Abstract
A novel proton-fueled molecular gate-like delivery system has been constructed for controlled cargo release using i-motif quadruplex DNA as caps onto pore outlets of mesoporous silica nanoparticles. Start from simple conformation changes, the i-motif DNA cap can open and close the pore system in smart response to pH stimulus. Importantly, the opening/closing and delivery protocol is highly reversible and a partial cargo delivery can be easily controlled at will. A pH-switchable nanoreactor has also been developed to validate the potential of our system for on-demand molecular transport. This proof of concept might open the door to a new generation of carrier materials and could also provide a general route to use other functional nucleic acids/peptide nucleic acids as capping agents in the fields of versatile controlled delivery nanodevices.
INTRODUCTION
Over the past two decades, nucleic acids have been recognized as an attractive building material for nanotechnology and materials science owning to their conformational polymorphism, programmable sequence-specific recognition and robust physicochemical nature. Many nucleic acids based, artificial structures/devices have been constructed and shown potential applications in miniaturized biosensors, microsurgery, drug delivery, nanorobotics and dynamic nanomaterials, etc. (1–5). Although these DNA nanodevices are promising, how to make them to perform further functions remains a big challenge in this field. Therefore, a new strategy is needed to overcome these problems for the development of simple DNA mechanical devices toward more sophisticated functions.
Here for the first time, we described the use of DNA as a biomolecule-based, proton-responsive cap system for MSN, and demonstrated the operability of this system with intelligent on-demand molecular transport. Because the unique feature of mesoporous silica nanoparticles (MSN), such as large load capacity, biocompatibility, high thermal stability and tunable pore structure, the development of functionalized MSNs as carrier vehicles in a stimuli-responsive capped/gated release mechanism have attracted great attention in basic discovery research as well as in biosensor, drug/gene delivery and detergent design, etc. (6–9). To date, the using of different kinds of pore blocking caps, such as nanoparticles (10–12), supermolecular assemblies (13,14) and large molecules (15,16) to keep compounds from leaching out of porous hosts and to permit their controlled release has taken chemistry to the frontier of nanoscience. Despite these burgeoning developments, many of the existing capping systems are offset by their poor applicability in aqueous solutions, irreversibility, the use of difficult-to-apply and/or complex stimuli and the toxicity of the capping agents used. In particular, regardless of very recent reported gated MSN that can be capped by certain antibodies or peptides (17–19), there is almost complete lack of MSN-based devices involving biomolecular caps.
An i-motif DNA which carries a piece of the human telomeric sequence was attached on the exterior of MCM-41 particles as a model system in this work. I-Motif DNA is a four-stranded DNA structure with stretches of cytosine base (20). It undergoes a precise structural change driven by a pH change with significant force (8–10 pN) (5,21–22). We then sought to take advantage of this unique feature to control the gate operation. The working principle of the system is illustrated in Figure 1. At low pH, the C residues are partially protonated and the DNA folds into the closed i-motif structure, the pores are capped by the quadruplex and the release of the cargo is strongly inhibited. When the pH is increased to basic, the C+ residues are deprotonated and the DNA unfolds to a single-stranded form, the silica nanopores are spontaneously unblocked which results in rapid delivery of the cargo from the pore voids into the aqueous solution. Thus, the interconversional cycles of the closed and open states of the gated system could be demonstrated by measuring the pH-dependent release of the loaded dye molecules.
Figure 1.
Schematic representation of proton-fueled release of guest molecules from the pores of MSN capped with i-motif DNA.
MATERIALS AND METHODS
Materials and instrumentation
Nanopure water (18.2 MΩ; Millpore Co., USA) was used in all experiments and to prepare all buffers. Tetraethylorthosilicate (TEOS), (3-aminopropyl)trimethoxysilane (APTES), 4-morpholineethanesulfonic acid (MES), rodamine B and 7-diethylamino-3-(4-maleimidophenyl)-4-methylcoumarin (CPM) were purchased from Sigma-Aldrich. N-cetyltrimethylammonium bromide (CTAB), 1-[3-(dimethylamino)propyl]-3-ethylcarbodiimide hydrochloride (EDC), trisodium citrate dihydrate and succinic anhydride were obtained from Alfa Aesar. N-hydroxysulfosucnimide sodium salt (sulfo-NHS) was purchased from Pierce Biotechnology. All the chemicals were used as received without further purification. The oligonucleotide used in this article was synthesized by Sangon Biotechnology Inc. (Shanghai, China). The sequence is as follows:
DNA-1: 5′-NH2-(CH2)6-CCCTAACCCTAACCCTAACCC-3′
DNA-2: 5′-NH2-(CH2)6-CTCTCACTCTCACTCTCCACC-3′
FT-IR analyses were carried out on a Bruker Vertex 70 FT-IR Spectrometer. X-ray measurements were performed on a Bruker D8 FOCUS Powder X-ray Diffractometer using Cu Kα radiation. Thermogravimetric analyses were carried out on a PerkinElmer Pyris Diamond TG/DTA Analyzer, using an oxidant atmosphere (Air) with a heating program consisting of a dynamic segment (10°С/min) from 373 to 1173 K. SEM images were obtained with a Hitachi S-4800 FE-SEM. N2 adsorption–desorption isotherms were recorded on a Micromeritics ASAP 2020M automated sorption analyzer. The samples were degassed at 150°С for 5 h. The specific surface areas were calculated from the adsorption data in the low pressure range using the BET model and pore size was determined following the BJH method. Solid-state 13C CP-MAS NMR spectra were obtained on Bruker AVANCE III 400 WB spectrometer equipped with a 4 mm standard bore CPMAS probe whose X channel was tuned to 100.62 MHz for 13C and the other channel was tuned to 400.18 MHz for broad band 1H decoupling, using a magnetic field of 9.39 T at 297 K. UV-vis spectroscopy was carried out with a JASCO V-550 UV/vis spectrometer. Fluorescence spectra were recorded with a JASCO FP-6500 spectrofluorometer.
Synthesis and chemical modification of the MSN surface
N-cetyltrimethylammonium bromide (CTAB, 1.00 g) was first dissolved in 50 ml of pure water by heating. After cooling to room temperature, aqueous ammonia (13 ml) and ethanol (75 ml) were added. The mixture was stirred for 15 min and TEOS (1.94 ml) added rapidly while stirring was continued. TEOS (30 µl) and APTES (30 µl) were introduced 30 min later. The mixture was allowed to stir for 2 h to give rise to white precipitates. The solid product was filtered, washed with deionized water and methanol, and dried in air. To remove the surfactant template (CTAB), the white powder was refluxed for 16 h in a solution of 1.00 ml of HCl (37%) and 80.00 ml of methanol followed by extensively washing with deionized water and methanol. The resulting surfactant-removed amine-functionalized MSN (MSN-NH2) was placed under high vacuum to remove the remaining solvent in the mesopores. The MSN-NH2 (50 mg) was reacted with succinic anhydride (1.00 g) in N,N-dimetylformamide solution (20 ml) under N2 gas for 8 h with continuous stirring. By doing so, carboxyl groups were formed onto the MSN surface for conjugation of DNA. After a thorough water wash, the carboxylated nanoparticles (MSN-COOH) were activated using EDC (10 mg/ml, 15 ml) and sulfo-NHS (10 mg/ml, 15 ml) in a MES buffer (pH 6.0) for 15 min at room temperature with continuous stirring. Twenty microliters of PBS buffer (100 mM, pH 7.4) was then added in the mixture, followed by the addition of DNA-1 or -2 (3 ml 98.3 µM) at room temperature with continuous stirring for 6 h and washing in PBS buffer (0.1 M, pH 7.4) to form the resultant DNA-conjugated nanoparticles (MSN-DNA or -cDNA). The unreacted DNA was purified with Vivaspin ultrafiltration spin column (MW 2 kDa). The quantification of left DNA was accomplished by UV-vis spectroscopy to be 104.1 nmol, which corresponded to an immobilization efficiency of ∼3.81µmol/g SiO2.
Rhodamine B loading and dye release experiments
The purified MSN-DNA was incubated in the phosphate-buffered saline (10 mM, 25 ml, pH 7.4) of rhodamine B (5 mg) for 24 h. The pH value of the suspension was adjusted to 5.0 by the addition of HCl. The solution was stirred for 16 h, followed by centrifuging and repeated washing with citrate buffer (25 mM, pH 5.0) to remove physisorbed rhodamine B molecules from the exterior surface of the material. All the washing solutions were collected, and the loading of rhodamine was calculated from the difference in the concentration of the initial and left dyes to be ∼40 µmol/g SiO2. Rhodamine-loaded MSN-DNA (10 mg) material was dispersed in 25 ml of citrate buffer at a certain pH value (pH 5.0 or 8.0). Aliquots were taken from the suspension and the delivery of rhodamine dye from the pore to the buffer solution was monitored via the absorbance band of the dye centered at 553 nm. As for partial cargo release, MSN-RhB (10 mg) was dispersed in 25 ml of citrate buffer at pH 5.0. Aliquots were taken from the suspension every 5 min over a range of 550 min, and the amount of cargo delivered by carrying out interconversion cycles of the closed and open states via pH variation.
CPM loading experiments
The purified MSN-DNA (10 mg) was incubated in dimethylsulfoxide (DMSO) of CPM (0.5 mg) at 20°C overnight, followed by centrifuging and washing with deionized water to afford CPM-loaded MSN-DNA.
Fluorescence detection of nanoreactor activity
CPM-loaded MSN-DNA (0.5 mg) material was dispersed in 2 ml of citrate buffer at a certain pH value (between pH 5.0 and 8.0). After the reaction with a model compound 6-mercapto-1-hexanol (50 µM) at 20°C overnight, the resultant nanoparticles were centrifuged and then dispersed in citrate buffer at corresponding pH. The pH-dependant nanoreactor activity was monitored via the fluorescence of thiol derivative of CPM (λex = 384 nm/λem = 410–650 nm) in the nanoreactor.
RESULTS AND DISCUSSION
Synthesis and characterization of MSN-DNA
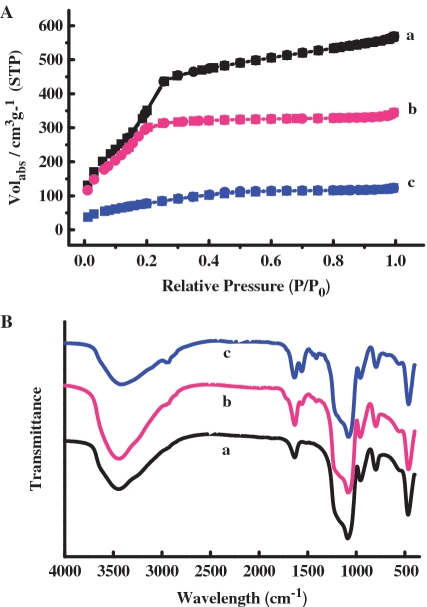
The MCM-41 particle was synthesized using a base-catalyzed sol-gel procedure (23) and MSN (400 nm in diameter) that contain hexagonally arranged pores were confirmed by SEM and X-ray diffraction (Supplementary Figures S1 and S2). The surface of MSN was then functionalized with amine groups by treatment with 3-aminopropyltriethoxysilane (APTES) to afford MSN-NH2. N2 adsorption–desorption isotherms of MSN-NH2 showed a typical Type IV curve with a specific surface area of 1476 m2 g−1, average pore diameter of 2.14 nm and a narrow pore distribution (Figure 2A and Table 1). The silica particle functionalized with a carboxylic group (MSN-COOH) was obtained by allowing MSN-NH2 to react with succinic anhydride in N,N-dimetylformamide (DMF). The resultant carboxyl unit on the surface was activated by 1-[3-(dimethylamino)propyl]-3-ethylcarbodiimide hydrochloride (EDC) and N-hydroxysulfosucnimide sodium salt (sulfo-NHS) in a MES buffer and subsequently treated with DNA-NH2 to obtain MSN-DNA.
Figure 2.
(A) Nitrogen sorption isotherms of the samples (a) MSN-NH2, (b) MSN-DNA and (c) MSN-RhB. (B) FTIR spectra of the samples (a) MSN-NH2, (b) MSN-COOH and (c) MSN-DNA.
Table 1.
BET specific surface values, pore volumes and pore sizes calculated from the N2 adsorption–desorption isotherms for selected materials
| SBET (m2g−1) | Pore volume (cm3g−1) | Pore size (nm) | |
|---|---|---|---|
| MSN-NH2 | 1476 | 0.79 | 2.14 |
| MSN-DNA | 1062 | 0.52 | 1.95 |
| MSN-RhB | 294 | 0.18 | – |
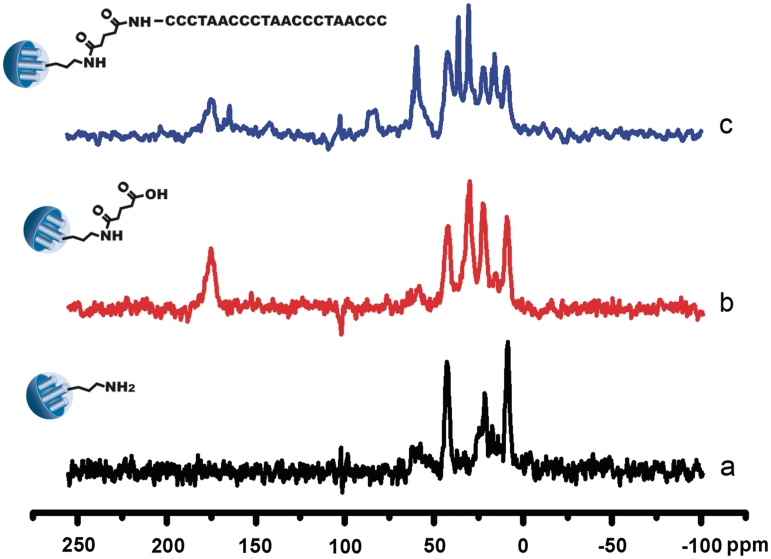
The surface functionalization of MSN was monitored by FTIR spectroscopy (Figure 2B). The emerging absorption band at around 1700 cm−1 in the sample MSN-COOH can be assigned to C=O stretching of the carboxyl groups contained within the attached succinic acid molecules. The efficient grafting of DNA onto mesoporous silica was validated by the appearance of an enhanced band at 1562 cm−1, which is characteristic of acylamide vibration. 13C CP-MAS NMR spectroscopy provides clear evidence for the successful incorporation of the functional groups (Figure 3). The MSN-NH2 nanoparticles had three peaks at 8.7 ppm (C1), 23.9 ppm (C2) and 43.0 ppm (C3). The resonances at 173.0 ppm and 175.4 ppm were typical positions for C=O of the acylamide and carboxyl groups. The grafting of DNA was also confirmed by the declined intensity of the resonance at 175.0 ppm and band shift to 164.9 ppm for C=O of the acylamide. The observed results agreed well with previous studies and were easily assigned (24,25). As shown in Figure 2A and Table 1, changes in pore volume and diameter were investigated by nitrogen sorption experiments. The decline in surface area and pore volume by ∼30% in the sample MSN-DNA is attributed to partial pore blocking effect induced by the large organic moieties on the outer shell of the MSN. On the other hand, practically similar pore sizes have been determined with value of 2.14 and 1.95 nm for samples MSN-NH2 and MSN-DNA, respectively. This result indicated that the internal surface of the mesoporous particles remained unfunctionalized and the residual porosity remaining in MSN-DNA had little effect on the dye loading. Quantification of the content of DNA anchored on MSN-DNA was accomplished by thermogravimetric studies (TGA) and UV-vis spectroscopy (Supplementary Figure S3).
Figure 3.
Solid state 13C CP-MAS NMR spectra of (a) MSN-NH2, (b) MSN-COOH and (c) MSN-DNA.
On-demand dye release
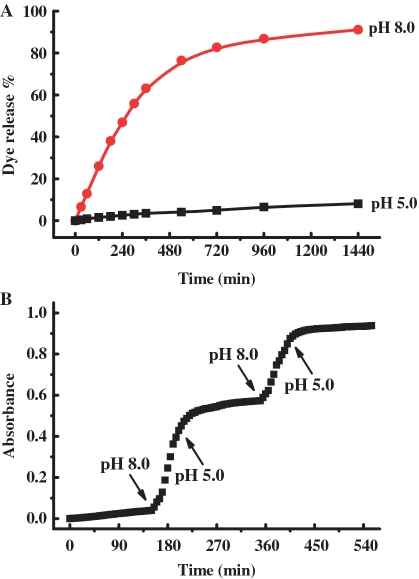
To investigate the proton-fueled gating behavior of the MSN-DNA system, rhodamine B was first loaded as guest molecule by soaking MSN-DNA in a phosphate-buffered saline (PBS) solution (pH 7.4). The pore was capped by i-motif quadruplex when the pH value of the solution was adjusted to five. The excess amount of dye was removed by centrifugation and repeated washing with PBS (pH 5.0). The resulting particles (denoted as MSN-RhB) were then dispersed the citrate buffer (25 ml, 25 mM, pH 5.0 or pH 8.0) to test their controlled release property. As can be seen in Figure 4A, a very clear and highly effective pH-operable gating effect was demonstrated by monitoring the absorbance maximum of rhodamine B (553 nm) as a function of time. When the pH value is adjusted to 8.0, 91% release is obtained after 24 h. However, only negligible release occurs at pH 5.0 under the same condition, indicating good capping efficiency and tunable release rate via pH change. The pH-dependent release rate is consistent with the mechanism of operation for the MSN system: release of guest molecules depends on the reversible conformational change between i-motif quadruplex and random coil DNA. Since the diameter of i-motif structure is 1.9 nm (26,27), we speculated that the gate-like structure formed at the closed state (pH 5.0) might be expected to be large enough to gate the ∼2-nm diameter pore and thus prevent rhodamine B molecules from escaping. Indeed, we note that the structural change of DNA could be extremely complicated on the surface. For example, a DNA strand may fold into more than one i-motif structure, while the i-motif could form by association of two or more single strands. Further studies will be required to elucidate the exact mechanisms involved, but these are beyond the scope of this brief communication. In contrast, when the folded four-stranded i-motif domain was denatured into single-stranded form with cross-sectional diameter of 0.6 nm at pH 8.0, the packing of ssDNA displayed poor coverage of the pore and consequently leaded to leakage of entrapped dye molecules. In comparison, the results of unfunctionalized MCM-41 and MSN-cDNA showed remarkable dye liberation at both pH 5.0 and 8.0 (Supplementary Figures S4–S6). These data clearly demonstrated that we were able to close the pore system of MSN with the i-motif quadruplex DNA, and to release the loaded molecules subsequently by unfolding the quadruplex DNA in smart response to pH stimulus. Compared to the fast capping/uncapping response on the basis of the conformational change of i-motif DNA, the relatively slow liberation processes observed is ascribed to the diffusion-controlled kinetics of dye release.
Figure 4.
(A) Release profiles of rhodamine B dye from MSN-RhB at pH 5.0 and 8.0. (B) Partial guest release profile of rhodamine B dye from MSN-RhB as function of pH variations.
One distinctive advantage of this system is that the reversible structural feature of i-motif DNA opened up new possibilities for a more sophisticated on-demand cargo delivery. As a proof of concept, a partial cargo release of trapped guest could be regulated with open-close cycles via alternating addition of H+ and OH−. As demonstrated in Figure 4B, the closed state at starting pH 5.0 strongly constrained the delivery of the cargo. On the contrary, a distinct release of the entrapped rhodamine dye was triggered in the open state as a result of the conformational change of i-motif DNA when the pH was suddenly changed to 8.0 by addition of OH− (see the arrow in Figure 4B). After 50 min the release of the entrapped dye was again restricted by lowering the pH to 5.0. This inhibition is somehow slower than the process when the delivery is triggered. At time 350 min, pH 8 was set back and further delivery of the entrapped dye occurred until pH 5.0 was again selected, inhibiting the release of the dye. The decrease of dye release in each open-close sequence was due to the reduced amount of dye to be delivered from the pores to the solution in each cycle. The results demonstrated that the interconversion protocol was reversible and that the delivery of the cargo in small portions could be operated at will owning to the high degree of cooperation between individual DNA molecules.
pH-switchable nanoreactor for controllable mass transport
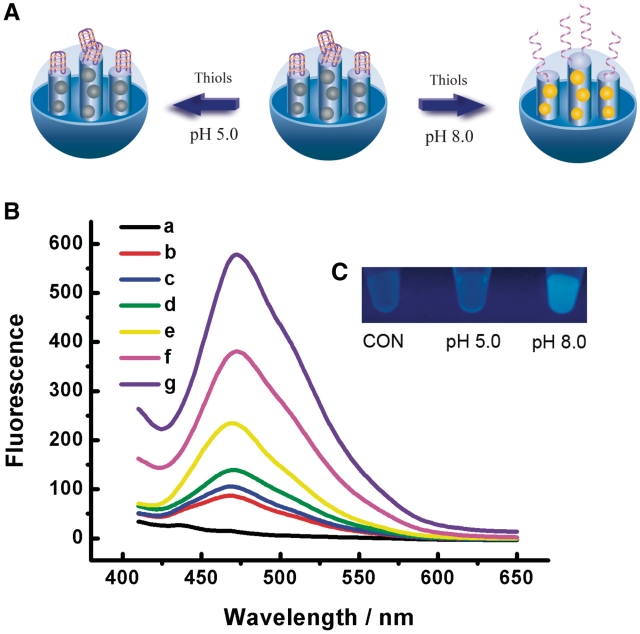
To realize the potential application of our system as pH-switchable nanoreactor with on-demand sensor-effector functionality, we took advantage of the large load capacity of mesoporous nanoparticles as the supporting matrix to incorporate hydrophobic molecules for chemical reactions in aqueous solution. A water-insoluble non-fluorescent probe 7-Diethylamino-3-(4-maleimidophenyl)-4-methylcoumarin (CPM) was first loaded in the DNA-capped mesoporous nanocontainer, and showed negligible release in solution once it was encapsulated owing to its poor solubibity. Only low molecular weight thiols were expected to diffuse into the pores due to the size-sieving ability of the mesoporous silica framework, and then react with CPM to give rise to highly fluorescent products as depicted in Figure 5A and Supplementary Figure S7. The nanoparticle showed significant blue luminescence at neutral conditions, while only faint fluorescence was found at acidic pH. Meanwhile, it is evident that the fluorescent signal intensity increased with enhanced pH value, which suggests that the gated system was able to change its state of activity by pH variation of the surrounding solution (Figure 5B and C). It was also found that thiol derivative of CPM, CPM-loaded MSN-cDNA and nanocontainers without DNA capping showed ignorable difference at acidic and basic conditions (Supplementary Figures S8–S10), indicating that the behavior of the nanoreactor depends on the conformational change of i-motif DNA on the mesoporous silica surface. Besides the application as nanoscale reactor, the specific recognition of CPM with thiol and unique pH-dependant conformational change of i-motif make it possible to immobilize the CPM-loaded nanocontainer on the solid support as a chip for sensing thiol or pH.
Figure 5.
(A) Illustration of the pH-switchable nanoreactor. (B) Fluorescence response of the nanoreactor activity in acidic and neutral environment: (a) MSN-DNA, (b) MSN-CPM before the addition of thiol, (c–g) MSN-CPM after the reaction of thiol from pH 5.0 to 7.0 with increments of 0.5 pH units. (C) Fluorescence images of the nanoreactors in the absence and presence of thiol. Only weak fluorescence could be observed with the addition of thiol at pH 5.0, whereas the container showed strong blue fluorescence at pH 8.0.
CONCLUSION
In summary, we have demonstrated the first example of a molecular gate-like functional system consisting of MSN material functionalized onto pore outlets with nucleic acids as caps. Starting from simple quadruplex/single strand conformation changes, the i-motif DNA cap can open and close the pore system in smart response to pH stimulus: the release of the cargo is inhibited at pH 5, whereas there is significant release of the guest molecule from the MPN at pH 8. Importantly, a partial cargo delivery can be easily controlled due to the reversible opening/closing protocol and the rapid structural switch of i-motif DNA, which opens the possibility of designing stimuli-induced pulsatile release supports. The operability of a pH-controlled nanoreactor for on-demand molecular transport makes the switchable nanocontainer promising for an easily producible, self-regulating, target-specific and stable carrier which can combine sensing and effector functionality on the nanoscale. Furthermore, the utility of nucleic acids in this system provides several critical advantages such as high biocompatibility, ease of synthesis and modification, straightforward operation and functional versatility. In contrast to the previous systems whose switching relies upon intermolecular interaction, the DNA-based nanocontainers can switch reversibly without the aid of other compounds, and thus make our system simple in design and facile in operation. This proof of concept might open the door to a new generation of carrier materials and could also provide a general route to use other functional nucleic acids/peptide nucleic acids as capping agents in the fields of versatile controlled delivery nanodevices.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
FUNDING
The National Natural Science Foundation of China (20831003, 90813001, 20833006 and 90913007); Chinese Academy of Sciences (CAS). Funding for open access charge: National Natural Science Foundation of China and Chinese Academy of Sciences.
Conflict of interest statement. None declared.
REFRENCES
- 1.Alberti P, Mergny JL. DNA duplex-quadruplex exchange as the basis for a nanomolecular machine. Proc. Natl Acad. Sci. USA. 2003;100:1569–1573. doi: 10.1073/pnas.0335459100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weizmann Y, Cheglakov Z, Pavlov V, Willner I. Autonomous fueled mechanical replication of nucleic acid templates for the amplified optical detection of DNA. Angew. Chem. Int. Ed. 2006;45:2238–2242. doi: 10.1002/anie.200503810. [DOI] [PubMed] [Google Scholar]
- 3.Simmel FC. Towards biomedical applications for nucleic acid nanodevices. Nanomedicine. 2007;2:817–830. doi: 10.2217/17435889.2.6.817. [DOI] [PubMed] [Google Scholar]
- 4.Miyoshi D, Karimata H, Wang Z, Koumoto K, Sugimoto N. Artificial G-wire switch with 2,2′-bipyridine units responsive to divalent metal ions. J. Am. Chem. Soc. 2007;129:5919–5925. doi: 10.1021/ja068707u. [DOI] [PubMed] [Google Scholar]
- 5.Sharma J, Chhabra R, Cheng A, Brownell J, Liu Y, Yan H. Control of self-assembly of DNA tubules through integration of gold nanoparticles. Science. 2009;323:112–116. doi: 10.1126/science.1165831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Descalzo AB, Martinez-Manez R, Sancenon R, Hoffmann K, Rurack K. The supramolecular chemistry of organic-inorganic hybrid materials. Angew. Chem. Int. Ed. 2006;45:5924–5948. doi: 10.1002/anie.200600734. [DOI] [PubMed] [Google Scholar]
- 7.Trewyn BG, Slowing, Giri S, Chen HT, Lin VSY. Synthesis and functionalization of a mesoporous silica nanoparticle based on the sol-gel process and applications in controlled release. Acc. Chem. Res. 2007;40:846–853. doi: 10.1021/ar600032u. [DOI] [PubMed] [Google Scholar]
- 8.Saha S, Leung KCF, Nguyen TD, Stoddart JF, Zink JI. Nanovalves. Adv. Funct. Mater. 2007;17:685–693. [Google Scholar]
- 9.Climent E, Marcos MD, Martinez-Manez R, Sancenon F, Soto J, Rurack K, Amoros P. The determination of methylmercury in real samples using organically capped mesoporous inorganic materials capable of signal amplification. Angew. Chem. Int. Ed. 2009;48:8519–8522. doi: 10.1002/anie.200904243. [DOI] [PubMed] [Google Scholar]
- 10.Lai CY, Trewyn BG, Jeftinija DM, Jeftinija K, Xu S, Jeftinija S, Lin VSY. A mesoporous silica nanosphere-based carrier system with chemically removable CdS nanoparticle caps for stimuli-responsive controlled release of neurotransmitters and drug molecules. J. Am. Chem. Soc. 2003;125:4451–4459. doi: 10.1021/ja028650l. [DOI] [PubMed] [Google Scholar]
- 11.Torney F, Trewyn BG, Lin VSY, Wang K. Mesoporous silica nanoparticles deliver DNA and chemicals into plants. Nat. Nanotechnol. 2007;2:295–300. doi: 10.1038/nnano.2007.108. [DOI] [PubMed] [Google Scholar]
- 12.Vivero-Escoto JL, Slowing, Wu CW, Lin VSY. Photoinduced intracellular controlled release drug delivery in human cells by gold-capped mesoporous silica nanosphere. J. Am. Chem. Soc. 2009;131:3462–3463. doi: 10.1021/ja900025f. [DOI] [PubMed] [Google Scholar]
- 13.Nguyen TD, Leung KCF, Liong M, Liu Y, Stoddart JF, Zink JI. Versatile supramolecular nanovalves reconfigured for light activation. Adv. Funct. Mater. 2007;17:2101–2110. [Google Scholar]
- 14.Hernandez R, Tseng HR, Wong JW, Stoddart JF, Zink JI. An operational supramolecular nanovalve. J. Am. Chem. Soc. 2004;126:3370–3371. doi: 10.1021/ja039424u. [DOI] [PubMed] [Google Scholar]
- 15.Radu DR, Lai CY, Wiench JW, Pruski M, Lin VSY. Gatekeeping layer effect: a poly(lactic acid)-coated mesoporous silica nanosphere-based fluorescence probe for detection of amino-containing neurotransmitters. J. Am. Chem. Soc. 2004;126:1640–1641. doi: 10.1021/ja038222v. [DOI] [PubMed] [Google Scholar]
- 16.Liu R, Zhao X, Wu T, Feng PY. Tunable redox-responsive hybrid nanogated ensembles. J. Am. Chem. Soc. 2008;130:14418–14419. doi: 10.1021/ja8060886. [DOI] [PubMed] [Google Scholar]
- 17.Thornton PD, Heise A. Highly specific dual enzyme-mediated payload release from peptide-coated silica particles. J. Am. Chem. Soc. 2010;132:2024–2028. doi: 10.1021/ja9094439. [DOI] [PubMed] [Google Scholar]
- 18.Schlossbauer A, Kecht J, Bein T. Biotin-avidin as a protease-responsive cap system for controlled guest release from colloidal mesoporous silica. Angew. Chem. Int. Ed. 2009;48:3092–3095. doi: 10.1002/anie.200805818. [DOI] [PubMed] [Google Scholar]
- 19.Climent E, Bernardos A, Martinez-Manez R, Maquieira A, Marcos MD, Pastor-Navarro N, Puchades R, Sancenon F, Soto J, Amoros P. Controlled delivery systems using antibody-capped mesoporous nanocontainers. J. Am. Chem. Soc. 2009;131:14075–14080. doi: 10.1021/ja904456d. [DOI] [PubMed] [Google Scholar]
- 20.Gueron M, Leroy JL. The i-motif in nucleic acids. Curr. Opin. Struct. Biol. 2000;10:326–331. doi: 10.1016/s0959-440x(00)00091-9. [DOI] [PubMed] [Google Scholar]
- 21.Shu WM, Liu DS, Watari M, Riener CK, Strunz T, Welland ME, Balasubramanian S, McKendry RA. DNA molecular motor driven micromechanical cantilever arrays. J. Am. Chem. Soc. 2005;127:17054–17060. doi: 10.1021/ja0554514. [DOI] [PubMed] [Google Scholar]
- 22.Chen C, Song GT, Ren JS, Qu XG. A simple and sensitive colorimetric pH meter based on DNA conformational switch and gold nanoparticle aggregation. Chem. Commun. 2008:6149–6151. doi: 10.1039/b812660g. [DOI] [PubMed] [Google Scholar]
- 23.Grun M, Lauer I, Unger KK. The synthesis of micrometer- and submicrometer-size spheres of ordered mesoporous oxide MCM-41. Adv. Mater. 1997;9:254–257. [Google Scholar]
- 24.Huh S, Wiench JW, Yoo JC, Pruski M, Lin VSY. Organic functionalization and morphology control of mesoporous silicas via a co-condensation synthesis method. Chem. Mater. 2003;15:4247–4256. [Google Scholar]
- 25.Liu R, Zhang Y, Zhao X, Agarwal A, Mueller LJ, Feng PY. pH-responsive nanogated ensemble based on gold-capped mesoporous silica through an acid-labile acetal linker. J. Am. Chem. Soc. 2010;132:1500–1501. doi: 10.1021/ja907838s. [DOI] [PubMed] [Google Scholar]
- 26.Phan AT, Gueron M, Leroy JL. The solution structure and internal motions of a fragment of the cytidine-rich strand of the human telomere. J. Mol. Biol. 2000;299:123–144. doi: 10.1006/jmbi.2000.3613. [DOI] [PubMed] [Google Scholar]
- 27.Kang CH, Berger I, Lockshin C, Ratliff R, Moyzis R, Rich A. Crystal structure of intercalated four-stranded d(C3T) at 1.4 A resolution Proc. Natl Acad. Sci. USA. 1994;91:11636–11640. doi: 10.1073/pnas.91.24.11636. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.