Abstract
The target of rapamycin (TOR) kinase is an evolutionarily conserved key regulator of eukaryotic cell growth and proliferation. Recently, it has been reported that inhibition of TOR signaling pathway can delay aging and extend lifespan in several eukaryotic organisms, but how lifespan extension is mediated by inhibition of TOR signaling is poorly understood. Here we report that rapamycin treatment and nitrogen starvation, both of which cause inactivation of TOR complex 1 (TORC1), lead to enhanced association of Sir2 with ribosomal DNA (rDNA) in Saccharomyces cerevisiae. TORC1 inhibition increases transcriptional silencing of RNA polymerase II-transcribed gene integrated at the rDNA locus and reduces homologous recombination between rDNA repeats that causes formation of toxic extrachromosomal rDNA circles. In addition, TORC1 inhibition induces deacetylation of histones at rDNA. We also found that Pnc1 and Net1 are required for enhancement of association of Sir2 with rDNA under TORC1 inhibition. Taken together, our findings suggest that inhibition of TORC1 signaling stabilizes the rDNA locus by enhancing association of Sir2 with rDNA, thereby leading to extension of replicative lifespan in S. cerevisiae.
INTRODUCTION
In eukaryotic cells, ribosomal DNA (rDNA) forms the basis of nucleolus where synthesis and processing of ribosomal RNA (rRNA) and ribosome assembly occur. In the budding yeast Saccharomyces cerevisiae, 100–200 copies of a 9.1-kb rDNA repeat exist as a tandem array on chromosome XII (1). Each rDNA repeat consists of the RNA polymerase (Pol) I-transcribed 35S rRNA gene and the non-transcribed spacer (NTS), which is divided by the Pol III-transcribed 5S rRNA gene into NTS1 and NTS2 (Figure 1B). Cells control the protein biosynthetic capacity by regulating transcription of rRNA genes and ribosomal protein genes and altering ribosome biosynthesis in response to nutrient availability (2–4). rRNA transcription is a critical initial step for ribosome biosynthesis and represents ∼60% of total transcription in rapidly growing yeast cells.
Figure 1.
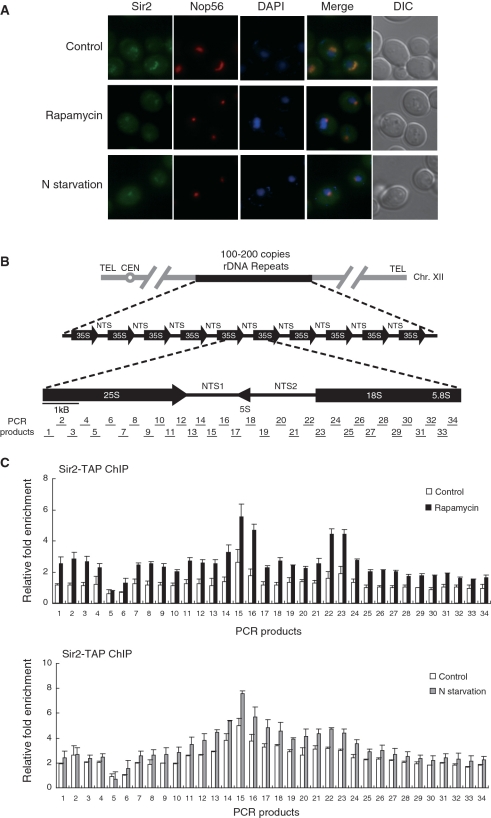
TORC1 inhibition leads to condensation of the nucleolus and enhancement of association of Sir2 with rDNA. (A) Rapamycin treatment and nitrogen starvation cause reduction of the nucleolar size and condensation of the Sir2-GFP signal. Subcellular localization of Sir2-GFP (green) and Nop56-RFP (red) was analyzed after treatment with 200 ng/ml rapamycin or incubation in nitrogen-depleted medium for 1 h. DAPI staining for visualization of the nucleus (blue) and differential interference contrast (DIC) images are also shown. (B) The structure of the tandemly repeating rDNA of S. cerevisiae is shown above, and a single 9.1-kb rDNA unit is shown expanded below. PCR products analyzed in the ChIP assays are indicated below the rDNA unit. (C) Rapamycin treatment and nitrogen starvation enhance association of Sir2 with rDNA. The degree of Sir2 binding to rDNA was measured using the ChIP assay after treatment with 200 ng/ml rapamycin (upper panel) or incubation in nitrogen-depleted medium for 1 h (lower panel). For control, cells were treated with DMSO only (upper panel) or grown in SC medium (lower panel). Relative fold enrichment refers to the relative ratio of PCR products amplified from immunoprecipitated DNA to products from whole-cell extract DNA. Values represent the average of three independent experiments and error bars indicate standard deviations.
Because of the highly repetitive nature, rDNA array is intrinsically unstable and an easy target for homologous recombination. A primary cause of aging in S. cerevisiae is known to be homologous recombination between rDNA repeats, which leads to formation of extrachromosomal rDNA circles that accumulate to toxic levels in mother cells (5). Under normal conditions, however, rDNA repeats remain relatively stable because rDNA recombination is negatively regulated through a mechanism referred to as rDNA silencing. In S. cerevisiae, Sir2 suppresses Pol II-dependent transcription at the rDNA locus (6). It has been reported that Sir2 can forestall aging by stabilizing the rDNA locus (5,7,8). Sir2 is an NAD+-dependent histone deacetylase and its activity is necessary for spreading of silencing complexes along the N-terminal tails of histones (9–11). Sir2 has been identified as a subunit of an rDNA silencing complex called RENT (regulator of nucleolar silencing and telophase exit) (12). Net1, another subunit of the RENT complex, recruits Sir2 to rDNA and is also required for rDNA silencing (13). Fob1 physically interacts with Net1 and Sir2 and has been shown to be required in rDNA silencing by recruiting Net1 and Sir2 to the NTS1 (6). It has also been shown that Net1 and Sir2 are associated with two regions of rDNA, the NTS1 and the NTS2/Pol I promoter (6). The NTS1 contains replication fork block and cis-element required for Fob1-dependent rDNA recombination (14–17) and the NTS2 overlaps with the Pol I transcription initiation region. In cells lacking Fob1, rDNA silencing is abolished specifically at the NTS1 (6).
The target of rapamycin (TOR) is a nutrient-responsive phosphatidylinositol kinase-related kinase highly conserved in all eukaryotes (18,19) and a key regulator of ribosome biogenesis in response to nutrient conditions (20–22). In S. cerevisiae, TOR kinases are encoded by TOR1 and TOR2, and they exist in two functionally distinct multi-protein complexes, TOR complex 1 (TORC1) and TOR complex 2 (TORC2) (23). An immunosuppressive and anticancer drug rapamycin specifically inhibits TORC1 and leads to a rapid decrease in the expression of ribosomal components, including 35S rRNA (by Pol I), ribosomal proteins (by Pol II) and 5S rRNA (by Pol III) (24–26). It has been reported that inhibition of TORC1 signaling by rapamycin can delay aging and extend lifespan in S. cerevisiae, Caenorhabditis elegans and Drosophila melanogaster (27–30). A recent study has shown that rapamycin can also extend lifespan in genetically heterogeneous mice (31).
The mechanism by which inhibition of TORC1 signaling delays aging and extends lifespan is poorly understood, but it has been proposed that it may involve alteration of ribosome assembly and translation (28,29,32,33). Whether Sir2 plays a role in rDNA stability and lifespan extension in S. cerevisiae under inhibition of TORC1 signaling is somewhat controversial; deletion of TOR1 has been shown to significantly increase lifespan but have no effect on the Sir2 activity (29), whereas a recent study has reported that TORC1 inhibition by rapamycin increases the Sir2 activity and stabilizes the rDNA locus (30).
To gain further insight into the relationship between TORC1 signaling and rDNA stability, we analyzed association of Sir2 with rDNA at high resolution under rapamycin treatment and nitrogen starvation. Here we report that association of Sir2 with rDNA increases under rapamycin treatment and nitrogen starvation, and inhibition of TORC1 signaling promotes transcriptional silencing of Pol II-transcribed gene at the rDNA locus and reduces homologous recombination between rDNA repeats. We also show that TORC1 inhibition induces deacetylation of histones at rDNA, and Pnc1 and Net1 are required for enhancement of association of Sir2 with rDNA under TORC1 inhibition. Our results propose a model in which TORC1 inhibition stabilizes the rDNA locus by enhancing Sir2 binding to rDNA and thus extends lifespan in yeast.
MATERIALS AND METHODS
Yeast strains, media and reagents
Yeast strains used in this study are listed in Supplementary Table S1. Yeast strains were genetically manipulated according to the one-step PCR-mediated gene targeting procedure as previously described (34). Strains for analyzing mURA3 silencing and loss of ADE2 in the rDNA locus have been described previously (35). Yeast transformation was performed using the lithium acetate method (36), and proper integration was confirmed by PCR. For overexpression of PNC1 and SIR2, the coding sequences for PNC1-GFP and SIR2-TAP were amplified by PCR and cloned into the XhoI and HindIII sites of the p416GPD vector (37). Rich medium (yeast extract, peptone, glucose; YPD) and synthetic complete (SC) medium lacking appropriate amino acids for selection were prepared as described earlier (38). Rapamycin (Tecoland) was stored in DMSO at a concentration of 2 mg/ml and used at a final concentration of 200 ng/ml. For rapamycin treatment, exponentially growing cells (OD600 = 1.0) in SC medium were treated with 200 ng/ml rapamycin for 1 h. For nitrogen starvation, cells grown to log phase (OD600 = 1.0) in SC medium were washed with distilled water and incubated in nitrogen-depleted medium (0.17% yeast nitrogen base without amino acids and ammonium sulfate, 2% glucose) for 1 h. For nicotinamide (Sigma) treatment, cells grown to log phase (OD600 = 1.0) in SC medium were treated with 5 mM nicotinamide and incubated for 1 h. All cultures were incubated at 30°C.
Fluorescence microscopy
After rapamycin treatment or nitrogen starvation, fluorescence microscopic analysis was performed on a Zeiss Axiovert 200 M inverted microscope as described earlier (39). Nuclei in live cells were stained with 4′,6-diamidino-2-phenylindole (DAPI).
Chromatin immunoprecipitation and quantitative real-time PCR analysis
Chromatin immunoprecipitation ChIP analysis was performed as previously described (6) with some modifications. After rapamycin treatment, nitrogen starvation or nicotinamide treatment, yeast cultures were crosslinked with 1% formaldehyde for 15 min and quenched with glycine at a final concentration of 125 mM for 5 min at room temperature. Cells were washed twice with cold phosphate-buffered saline. Cell pellets were resuspended in 500 µl of lysis buffer (50 mM HEPES–NaOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, 1 μg/ml leupeptin and 1 μg/ml pepstatin) and bead-beated with 0.5 mm glass beads ten times for 1 min at 4°C. Samples were incubated on ice for 2 min between bead beatings. Cell lysates were sonicated five times for 15 s with amplitude set at 20% and centrifuged twice at 16 000g for 10 min. For TAP ChIP experiments, 10 µl of 50% slurry of pre-washed IgG–agarose beads (Amersham Biosciences) was incubated with 200 µl of lysate at 4°C for 3 h. For acetylated histone H3 ChIP experiments, 200 µl of lysate was incubated at 4°C overnight with anti-acetyl Lys9/18 histone H3 (07-593; Upstate Biotechnology) and then incubated with Protein A-agarose beads (Sigma) at 4°C for 3 h. Beads were washed twice in lysis buffer, once with washing buffer I (50 mM HEPES–NaOH, pH 7.5, 500 mM NaCl, 1 mM EDTA, 1% Triton X-100 and 0.1% sodium deoxycholate), once with washing buffer II (10 mM Tris–HCl, pH 8.0, 0.25 M LiCl, 0.5% NP-40, 0.5% sodium deoxycholate and 1 mM EDTA) and once with TE buffer at room temperature. Beads were eluted by adding 100 µl of elution buffer (50 mM Tris–HCl, pH 8.0, 10 mM EDTA and 1% SDS) and incubating at 65°C for 15 min. Eluate was transferred to a fresh tube and pooled with 150 µl of TE containing 0.67% SDS. For input DNA, 200 µl of elution buffer was added to 50 µl of lysate. Input and ChIP samples were incubated at 65°C overnight. All samples were combined with 250 µl of TE, 5 µg of glycogen and 100 µg of proteinase K, and incubated at 37°C for 2 h. After addition of 55 µl of 4 M LiCl, samples were extracted once with phenol:chloroform:isoamyl alcohol and once with chloroform. Precipitated and washed DNA was resuspended in 100 µl of TE with 10 µg of RNase A and incubated at 37°C for 1 h. ChIP samples were analyzed by quantitative real-time PCR using SYBR Green. Primers used for PCR were the same as previously described (6). Relative fold enrichment was determined by calculating the ratio of rDNA to CUP1, an internal control, as follows: [rDNA(IP)/CUP1(IP)]/[rDNA(input)/CUP1(input)]. Each set of experiments was performed at least three times.
Western blot analysis
Whole-cell extracts were run on SDS–polyacrylamide gel electrophoresis. Western blot analysis was performed by standard methods using HRP-conjugated anti-mouse IgG antibody (A9044; Sigma) for detection of TAP-tagged proteins. Hexokinase was used as a loading control and detected by anti-hexokinase antibody (H2035-02; United States Biological).
Immunoprecipitation reactions
Reactions were performed as described earlier (13). For immunoprecipitation of Sir2-Myc, mouse anti-Myc antibody (sc-40; Santa Cruz Biotechnology) was added to the cell extract and incubated at 4°C for 2 h. Protein A-agarose (Sigma) was then added to the reaction mixture and incubated at 4°C for 1 h. The immune complexes were collected and loaded on SDS–polyacrylamide gels for western blot analysis. Proteins were detected using HRP-conjugated anti-HA antibody (sc-7392; Santa Cruz Biotechnology) and rabbit anti-Myc antibody (06-549; Upstate Biotechnology). Hexokinase was used as a loading control and detected by anti-hexokinase antibody (H2035-02; United States Biological).
Quantification of mURA3 mRNA
Total RNA was isolated from yeast cells using the RNeasy MiniKit (Qiagen). cDNA for reverse transcription–PCR was generated using the ProtoScript First Strand cDNA Synthesis Kit (New England Biolabs). The mURA3 silencing reporter gene, which contains the TRP1 promoter instead of the URA3 promoter, has been described previously (40). The amount of mURA3 and ACT1 mRNA was analyzed by quantitative real-time reverse transcription–PCR using the Applied Biosystems 7300 Real-Time PCR system. Amplification efficiencies were validated and normalized against ACT1, and fold increases were calculated using the 2−ΔΔCT method (41). Primers used for amplification of mURA3 were 5′-CTGTTGACATTGCGAAGAGC-3′ and 5′-TCTCCCTTGTCATCTAAACC-3′, and those for ACT1 were 5′-TGACTGACTACTTGATGAAG-3′ and 5′-TGCATTTCTTGTTCGAAGTC-3′. All reactions were carried out in triplicate.
rDNA recombination assay
rDNA recombination rates were determined by measuring the frequency of loss of ADE2 integrated at the rDNA locus of strain DMY3010 as previously described (7). Exponentially growing cells (OD600 = 1.0) in SC medium were treated with or without 200 ng/ml rapamycin for 1 h, sonicated briefly to prevent aggregation and spread on SC plates containing 5 mM nicotinamide or not. Colonies were allowed to grow for 2 days at 30°C and then placed at 4°C for 2 days to enhance color development. rDNA recombination rate was calculated by dividing the number of half-red/half-white colonies by the total number of colonies. Entirely red colonies were excluded from all calculations. More than 20 000 colonies were examined for each assay.
Analysis of replicative lifespan
Analysis of replicative lifespan was carried out by micromanipulation as previously described (42), using a Zeiss Tetrad Microscope. Lifespan analysis was performed on YPD plates with or without 10 ng/ml rapamycin or 5 mM nicotinamide. Cells that never budded were excluded from calculation. For statistical analysis, lifespan data sets were compared by a two-tailed Wilcoxon Rank-Sum test. Lifespan was determined for 40 cells in each experiment.
RESULTS
Rapamycin treatment and nitrogen starvation cause condensation of the nucleolus and enhance association of Sir2 with rDNA
The yeast nucleolus normally appears as a crescent shape localized at the periphery of the nucleus and is a dynamic structure that can be altered by environmental condition or stress. When TORC1 signaling is inhibited, nucleolar size is rapidly reduced and rDNA undergoes condensation (43). To examine the subcellular localization of rDNA silencing factor Sir2 under TORC1 inhibition, we performed fluorescence microscopic analysis on cells in which the endogenous SIR2 gene was modified to produce C-terminal GFP fusion protein. For comparison, we also analyzed the localization of Nop56-RFP, a well-characterized nucleolar marker (44). Similar to Nop56-RFP, Sir2-GFP formed a crescent-shaped structure and did not colocalize with the DNA-specific dye DAPI, which is typically excluded from the nucleolus during normal growth (Figure 1A). However, rapamycin treatment and nitrogen starvation, conditions inhibiting TORC1 signaling, caused reduction of the nucleolar size and condensation of the fluorescent signal of Sir2-GFP. This morphological change of the nucleolus under TORC1 inhibition has been reported to be accompanied by release of Pol I components from the nucleolus (43). Our results indicate that, unlike Pol I components, Sir2 remains stably associated with rDNA under TORC1 inhibition.
To gain insight into the relationship between rDNA silencing factor Sir2 and TORC1 signaling, we analyzed the binding of Sir2 to rDNA under rapamycin treatment or nitrogen starvation. To obtain a high-resolution map for this binding, we performed the ChIP experiment followed by quantitative real-time PCR analysis using a panel of 68 primers, which were designed to amplify fragments of ∼0.25 kb in length that span an entire 9.1-kb rDNA unit (6). The structure of an rDNA repeat unit and the PCR products analyzed in the ChIP assay are shown in Figure 1B. We constructed yeast strains in which the endogenous SIR2 gene was modified to express C-terminal TAP fusion protein. After rapamycin treatment or nitrogen starvation, Sir2-TAP was immunoprecipitated using IgG-agarose beads from extracts containing sheared chromatin. Whole-cell extract chromatin and immunoprecipitated chromatin were used as template DNA for quantitative real-time PCR. Consistent with the previous report (6), Sir2 bound highly to the NTS1 and NTS2/18S regions (Figure 1C). Remarkably, association of Sir2 with rDNA increased over the entire rDNA locus under rapamycin treatment and nitrogen starvation compared to normal condition. This result indicates that TORC1 inhibition by rapamycin treatment or nitrogen starvation enhances association of Sir2 with rDNA array.
To test a possibility that enhanced association of Sir2 with rDNA might be due to increased expression of Sir2 under TORC1 inhibition, we examined the expression level of Sir2 under rapamycin treatment or nitrogen starvation. Western blot analysis of whole-cell extracts showed that the protein level of Sir2 did not increase under rapamycin treatment or nitrogen starvation (data not shown). This result indicates that the enrichment of Sir2 at the rDNA region under TORC1 inhibition is not due to increased protein level of Sir2. Therefore, we conclude that inhibition of TORC1 signaling promotes interaction between Sir2 and rDNA.
Inhibition of TORC1 signaling increases rDNA silencing in a Sir2-dependent manner
Sir2 is a well-known rDNA silencing factor that suppresses Pol II-dependent transcription at the rDNA locus in S. cerevisiae (6). Enhanced association of Sir2 with rDNA under rapamycin treatment or nitrogen starvation raised a possibility that inhibition of TORC1 signaling might increase rDNA silencing. To check this possibility, we performed a spot assay using yeast strains carrying the mURA3 silencing reporter gene inserted either inside the NTS1 region of the rDNA locus (RDN1-NTS1::mURA3) or outside the rDNA array (leu2::mURA3) (35). Cells were 10-fold serially diluted and spotted on SC medium or SC medium containing 5-fluoroorotic acid (5-FOA), each with or without rapamycin, to monitor silencing of mURA3. Consistent with the previous observations (6,40), the mURA3 reporter gene was efficiently silenced at the NTS1, as indicated by good growth on medium containing 5-FOA (Supplementary Figure S1A, upper panel). In the presence of rapamycin, overall cell growth was severely impaired on SC medium, but cells carrying mURA3 within the NTS1 showed enhanced growth on SC medium containing 5-FOA (Supplementary Figure S1A, lower panel). This observation suggests that TORC1 inhibition by rapamycin treatment increases silencing of mURA3 at rDNA. Additionally, to investigate the effect of TORC1 inhibition on rDNA silencing more directly, we deleted TOR1 in strains carrying the mURA3 silencing reporter gene inserted either inside the NTS1 region of the rDNA locus (RDN1-NTS1::mURA3) or outside the rDNA array (leu2::mURA3). Cells were 10-fold serially diluted and spotted on SC medium as a plating control and on medium without uracil or with 5-FOA to monitor silencing of mURA3. As expected, the reporter gene was efficiently silenced at the NTS1 in wild-type cells, as indicated by poor growth on medium lacking uracil or by good growth on medium containing 5-FOA (Supplementary Figure S1B). In tor1Δ cells, the reporter gene was further silenced at the NTS1, as indicated by poorer growth on medium lacking uracil or by better growth on medium containing 5-FOA compared to wild-type cells. These results indicate that inhibition of TORC1 signaling increases rDNA silencing.
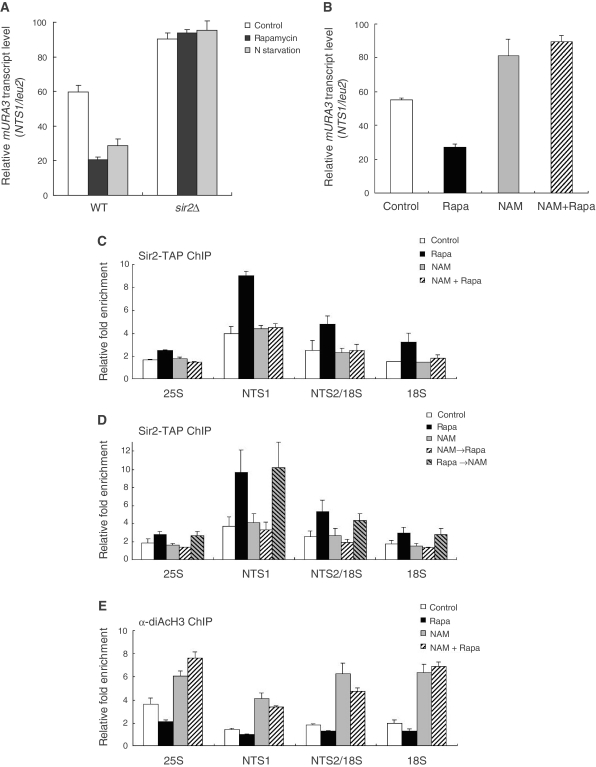
To further examine rDNA silencing under TORC1 inhibition, we analyzed transcription of the mURA3 silencing reporter gene integrated at the rDNA locus. We performed a real-time reverse transcription–PCR analysis to directly measure the transcript levels of the Pol II-transcribed mURA3 gene inserted either inside the NTS1 region of the rDNA locus (RDN1-NTS1::mURA3) or outside the rDNA array (leu2::mURA3). In normally growing wild-type cells, the reporter mURA3 gene was efficiently silenced at the rDNA locus (>40%) compared to outside rDNA (Figure 2A). As expected, silencing of the reporter gene at the rDNA locus was almost completely abolished in normally growing sir2Δ cells. Interestingly, in wild-type cells under rapamycin treatment or nitrogen starvation, silencing of the mURA3 gene at the rDNA locus increased considerably (>70%) compared to outside rDNA. However, the relative transcript levels of mURA3 in sir2Δ cells under rapamycin treatment and nitrogen starvation were similar to those in normally growing sir2Δ cells. These results indicate that inhibition of TORC1 signaling increases rDNA silencing, which is dependent on the presence of Sir2.
Figure 2.
Inhibition of TORC1 signaling enhances rDNA silencing and histone deacetylation at rDNA in a Sir2-dependent manner. (A) Rapamycin treatment and nitrogen starvation promotes transcriptional silencing of the mURA3 reporter gene at the rDNA locus in a Sir2-dependent manner. Total RNA was extracted from wild-type (WT) and sir2Δ cells after treatment with 200 ng/ml rapamycin or incubation in nitrogen-depleted medium for 1 h. Quantitative real-time reverse transcription–PCR analysis was performed to measure the transcript levels of the mURA3 reporter gene inserted either inside the NTS1 region or outside the rDNA array. Amplification efficiencies were validated and normalized against ACT1. Relative mURA3 transcript levels were calculated as the ratio of the normalized transcript level of the mURA3 reporter gene inside the NTS1 region to that outside the rDNA array. (B) Nicotinamide abolishes silencing of the mURA3 reporter gene at the rDNA locus. Total RNA was extracted from wild-type cells after treatment with 200 ng/ml rapamycin (Rapa), 5 mM nicotinamide (NAM), or both (NAM + Rapa) for 1 h. Relative mURA3 transcript levels were calculated as described above. (C) Association of Sir2 with rDNA is not enhanced by rapamycin under nicotinamide treatment. The degree of Sir2 binding to four representative regions in the rDNA (25S, NTS1, NTS2/18S and 18S regions) was measured using the ChIP assay after treatment with 200 ng/ml rapamycin (Rapa), 5 mM nicotinamide (NAM), or both (NAM + Rapa) for 1 h. (D) Nicotinamide does not disturb the already-established association of Sir2 with rDNA. The degree of Sir2 binding to rDNA was measured using the ChIP assay after the following treatments: 200 ng/ml rapamycin for 1 h (Rapa), 5 mM nicotinamide for 1 h (NAM), 5 mM nicotinamide for 1 h and then 200 ng/ml rapamycin for 1 h (NAM→Rapa), 200 ng/ml rapamycin for 1 h and then 5 mM nicotinamide for 1 h (Rapa→NAM). (E) The acetylation level of histone H3 at rDNA is reciprocally proportional to the enzymatic activity of Sir2. The acetylation level of histone H3 associated with the rDNA regions was measured using the ChIP assay after treatment with 200 ng/ml rapamycin (Rapa), 5 mM nicotinamide (NAM), or both (NAM + Rapa) for 1 h. For control, cells were treated with DMSO only. Values represent the average of three independent experiments and error bars indicate standard deviations.
To confirm that increase of rDNA silencing under TORC1 inhibition is mediated by Sir2, we measured the transcript levels of the mURA3 reporter gene under treatment of nicotinamide, a well-known inhibitor of the enzymatic activity of Sir2 (45,46). Consistent with the previous report (45,46), nicotinamide inhibited the histone deacetylase activity of Sir2 in vitro (Supplementary Figure S2A). As expected, nicotinamide treatment considerably abolished silencing of the reporter gene at the rDNA locus (Figure 2B; see also Supplementary Figure S2B), demonstrating that Sir2 plays critical roles in rDNA silencing. Interestingly, when nicotinamide and rapamycin were co-treated to cells, silencing of the reporter gene at the rDNA locus was also abolished, suggesting that the inhibitory effect of nicotinamide on rDNA silencing overrides the stimulatory effect of TORC1 inhibition on rDNA silencing. When the expression level of Sir2 was measured under nicotinamide treatment, no difference was observed whether nicotinamide was treated or not (data not shown), indicating that nicotinamide does not interfere with the expression of Sir2 and, therefore, the effect of nicotinamide on rDNA silencing results from inhibition of the Sir2 activity.
We next analyzed the effect of nicotinamide on association of Sir2 with rDNA. For this analysis, we performed the ChIP experiment for four representative regions in the rDNA locus—25S, NTS1, NTS2/18S and 18S regions (corresponding to the PCR products 3, 15, 23 and 30, respectively; see Figure 1B). Under nicotinamide treatment, the degree of binding of Sir2 to rDNA was essentially unchanged whether rapamycin was co-treated or not (Figure 2C). This result correlates well with the observation that silencing of the mURA3 reporter gene at the NTS1 region of the rDNA locus was relieved by nicotinamide even under rapamycin treatment (Figure 2B). The observation that the degree of Sir2 binding to rDNA was neither decreased nor increased by nicotinamide raised a possibility that, although nicotinamide inhibits the activity of Sir2, it may not affect the already-established association of Sir2 with rDNA. To check this possibility, we assayed whether enhanced association of Sir2 with rDNA by rapamycin would be maintained upon later treatment of nicotinamide. Interestingly, we observed that enhanced association of Sir2 with rDNA by rapamycin was not affected by later treatment of nicotinamide (Figure 2D), indicating that nicotinamide does not disturb the already-established association of Sir2 with rDNA. However, silencing of the mURA3 reporter gene at the NTS1 region of the rDNA locus was still abolished under this condition (Supplementary Figure S2C). When nicotinamide was treated before rapamycin, enhancement of association of Sir2 with rDNA was not observed (Figure 2D), suggesting that, upon inactivation by nicotinamide, rDNA-unbound form of Sir2 does not respond to rapamycin and is no longer recruited to rDNA.
We also measured the acetylation level of histone H3 associated with the rDNA regions. Based on the fact that Sir2 is a histone deacetylase (9,10), we expected that the acetylation level of histone H3 at rDNA would be reciprocally proportional to the enzymatic activity of Sir2. As expected, the acetylation level of H3 was lowest in the NTS1 region where the degree of binding of Sir2 to rDNA was highest whether rapamycin was treated or not (Figure 2E). When cells were treated with nicotinamide, the H3 acetylation level increased significantly in all regions of the rDNA locus, correlating with the previous report that deletion of SIR2 causes an increase in the H3 acetylation level throughout the rDNA regions (6). This effect of nicotinamide on the H3 acetylation level was still observed even under rapamycin treatment. Taken together, these results indicate that Sir2 is crucial for increasing rDNA silencing under inhibition of TORC1 signaling and nicotinamide negatively regulates rDNA silencing.
Inhibition of TORC1 signaling promotes rDNA stability and extends lifespan in yeast
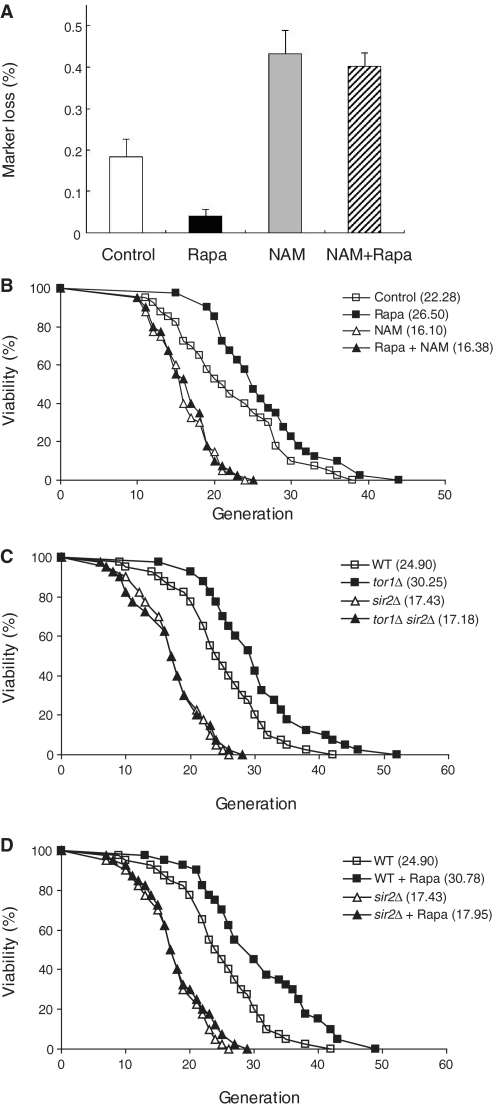
Based on the results that inhibition of TORC1 signaling enhances association of Sir2 with rDNA and increases rDNA silencing, we examined if TORC1 inhibition could reduce rDNA recombination by promoting rDNA stability. To measure the rDNA recombination rate under TORC1 inhibition, the frequency of loss of the ADE2 marker gene integrated at the rDNA locus was monitored under rapamycin treatment. Colonies in which the ADE2 marker has been lost accumulate a red pigment, while colonies that maintain and express ADE2, which is weakly silenced in rDNA, remain white. A marker loss event during the first division after plating results in half-red/half-white colonies. Thus, rDNA recombination rate can be determined by dividing the number of half-red/half-white colonies by the total number of colonies (7,35). In accordance with the observation that TORC1 inhibition increases rDNA silencing, cells treated with rapamycin showed a decreased rate of marker loss compared to control cells (Figure 3A). This result demonstrates that TORC1 inhibition suppresses rDNA recombination and promotes rDNA stability. In contrast, nicotinamide treatment increased the rate of rDNA recombination. rDNA recombination was still increased when cells were co-treated with rapamycin and nicotinamide, indicating that enzymatically active Sir2 is critical for suppression of rDNA recombination under inhibition of TORC1 signaling. Taken together with the previous data, this result suggests that inhibition of TORC1 signaling suppresses rDNA recombination and promotes rDNA stability by enhancing association of Sir2 with rDNA.
Figure 3.
TORC1 inhibition promotes rDNA stability and extends lifespan in yeast. (A) Inhibition of TORC1 signaling suppresses rDNA recombination. rDNA recombination is represented by the rate of loss of the ADE2 marker gene integrated at the rDNA locus, which was calculated as the ratio of half-sectored colonies to the total number of colonies (‘Materials and Methods’ section). Exponentially growing cells were treated with or without 200 ng/ml rapamycin for 1 h and plated on SC plates containing 5 mM nicotinamide or not. After color development, the number of half-red/half-white colonies was counted and divided by the total number of colonies. Entirely red colonies have lost the marker prior to plating and thus were excluded from the total number of colonies. Values represent the average of three independent experiments and error bars indicate standard deviations. (B) Rapamycin extends replicative lifespan of cells, whereas nicotinamide accelerates aging of cells. The P-values for 10 ng/ml rapamycin (Rapa), 5 mM nicotinamide (NAM) and both treatment (Rapa + NAM) versus DMSO only (Control) are 3.7 × 10−3, 3.3 × 10−6 and 7.8 × 10−6, respectively. (C) Deletion of TOR1 fails to increase the lifespan of sir2Δ cells. The P-values for tor1Δ, sir2Δ, and tor1Δ sir2Δ cells versus wild-type cells (WT) are 5.8 × 10−4, 2.7 × 10−7, and 4.3 × 10−7, respectively. (D) Rapamycin fails to increase the lifespan of sir2Δ cells. The P-values for wild-type cells treated with 10 ng/ml rapamycin (WT + Rapa), sir2Δ, and sir2Δ cells treated with 10 ng/ml rapamycin (sir2Δ + Rapa) versus wild-type cells (WT) are 4.8 × 10−4, 2.7 × 10−7 and 2.3 × 10−6, respectively. Replicative lifespan was determined by scoring the number of daughter cells produced by each mother cell. Mean lifespans are shown in parentheses.
It has been reported that rDNA recombination leads to the formation of extrachromosomal rDNA circles that are toxic to cells and induce cell aging (5), and TORC1 inhibition increases replicative lifespan in yeast (29,30). Our result showing that TORC1 inhibition suppresses rDNA recombination and promotes rDNA stability provides a clue as to why TORC1 inhibition increases replicative lifespan in yeast. To confirm that replicative lifespan extension by TORC1 inhibition is closely related to increased rDNA silencing and rDNA stability resulting from enhanced association of Sir2 with rDNA, we performed the replicative lifespan analysis under treatment of rapamycin, nicotinamide, or both. As shown in Figure 3B, rapamycin significantly extended replicative lifespan of cells. In contrast, nicotinamide accelerated aging of cells. When rapamycin and nicotinamide were co-treated to cells, nicotinamide abolished the stimulatory effect of rapamycin on lifespan extension. These results correlate well with the effects of rapamycin and nicotinamide on rDNA silencing (Figure 2B) and rDNA stability (Figure 3A). Consistent with the previous report (7), deletion of SIR2 caused reduction in replicative lifespan of cells (Figure 3C and D). In sir2Δ cells, replicative lifespan was not increased under TORC1 inhibition by TOR1 deletion (Figure 3C) or rapamycin treatment (Figure 3D). Taken together, our data suggest that inhibition of TORC1 signaling increases rDNA silencing and rDNA stability by enhancing association of Sir2 with rDNA, thereby leading to extension of replicative lifespan in yeast.
Overexpression of PNC1 and SIR2 enhances association of Sir2 with rDNA and decreases the acetylation level of histone H3 at rDNA
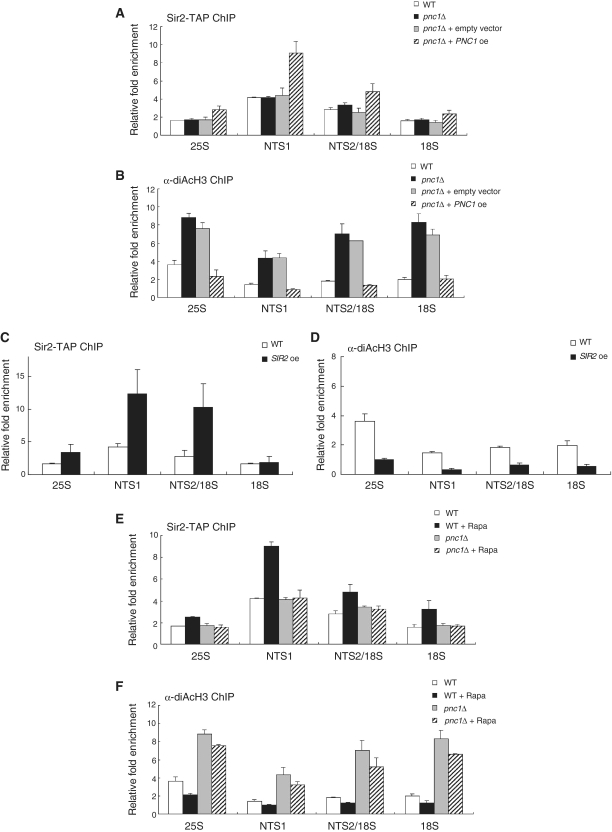
The PNC1 gene encodes a nicotinamidase that converts nicotinamide to nicotinic acid as part of the NAD+ salvage pathway (47). Therefore, deletion of PNC1 results in an elevated level of nicotinamide in cells that can inhibit the Sir2 activity. It has been shown that calorie restriction and other mild stresses extend yeast lifespan by increasing expression of PNC1 (46,48). A recent study has reported that inhibition of TORC1 signaling by rapamycin also promotes expression of PNC1 (30). Our results showing that rapamycin enhances association of Sir2 with rDNA and lowers the H3 acetylation level at rDNA raised a possibility that overexpression of PNC1 would have the same effect of rapamycin on rDNA. To check this possibility, we analyzed the degree of Sir2 binding to rDNA and the H3 acetylation level at rDNA in cells where PNC1 was deleted or overexpressed. In pnc1Δ cells, the degree of Sir2 binding to rDNA was essentially unchanged (Figure 4A), but the H3 acetylation level at rDNA increased considerably compared to wild-type cells (Figure 4B). In contrast, overexpression of PNC1 increased binding of Sir2 to rDNA (Figure 4A) and lowered the H3 acetylation level at rDNA (Figure 4B). Whether PNC1 was deleted or overexpressed, the protein level of Sir2 was not changed (data not shown). This result demonstrates that overexpression of PNC1 and rapamycin treatment exert the same effect on rDNA and suggests that enhancement of association of Sir2 with rDNA is a common mechanism underlying extension of lifespan in yeast by calorie restriction, other mild stresses, or TORC1 inhibition.
Figure 4.
Pnc1 is required for enhancement of Sir2 binding to rDNA under TORC1 inhibition. (A) PNC1 overexpression enhances association of Sir2 with rDNA. The degree of Sir2 binding to rDNA was measured using the ChIP assay in wild-type, pnc1Δ, pnc1Δ cells containing an empty vector and pnc1Δ cells expressing Pnc1-GFP on the p416GPD vector. (B) PNC1 overexpression lowers the H3 acetylation level at rDNA. The acetylation level of histone H3 at the rDNA regions was measured using the ChIP assay in wild-type, pnc1Δ, pnc1Δ cells containing an empty vector and pnc1Δ cells expressing Pnc1-GFP on the p416GPD vector. (C) SIR2 overexpression enhances association of Sir2 with rDNA. The degree of Sir2 binding to rDNA was measured using the ChIP assay in wild-type and sir2Δ cells expressing Sir2-TAP on the p416GPD vector. (D) SIR2 overexpression lowers the H3 acetylation level at rDNA. The acetylation level of histone H3 at the rDNA regions was measured using the ChIP assay in wild-type and sir2Δ cells expressing Sir2-TAP on the p416GPD vector. (E) Rapamycin does not enhance association of Sir2 with rDNA in pnc1Δ cells. The degree of Sir2 binding to rDNA was measured using the ChIP assay in wild-type and pnc1Δ cells after treatment with or without 200 ng/ml rapamycin for 1 h. (F) Rapamycin does not lower the H3 acetylation level at rDNA in pnc1Δ cells. The acetylation level of histone H3 at the rDNA regions was measured using the ChIP assay in wild-type and pnc1Δ cells after treatment with or without 200 ng/ml rapamycin for 1 h. Values represent the average of three independent experiments and error bars indicate standard deviations.
It is well established that increased dosage of the SIR2 gene extends lifespan in several organisms (7,49,50). To examine whether association of Sir2 with rDNA would be enhanced by overexpression of SIR2, we measured the degree of Sir2 binding to rDNA and the H3 acetylation level at rDNA in cells where SIR2 was overexpressed. For this experiment, we used sir2Δ cells with ectopically expressing TAP-tagged Sir2 to exclude the interfering effect of untagged Sir2 on the ChIP assay. In cells overexpressing Sir2, association of Sir2 with rDNA was considerably enhanced throughout the rDNA region (Figure 4C). Correlating with this observation, the H3 acetylation level at rDNA was significantly lowered in those cells (Figure 4D). This result corroborates the notion that enhancement of association of Sir2 with rDNA is a common mechanism underlying extension of lifespan in yeast by calorie restriction, TORC1 inhibition, or SIR2 overexpression.
Upon loss of PNC1, rapamycin cannot increase association of Sir2 with rDNA and histone deacetylation at rDNA
We have shown that inhibition of TORC1 signaling enhances association of Sir2 with rDNA and increases histone deacetylation at rDNA. Recently, it has been reported that TORC1 inhibition extends lifespan by relocalizing two transcription factors, Msn2 and Msn4, from the cytoplasm to the nucleus, where they activate expression of Pnc1 (30). Increased Pnc1 level would then lead to reduction of intracellular nicotinamide and consequent increase of the Sir2 activity. Taking into account these facts, we hypothesized that increased expression of Pnc1 would be a prerequisite for enhancement of association of Sir2 with rDNA under TORC1 inhibition. To examine this hypothesis, we measured the degree of Sir2 binding to rDNA and the H3 acetylation level at rDNA in pnc1Δ cells under rapamycin treatment. When pnc1Δ cells were treated with rapamycin, we could not observe any enhancement of Sir2 binding to rDNA (Figure 4E). Correlating with this result, rapamycin could not lower the H3 acetylation level at rDNA in pnc1Δ cells (Figure 4F). Deletion of PNC1 or treatment of rapamycin did not change the protein level of Sir2 (data not shown). The patterns of Sir2 binding to rDNA and histone deacetylation at rDNA in pnc1Δ cells under rapamycin treatment were very similar to those observed in cells co-treated with nicotinamide and rapamycin (Figure 2C and E). These results suggest that reduction of nicotinamide by increased Pnc1 is required for enhancement of Sir2 binding to rDNA and increase of histone deacetylation at rDNA under TORC1 inhibition.
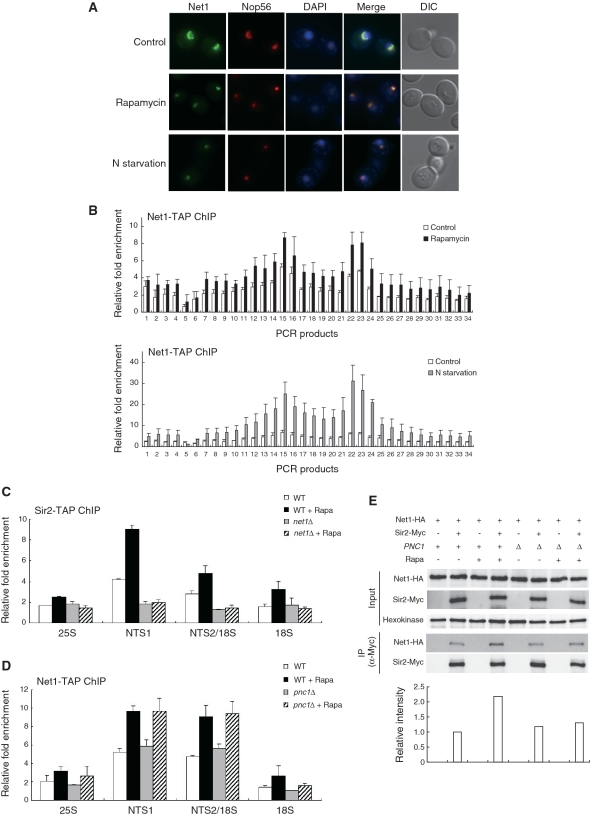
Net1 is required for enhancement of association of Sir2 with rDNA under TORC1 inhibition
In yeast, Sir2 is found as a subunit of an rDNA silencing complex called RENT (12). This complex is tethered to the nucleolus by its core subunit Net1. Net1 is required for association of Sir2 with rDNA, but association of Net1 with rDNA is independent of Sir2 (13). Since association of Sir2 with rDNA is dependent on Net1, we asked whether Net1 would also be required for enhanced association of Sir2 with rDNA under TORC1 inhibition. To test this possibility, we first analyzed the subcellular localization of Net1 under TORC1 inhibition. Rapamycin treatment and nitrogen starvation led to condensation of the fluorescent signal of Net1-GFP (Figure 5A). This observation suggests that, similar to Sir2, Net1 remains stably associated with rDNA under TORC1 inhibition. Moreover, association of Net1 with rDNA increased over the entire rDNA locus under rapamycin treatment and nitrogen starvation (Figure 5B). The protein level of Net1 was not changed under these conditions (data not shown). Together, these results indicate that TORC1 inhibition enhances association of not only Sir2 but also Net1 with rDNA array.
Figure 5.
Net1 is required for enhancement of association of Sir2 with rDNA under TORC1 inhibition. (A) Rapamycin treatment and nitrogen starvation cause condensation of the Net1-GFP signal. Subcellular localization of Net1-GFP (green) and Nop56-RFP (red) was analyzed after treatment with 200 ng/ml rapamycin or incubation in nitrogen-depleted medium for 1 h. DAPI staining for visualization of the nucleus (blue) and DIC images are also shown. (B) Rapamycin treatment and nitrogen starvation enhance association of Net1 with rDNA. The degree of Net1 binding to rDNA was measured using the ChIP assay after treatment with 200 ng/ml rapamycin (upper panel) or incubation in nitrogen-depleted medium for 1 h (lower panel). For control, cells were treated with DMSO only (upper panel) or grown in SC medium (lower panel). (C) Rapamycin does not enhance association of Sir2 with rDNA in net1Δ cells. The degree of Sir2 binding to rDNA was measured using the ChIP assay in wild-type and net1Δ cells after treatment with or without 200 ng/ml rapamycin for 1 h. (D) Association of Net1 with rDNA is not affected by Pnc1. The degree of Net1 binding to rDNA was measured using the ChIP assay in wild-type and pnc1Δ cells after treatment with or without 200 ng/ml rapamycin for 1 h. Values represent the average of three independent experiments and error bars indicate standard deviations. (E) TORC1 inhibition enhances interaction between Net1 and Sir2. Interaction between Net1 and Sir2 was analyzed by coimmunoprecipitation in wild-type and pnc1Δ cells after treatment with or without 200 ng/ml rapamycin for 1 h. Hexokinase was used as a loading control. The levels of Net1-HA coimmunoprecipitated with Sir2-Myc were quantified and indicated below the respective lanes.
We next examined the degree of binding of Sir2 to rDNA under rapamycin treatment in cells where NET1 was deleted. Interestingly, when net1Δ cells were treated with rapamycin, we could not observe any enhancement of Sir2 binding to rDNA (Figure 5C). Whether NET1 was deleted or not, the protein level of Sir2 was not altered (data not shown). This result demonstrates that Net1 is required for enhancement of Sir2 binding to rDNA under TORC1 inhibition.
As shown in Figure 4A, overexpression of Pnc1 enhances Sir2 binding to rDNA. To determine whether the Pnc1 activity would influence association of Net1 with rDNA, we examined the degree of Net1 binding to rDNA under rapamycin treatment in cells where PNC1 was deleted. There was little difference in the degree of Net1 binding to rDNA between wild-type and pnc1Δ cells under normal condition (Figure 5D). When rapamycin was treated, association of Net1 with rDNA was considerably enhanced in pnc1Δ cells as well as in wild-type cells. This result indicates that the Pnc1 activity does not affect association of Net1 with rDNA whether rapamycin is present or not. This binding pattern of Net1 is contrasting to that of Sir2, which does not show any enhancement of binding to rDNA in pnc1Δ cells even when rapamycin is present (Figure 4E).
We next asked whether rapamycin would influence interaction between Net1 and Sir2. To examine interaction between Net1 and Sir2, we constructed yeast strains in which the endogenous NET1 and SIR2 genes were modified to express C-terminal HA and Myc fusion proteins, respectively. Remarkably, coprecipitation of Net1 and Sir2 was significantly increased under rapamycin treatment (Figure 5E). This observation indicates that TORC1 inhibition enhances interaction between Net1 and Sir2. In pnc1Δ cells, however, coprecipitation of Net1 and Sir2 was only marginally increased under rapamycin treatment, indicating that enhancement of interaction between Net1 and Sir2 under TORC1 inhibition is dependent on the Pnc1 activity and only the active form of Sir2 can be bound to Net1 under TORC1 inhibition. Taken together with the previous data, these results suggest that enhancement of Sir2 binding to rDNA under TORC1 inhibition is due to increased association of Net1 with rDNA and enhanced interaction between Net1 and Sir2.
DISCUSSION
Inhibition of TORC1 signaling by rapamycin can delay aging and extend lifespan in several organisms. However, the mechanism by which inhibition of TORC1 signaling delays aging and extends lifespan is poorly understood. A recent study has shown that calorie restriction and TORC1 inhibition promote the activity of Sir2 by relocalizing the transcription factors Msn2 and Msn4 from the cytoplasm to the nucleus, where they increase expression of PNC1 encoding a nicotinamidase that depletes cellular nicotinamide, a physiological inhibitor of Sir2 (30). Taken together with the fact that increase of the Sir2 activity extends lifespan in yeast (7), it is likely that extension of lifespan by TORC1 inhibition is due to the promoted activity of Sir2. Our results support this notion and provide insight into the molecular mechanism underlying promotion of the Sir2 activity by TORC1 inhibition. We have shown that inhibition of TORC1 signaling enhances association of Sir2 with rDNA (Figure 1), promotes transcriptional silencing of Pol II-transcribed gene at the rDNA locus and induces deacetylation of histones at rDNA (Figure 2). Furthermore, we have shown that TORC1 inhibition reduces homologous recombination between rDNA repeats and extends replicative lifespan (Figure 3), and Pnc1 and Net1 are required for enhancement of Sir2 binding to rDNA under TORC1 inhibition (Figures 4 and 5). Our results provide evidence that TORC1 inhibition stabilizes the rDNA locus and increases rDNA silencing by enhancing association of Sir2 with rDNA, thereby leading to lifespan extension in yeast.
It is well known that accumulation of extrachromosomal rDNA circles is toxic to cells and induces replicative aging in yeast (5). Extrachromosomal rDNA circles are formed by homologous recombination within the rDNA. The replication fork block protein Fob1 promotes rDNA recombination and formation of extrachromosomal rDNA circles (16,17). Therefore, deletion of FOB1 extends replicative lifespan in yeast (51). We observed that inhibition of TORC1 by rapamycin resulted in an additive increase of replicative lifespan when combined with deletion of FOB1 (Supplementary Figure S3), which is consistent with the previous report that deletion of TOR1 increases replicative lifespan additively with deletion of FOB1 (29). This observation suggests that TORC1 inhibition may contribute to lifespan extension by additional mechanisms other than promotion of rDNA silencing and rDNA stability by enhancing association of Sir2 with rDNA. Recent studies have reported that Sir2 promotes replicative lifespan in yeast by additional non-rDNA mechanisms. It has been shown that Sir2 is required for asymmetric retention of damaged cytoplasmic proteins in the mother cell, resulting in an enhanced capacity to respond to oxidative stress in the daughter cell (52,53). Another non-rDNA function of Sir2 suggested to contribute to longevity is maintenance of telomeric chromatin. Dang et al. (54) showed that Sir2 abundance declines in replicatively old yeast cells and that this loss of Sir2 is accompanied by a decrease in histone acetylation and histone abundance near telomeres. Whether TORC1 signaling modulates these non-rDNA functions of Sir2 is not known yet. Further studies are needed to establish if TORC1 inhibition contributes to lifespan extension by regulating these non-rDNA functions of Sir2, in addition to by promoting rDNA silencing and rDNA stability.
Recently, it has been reported that deletion of genes encoding many of 60S subunit ribosomal proteins are each sufficient to significantly increase lifespan in yeast (55). Deletion of some ribosomal protein genes results in lifespan extension exceeding 50% (e.g. 90.9 and 60.9% in rpl22aΔ and rpl13aΔ cells, respectively). Deletion of some other ribosomal proteins has no effect on lifespan or rather results in lifespan shortening (e.g. −8.8% in rpl35bΔ cells). To test a possibility that lifespan extension by depletion of 60S ribosomal proteins might has any relation to enhanced association of Sir2 with rDNA, we examined the degree of Sir2 binding to rDNA in rpl22aΔ and rpl13aΔ cells. These mutant cells did not show significant change in Sir2 binding to rDNA compared to that of wild-type or rpl35bΔ cells (Supplementary Figure S4). In addition, the protein level of Sir2 was not changed in these mutant cells (data not shown). This observation indicates that depletion of 60S ribosomal proteins extends lifespan by mechanisms different from promotion of rDNA silencing and rDNA stability by Sir2 and suggests that multiple pathways act in parallel to promote lifespan in yeast.
Saccharoymyces cerevisiae contains four additional proteins homologous to Sir2 (Hst1, Hst2, Hst3 and Hst4), some of which have been reported to have functional redundancy with Sir2. For example, Hst2 can compensate for the lack of Sir2 during calorie restriction (56). Nicotinamide has been shown to inhibit not only Sir2 but also Hst1 and Hst2 (46,57). Under the experimental conditions and with the strains used in this study, we have observed that nicotinamide treatment abolishes silencing of the reporter mURA3 gene at the rDNA locus to an extent similar to that of deletion of SIR2 (Figure 2A and B). This observation suggests that Sir2 plays a major role in rDNA silencing while Hst1 and Hst2 make a minor contribution to rDNA silencing. Consistent with this notion, the ChIP assay revealed that Hst1 and Hst2 were not associated with the rDNA regions whether rapamycin was treated or not (data not shown). A previous study has also reported that deletion of HST1 and HST2 has a very minor effect on rDNA silencing (58). In contrast, deletion of HST2 has been shown to significantly increase the rDNA recombination rate (56). Since functional redundancy between Sir2 and Hst proteins in rDNA silencing remains controversial, further investigation and comparison of the action mechanisms of Sir2 and Hst proteins should be useful in resolving the controversy over functional redundancy between them.
Intriguingly, we observed that the rDNA-binding pattern of Net1 under nitrogen starvation is slightly different from that of Sir2 under the same condition (Figures 1C and 5B). Under nitrogen starvation, Net1 most strongly bound to the NTS2/18S region, whereas Sir2 most strongly bound to the NTS1 region. It is widely accepted that rapamycin treatment induces similar responses in eukaryotic cells as under condition of nitrogen depletion (59). However, it is reasonable to assume that the response of cells to nitrogen depletion would be slightly different from that to rapamycin treatment. This is because rapamycin directly exerts its inhibitory effect on TORC1 via Fpr1 but not on the upstream effectors of TORC1 (60), whereas nitrogen depletion shuts down amino acid signaling (61), which may affect the upstream effectors of TORC1. Stp1, a well-known transcription factor that regulates the expression of several amino acid permease genes, represents a clear example of the differences between response of cells to nitrogen depletion and that to rapamycin treatment (62). It is presumable that some unknown upstream effectors of TORC1 may differentially influence the rDNA-binding activity of Net1 and Sir2.
Since Sir2 was shown to extend lifespan of budding yeast by repressing genome instability (5,7), Sir2-like proteins, or sirtuins, have been found in organisms ranging from bacteria to humans (63,64). Mammals contain seven sirtuins, SIRT1-7 (64). Human SIRT1 has been shown to deacetylate H3 at rDNA repeats (65). Recently, it has been reported that nuclear SIRT1 is recruited to the nucleolar compartment dynamically (66) and is associated with chromatin throughout the rDNA locus (67). Moreover, a nucleolar sirtuin SIRT7 associates with the rDNA locus, interacts with RNA Pol I and activates rRNA transcription (68). Presumably, enhancement of association of Sir2 and some sirtuins with rDNA may be a common mechanism underlying increase of rDNA stability and extension of lifespan among yeast and higher organisms. It remains to be determined whether TORC1 inhibition by rapamycin would also enhance association of sirtuins with rDNA, thereby stabilizing the rDNA locus and increasing rDNA silencing in higher organisms. Recent studies have shown that mammalian nicotinamide phosphoribosyltransferase (Nampt/PBEF/Visfatin), a putative functional ortholog of Pnc1 that is required for conversion of nicotinamide to NAD+, positively regulates sirtuin activity (69,70). It would also be interesting to check if association of sirtuin proteins with rDNA is enhanced in mammalian cells overexpressing nicotinamide phosphoribosyltransferase.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
21C Frontier Functional Proteomics Project (FPR08A1-060); 21C Frontier Microbial Genomics and Application Center Program (MG-11-2008-09-004-00); SRC/ERC Program of MOST/KOSEF (R11-2005-009-05001-0), Republic of Korea. Funding for open access charge: The Brain Korea 21 Project from the Ministry of Education, Science and Technology, Republic of Korea.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Danesh Moazed and Masayasu Nomura for providing strains. The authors also thank members of the Huh lab for helpful discussions.
REFERENCES
- 1.Johnston M, Hillier L, Riles L, Albermann K, Andre B, Ansorge W, Benes V, Bruckner M, Delius H, Dubois E, et al. The nucleotide sequence of Saccharomyces cerevisiae chromosome XII. Nature. 1997;387:87–90. [PMC free article] [PubMed] [Google Scholar]
- 2.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 3.Moss T. At the crossroads of growth control; making ribosomal RNA. Curr. Opin. Genet. Dev. 2004;14:210–217. doi: 10.1016/j.gde.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Rudra D, Warner JR. What better measure than ribosome synthesis? Genes Dev. 2004;18:2431–2436. doi: 10.1101/gad.1256704. [DOI] [PubMed] [Google Scholar]
- 5.Sinclair DA, Guarente L. Extrachromosomal rDNA circles–a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 6.Huang J, Moazed D. Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes Dev. 2003;17:2162–2176. doi: 10.1101/gad.1108403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fritze CE, Verschueren K, Strich R, Easton Esposito R. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 1997;16:6495–6509. doi: 10.1093/emboj/16.21.6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 10.Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl Acad. Sci. USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, et al. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl Acad. Sci. USA. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shou W, Seol JH, Shevchenko A, Baskerville C, Moazed D, Chen ZW, Jang J, Charbonneau H, Deshaies RJ. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- 13.Straight AF, Shou W, Dowd GJ, Turck CW, Deshaies RJ, Johnson AD, Moazed D. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell. 1999;97:245–256. doi: 10.1016/s0092-8674(00)80734-5. [DOI] [PubMed] [Google Scholar]
- 14.Lin YH, Keil RL. Mutations affecting RNA polymerase I-stimulated exchange and rDNA recombination in yeast. Genetics. 1991;127:31–38. doi: 10.1093/genetics/127.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi T, Horiuchi T. A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells. 1996;1:465–474. doi: 10.1046/j.1365-2443.1996.d01-256.x. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi T, Heck DJ, Nomura M, Horiuchi T. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 1998;12:3821–3830. doi: 10.1101/gad.12.24.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johzuka K, Horiuchi T. Replication fork block protein, Fob1, acts as an rDNA region specific recombinator in S. cerevisiae. Genes Cells. 2002;7:99–113. doi: 10.1046/j.1356-9597.2001.00508.x. [DOI] [PubMed] [Google Scholar]
- 18.Hunter T. When is a lipid kinase not a lipid kinase? When it is a protein kinase. Cell. 1995;83:1–4. doi: 10.1016/0092-8674(95)90225-2. [DOI] [PubMed] [Google Scholar]
- 19.Keith CT, Schreiber SL. PIK-related kinases: DNA repair, recombination, and cell cycle checkpoints. Science. 1995;270:50–51. doi: 10.1126/science.270.5233.50. [DOI] [PubMed] [Google Scholar]
- 20.Schneper L, Duvel K, Broach JR. Sense and sensibility: nutritional response and signal integration in yeast. Curr. Opin. Microbiol. 2004;7:624–630. doi: 10.1016/j.mib.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 21.Dennis PB, Fumagalli S, Thomas G. Target of rapamycin (TOR): balancing the opposing forces of protein synthesis and degradation. Curr. Opin. Genet. Dev. 1999;9:49–54. doi: 10.1016/s0959-437x(99)80007-0. [DOI] [PubMed] [Google Scholar]
- 22.Schmelzle T, Hall MN. TOR, a central controller of cell growth. Cell. 2000;103:253–262. doi: 10.1016/s0092-8674(00)00117-3. [DOI] [PubMed] [Google Scholar]
- 23.Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 24.Mahajan PB. Modulation of transcription of rRNA genes by rapamycin. Int. J. Immunopharmacol. 1994;16:711–721. doi: 10.1016/0192-0561(94)90091-4. [DOI] [PubMed] [Google Scholar]
- 25.Zaragoza D, Ghavidel A, Heitman J, Schultz MC. Rapamycin induces the G0 program of transcriptional repression in yeast by interfering with the TOR signaling pathway. Mol. Cell. Biol. 1998;18:4463–4470. doi: 10.1128/mcb.18.8.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Powers T, Walter P. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol. Biol. Cell. 1999;10:987–1000. doi: 10.1091/mbc.10.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- 28.Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr. Biol. 2004;14:885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 30.Medvedik O, Lamming DW, Kim KD, Sinclair DA. MSN2 and MSN4 link calorie restriction and TOR to sirtuin-mediated lifespan extension in Saccharomyces cerevisiae. PLoS Biol. 2007;5:e261. doi: 10.1371/journal.pbio.0050261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Syntichaki P, Troulinaki K, Tavernarakis N. eIF4E function in somatic cells modulates ageing in Caenorhabditis elegans. Nature. 2007;445:922–926. doi: 10.1038/nature05603. [DOI] [PubMed] [Google Scholar]
- 33.Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- 34.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 35.Huang J, Brito IL, Villen J, Gygi SP, Amon A, Moazed D. Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer. Genes Dev. 2006;20:2887–2901. doi: 10.1101/gad.1472706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 37.Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 38.Sherman F. Getting started with yeast. Methods Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- 39.Sung MK, Huh WK. Bimolecular fluorescence complementation analysis system for in vivo detection of protein-protein interaction in Saccharomyces cerevisiae. Yeast. 2007;24:767–775. doi: 10.1002/yea.1504. [DOI] [PubMed] [Google Scholar]
- 40.Smith JS, Boeke JD. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- 41.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 42.Park PU, McVey M, Guarente L. Separation of mother and daughter cells. Methods Enzymol. 2002;351:468–477. doi: 10.1016/s0076-6879(02)51865-6. [DOI] [PubMed] [Google Scholar]
- 43.Tsang CK, Bertram PG, Ai W, Drenan R, Zheng XF. Chromatin-mediated regulation of nucleolar structure and RNA Pol I localization by TOR. EMBO J. 2003;22:6045–6056. doi: 10.1093/emboj/cdg578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 45.Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J. Biol. Chem. 2002;277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- 46.Gallo CM, Smith DL, Jr, Smith JS. Nicotinamide clearance by Pnc1 directly regulates Sir2-mediated silencing and longevity. Mol. Cell. Biol. 2004;24:1301–1312. doi: 10.1128/MCB.24.3.1301-1312.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ghislain M, Talla E, Francois JM. Identification and functional analysis of the Saccharomyces cerevisiae nicotinamidase gene, PNC1. Yeast. 2002;19:215–224. doi: 10.1002/yea.810. [DOI] [PubMed] [Google Scholar]
- 48.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Cohen H, Lin SS, Manchester JK, Gordon JI, Sinclair DA. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J. Biol. Chem. 2002;277:18881–18890. doi: 10.1074/jbc.M111773200. [DOI] [PubMed] [Google Scholar]
- 49.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 50.Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- 51.Defossez PA, Prusty R, Kaeberlein M, Lin SJ, Ferrigno P, Silver PA, Keil RL, Guarente L. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol. Cell. 1999;3:447–455. doi: 10.1016/s1097-2765(00)80472-4. [DOI] [PubMed] [Google Scholar]
- 52.Aguilaniu H, Gustafsson L, Rigoulet M, Nystrom T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 2003;299:1751–1753. doi: 10.1126/science.1080418. [DOI] [PubMed] [Google Scholar]
- 53.Erjavec N, Nystrom T. Sir2p-dependent protein segregation gives rise to a superior reactive oxygen species management in the progeny of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2007;104:10877–10881. doi: 10.1073/pnas.0701634104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, Kaeberlein M, Kennedy BK, Berger SL. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, Dang N, Johnston ED, Oakes JA, Tchao BN, et al. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell. 2008;133:292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lamming DW, Latorre-Esteves M, Medvedik O, Wong SN, Tsang FA, Wang C, Lin SJ, Sinclair DA. HST2 mediates SIR2-independent life-span extension by calorie restriction. Science. 2005;309:1861–1864. doi: 10.1126/science.1113611. [DOI] [PubMed] [Google Scholar]
- 57.Landry J, Slama JT, Sternglanz R. Role of NAD(+) in the deacetylase activity of the SIR2-like proteins. Biochem. Biophys. Res. Commun. 2000;278:685–690. doi: 10.1006/bbrc.2000.3854. [DOI] [PubMed] [Google Scholar]
- 58.Perrod S, Cockell MM, Laroche T, Renauld H, Ducrest AL, Bonnard C, Gasser SM. A cytosolic NAD-dependent deacetylase, Hst2p, can modulate nucleolar and telomeric silencing in yeast. EMBO J. 2001;20:197–209. doi: 10.1093/emboj/20.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rohde J, Heitman J, Cardenas ME. The TOR kinases link nutrient sensing to cell growth. J. Biol. Chem. 2001;276:9583–9586. doi: 10.1074/jbc.R000034200. [DOI] [PubMed] [Google Scholar]
- 60.Koltin Y, Faucette L, Bergsma DJ, Levy MA, Cafferkey R, Koser PL, Johnson RK, Livi GP. Rapamycin sensitivity in Saccharomyces cerevisiae is mediated by a peptidyl-prolyl cis-trans isomerase related to human FK506-binding protein. Mol. Cell Biol. 1991;11:1718–1723. doi: 10.1128/mcb.11.3.1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Forsberg H, Gilstring CF, Zargari A, Martinez P, Ljungdahl PO. The role of the yeast plasma membrane SPS nutrient sensor in the metabolic response to extracellular amino acids. Mol. Microbiol. 2001;42:215–228. doi: 10.1046/j.1365-2958.2001.02627.x. [DOI] [PubMed] [Google Scholar]
- 62.Shin CS, Kim SY, Huh WK. TORC1 controls degradation of the transcription factor Stp1, a key effector of the SPS amino-acid-sensing pathway in Saccharomyces cerevisiae. J. Cell Sci. 2009;122:2089–2099. doi: 10.1242/jcs.047191. [DOI] [PubMed] [Google Scholar]
- 63.Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun. 1999;260:273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- 64.Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 65.Liou GG, Tanny JC, Kruger RG, Walz T, Moazed D. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell. 2005;121:515–527. doi: 10.1016/j.cell.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 66.Espada J, Ballestar E, Santoro R, Fraga MF, Villar-Garea A, Nemeth A, Lopez-Serra L, Ropero S, Aranda A, Orozco H, et al. Epigenetic disruption of ribosomal RNA genes and nucleolar architecture in DNA methyltransferase 1 (Dnmt1) deficient cells. Nucleic Acids Res. 2007;35:2191–2198. doi: 10.1093/nar/gkm118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Murayama A, Ohmori K, Fujimura A, Minami H, Yasuzawa-Tanaka K, Kuroda T, Oie S, Daitoku H, Okuwaki M, Nagata K, et al. Epigenetic control of rDNA loci in response to intracellular energy status. Cell. 2008;133:627–639. doi: 10.1016/j.cell.2008.03.030. [DOI] [PubMed] [Google Scholar]
- 68.Ford E, Voit R, Liszt G, Magin C, Grummt I, Guarente L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 2006;20:1075–1080. doi: 10.1101/gad.1399706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J. Biol. Chem. 2004;279:50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- 70.van der Veer E, Ho C, O'Neil C, Barbosa N, Scott R, Cregan SP, Pickering JG. Extension of human cell lifespan by nicotinamide phosphoribosyltransferase. J. Biol. Chem. 2007;282:10841–10845. doi: 10.1074/jbc.C700018200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.