in hormone-dependent LNCaP cell growth (Figure 7A) compared to control cells indicating a pro-proliferative effect of SET9 on the AR signalling cascade. A similar result was achieved using doxycycline-induced SET9 knockdown in LNCaP cells (data not shown). Further analysis revealed that the reduction in LNCaP cell number upon SET9 knockdown was, in part, due to a small, but significant increase in apoptosis as measured by caspase 3 expression (Figure 7B, left panel). This finding was unexpected considering SET9-null mouse embryonic fibroblasts (MEFs) demonstrated reduced apoptosis in response to the DNA damaging agent doxorubicin which was found to be the result of attenuated p53 activity (48). To test the effect of DNA damage on cell apoptosis with and without SET9-depletion in our system, LNCaP cells transiently transfected with either SET9 or N/S siRNAs were treated with 0.5 µM doxorubicin for 24-hours prior to caspase 3 expression analysis. As expected, control LNCaP cells showed a robust increase in apoptosis from ∼9–40% in the presence of doxorubicin (Figure 7B, middle panel). Interestingly, doxorubicin-induced apoptosis was enhanced significantly by SET9 knockdown (Figure 7B, middle panel, compare bars 1 and 2) suggesting that SET9 function in LNCaP cells is anti-apoptotic as opposed to pro-apoptotic in MEFs (48) and U2OS cells (Supplementary Figure S9A) [as described in ref. (31)]. Furthermore, although p53 knockdown had no effect on doxorubicin-induced LNCaP cells apoptosis, dual depletion of SET9 and p53 in LNCaP cells abrogated the effects of SET9 knockdown alone (Figure 7B, right panel, compare bars 2 and 4 and Supplementary Figure S9B) indicating that p53 remains functional in the absence of SET9 to facilitate cell death.
Figure 7.
SET9 regulates LNCaP cell proliferation, apoptosis and is aberrantly expressed in prostate cancer. (A) WST-1 proliferation assays were conducted in SET9-depleted and non-depleted LNCaP cells grown in steroid-depleted media supplemented with 10 nM DHT for 72 h. Data represents the mean of three independent experiments. (B) LNCaP cells grown in serum-containing media and transiently transfected with non-silencing (N/S), SET9 and/or p53 siRNAs were treated with and without 0.5 µM doxorubicin for 24-h prior to caspase 3 analysis. Data represents the mean of three independent repeats ± standard error (asterisk represents statistical significance <0.05). (C) Representative SET9 staining of prostate tissue using an anti-SET9 antibody in immunohistochemistry (left panel) and table summarizing nuclear and stromal staining pattern of SET9 in cancer and normal tissue (right panel).
Considering the involvement of SET9 in LNCaP cell proliferation and apoptosis, SET9, like other AR co-regulators including TIP60 (49) and hPIRH2 (50), may be aberrantly expressed in prostate cancer (CaP). To examine expression in human prostate tumours, paraffin-embedded prostate tissue samples, retrieved by transurethral resection and incorporated into a tissue micro-array (TMA), were assayed by immunohistochemistry using an anti-SET9 antibody. Out of 76 cases of CaP and 24 benign prostate controls, 90.5% of malignant tissue demonstrated nuclear SET9 staining (67/74) which is in contrast to only 33.3% for the benign prostatic samples (8/24). Representative images are shown in Figure 7C. Nuclear SET9 expression was found to be significantly up-regulated in the malignant epithelium compared to benign tissue (Mann–Whitney U-test, P < 0.0001). SET9 immunoreactivity was localized to the stromal tissue in 66.6% of benign samples (16/24), compared to only 4.1% of prostate cancer samples (3/74) indicating SET9 stromal staining is significantly down-regulated in malignant tissue compared to BPH samples (Mann–Whitney U-test, P < 0.0001) (Figure 7C, right panel). There was no correlation demonstrated between SET9 nuclear immunoreactivity and Gleason grade (Kruskal–Wallis, P = 0.892), presence of bony metastases at diagnosis (Mann–Whitney U-test, P = 0.943) or development of bony metastases subsequent to diagnosis (Mann–Whitney U-test, P = 0.686). There was also no correlation between SET9 stromal immunoreactivity and Gleason grade (Kruskal–Wallis, P = 0.767). In all, this data demonstrates that SET9 nuclear expression is up-regulated in epithelial cells and concurrently down-regulated in stromal cells of prostate cancer tissue compared to benign prostate tissues. Furthermore, SET9 was shown to be pro-proliferative and anti-apoptotic in LNCaP cells suggesting deregulation of SET9 expression may drive uncontrolled cell growth in cancer.
DISCUSSION
The expanding repertoire of AR co-regulator proteins identified over the past decade has provided an indication to the mechanistic complexity required for acute transcriptional control of the receptor. Additional layers of regulation have been attributed to various post-translational modifications of the AR, including phosphorylation (4), acetylation (9) and ubiquitylation (42) which control, for example, transcriptional output and stability of the receptor. Importantly, deregulated activity of several co-regulator proteins has been postulated to facilitate the transition from androgen-dependent CaP to an untreatable castrate-resistant phenotype suggesting future therapies for prostate malignancy may target components of the AR transcriptome (5). Our current studies identify SET9 as a novel AR co-regulator that directly methylates lysine 632 within the hinge domain of the receptor to enhance AR-mediated transcription. This activation signal functions to facilitate both inter-domain communication between the N- and C- termini of the receptor and AR-promoter association without effecting protein stability or nuclear translocation. That SET9 is pro-proliferative and anti-apoptotic in LNCaP prostate cancer cells and is aberrantly expressed in CaP suggest an important role for SET9 in CaP progression and may constitute a novel therapeutic target.
SET9 was initially characterized as a histone H3K4 mono-methyltransferase (28). However, its inability to directly methylate mono- and oligonucleosomes in vitro suggested a more prevalent role for SET9 in non-histone protein modification (51). Indeed, over the past few years, several in vivo targets of SET9-mediated methylation have been identified, including p53 (31), ER (33), RelA (34,35) and DNMT1 (41). In all cases, methylation has provided an additional layer of regulation by controlling transcriptional activity and/or stability of these proteins. We find that SET9 directly methylates the AR predominantly at lysine 632 in vitro and in vivo. Interaction between AR and SET9 in LNCaP cells is enhanced by androgen (Figure 1A) and this correlated with methylation of the receptor, as detected using a pan-methyl-lysine antibody (Figure 2B) indicating a positive correlation between the level of methylation and activation status of the receptor. Establishing a role for SET9-mediated AR methylation in vivo was achieved by depleting SET9 levels in LNCaP cells. Although knockdown was not completely efficient by SET9 siRNA, AR methylation was markedly reduced indicating that SET9 is important for receptor modification (Figure 2C and Supplementary Figure S1). Knockdown of other HMT enzymes, including SET8, in LNCaP cells failed to affect AR methylation (data not shown) suggesting discriminate receptor modification by SET9 in vivo. This notion was confirmed in HEK293T cells in which AR methylation was shown to be up-regulated by ectopic expression of wild-type SET9, but not the methylase-dead SET9H297A mutant. Importantly, SET9H297A, acting as a dominant negative, completely diminished methylation of the AR below basal levels indicating that SET9 is the sole methyltransferase for AR modification in this cell type.
In vitro interaction of SET9 with an AR fragment encompassing the DBD/H/LBD domains catalysed receptor methylation that we hypothesised to be within the KLKK motif of the hinge region due to its similarity with the KSKK and RSKK motifs of p53 and ER, respectively. Using a combination of in vitro peptide methylation assays and mass spectrometry, we showed that lysine 632 was predominantly methylated in favour of lysines 630 and 633 within the KLKK motif and the downstream lysine 638. Indeed, substitution of lysine 632 in the context of the 18-mer peptide markedly reduced peptide methylation by SET9 confirming specificity of SET9 for the single lysine 632 site. Moreover, lysine 632 was found to be mono-methylated by SET9 which is consistent with the catalytic mechanism of the methyltransferase (52).
This finding is in contrast to a recent study investigating ER methylation that demonstrated a 21-mer AR peptide containing the KLKK motif (AR-624–644) was not methylated in vitro by SET9, while peptides containing the RSKK (ER) and KSKK (mineralocorticoid receptor) sequences were modified. Several in vitro methylation experiments have suggested that the consensus sequence for SET9 mediated methylation is [R/K][S/T/A]K (where K is the substrate lysine) and the failure for SET9 to modify the AR peptide was due to the presence of a leucine adjacent to the target lysine which reduces the ability for SET9 to directly modify substrate lysines (52). However, RelA has recently been identified as a SET9 methylation target which does not conform to the consensus sequence; RYK in the N-terminus (Ea and Baltimore, 2009) and TMKK in the C-terminus (34), suggesting flexibility to the catalytic activity of SET9 for substrate modification. Therefore, our data demonstrating AR methylation likely represents the ability for SET9 to methylate proteins containing moderately or poorly conserved methylation consensus sequences and the differences between our in vitro experiments and those of Subramanian et al. (33) may be a reflection of variation to experimental procedure.
Using SET9 knockdown and over-expression studies, we have shown that SET9 is a co-activator for the AR (Figure 4A and B). Moreover, up-regulation of receptor activity is dependent upon the HMT activity of SET9 and the target methylation site lysine 632 within the AR (Figure 4B and C) suggesting methylation of the receptor is inherently linked with AR activation. Unlike p53, ER and RelA, however, the stability of the AR was not affected by SET9 (Figure 5A and B) nor was there an impact on nuclear shuttling of the active receptor upon SET9 knockdown or removal of lysine 632 (Figure 5C and D). Instead, we found that SET9, but not the methylase-inactive mutant SET9H297A, facilitated the N–C terminal interaction of the receptor (Figure 5E), suggesting for the first time a role for methylation in controlling allosteric changes within a protein. Inter-domain communication between the N- and C-termini of the AR is important for facilitating the full transcriptional activity of the receptor and has recently been shown to be negatively regulated by a region of the hinge domain containing residues 629–636 (53), encompassing lysine 632. We speculate that methylation of lysine 632 within this ‘inhibitory region’ of the hinge domain may overcome the repressive effect of this short sequence on the N–C terminal interaction, via an as yet undefined mechanism, to up-regulate transcriptional activity of the receptor.
Using ChIP analysis, we found SET9 associated with the proximal and distal regions of the inactive androgen-regulated PSA promoter; a finding similar to that described for SET9 association at the quiescent IL-8 promoter (34). Upon hormone stimulation, we showed a dynamic dissociation-association profile for SET9 that, again, resembled the kinetics of SET9 during IL-8 activation. Interestingly, upon SET9 knockdown, we found that mono-methylation of H3K4 at the distal promoter element ARE III was significantly reduced suggesting that, contrary to the notion of SET9 as a non-HMT, SET9 is important for methylating histone H3K4 in the context of the chromatinized PSA promoter. This finding is in-line with a role for SET9 in regulating mono-methylation of H3K4 at both MCP-1 and TNF-α promoter elements (54). We also demonstrated reduced AR recruitment to the distal and proximal regions of the PSA promoter indicating the importance of SET9 in facilitating AR association to target genes. Whether this effect is a result of failure of the AR to mediate the N–C terminal interaction, a process required for androgen-dependent expression of PSA (55), or due to a reduction in the histone marks required for AR recruitment is still unknown, but is likely to be a contribution by both SET9-mediated activities. As shown in Figure 8, we speculate that SET9-mediated H3K4 mono-methylation within the context of the inactive promoter creates a chromatin landscape primed to respond to AR activation and that upon androgen binding, direct interaction with SET9 and receptor methylation facilitates AR folding and recruitment to target genes. Indeed, our data demonstrating up-regulation of the endogenous PSA gene in PC3 cells in the presence of ectopically expressed wild-type AR, but not AR632, implicated a role for direct receptor methylation in regulating receptor activity and this event combined with the presence of positive-acting methylation marks at target genes is likely to contribute to powerful transcriptional activation. Preliminary data have shown that SET9 depletion up-regulates both repressive H3K9 methylation marks and HDAC1 association at the distal and proximal PSA promoter elements, suggesting that SET9 functions to retain androgen-regulated genes in a state of readiness for transcriptional activation (Gaughan and Robson, unpublished data).
The AR is a target for numerous post-translational modifications, including phosphorylation (56), ubiquitylation (42,57) and acetylation (9,58). Lysines 630, 632 and 633 of the hinge domain are targets for acetylation by the histone acetyltransferases (HATs) p300, PCAF (58) and TIP60 (42) which is required for optimal transcriptional activity of the AR. Our finding that lysine 632 of the receptor is methylated and is required for SET9-mediated co-activation, suggests potential interplay between these modifications during transcription of AR target genes. It might be interesting to establish if methylation precedes, and is required for, acetylation of the AR, a process that has been described for p53 (48). Preliminary data has found that PCAF interaction with the AR is reduced upon removal of the AR methylation site and co-operativity between SET9 and PCAF for AR co-activation is dependent upon the HMT activity of SET9 suggesting a role for methylation in facilitating PCAF-AR interaction and subsequent co-activation by PCAF (Gaughan and Robson, unpublished data). Additional studies to further dissect interaction between acetylation and methylation would be greatly facilitated by antibodies to acetylated and methylated AR; both of which are currently in production.
In keeping with the role of SET9 as an AR co-activator, our data demonstrate that SET9 depletion reduces androgen-dependent growth of AR-expressing LNCaP cells by ∼40% (Figure 7A) which is similar to other known AR co-regulators, including hPIRH2 (50). For the first time, SET9 tissue expression has been analysed by immunohistochemistry. Importantly, our analysis of prostate tissue demonstrated up-regulated SET9 expression in cancer compared to normal (Figure 7C), suggesting deregulated activity of the methylase in prostate malignancy. We hypothesise that up-regulated expression of SET9 in the nucleus of CaP tissue, irrespective of tumour grade or presence of bone metastasis, enhances nuclear AR function to drive uncontrolled prostate cell proliferation. It is possible that SET9 deregulation in CaP impacts on pathways other than AR signalling, such as the p53 and NF-κB cascades. Interestingly, we show that SET9 is anti-apoptotic in LNCaP cells (Figure 7B) which is in contrast to studies in the osteosarcoma cell line U2OS (31) and MEFs derived from SET9 null animals (48) that demonstrate SET9 is pro-apoptotic as a consequence of up-regulating the p53 pathway. It is interesting to speculate that the balance of SET9 activity in specific cancer types is tipped towards facilitating the activity of pro-proliferative factors, such as the AR, at the expense of the anti-proliferative and pro-apoptotic agents like p53.
In summary, our data highlight a new mode of transcriptional regulation of the AR that adds methylation to the list of post-translational modifications of the receptor. Investigating interplay between these various modifications will provide a more comprehensive understanding of AR control and new therapeutic targets for CaP therapy.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Cancer Research UK (to J.S.) and Association for International Cancer Research (to K.A.). Funding for open access charge: Cancer Research UK Association for International Cancer Research.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank Prof. Danny Reinberg for SET/SET9H297A mammalian expression vectors.
REFERENCES
- 1.Culig Z. Role of the androgen receptor axis in prostate cancer. Urology. 2003;62:21–26. doi: 10.1016/s0090-4295(03)00698-8. [DOI] [PubMed] [Google Scholar]
- 2.Culig Z, Hobisch A, Bartsch G, Klocker H. Expression and function of androgen receptor in carcinoma of the prostate. Microsc. Res. Tech. 2000;51:447–455. doi: 10.1002/1097-0029(20001201)51:5<447::AID-JEMT7>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 3.Culig Z, Klocker H, Bartsch G, Steiner H, Hobisch A. Androgen receptors in prostate cancer. J. Urol. 2003;170:1363–1369. doi: 10.1097/01.ju.0000075099.20662.7f. [DOI] [PubMed] [Google Scholar]
- 4.Edwards J, Bartlett JM. The androgen receptor and signal-transduction pathways in hormone-refractory prostate cancer. Part 1: Modifications to the androgen receptor. BJU Int. 2005;95:1320–1326. doi: 10.1111/j.1464-410X.2005.05526.x. [DOI] [PubMed] [Google Scholar]
- 5.Edwards J, Bartlett JM. The androgen receptor and signal-transduction pathways in hormone-refractory prostate cancer. Part 2: Androgen-receptor cofactors and bypass pathways. BJU Int. 2005;95:1327–1335. doi: 10.1111/j.1464-410X.2005.05527.x. [DOI] [PubMed] [Google Scholar]
- 6.Culig Z, Comuzzi B, Steiner H, Bartsch G, Hobisch A. Expression and function of androgen receptor coactivators in prostate cancer. J. Steroid Biochem. Mol. Biol. 2004;92:265–271. doi: 10.1016/j.jsbmb.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Wang L, Hsu CL, Chang C. Androgen receptor corepressors: an overview. Prostate. 2005;63:117–130. doi: 10.1002/pros.20170. [DOI] [PubMed] [Google Scholar]
- 8.Kang Z, Janne OA, Palvimo JJ. Coregulator recruitment and histone modifications in transcriptional regulation by the androgen receptor. Mol. Endocrinol. 2004;18:2633–2648. doi: 10.1210/me.2004-0245. [DOI] [PubMed] [Google Scholar]
- 9.Gaughan L, Logan IR, Cook S, Neal DE, Robson CN. Tip60 and histone deacetylase 1 regulate androgen receptor activity through changes to the acetylation status of the receptor. J. Biol. Chem. 2002;277:25904–25913. doi: 10.1074/jbc.M203423200. [DOI] [PubMed] [Google Scholar]
- 10.Sims RJ, 3rd, Nishioka K, Reinberg D. Histone lysine methylation: a signature for chromatin function. Trends Genet. 2003;19:629–639. doi: 10.1016/j.tig.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 11.Ng SS, Yue WW, Oppermann U, Klose RJ. Dynamic protein methylation in chromatin biology. Cell Mol. Life Sci. 2009;66:407–422. doi: 10.1007/s00018-008-8303-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Margueron R, Trojer P, Reinberg D. The key to development: interpreting the histone code? Curr. Opin. Genet. Dev. 2005;15:163–176. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 13.Wood A, Schneider J, Shilatifard A. Cross-talking histones: implications for the regulation of gene expression and DNA repair. Biochem Cell Biol. 2005;83:460–467. doi: 10.1139/o05-116. [DOI] [PubMed] [Google Scholar]
- 14.Daniel JA, Pray-Grant MG, Grant PA. Effector proteins for methylated histones: an expanding family. Cell Cycle. 2005;4:919–926. doi: 10.4161/cc.4.7.1824. [DOI] [PubMed] [Google Scholar]
- 15.Fischle W, Wang Y, Allis CD. Binary switches and modification cassettes in histone biology and beyond. Nature. 2003;425:475–479. doi: 10.1038/nature02017. [DOI] [PubMed] [Google Scholar]
- 16.Vakoc CR, Sachdeva MM, Wang H, Blobel GA. A profile of histone lysine methylation across transcribed mammalian chromatin. Mol. Cell Biol. 2006;26:9185–9195. doi: 10.1128/MCB.01529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peterson CL, Laniel MA. Histones and histone modifications. Curr. Biol. 2004;14:R546–R551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 18.Strahl BD, Ohba R, Cook RG, Allis CD. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc. Natl Acad. Sci. USA. 1999;96:14967–14972. doi: 10.1073/pnas.96.26.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J. Biol. Chem. 2005;280:17732–17736. doi: 10.1074/jbc.M500796200. [DOI] [PubMed] [Google Scholar]
- 20.Heard E, Rougeulle C, Arnaud D, Avner P, Allis CD, Spector DL. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell. 2001;107:727–738. doi: 10.1016/s0092-8674(01)00598-0. [DOI] [PubMed] [Google Scholar]
- 21.Chen H, Ke Q, Kluz T, Yan Y, Costa M. Nickel ions increase histone H3 lysine 9 dimethylation and induce transgene silencing. Mol. Cell Biol. 2006;26:3728–3737. doi: 10.1128/MCB.26.10.3728-3737.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rougeulle C, Chaumeil J, Sarma K, Allis CD, Reinberg D, Avner P, Heard E. Differential histone H3 Lys-9 and Lys-27 methylation profiles on the X chromosome. Mol. Cell Biol. 2004;24:5475–5484. doi: 10.1128/MCB.24.12.5475-5484.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karachentsev D, Sarma K, Reinberg D, Steward R. PR-Set7-dependent methylation of histone H4 Lys 20 functions in repression of gene expression and is essential for mitosis. Genes Dev. 2005;19:431–435. doi: 10.1101/gad.1263005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P, Tempst P, Steward R, et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol. Cell. 2002;9:1201–1213. doi: 10.1016/s1097-2765(02)00548-8. [DOI] [PubMed] [Google Scholar]
- 25.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 26.Wissmann M, Yin N, Muller JM, Greschik H, Fodor BD, Jenuwein T, Vogler C, Schneider R, Gunther T, Buettner R, et al. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat. Cell Biol. 2007;9:347–353. doi: 10.1038/ncb1546. [DOI] [PubMed] [Google Scholar]
- 27.Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 28.Nishioka K, Chuikov S, Sarma K, Erdjument-Bromage H, Allis CD, Tempst P, Reinberg D. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 2002;16:479–489. doi: 10.1101/gad.967202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H, Cao R, Xia L, Erdjument-Bromage H, Borchers C, Tempst P, Zhang Y. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol. Cell. 2001;8:1207–1217. doi: 10.1016/s1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- 30.Wilson JR, Jing C, Walker PA, Martin SR, Howell SA, Blackburn GM, Gamblin SJ, Xiao B. Crystal structure and functional analysis of the histone methyltransferase SET7/9. Cell. 2002;111:105–115. doi: 10.1016/s0092-8674(02)00964-9. [DOI] [PubMed] [Google Scholar]
- 31.Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, McKinney K, Tempst P, Prives C, Gamblin SJ, et al. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 32.Kouskouti A, Scheer E, Staub A, Tora L, Talianidis I. Gene-specific modulation of TAF10 function by SET9-mediated methylation. Mol. Cell. 2004;14:175–182. doi: 10.1016/s1097-2765(04)00182-0. [DOI] [PubMed] [Google Scholar]
- 33.Subramanian K, Jia D, Kapoor-Vazirani P, Powell DR, Collins RE, Sharma D, Peng J, Cheng X, Vertino PM. Regulation of estrogen receptor alpha by the SET7 lysine methyltransferase. Mol. Cell. 2008;30:336–347. doi: 10.1016/j.molcel.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang XD, Huang B, Li M, Lamb A, Kelleher NL, Chen LF. Negative regulation of NF-kappaB action by Set9-mediated lysine methylation of the RelA subunit. Embo J. 2009;28:1055–1066. doi: 10.1038/emboj.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ea CK, Baltimore D. Regulation of NF-kappaB activity through lysine monomethylation of p65. Proc. Natl Acad. Sci. USA. 2009;106:18972–18977. doi: 10.1073/pnas.0910439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brady ME, Ozanne DM, Gaughan L, Waite I, Cook S, Neal DE, Robson CN. Tip60 is a nuclear hormone receptor coactivator. J. Biol. Chem. 1999;274:17599–17604. doi: 10.1074/jbc.274.25.17599. [DOI] [PubMed] [Google Scholar]
- 37.Gaughan L, Brady ME, Cook S, Neal DE, Robson CN. Tip60 is a co-activator specific for class I nuclear hormone receptors. J. Biol. Chem. 2001;276:46841–46848. doi: 10.1074/jbc.M103710200. [DOI] [PubMed] [Google Scholar]
- 38.Clark EL, Coulson A, Dalgliesh C, Rajan P, Nicol SM, Fleming S, Heer R, Gaughan L, Leung HY, Elliott DJ, et al. The RNA helicase p68 is a novel androgen receptor coactivator involved in splicing and is overexpressed in prostate cancer. Cancer Res. 2008;68:7938–7946. doi: 10.1158/0008-5472.CAN-08-0932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davies P, Watt K, Kelly SM, Clark C, Price NC, McEwan IJ. Consequences of poly-glutamine repeat length for the conformation and folding of the androgen receptor amino-terminal domain. J. Mol. Endocrinol. 2008;41:301–314. doi: 10.1677/JME-08-0042. [DOI] [PubMed] [Google Scholar]
- 40.Duff J, McEwan IJ. Mutation of histidine 874 in the androgen receptor ligand-binding domain leads to promiscuous ligand activation and altered p160 coactivator interactions. Mol. Endocrinol. 2005;19:2943–2954. doi: 10.1210/me.2005-0231. [DOI] [PubMed] [Google Scholar]
- 41.Esteve PO, Chin HG, Benner J, Feehery GR, Samaranayake M, Horwitz GA, Jacobsen SE, Pradhan S. Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proc. Natl Acad. Sci. USA. 2009;106:5076–5081. doi: 10.1073/pnas.0810362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gaughan L, Logan IR, Neal DE, Robson CN. Regulation of androgen receptor and histone deacetylase 1 by Mdm2-mediated ubiquitylation. Nucleic Acids Res. 2005;33:13–26. doi: 10.1093/nar/gki141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He B, Kemppainen JA, Wilson EM. FXXLF and WXXLF sequences mediate the NH2-terminal interaction with the ligand binding domain of the androgen receptor. J. Biol. Chem. 2000;275:22986–22994. doi: 10.1074/jbc.M002807200. [DOI] [PubMed] [Google Scholar]
- 44.He B, Wilson EM. The NH(2)-terminal and carboxyl-terminal interaction in the human androgen receptor. Mol. Genet. Metab. 2002;75:293–298. doi: 10.1016/S1096-7192(02)00009-4. [DOI] [PubMed] [Google Scholar]
- 45.Hsu CL, Chen YL, Ting HJ, Lin WJ, Yang Z, Zhang Y, Wang L, Wu CT, Chang HC, Yeh S, et al. Androgen receptor (AR) NH2- and COOH-terminal interactions result in the differential influences on the AR-mediated transactivation and cell growth. Mol. Endocrinol. 2005;19:350–361. doi: 10.1210/me.2004-0190. [DOI] [PubMed] [Google Scholar]
- 46.Wang Q, Carroll JS, Brown M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol. Cell. 2005;19:631–642. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 47.Lin B, Wang J, Hong X, Yan X, Hwang D, Cho JH, Yi D, Utleg AG, Fang X, Schones DE, et al. Integrated expression profiling and ChIP-seq analyses of the growth inhibition response program of the androgen receptor. PLoS One. 2009;4:e6589. doi: 10.1371/journal.pone.0006589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kurash JK, Lei H, Shen Q, Marston WL, Granda BW, Fan H, Wall D, Li E, Gaudet F. Methylation of p53 by Set7/9 mediates p53 acetylation and activity in vivo. Mol. Cell. 2008;29:392–400. doi: 10.1016/j.molcel.2007.12.025. [DOI] [PubMed] [Google Scholar]
- 49.Halkidou K, Gnanapragasam VJ, Mehta PB, Logan IR, Brady ME, Cook S, Leung HY, Neal DE, Robson CN. Expression of Tip60, an androgen receptor coactivator, and its role in prostate cancer development. Oncogene. 2003;22:2466–2477. doi: 10.1038/sj.onc.1206342. [DOI] [PubMed] [Google Scholar]
- 50.Logan IR, Gaughan L, McCracken SR, Sapountzi V, Leung HY, Robson CN. Human PIRH2 enhances androgen receptor signaling through inhibition of histone deacetylase 1 and is overexpressed in prostate cancer. Mol. Cell Biol. 2006;26:6502–6510. doi: 10.1128/MCB.00147-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang J, Berger SL. The emerging field of dynamic lysine methylation of non-histone proteins. Curr. Opin. Genet. Dev. 2008;18:152–158. doi: 10.1016/j.gde.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 52.Couture JF, Collazo E, Hauk G, Trievel RC. Structural basis for the methylation site specificity of SET7/9. Nat. Struct. Mol. Biol. 2006;13:140–146. doi: 10.1038/nsmb1045. [DOI] [PubMed] [Google Scholar]
- 53.Haelens A, Tanner T, Denayer S, Callewaert L, Claessens F. The hinge region regulates DNA binding, nuclear translocation, and transactivation of the androgen receptor. Cancer Res. 2007;67:4514–4523. doi: 10.1158/0008-5472.CAN-06-1701. [DOI] [PubMed] [Google Scholar]
- 54.Li Y, Reddy MA, Miao F, Shanmugam N, Yee JK, Hawkins D, Ren B, Natarajan R. Role of the histone H3 lysine 4 methyltransferase, SET7/9, in the regulation of NF-kappaB-dependent inflammatory genes. Relevance to diabetes and inflammation. J. Biol. Chem. 2008;283:26771–26781. doi: 10.1074/jbc.M802800200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He B, Lee LW, Minges JT, Wilson EM. Dependence of selective gene activation on the androgen receptor NH2- and COOH-terminal interaction. J. Biol. Chem. 2002;277:25631–25639. doi: 10.1074/jbc.M202809200. [DOI] [PubMed] [Google Scholar]
- 56.Wang G, Sadar MD. Amino-terminus domain of the androgen receptor as a molecular target to prevent the hormonal progression of prostate cancer. J. Cell Biochem. 2006;98:36–53. doi: 10.1002/jcb.20802. [DOI] [PubMed] [Google Scholar]
- 57.Lin HK, Wang L, Hu YC, Altuwaijri S, Chang C. Phosphorylation-dependent ubiquitylation and degradation of androgen receptor by Akt require Mdm2 E3 ligase. Embo J. 2002;21:4037–4048. doi: 10.1093/emboj/cdf406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fu M, Wang C, Reutens AT, Wang J, Angeletti RH, Siconolfi-Baez L, Ogryzko V, Avantaggiati ML, Pestell RG. p300 and p300/cAMP-response element-binding protein-associated factor acetylate the androgen receptor at sites governing hormone-dependent transactivation. J. Biol. Chem. 2000;275:20853–20860. doi: 10.1074/jbc.M000660200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.