Abstract
Myogenic microRNAs are important regulators of muscle development and differentiation. To better understand the roles of chromatin-modifying and remodeling enzymes in the activation of myogenic microRNA expression, we have functionally analyzed two different protein arginine methyltransferases, Prmt5 and Prmt4, both of which have previously been implicated in the regulation of myogenic mRNA expression. Both Prmts are required for myogenic microRNA induction during differentiation. Prmt5 is indirectly required due to the necessity of Prmt5 for expression of the transcriptional regulator, myogenin, as ectopic expression of myogenin eliminates Prmt5 dependency. By contrast, Prmt4 binds to the upstream regulatory regions of myogenic microRNAs and is required for dimethylation of the Prmt4 substrate, H3R17, at microRNA regulatory sequences. Deletion of Prmt4 does not alter MyoD binding at myogenic microRNA regulatory sequences but prevents the binding of both myogenin and the Brg1 ATPase that catalyzes SWI/SNF-dependent chromatin remodeling, resulting in an inhibition of microRNA expression.
INTRODUCTION
microRNAs (miRNAs) are ∼22-nt RNAs that post-transcriptionally regulate gene expression and that are required for organismal development (1–3). They function by binding to complementary sequences, generally in the 3′ untranslated regions of mRNAs, leading to messenger RNA (mRNA) degradation or inhibition of translation (4). Considerable effort has been expended to understand the specific targets and functions of individual miRNAs; however, the mechanisms by which miRNA expression is regulated have not been as well studied.
Given the requirement for miRNAs and the miRNA processing enzymes in most aspects of development and cell differentiation, we have focused our efforts on understanding the induction of developmentally regulated miRNAs. Cardiac and skeletal muscle development is marked by the induction and function of several miRNA molecules. miR-1 inhibits muscle growth and promotes differentiation by inhibiting the expression of a Hand family transcriptional activator that promotes cell growth and proliferation and by targeting histone deacetylase 4, which is an inhibitor of muscle differentiation (5,6). miR-206 also promotes myogenic differentiation by targeting the DNA polymerase-α, resulting in DNA synthesis inhibition (7). By contrast, miR-133 promotes growth and inhibits differentiation by targeting the serum response factor, which is a key activator of myogenesis (5). Other miRNAs have been implicated in cardiac and skeletal muscle disease (8–12).
Studies examining the regulation of myogenic miRNAs in skeletal muscle have indicated a role for known myogenic transcriptional regulatory proteins. Several E-boxes are present in the regions upstream of myogenic miRNA genes and have been shown by chromatin immunoprecipitation (ChIP) to be occupied by MyoD and myogenin (13–15). Other studies demonstrated that the MyoD binding sites and myocyte enhancer factor 2 (Mef2) sites present in myogenic miRNA regulatory sequences mediate miRNA expression (6,16). In addition, the Twist transcriptional regulator was identified as a regulator of a myogenic miRNAs in Drosophila (17). More recently, the expression of several myogenic miRNAs was shown to be dependent upon the chromatin remodeling function of the Brg1 ATPase that is the catalytic subunit of some SWI/SNF chromatin remodeling enzymes (13). More detailed analysis of how myogenic miRNAs expression is regulated is lacking.
Post-translational modifications of histones can alter nucleosomes, cause conformational changes in chromatin structure, lead to recruitment of proteins or protein complexes and contribute to the regulation of transcription. The histone-modifying enzymes, together with ATP-dependent chromatin remodeling enzymes and transcription factors, cooperate to modulate RNA polymerase II function. Histone modifiers comprise a large family of enzymes that add or remove a variety of moieties from histone proteins. These modifications include acetylation, deacetylation, methylation, demethylation, phosphorylation, ubiquitylation, sumoylation, poly(ADP-ribosylation) and biotinylation (18–24). The requirement for individual enzymes and the interplay and functional relationships between different enzymes is an area of intense interest.
Among the histone-modifying enzymes are the protein arginine methyltransferases (Prmts), which asymmetrically (type I) or symmetrically (type II) dimethylate arginine residues in substrate proteins (25). Nine Prmts have been identified in mammals (26). Prmts have roles in multiple cellular processes such as regulation of cell signaling, cytokine production, differentiation, transcription, RNA processing and nucleo-cytoplasmic transport (26). Investigation of cellular differentiation, in particular the differentiation of skeletal muscle, has led to identification of roles for two Prmts in myogenesis. Prmt5 is a type II Prmt that dimethylates histone 3 at arginine 8 (H3R8) and histone H4 at arginine 3 (27,28). The induction of myogenin requires Prmt5, and H3R8 dimethylation at the myogenin promoter is dependent upon Prmt5 (21). Prmt4, also called coactivator-associated arginine methyltransferase 1 (Carm1), and hereafter referred to as Carm1/Prmt4, asymmetrically dimethylates H3R17 and H3R26 (29,30). Carm1/Prmt4 binds to genes expressed at late times of skeletal muscle differentiation and is required for the expression of these genes (20,31). Interestingly, both Prmt5 and Carm1/Prmt4 can associate with SWI/SNF chromatin remodeling enzymes (28,32,33), suggesting functional links between these different classes of enzymes. Indeed deficiency of either Prmt5 or Carm1/Prmt4 leads not only to an absence of arginine dimethylated histones at target promoters but, in both cases, also results in an inhibition of SWI/SNF enzyme binding at target genes. Consequently, the target genes do not undergo chromatin remodeling and are not transcriptionally induced (20,21).
In the current study, we asked whether Prmt5 and Carm1/Prmt4 are involved in the induction of myogenic miRNA expression during differentiation. The results indicate that while both Prmts are required for myogenic miRNA expression, the mechanism by which each enzyme functions is distinct. Prmt5 is indirectly needed for myogenic miRNA expression via its requirement for myogenin expression. By contrast, Carm1/Prmt4 binds to myogenic miRNA regulatory sequences, modifies histones in these regions, and is required for the binding of both the Brg1 ATPase of SWI/SNF chromatin remodeling enzymes and myogenin. The results demonstrate a role for multiple Prmts in the induction of myogenic miRNAs during differentiation and reinforce the idea that specific Prmts and ATP-dependent chromatin remodeling enzymes can cooperate during gene activation.
MATERIALS AND METHODS
Cell culture, retrovirus generation and myogenic differentiation
NIH 3T3 cells were derived by passaging cells from a mouse embryo (strain NIH/Swiss) using a classical 3T3 protocol, resulting in an immortalized mouse embryo fibroblast (MEF) line (34). These were purchased from ATCC. Prmt5 antisense cells are NIH 3T3 fibroblasts that constitutively express a Prmt5 antisense vector. Clone 12 (C12) was previously described (28). Clone 1 (C1) is another independently derived antisense clone that has not previously been published but that phenotypically is identical to previously published Prmt5 antisense clonal cell lines (21,28). The wild-type and Carm1/Prmt4 null immortalized MEFs were generated by passaging cells from a mouse embryo (mixed strain TC-1/Black Swiss) using a classical 3T3 protocol (35). All cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% calf serum, penicillin and streptomycin. Prmt5 antisense cells were cultured in the presence of 2.5 ug/ml of puromycin. The pBABE-MyoD retroviral construct (21) was transfected into BOSC23 cells (36) for generation of retroviral particles, infection and differentiation as described (21,37). To induce myogenic differentiation with myogenin and Mef2D1b, a blasticidin resistant pBABE-myogenin construct and neomycin-resistant pBABE-Mef2D1b construct were used to generate retrovirus. Cells were infected with the myogenin encoding virus, subjected to drug selection, then infected with the Mef2D1b retrovirus, and again subjected to drug selection to ensure efficient double infection prior to differentiation. Samples for RNA and ChIP were collected at different time points of differentiation.
RNA isolation, reverse transcription-polymerase chain reaction and northern blots
Total RNA was isolated from cells using TRIzol (Invitrogen) reagent according to the manufacturer’s instructions. Five-hundred nanograms total RNA was used for reverse transcription reactions to generate cDNA using superscript III (Invitrogen) reverse transcriptase enzymes as previously described (38). Quantitative polymerase chain reaction (PCR) was performed with SYBR green master mix (ABI) according to the manufacturer’s protocol using MyoD, Mef2D and myogenin primers described (39,40). Primers for Acta1 and primary miRNA transcripts were published (13). Primers for SRF were 5′- atgccccatcccttaaaatccctttgg-3′ and 5′-ggatacaggtggtttaggctggctctgac-3′. Amplification and quantification were performed using an ABI StepOne Plus System. Values for each gene or miRNA were normalized to levels of EF1alpha mRNA. Primers for EF1alpha were described (39). Northern blots were performed exactly as described using the γ32P-radiolabeled probes described for miR1-1, miR-133a, miR-29a and U6 (13). The probe sequence for miR-143 was 5′-gagctacagtgcttcatctca-3′.
Western blots
Western blots to detect Prmt5, MyoD and Carm1/Prmt4 were performed on whole cell extracts as described (41) using rabbit polyclonal antisera against Prmt5 (Santa Cruz sc-22132), MyoD (Santa Cruz sc-32758) and Prmt4 (Millipore 09-818).
ChIP
ChIP assays were carried out as previously described (21). Analysis of immunoprecipitated DNA was performed by quantitative real-time PCR using SYBR green master mix (ABI) on an ABI StepOne Plus RT-PCR system. Primers used for ChIP assays were described (13). The antibodies used for the immunoprecipitation step included rabbit polyclonal antisera or partially purified antibodies against Prmt5 (32), Carm1/Prmt4 (Millipore 09-818), dimethylated H3R17 (Millipore 07-214), MyoD (Santa Cruz sc-304), myogenin (Santa Cruz sc-576) and Brg1 (41). IgG (Millipore 12-370) was used as a control.
RESULTS
Kinetics of miRNA expression
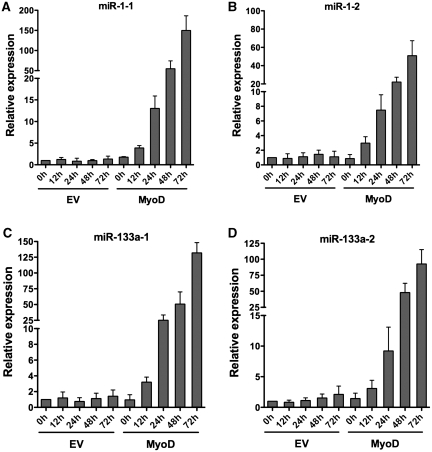
miRNA-1 and miRNA-133a are skeletal and cardiac muscle-specific miRNAs. Expression of these miRNAs is induced during myogenesis (5,6). To further understand the kinetics of induction of these miRNAs during skeletal muscle differentiation, we utilized a well-defined culture model for myogenesis (42). NIH 3T3 fibroblasts were differentiated along the skeletal muscle lineage by introducing a retrovirus encoding MyoD, allowing the cells to become confluent and inducing differentiation via exposure to a low serum media. Primary transcripts of miR-1-1, miR-1-2, miR133a-1 and miR-133a-2 were induced by 12 h post-differentiation in the MyoD-differentiated cells, and the expression levels continued to increase with time post-differentiation. Empty vector infected cells did not express these miRNAs (Figure 1A–D).
Figure 1.
Kinetics of myogenic miRNA expression during myogensis. (A–D) qPCR analyses of miR-1-1, miR-1-2, miR-133a-1 and miR-133a-2 primary transcripts in MyoD-differentiated NIH 3T3 cells. Relative expression was analyzed at the indicated times post-differentiation. The expression level at time 0 in the empty vector (EV) control was normalized to 1. Results are the average from three independent experiments± standard deviation. h, hours.
Prmt5 is indirectly required for the expression of miR-1 and miR-133a
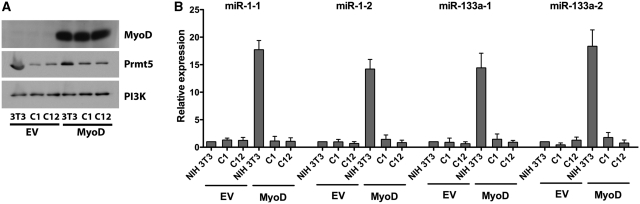
Prmt5 and Carm1/Prmt4 play important roles in the regulation of myogenic gene expression and differentiation. Although Prmt5 is required for the expression of the myogenin gene, which is expressed at early times post-differentiation, it is not directly required for activation of myogenic late genes (20,21). Conversely, Carm1/Prmt4 is required for the expression of late genes but is dispensable for the expression of myogenin during myogenic differentiation (20). To investigate whether Prmt5 plays a role in the regulation of myogenic miRNA expression, we differentiated NIH 3T3 and Prmt5 antisense (AS) cell line clones C12 and C1 following the introduction of retrovirus encoding MyoD or an empty vector control. We confirmed the knockdown of Prmt5 by western blot in both the C1 and C12 Prmt5 AS lines and the equivalent expression of MyoD in the cells infected with the MyoD retrovirus (Figure 2A). The expression of miR-1 and miR-133a primary transcripts at 24 h post-differentiation was completely inhibited in both of the Prmt5 AS cell lines, indicating that Prmt5 is required for the induction of myogenic miRNA expression (Figure 2B).
Figure 2.
Myogenic miRNA expression is compromised in Prmt5 antisense cell lines. (A) Immunoblot showing the Prmt5 and MyoD protein levels in vector- and MyoD-infected NIH 3T3 and Prmt5 antisense cell lines C1 and C12 at the onset of differentiation. The same blot was stripped and probed with PI3 Kinase (PI3K) antibody as a loading control. (B) qPCR analyses of primary transcripts of miR-1 and miR-133a upon MyoD-mediated differentiation of NIH 3T3 cells and the Prmt5 antisense cell lines, C1 and C12, along with the empty vector (EV) control. Expression levels were monitored 24 h post-differentiation. The expression of each miRNA in NIH 3T3 cells infected and differentiated with the empty vector retrovirus was normalized to 1. Results are the average of three independent experiments ± standard deviation.
To investigate whether the regulation of miRNA expression by Prmt5 was direct, we performed ChIP for Prmt5 and dimethylated histone 3 arginine 8 (diMeH3R8), a Prmt5-mediated histone modification (28), at miRNA regulatory regions in MyoD-differentiated cells. As previously reported (21), we found that Prmt5 and diMeH3R8 localized to the myogenin promoter in differentiated cells, but we could not detect the binding of Prmt5 or diMeH3R8 on any of the previously characterized myogenic regulatory regions upstream of miR-1 or miR-133a (data not shown). This suggests that the requirement for Prmt5 during myogenic miRNA induction is indirect. As Prmt5 is directly required for myogenin expression during differentiation (21), we hypothesized that the failure to induce myogenic miRNAs was due to the lack of myogenin expression when Prmt5 levels were reduced. To address this possibility, we took advantage of our previous findings that these fibroblasts could be differentiated into myotubes in the absence of MyoD by simultaneously expressing myogenin and Mef2D1b (39). Mef2D1b is a muscle-specific isoform of Mef2D that cooperates with myogenin to induce differentiation but that is lacking in these fibroblasts (39,43,44). If the lack of myogenic miRNA expression in Prmt5 AS cells is due to the lack of myogenin, then one would expect that introducing myogenin during the differentiation process would restore the ability of the differentiating cells to express the myogenic miRNAs.
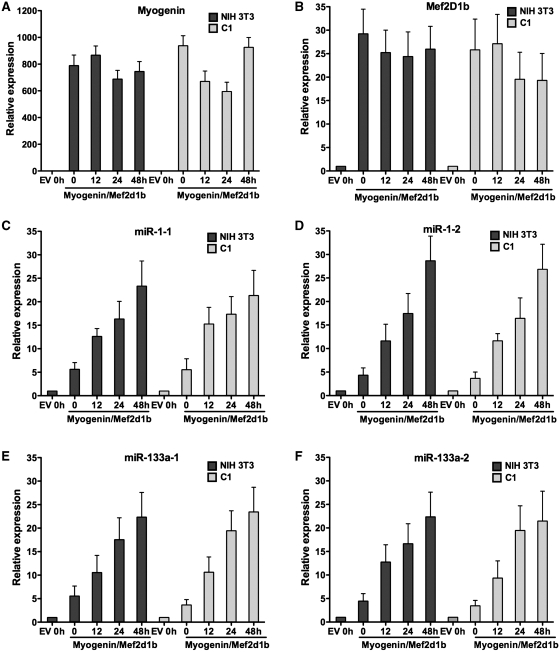
NIH 3T3 and C1 Prmt5 AS cells were infected with retroviruses encoding myogenin and Mef2D1b in a sequential manner and differentiated. We could detect the expression of myogenin and Mef2D1b in both the NIH 3T3 and the C1 cells from the onset of differentiation throughout the differentiation time course (Figure 3A and B), demonstrating that each of the cell lines at each of the time points assayed were equivalently expressing myogenin and Mef2D1b. We then tested the expression of primary miR-1 and miR-133a transcripts and observed that all four were robustly expressed in a manner independent of the presence of Prmt5 (Figure 3C–F). These results demonstrate that loss of miRNA expression in Prmt5 AS cell lines could be complemented by expressing myogenin and Mef2D1b, indicating that the Prmt5 requirement for myogenic miRNA expression is indirect via the induction of myogenin. In this experiment, we also noted that miRNA expression was induced earlier when cells were differentiated by myogenin and Mef2D1b than when they were differentiated by MyoD (compare Figures 1 and 3C–F). This observation argues that myogenic miRNAs may be targets for activation by myogenin because the onset of myogenic microRNA expression correlated with the introduction of myogenin. This observation is also consistent with a previous report indicating that myogenin can bind to E boxes upstream of these myogenic miRNA sequences (14).
Figure 3.
Expression of myogenin and Mef2D1b complements the loss of myogenic miRNA expression in a Prmt5 AS cell line. (A and B) Relative expression of myogenin and Mef2D1b upon differentiation in NIH 3T3 and in the Prmt5 AS cell line, C1, infected with retrovirus encoding myogenin and Mef2D1b. (C–F) qPCR analyses of primary transcripts of miR-1 and miR-133a in NIH 3T3 and C1 cells expressing myogenin and Mef2D1b at various times post-differentiation. The data represent the average of three independent experiments ± standard deviation. Expression at Time 0 in the empty vector (EV) control is normalized to 1. h, hours.
Carm1/Prmt4 is required for myogenic miRNA expression and interacts with miRNA regulatory sequences
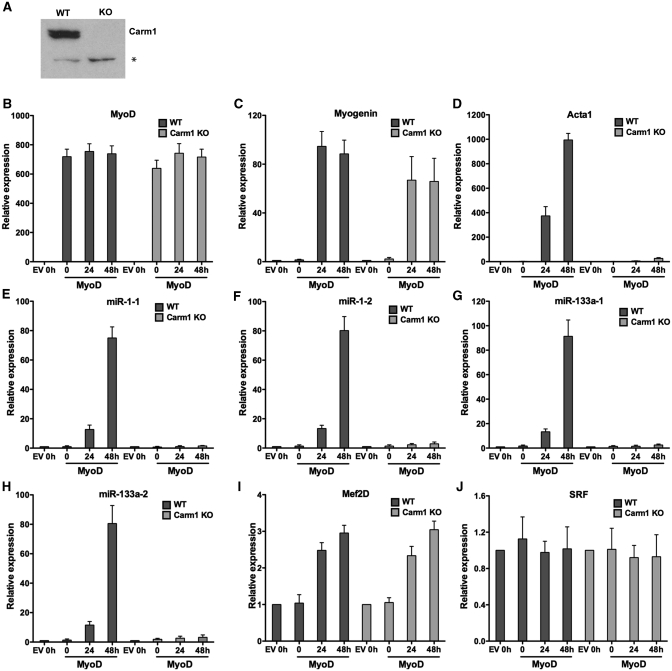
Carm1/Prmt4 is an arginine methyltransferase that methylates arginines 17 and 26 on histone H3 (29,30). Carm1/Prmt4 is required for the expression of late myogenic genes but is dispensable for the expression of myogenin, which is expressed at early times post-differentiation (20). To determine whether Carm1/Prmt4 contributes to myogenic miRNA expression, we utilized cell lines that were derived from wild type and Carm1/Prmt4-deficient mouse embryos via a 3T3 passaging protocol (35). An immunoblot with Carm1/Prmt4 antibody confirmed the absence of Carm1/Prmt4 protein in the knockout (KO) fibroblasts (Figure 4A). We then tested whether Carm1/Prmt4 was required for the proper expression of myogenic miRNA genes. We differentiated wild type and Carm1/Prmt4 KO fibroblasts by expressing MyoD or the empty vector as a control. Cells infected with the MyoD encoding retrovirus expressed similar levels of MyoD (Figure 4B). Differentiated Carm1/Prmt4 KO fibroblasts expressed normal levels of myogenin but failed to express late marker gene Acta1, in agreement with previous results [(20); Figure 4C and D]. When tested for the expression of primary transcripts of miR-1 and miR-133a, no or minimal miRNA induction was observed in Carm1/Prmt4 KO cells at all times post-differentiation, whereas normal induction of miRNA expression was observed in the wild-type fibroblasts upon myogenic differentiation (Figure 4E–H). Analysis of other myogenic regulatory protein such as Mef2D and SRF showed that the absence of Prmt4/Carm1 did not alter the expression of these genes (Figure 4I and J). These results show that Carm1/Prmt4 is required for miR-1 and miR-133a expression during myogenesis.
Figure 4.
Carm1/Prmt4 is required for the induction of myogenic miRNA expression during myogenesis. (A) Immunoblot demonstrating the absence of Carm1/Prmt4 in the knockout (KO) cells. A cross-reacting band (asterisk) demonstrates equal loading between gel lanes. (B–D) Relative expression of MyoD, myogenin and Acta1 mRNAs in the wild-type (WT) and Carm1/Prmt4 KO cells differentiated along the myogenic pathway by expressing MyoD or the empty vector (EV) control. (E–H) Relative expression of primary transcripts of miR-1-1, miR-1-2, miR-133a-1 and miR-133a-2 at various times post-differentiation in the WT and Carm1/Prmt4 knockout (KO) cells. (I and J) Relative expression of Mef2D and SRF mRNAs at various times post-differentiation in the WT and Carm1/Prmt4 knockout (KO) cells. The data represent the average of three independent experiments ± standard deviation. The expression at time 0 in empty vector infected cells was normalized to 1. h, hours.
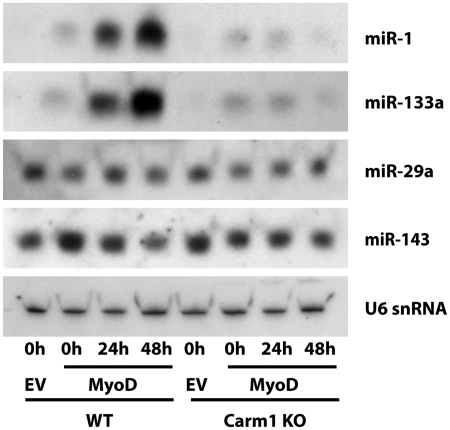
To determine whether Carm1/Prmt4 is required for the accumulation of mature microRNAs, we performed northern blots. Figure 5 shows differentiation-dependent accumulation of the mature forms of miR-1 and miR-133 that is severely reduced in the absence of Carm1/Prmt4. By contrast, the widely expressed microRNAs miR-29a and miR-143 were unaffected by differentiation or by the absence of Carm1/Prmt4. These data indicate that the accumulation of mature myogenic microRNAs, like the accumulation of myogenic microRNA primary transcripts, is dependent on Carm1/Prmt4.
Figure 5.
Carm1/Prmt4 is required for the accumulation of mature myogenic miRNAs during myogenesis. Duplicate Northern blots were prepared using RNA isolated from the indicated samples at the indicated times. Levels of the myogenic microRNAs, miR-1 and miR-133a, and the widely expressed microRNAs, miR-29a and miR-143, were determined. U6 snRNA was probed as a loading control. Blot 1 was sequentially probed for miR-1, miR-29a and U6. Blot 2 was sequentially probed for miR-133a, miR-143 and U6. The differences in the two U6 blots were negligible; only the U6 image from Blot 1 is shown.
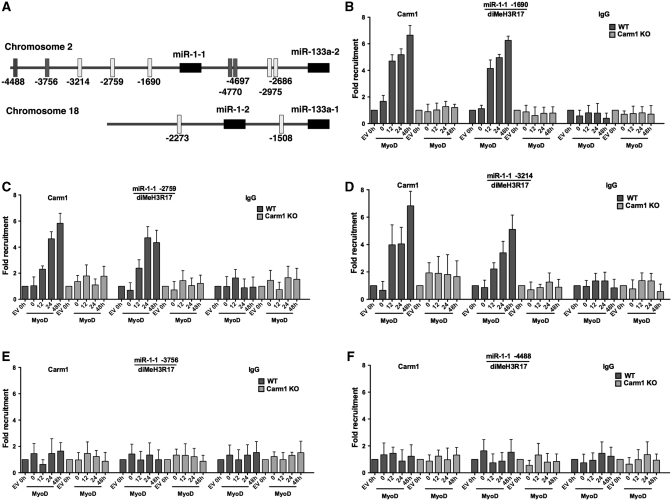
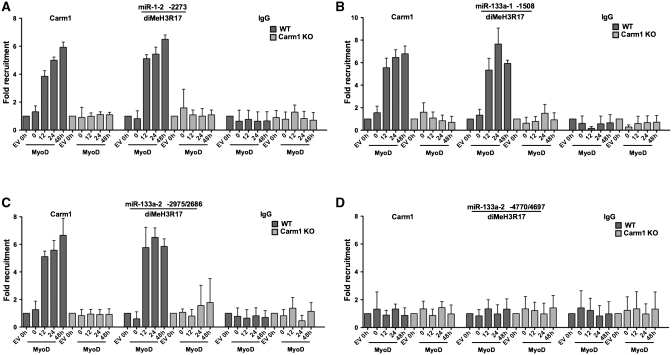
We next addressed whether Carm1/Prmt4 binds to the upstream regulatory regions of miRNA genes and whether the Carm1/Prmt4 substrate, dimethylated H3R17, is also present at these sequences. A schematic diagram illustrates the positions of E boxes upstream of the miR-1-1, miR-133a-2, miR-1-2 and miR-133a-1 stem–loop sequences (Figure 6A). Open rectangles indicate E box containing regions that we previously showed were bound by both MyoD and the Brg1 ATPase that is the enzymatic subunit of the SWI/SNF ATP-dependent chromatin remodeling enzyme. Solid rectangles indicate E box containing sequences that we previously showed were not bound by either MyoD or Brg1 [Figure 6A; (13)]. ChIP experiments for Carm1/Prmt4 and diMeH3R17 at each of these putative miRNA regulatory regions in MyoD-differentiated wild-type and Carm1/Prmt4 KO fibroblasts showed that Carm1/Prmt4 binds to the proximal three E box containing regions of the miR-1-1 upstream region but not to the two more distal E-boxes (Figure 6B–F). Carm1/Prmt4 also interacted with the proximal E box containing sequences of the miR-1-2, miR-133a-1 and miR-133a-2 upstream regions. However, no binding was observed at the distal E box containing sequences upstream of miR-133a-2 (Figure 7A–D). Carm1/Prmt4 binding at these regions correlated precisely with the presence of diMeH3R17, and the presence of diMeH3R17 was entirely Prmt4/Carm1 dependent (Figures 6B–F and 7A–D). Binding of both Carm1/Prmt4 and diMeH3R17 was first observed at 12 h post-differentiation, consistent with the onset of miRNA expression (Figure 1) and was maintained through the 48-h post-differentiation time point. These results demonstrate that Carm1/Prmt4 and diMeH3R17 are present in the upstream regulatory regions of myogenic miRNAs, miR-1 and miR-133a, that dimethylation of H3R17 at these sequences required Carm1/Prmt4, and that the onset of Carm1/Prmt4 and diMeH3R17 binding occurred at the time of myogenic miRNA gene induction. In combination with studies from our previous work (13), we have demonstrated that each of the sites bound by Carm1/Prmt4 and diMeH3R17 also are bound by MyoD and by Brg1.
Figure 6.
Prmt4/Carm1 binds and dimethylates H3R17 at regulatory regions upstream of myogenic miR-1-1 miRNA. (A) A schematic diagram showing the location of consensus E-boxes, the cis elements that mediate the interaction of myogenic regulatory factors, upstream of miR-1 and miR-133a genes. This schematic diagram is a modified version of the diagram published in Figure 6 of ref. (13), that was amended with permission from the American Society for Microbiology. (B–F) ChIP experiments demonstrating the interaction of Carm1/Prmt4 and the presence of dimethylated (diMe) H3R17 at the three proximal E-boxes but not at the two distal E-boxes of miR-1-1 in WT but not in the Carm1/Prmt4 KO cells at various times during myogenic differentiation. No binding of IgG was found at any of the E-boxes at any time point. The binding at time 0 in the empty vector (EV) control in WT and Carm1/Prmt4 KO cells was normalized to 1. Data represent the average of three independent experiments ± standard deviation. h, hours.
Figure 7.
Carm1/Prmt4 binding and the presence of dimethylated (diMe)H3R17 at miR-1-2, miR-133a-1 and miR-133a-2 regulatory regions. ChIP experiments show the presence of Carm1, and diMeH3R17 at E-box containing sequences upstream of miR-1-2 (A) and miR-133a-1 (B) and proximal E-boxes of miR-133a-2 (C) but not the distal E-boxes of miR-133a-2 (D) at various times following MyoD mediated differentiation in the WT but not the Carm1/Prmt4 KO cells. Values for empty vector (EV) infected cells at time 0 of differentiation were normalized to 1. Data represent the average of three independent experiments ± standard deviation. h, hours.
Carm1/Prmt4 is required for the binding of myogenin and Brg1 at miRNA regulatory regions
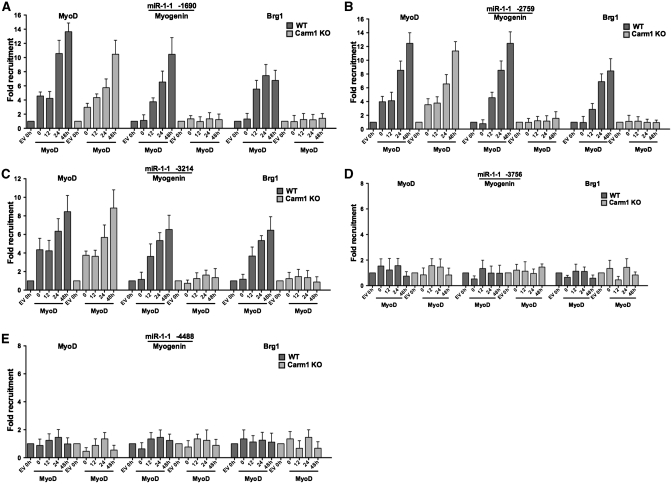
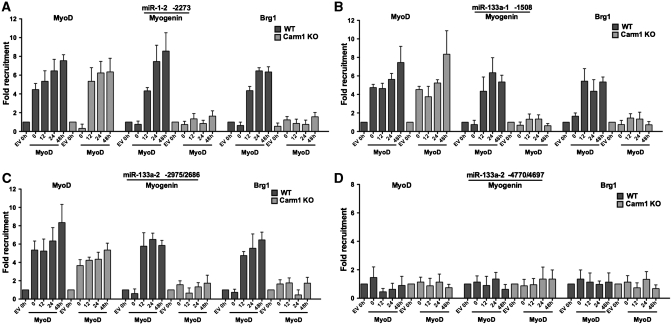
Brg1-based SWI/SNF chromatin remodeling enzyme function is required for the expression of miR-1 and miR-133a primary transcripts during myogenesis (13). In addition, MyoD has been shown to interact with upstream regulatory regions of miR-1 and miR-133a in differentiated C2C12 myoblasts (14) as well as in MyoD-differentiated fibroblasts and in skeletal muscle tissue (13). The myogenin factor also binds to some of these sequences in differentiated C2C12 myoblasts (14). To address the relationship between Carm1/Prmt4, the dimethylation of H3R17, and the binding of Brg1, MyoD and myogenin, we carried out ChIP assays on wild-type and Carm1/Prmt4 KO cells that were differentiated by MyoD or the empty vector. Consistent with our previous report (13), MyoD was found bound to the proximal but not distal regulatory regions of miR-1 and miR-133a in wild-type cells both at the onset of and during differentiation (Figures 8 and 9). Binding of MyoD at these sites was not affected in the Carm1/Prmt4 KO cells (Figures 8 and 9), indicating that MyoD could access these E boxes in the absence of Prmt4 and in the absence of dimethylated H3R17. By contrast, myogenin binding was not observed at any region upstream of the microRNAs at the onset of differentiation. However, myogenin did bind to the same sequences bound by MyoD, Carm1/Prmt4 and diMeH3R17 at 12 h post-differentiation and beyond in MyoD-differentiated cells (Figures 8 and 9). Intriguingly, myogenin binding was completely inhibited in the absence of Carm1/Prmt4 (Figures 8 and 9). This was not due to a change in myogenin expression in the Carm1/Prmt4-deficient cells (Figure 4C), suggesting instead that the local chromatin environment is insufficient to permit myogenin binding to the miRNA regulatory regions of miR-1 and miR-133a in the absence of Carm1/Prmt4.
Figure 8.
The myogenic regulatory factors MyoD and myogenin and the chromatin remodeling enzyme Brg1 interact with the proximal E-boxes containing sequences upstream of miR-1-1. ChIP experiments demonstrated the binding of MyoD, myogenin and Brg1 at regions containing the proximal E-boxes (A–C) but not to the distal E-boxes (D and E) of miR-1-1 at the indicated times in MyoD differentiated WT and Carm1/Prmt4 KO cells. Values at time 0 in the empty vector (EV) control were normalized to 1. The experiment was repeated three times, and the data represent the average of three experiments ± standard deviation. h, hours.
Figure 9.
Interaction of MyoD, myogenin and Brg1 at upstream regulatory regions of miR-1-2, miR-133a-1 and miR-133a-2. ChIP experiments demonstrate the interaction of MyoD, myogenin and Brg1 at sequences containing the E-box regulatory elements upstream of miR-1-2 (A), miR-133a-1 (B) and the proximal (C) but not the distal E-boxes (D) of miR-133a-2 at the indicated times following MyoD-mediated differentiation of WT and Carm1/Prmt4 KO cells. The values from the empty vector (EV) control at time 0 in both WT and Carm1/Prmt4 KO cells were normalized to 1. The results are the average of three independent experiments ± standard deviation. h, hours.
Additional ChIP experiments revealed binding of Brg1 to the proximal regulatory regions but not to the distal E-boxes of miR-1-1 in the wild-type MyoD-differentiated cells (Figure 8A–E). Brg1 was also found to interact with regulatory regions upstream of miR-1-2 and miR-133a-1 and to the proximal but not distal regions of miR-133a-2 upon myogenic differentiation. No Brg1 binding was observed at any of these sites in the Carm1/Prmt4 KO cells at any time (Figure 9A–D). These results demonstrate that Carm1/Prmt4 is required for the binding of myogenin and Brg1 and suggest that the requirement for Carm1/Prmt4 for the proper expression of myogenic miRNAs during myogenic differentiation is based on its ability to facilitate Brg1 and myogenin binding to the miRNA regulatory sequences.
DISCUSSION
An increasing number of miRNAs, both muscle-specific and ubiquitously or widely expressed, have been shown to affect muscle cell proliferation, development and differentiation (45–48). In addition, there is a growing appreciation that specific miRNAs are involved in the onset of disease states affecting all three types of muscle (8,10). miRNAs, therefore, play an important and essential role in the regulation of muscle formation and function, and defining the principles controlling the expression of these miRNAs is critical to better understanding muscle biology and muscle disease.
We have focused on the transcriptional regulation of expression of several miRNAs that are induced during muscle differentiation and are largely muscle specific in their expression. The miRNAs are critical regulators of myoblast proliferation and differentiation (5). Previous studies have implicated MyoD, myogenin, Mef2 and the respective binding sites for these factors in myogenic miRNA regulatory sequences in the induction of miRNA expression (5,14,16). Thus, the data support the idea that myogenic, DNA-binding transcription factors that control myogenic mRNA expression also control myogenic miRNA expression.
Considerable effort has been made to understand how epigenetic regulators influence myogenic mRNA expression. These studies have revealed roles for numerous histone-modifying enzymes, specific histone modifications and ATP-dependent chromatin remodeling enzymes (49–51). Whether such chromatin modifiers also contribute myogenic miRNA expression has only recently begun to be investigated. Recently, we used morpholinos to reduce Brg1 levels in developing zebrafish and observed that ∼40% had altered tail development with morphologically altered somite structure. In addition, these embryos had altered sarcomeric actin organization in the tissue. Analysis of myogenic miRNA expression in these altered tissues showed that there was a significant decrease in expression relative to the controls (13). This phenotype was remarkably similar to that observed by others when the microRNA processing enzyme, Dicer, was mutated or when the levels of myogenic miRNAs miR-1 and miR-133 were reduced by morpholino injection (52). Together, the two studies suggest that Brg1 and miRNAs are part of the same regulatory pathway. Subsequent studies in Brg1-deficient myoblasts, in primary skeletal muscle tissue and in a cell culture model for skeletal muscle differentiation indicated that Brg1 is not only required for skeletal muscle mRNA expression and differentiation, as previously reported (37,53,54), but also directly required for myogenic miRNA expression (13). To date, no other chromatin modifiers or remodeling enzymes have been shown to contribute to myogenic miRNA regulation.
Given the recently demonstrated roles for the arginine methyltransferases Prmt5 and Carm1/Prmt4 in myogenic mRNA expression, we asked whether these enzymes are involved in myogenic miRNA regulation. We previously showed that Prmt5 is directly required for expression of the myogenin gene (21), but despite binding to regulatory sequences controlling myogenic genes expressed at late times of myogenesis in cultured cells, it was not required for the expression of these genes. Carm1/Prmt4, in contrast, bound to late gene regulatory sequences and was required for expression during differentiation in culture (20). Our studies of Prmt5 and Carm1/Prmt4 function during myogenic miRNA expression revealed that the requirements for these arginine methyltransferases were similar to their functions in regulating myogenic late gene mRNA expression. Thus, there is conservation between Prmt5 and Carm1/Prmt4 function in the regulation of myogenic miRNAs and in the regulation of a subset of myogenic mRNAs. It is important to note, however, that despite the similarity to the regulation of myogenic late gene mRNAs, this does not indicate that the myogenic miRNAs should be considered ‘late’ expressing genes. In vivo analysis of the expression of transgene constructs controlled by regulatory sequences of several myogenic miRNAs indicates that they can be detected in somites at E9.5 and at later stages of development (6,16).
The ChIP experiments performed in the Carm1/Prmt4-deficient cells revealed several mechanistic insights into how Carm1/Prmt4 functions at myogenic miRNA regulatory sequences. First, incorporation of the Carm1/Prmt4 substrate, dimethylated H3R17, at these sequences is absolutely dependent upon the presence of Carm1/Prmt4 (Figures 6 and 7). Second, the subset of E box containing sequences that showed Carm1/Prmt4 binding correlated exactly with those sequences previously found to bind both MyoD and the Brg1 SWI/SNF ATPase (13). This suggests that factors regulating myogenic miRNA expression are functioning at the same cis-acting sequences. Third, cells lacking Carm1/Prmt4 not only were deficient for the H3R17 modification, but also failed to target the Brg1 ATPase of SWI/SNF chromatin remodeling enzymes to the regulatory sequences (Figures 8 and 9). We have previously demonstrated that chromatin remodeling at these miRNA sequences and microRNA expression are entirely dependent upon Brg1 (13). The absence of Brg1 indicates that these sequences cannot undergo chromatin remodeling and are therefore not accessible for active gene expression. The mechanism(s) by which Carm1/Prmt4 directly or indirectly facilitates Brg1 binding remains to be determined, but we can speculate that Carm1/Prmt4 mediated histone modifications likely contribute to targeting the ATP-dependent chromatin remodeler.
The last significant conclusion from the ChIP experiments with Carm1/Prmt4 deficient cells is the novel observation that MyoD binding to these sequences was unaffected while myogenin binding was abolished (Figures 8 and 9). The data presented here and in our prior study (13) indicate that MyoD binding to these sequences is independent of Brg1 function and the presence of Carm1/Prmt4. Thus, MyoD binding does not require these particular enzymes or the chromatin structural modifications that they mediate. Whether different chromatin-modifying or remodeling enzymes are required for MyoD binding at these sequences is not known. By contrast, myogenin binding is completely dependent on Carm1/Prmt4. Despite the sequence similarities between the MyoD and myogenin proteins, it has long been established that MyoD is intrinsically better at functioning in a chromatin environment than is myogenin (55,56). How the structural differences between MyoD and myogenin relate to the Carm1/Prmt4 dependence of binding at miRNAs regulatory sequences is an obvious topic for further work. Nevertheless, the presence of MyoD at these sequences in both wild-type and Carm1/Prmt4-deficient cells suggests that MyoD binding is not sufficient for myogenic miRNA expression. Instead, the binding of myogenin correlates with transcriptional competence, suggesting that it is myogenin that is facilitating transcriptional activation. Previous cooperativity between Carm1/Prmt4 and both myogenin and Mef2 has been reported (20,31), supporting the idea that Carm1/Prmt4 acts as a co-activator for myogenin and Mef2 to promote myogenic miRNA expression.
The functional interrelationships between two distinct types of Prmts and the Brg1 chromatin-remodeling enzyme during differentiation-mediated induction of myogenic miRNAs suggest that regulation of these myogenic miRNAs will be as complex as the regulation of myogenic mRNAs. It is therefore likely that histone acetyltransferases, lysine methyltransferases and other histone-modifying enzymes will also be involved in myogenic miRNA expression. Because histone modifications are dynamically regulated, there also will likely be roles for deacetylating and demethylating enzymes. How the activities of multiple chromatin modifiers are integrated with the functions of SWI/SNF enzymes and gene specific transcription factors to regulate myogenic miRNA expression will be an interesting topic for future studies.
Finally, the similarity in the regulation of the miR-1 and miR-133 myogenic microRNAs is interesting because miR-1 and miR-133 have opposing functions; miR-1 promotes differentiation, whereas miR-133 promotes myoblast proliferation (5). We showed that differentiation-dependent kinetics of induction of miR-1-1, miR-1-2, miR-133a-1 and miR-133a-2 are equivalent in MyoD-differentiated fibroblasts and that the four myogenic microRNAs are also induced with similar kinetics in myogenin/Mef2D1b-differentiated fibroblasts (Figures 1 and 3). This is consistent with the work of others who have seen differentiation-dependent increases in miR-1 and miR-133 expression in C2C12 cells (5,7,14,57) and with data from transgenic animal studies indicating that the intragenic regions between the two miR-1 and miR-133 clusters both drive reporter gene expression in the heart and somites with similar kinetics (6,16). The reason that the induction and regulation of two microRNAs that promote opposite developmental outcomes is similar is not known. However, others have speculated that differential regulation of the miRNAs at steps downstream of the transcription of these microRNAs might promote their differences in function, as might the obvious differences in the functions of the proteins encoded by their target mRNAs (16,58,59). In addition, despite the similarities in regulation by chromatin-modifying and remodeling enzymes documented here and previously (13), it remains possible that subtle, yet uncharacterized, differences in regulatory mechanisms or in the timing of microRNA expression could promote differences in miR-1 and miR-133 function.
FUNDING
National Institutes of Health (grants GM56244 to A.N.I. and CA116093 to S.S.). A.N.I. is a member of the University of Massachusetts Medical School Diabetes Endocrine Research Center supported by National Institutes of Health grant DK32520. Funding for open access charge: National Institutes of Health R01 (GM56244 to A.N.I.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Silvana Konda for retrovirus preparation, Dr. Mark Bedford for providing the Carm1/Prmt4 deficient cell line and its wild-type counterpart, and Qiong Wu for reviewing the manuscript.
REFERENCES
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bushati N, Cohen SM. microRNA functions. Annu. Rev. Cell Dev. Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 3.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Na.t Rev. Mol. Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 4.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 5.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 7.Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J. Cell. Biol. 2006;174:677–687. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen JF, Callis TE, Wang DZ. microRNAs and muscle disorders. J. Cell. Sci. 2009;122:13–20. doi: 10.1242/jcs.041723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisenberg I, Alexander MS, Kunkel LM. miRNAS in normal and diseased skeletal muscle. J. Cell. Mol. Med. 2009;13:2–11. doi: 10.1111/j.1582-4934.2008.00524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams AH, Liu N, van Rooij E, Olson EN. MicroRNA control of muscle development and disease. Curr. Opin. Cell. Biol. 2009;21:461–469. doi: 10.1016/j.ceb.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Small EM, Frost RJ, Olson EN. MicroRNAs add a new dimension to cardiovascular disease. Circulation. 2010;121:1022–1032. doi: 10.1161/CIRCULATIONAHA.109.889048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Latronico MV, Condorelli G. MicroRNAs and cardiac pathology. Nat. Rev. Cardiol. 2009;6:419–429. doi: 10.1038/nrcardio.2009.56. [DOI] [PubMed] [Google Scholar]
- 13.Mallappa C, Nasipak BT, Etheridge L, Androphy EJ, Jones SN, Sagerstrom CG, Ohkawa Y, Imbalzano AN. Myogenic microRNA expression requires ATP-dependent chromatin remodeling enzyme function. Mol. Cell. Biol. 2010;30:3176–3186. doi: 10.1128/MCB.00214-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc. Natl Acad. Sci. USA. 2006;103:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenberg MI, Georges SA, Asawachaicharn A, Analau E, Tapscott SJ. MyoD inhibits Fstl1 and Utrn expression by inducing transcription of miR-206. J. Cell. Biol. 2006;175:77–85. doi: 10.1083/jcb.200603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu N, Williams AH, Kim Y, McAnally J, Bezprozvannaya S, Sutherland LB, Richardson JA, Bassel-Duby R, Olson EN. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc. Natl Acad. Sci. USA. 2007;104:20844–20849. doi: 10.1073/pnas.0710558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sokol NS, Ambros V. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes Dev. 2005;19:2343–2354. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr. Opin. Cell. Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 19.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dacwag CS, Bedford MT, Sif S, Imbalzano AN. Distinct protein arginine methyltransferases promote ATP-dependent chromatin remodeling function at different stages of skeletal muscle differentiation. Mol. Cell. Biol. 2009;29:1909–1921. doi: 10.1128/MCB.00742-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dacwag CS, Ohkawa Y, Pal S, Sif S, Imbalzano AN. The protein arginine methyltransferase Prmt5 is required for myogenesis because it facilitates ATP-dependent chromatin remodeling. Mol. Cell. Biol. 2007;27:384–394. doi: 10.1128/MCB.01528-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 23.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 24.Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat. Struct. Mol. Biol. 2007;14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- 25.Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol. Cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 26.Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol. Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pollack BP, Kotenko SV, He W, Izotova LS, Barnoski BL, Pestka S. The human homologue of the yeast proteins Skb1 and Hsl7p interacts with Jak kinases and contains protein methyltransferase activity. J. Biol. Chem. 1999;274:31531–31542. doi: 10.1074/jbc.274.44.31531. [DOI] [PubMed] [Google Scholar]
- 28.Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol. Cell. Biol. 2004;24:9630–9645. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bauer UM, Daujat S, Nielsen SJ, Nightingale K, Kouzarides T. Methylation at arginine 17 of histone H3 is linked to gene activation. EMBO Rep. 2002;3:39–44. doi: 10.1093/embo-reports/kvf013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 31.Chen SL, Loffler KA, Chen D, Stallcup MR, Muscat GE. The coactivator-associated arginine methyltransferase is necessary for muscle differentiation: CARM1 coactivates myocyte enhancer factor-2. J. Biol. Chem. 2002;277:4324–4333. doi: 10.1074/jbc.M109835200. [DOI] [PubMed] [Google Scholar]
- 32.Pal S, Yun R, Datta A, Lacomis L, Erdjument-Bromage H, Kumar J, Tempst P, Sif S. mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol. Cell. Biol. 2003;23:7475–7487. doi: 10.1128/MCB.23.21.7475-7487.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu W, Cho H, Kadam S, Banayo EM, Anderson S, Yates JR, III, Emerson BM, Evans RM. A methylation-mediator complex in hormone signaling. Genes Dev. 2004;18:144–156. doi: 10.1101/gad.1141704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jainchill JL, Aaronson SA, Todaro GJ. Murine sarcoma and leukemia viruses: assay using clonal lines of contact-inhibited mouse cells. J. Virol. 1969;4:549–553. doi: 10.1128/jvi.4.5.549-553.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yadav N, Lee J, Kim J, Shen J, Hu MC, Aldaz CM, Bedford MT. Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proc. Natl Acad. Sci. USA. 2003;100:6464–6468. doi: 10.1073/pnas.1232272100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl Acad. Sci. USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de la Serna IL, Carlson KA, Imbalzano AN. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat. Genet. 2001;27:187–190. doi: 10.1038/84826. [DOI] [PubMed] [Google Scholar]
- 38.Rawls A, Morris JH, Rudnicki M, Braun T, Arnold HH, Klein WH, Olson EN. Myogenin's functions do not overlap with those of MyoD or Myf-5 during mouse embryogenesis. Dev. Biol. 1995;172:37–50. doi: 10.1006/dbio.1995.0004. [DOI] [PubMed] [Google Scholar]
- 39.Ohkawa Y, Marfella CG, Imbalzano AN. Skeletal muscle specification by myogenin and Mef2D via the SWI/SNF ATPase Brg1. EMBO J. 2006;25:490–501. doi: 10.1038/sj.emboj.7600943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valdez MR, Richardson JA, Klein WH, Olson EN. Failure of Myf5 to support myogenic differentiation without myogenin, MyoD, and MRF4. Dev. Biol. 2000;219:287–298. doi: 10.1006/dbio.2000.9621. [DOI] [PubMed] [Google Scholar]
- 41.de La Serna IL, Carlson KA, Hill DA, Guidi CJ, Stephenson RO, Sif S, Kingston RE, Imbalzano AN. Mammalian SWI-SNF complexes contribute to activation of the hsp70 gene. Mol. Cell. Biol. 2000;20:2839–2851. doi: 10.1128/mcb.20.8.2839-2851.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 43.Martin JF, Miano JM, Hustad CM, Copeland NG, Jenkins NA, Olson EN. A Mef2 gene that generates a muscle-specific isoform via alternative mRNA splicing. Mol. Cell. Biol. 1994;14:1647–1656. doi: 10.1128/mcb.14.3.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Russo S, Tomatis D, Collo G, Tarone G, Tato F. Myogenic conversion of NIH3T3 cells by exogenous MyoD family members: dissociation of terminal differentiation from myotube formation. J. Cell. Sci. 1998;111(Pt 6):691–700. doi: 10.1242/jcs.111.6.691. [DOI] [PubMed] [Google Scholar]
- 45.Cardinali B, Castellani L, Fasanaro P, Basso A, Alema S, Martelli F, Falcone G. Microrna-221 and microrna-222 modulate differentiation and maturation of skeletal muscle cells. PLoS One. 2009;4:e7607. doi: 10.1371/journal.pone.0007607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Small EM, O'Rourke JR, Moresi V, Sutherland LB, McAnally J, Gerard RD, Richardson JA, Olson EN. Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proc. Natl Acad. Sci. USA. 2010;107:4218–4223. doi: 10.1073/pnas.1000300107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sarkar S, Dey BK, Dutta A. MiR-322/424 and -503 are induced during muscle differentiation and promote cell cycle quiescence and differentiation by down-regulation of Cdc25A. Mol. Biol. Cell. 2010;21:2138–2149. doi: 10.1091/mbc.E10-01-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Townley-Tilson WH, Callis TE, Wang D. MicroRNAs 1, 133, and 206: critical factors of skeletal and cardiac muscle development, function, and disease. Int. J. Biochem. Cell. Biol. 2010;42:1252–1255. doi: 10.1016/j.biocel.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de la Serna IL, Ohkawa Y, Imbalzano AN. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat. Rev. Genet. 2006;7:461–473. doi: 10.1038/nrg1882. [DOI] [PubMed] [Google Scholar]
- 51.Saccone V, Puri PL. Epigenetic regulation of skeletal myogenesis. Organogenesis. 2010;6:48–53. doi: 10.4161/org.6.1.11293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mishima Y, Abreu-Goodger C, Staton AA, Stahlhut C, Shou C, Cheng C, Gerstein M, Enright AJ, Giraldez AJ. Zebrafish miR-1 and miR-133 shape muscle gene expression and regulate sarcomeric actin organization. Genes Dev. 2009;23:619–632. doi: 10.1101/gad.1760209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de la Serna IL, Ohkawa Y, Berkes CA, Bergstrom DA, Dacwag CS, Tapscott SJ, Imbalzano AN. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol. Cell. Biol. 2005;25:3997–4009. doi: 10.1128/MCB.25.10.3997-4009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simone C, Forcales SV, Hill DA, Imbalzano AN, Latella L, Puri PL. p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat. Genet. 2004;36:738–743. doi: 10.1038/ng1378. [DOI] [PubMed] [Google Scholar]
- 55.Bergstrom DA, Tapscott SJ. Molecular distinction between specification and differentiation in the myogenic basic helix-loop-helix transcription factor family. Mol. Cell. Biol. 2001;21:2404–2412. doi: 10.1128/MCB.21.7.2404-2412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gerber AN, Klesert TR, Bergstrom DA, Tapscott SJ. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: a mechanism for lineage determination in myogenesis. Genes Dev. 1997;11:436–450. doi: 10.1101/gad.11.4.436. [DOI] [PubMed] [Google Scholar]
- 57.Boutz PL, Chawla G, Stoilov P, Black DL. MicroRNAs regulate the expression of the alternative splicing factor nPTB during muscle development. Genes Dev. 2007;21:71–84. doi: 10.1101/gad.1500707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Callis TE, Chen JF, Wang DZ. MicroRNAs in skeletal and cardiac muscle development. DNA Cell Biol. 2007;26:219–225. doi: 10.1089/dna.2006.0556. [DOI] [PubMed] [Google Scholar]
- 59.van Rooij E, Liu N, Olson EN. MicroRNAs flex their muscles. Trends Genet. 2008;24:159–166. doi: 10.1016/j.tig.2008.01.007. [DOI] [PubMed] [Google Scholar]