Abstract
Wybutosine (yW) is a hypermodified nucleoside found in position 37 of tRNAPhe, and is essential for correct phenylalanine codon translation. yW derivatives widely exist in eukaryotes and archaea, and their chemical structures have many species-specific variations. Among them, its hydroxylated derivative, hydroxywybutosine (OHyW), is found in eukaryotes including human, but the modification mechanism remains unknown. Recently, we identified a novel Jumonji C (JmjC)-domain-containing protein, TYW5 (tRNA yW-synthesizing enzyme 5), which forms the OHyW nucleoside by carbon hydroxylation, using Fe(II) ion and 2-oxoglutarate (2-OG) as cofactors. In this work, we present the crystal structures of human TYW5 (hTYW5) in the free and complex forms with 2-OG and Ni(II) ion at 2.5 and 2.8 Å resolutions, respectively. The structure revealed that the catalytic domain consists of a β-jellyroll fold, a hallmark of the JmjC domains and other Fe(II)/2-OG oxygenases. hTYW5 forms a homodimer through C-terminal helix bundle formation, thereby presenting a large, positively charged patch involved in tRNA binding. A comparison with the structures of other JmjC-domain-containing proteins suggested a mechanism for substrate nucleotide recognition. Functional analyses of structure-based mutants revealed the essential Arg residues participating in tRNA recognition by TYW5. These findings extend the repertoire of the tRNA modification enzyme into the Fe(II)/2-OG oxygenase superfamily.
INTRODUCTION
Over 100 RNA modifications have been found throughout the three phylogenic domains (1), and these modified nucleosides have significant biological roles in maintaining life. Most of these modifications exist in transfer RNA (tRNA), and they confer various chemical properties to the RNA residues. Wybutosine (yW) and its derivatives are highly modified nucleosides characterized by a tricyclic base and a bulky side chain (2,3) (Figure 1). These nucleosides are located at position 37, adjacent to the 3′ position of the anticodon, of eukaryotic and archaeal tRNAPhe. Recent studies have shown that the yW base is important for the maintenance of translational fidelity. Whereas yW does not affect the phenylalanylation of tRNAPhe under physiological conditions, tRNAPhe lacking the yW modification enhances codon frame shifting (4). Moreover, the bulky side chain of the yW base significantly suppresses the −1 frame shifting at a phenylalanine codon (5). Therefore, the bulky and hydrophobic nature of yW may stabilize the codon–anticodon pairing in the ribosome through a base-stacking interaction, thereby ensuring the correct reading frame (6).
Figure 1.
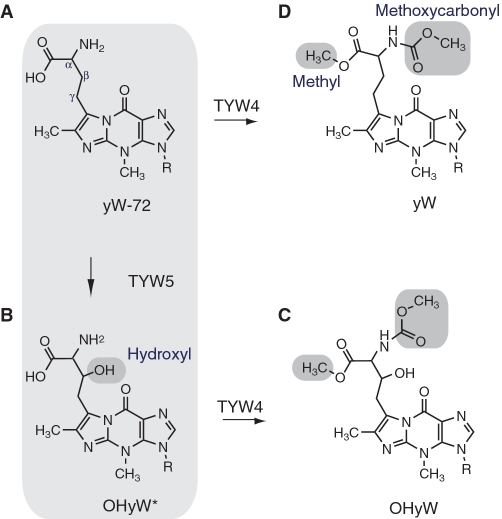
Biosynthetic pathway of wybutosine. The chemical structures of (A) 7-(α-amino-α-carboxypropyl)wyosine (yW-72), (B) undermodified hydroxywybutosine (OHyW*), (C) hydroxywybutosine (OHyW) and (D) wybutosine (yW) are shown.
Another distinctive feature of the yW modification is the existence of a wide variety of derivatives. At least ten derivatives reportedly exist in different phylogenetic groups (7). Among them, hydroxywybutosine (OHyW), undermodified hydroxywybutosine (OHyW*) and peroxywybutosine (o2yW) are hydroxylated yW derivatives found in higher eukaryotes, including human. These derivatives have a hydroxyl group or hydroperoxide group at the β position of the bulky chain of the yW base.
The biosynthetic pathway of yW has been well characterized in yeast (12). At first, the guanosine residue at position 37 is converted into the distinctive tricyclic structure by tRNA G37 methyltransferase, Trm5 (8) and the [4Fe-4S] cluster-containing radical SAM enzyme, TYW1 (9,10). Subsequently, aminocarboxyl transferase TYW2 (11) and methyltransferase TYW3 append the exocyclic groups in an Ado-Met-dependent manner to form yW-72 (yW minus 72 Da) (Figure 1). Finally, TYW4 attaches both the methyl and methoxycarbonyl groups to the aminocarboxyl side chain, to complete yW formation (12). However, since yeast tRNAPhe lacks OHyW, the biosynthetic mechanism for the hydroxylated yW derivatives remained elusive for over 30 years.
Recently, our group found that the biosynthesis of OHyW is catalyzed by a novel Jumonji C (JmjC)-domain-containing protein (13), TYW5 (tRNA yW-synthesizing enzyme 5). The JmjC domain was originally discovered in histone demethylases (14), which have important roles in the epigenetic regulation of gene expression, to control cell fate and identity. The JmjC-domain-containing histone demethylases catalyze the removal of the methyl group attached to the lysine ε-amino group in histones, through an oxidative reaction that requires Fe(II) and 2-oxoglutarate (2-OG) as cofactors. The JmjC domain is also found in other proteins, such as factor inhibiting hypoxia inducible factor 1 (FIH), which hydroxylates the β carbon of the Asn side chain in the hypoxia inducible factor (Hif) 1α transcription factor, thereby regulating the hypoxic response (15). Furthermore, these JmjC-domain-containing proteins belong to a large superfamily of Fe(II)/2-OG dependent oxygenases, which have a variety of roles in nutrient acquisition, antibiotic synthesis and alkylated nucleic acid repair (16). Among the JmjC-domain-containing protein family members, the structure of FIH shows the highest sequence similarity to TYW5 (26% identity to human TYW5). TYW5 also shares functional similarity to FIH (i.e. hydroxylates the β carbon of the amino-acid derived side chain of yW), and thus is considered to catalyze hydroxylation by a similar mechanism to that of FIH. Actually, our functional analysis demonstrated that TYW5 catalyzes Fe(II)/2-OG-dependent hydroxylation (17). Furthermore, we found that TYW5 hydroxylates the precursor of yW, yW-72 (Figure 1A), but not yW (Figure 1D), suggesting that the hydroxylation by TYW5 occurs prior to the modification by TYW4. Consistent with these findings, a recent bioinformatics analysis also predicted that TYW5 is involved in OHyW formation (18). However, the tRNA recognition manner and the hydroxylation mechanism by TYW5 have remained elusive.
To address these questions, we solved the crystal structures of human TYW5 (hTYW5) in the free and complex forms with 2-OG and Ni(II) ion at 2.5 and 2.8 Å resolutions, respectively. The structures revealed that hTYW5 consists of a C-terminal helix bundle and an N-terminal catalytic domain, which shares a similar β-jellyroll fold with the other JmjC-domain-containing proteins and the Fe(II)/2-OG dependent oxygenases. Based on a structural comparison with the active site of FIH, we found that two Arg residues, specifically conserved among the TYW5s, are located in the active site. The structure-based mutational analysis revealed that these two Arg residues are essential for tRNA recognition. In addition, homodimer formation through the C-terminal helix bundle is crucial for the hydroxylation activity. These results provide structural insights into the mechanism of Fe(II)/2-OG dependent hydroxylation and tRNA recognition by TYW5.
MATERIALS AND METHODS
Cloning, expression and purification of human TYW5 proteins
The gene encoding human TYW5 was cloned into the NdeI/BamHI sites of a modified pET28a vector (Novagen), with the thrombin site replaced by the PreScission Protease site (GE Healthcare). The protein was overexpressed in the Escherichia coli C41(DE3) strain (Lucigen) transformed with the pRARE plasmid (Novagen). Escherichia coli cells were grown to an OD600 = 0.8, and then the expression was induced by 0.5 mM isopropyl β-d-thiogalactopyranoside (IPTG), followed by cultivation for 18 h at 20°C. The cells were harvested by centrifugation, resuspended in buffer (50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 10 mM imidazole, 1 mM PMSF and 5 mM 2-mercaptoethanol), sonicated, and then centrifuged. The supernatant was applied to a Ni–NTA Superflow column (Qiagen). The eluted fractions were collected and dialyzed against 50 mM Tris–HCl, pH 8.0, containing 150 mM NaCl and 1 mM DTT, and then applied to a Resource Q column (GE Healthcare). The flow-through fractions, containing the enzyme activity, were collected, concentrated by ultrafiltration, and loaded onto a HiLoad 16/60 Superdex 200 column (GE Healthcare) equilibrated in 20 mM Tris–HCl buffer (pH 8.0), containing 300 mM NaCl and 1 mM DTT. The purified hTYW5 fractions eluted from the gel-filtration column were collected and concentrated to 10 mg ml−1 with an Amicon Ultra-4 filter (Millipore). The protein purity was assessed by SDS–PAGE. To obtain the selenomethionine-labeled proteins, the methionine-auxotroph E. coli strain B834(DE3) CodonPlus was transformed with the same plasmid. The cells were cultivated in M9 minimum medium containing 50 µg ml−1 selenomethionine, and the protein was purified in the same manner as native hTYW5.
Crystallization and data collection
The native crystals were obtained by the sitting-drop vapor diffusion method. The drops were prepared by mixing 1 µl of a 10 mg ml−1 TYW5 solution and 1 µl of the reservoir solution, containing 200 mM potassium thiocyanate and 10% (w/v) PEG 3350. Crystallization was performed at 20°C by the sitting drop vapor diffusion method. Crystals suitable for structural analysis appeared within 3 days. The selenomethionine-labeled crystals were obtained by the micro-seeding technique, using the native crystal as a seed, under the same crystallization conditions as for the native protein. Crystals were transferred stepwise to the harvesting solution, 240 mM potassium thiocyanate and 12% (w/v) PEG 3350, containing 30% (v/v) glycerol as a cryoprotectant and were flash-cooled in a liquid nitrogen stream at 100 K. To obtain the cofactor-bound crystals, the hTYW5 crystals were soaked for 3 h in a harvest solution, containing 1 mM NiCl2, 10mM 2-OG, 240 mM potassium thiocyanate and 12% (w/v) PEG 3,350.
Data collection, structure determination and refinement
X-ray diffraction data were collected at 100 K on beamline BL41XU at SPring-8 and beamline NW12A at the Photon Factory Advanced Ring (Tsukuba, Japan). Diffraction data were processed with the HKL2000 program (HKL Research). The structure was solved by the MAD method, using the selenomethionine-labeled crystal. The 16 selenium sites were initially located by the programs SHELXC and SHELXD (19), and the initial phases were calculated by using SHARP/autoSHARP (20), followed by automated model building using RESOLVE (21). The resultant model was refined by using the 2.80-Å resolution data. The model was further manually built with COOT (22) and refined with PHENIX (23). Molecular graphics were illustrated with CueMol (http://www.cuemol.org/). Statistics on data collection, phasing and refinement are provided in Table 1 and 2.
Table 1.
Data collection and phasing statistics
| Data collection statistics | SeMet |
|||
|---|---|---|---|---|
| X-ray source | SPring-8 BL41XU |
|||
| Wavelength (Å) | Peak | Edge | Remote high | Remote low |
| 0.97911 | 0.97937 | 0.96414 | 0.99509 | |
| Unit cell dimensions (Å, °) | a = 165, b = 165, c = 105 α = β = γ = 90 | |||
| Resolution (Å) | 50–2.5 (2.95–2.49) | 50–2.9 (2.95–2.90) | 50–2.9 (2.95–2.90) | 50–2.9 (2.95–2.90) |
| Unique reflections | 33 018 (1592) | 32 694 (1390) | 32 644 (1454) | 32 735 (1366) |
| Redundancy | 20.9 (9.1) | 10.2 (3.6) | 10.5 (4.2) | 10.3 (8.3) |
| Completeness (%) | 99.6 (98.9) | 98.6 (86.2) | 98.7 (90.3) | 98.3 (83.6) |
| I/σ(I) | 57.2 (3.4) | 37.6 (2.0) | 37.8 (2.1) | 37.7 (2.0) |
| Rsym | 0.078 (0.411) | 0.064 (0.403) | 0.065 (0.396) | 0.063 (0.400) |
| Phasing statistics | ||||
| Number of Se sites | 16 | |||
| Phasing power | ||||
| Iso (cen./acen.) | – | 1.240/1.378 | 1.247/1.310 | 1.334/1.401 |
| Ano | 0.574 | 2.202 | 0.897 | 0.129 |
| Rcullis | ||||
| Iso (cen./acen.) | – | 0.658/0.675 | 0.582/0.647 | 0.567/0.638 |
| Ano | 0.933 | 0.560 | 0.857 | 0.996 |
| Mean FOM | ||||
| Cen./Acen. | 0.44079/0.49979 | |||
The numbers in parentheses are for the last shell.

Table 2.
Structure refinement statistics
| Data collection statistics | Apo form | Cofactor bound form |
|---|---|---|
| X-ray source | PF NW12 | SPring-8 BL41XU |
| Wavelength (Å) | 1.0 | 1.48505 |
| Resolution (Å) | 50–2.5 (2.54–2.5) | 50–2.8 (2.85–2.8) |
| Unique reflections | 49 257 (2349) | 36 950 (1776) |
| Redundancy | 9.0 (4.3) | 19.0 (5.7) |
| Completeness (%) | 97.6 (94.9) | 99.6 (98.1) |
| I/σ(I) | 28.0 (2.6) | 56.4 (2.5) |
| Rsym | 0.075 (0.392) | 0.082 (0.480) |
| Refinement statistics | ||
| Resolution (Å) | 50–2.5 | 50–2.8 |
| No. of reflections (all/test) | 49 227/2462 | 36782/1840 |
| Rwork/Rfree | 0.2196/0.2812 | 0.2349/0.2993 |
| Number of atoms | ||
| Non-hydrogen | 9288 | 9057 |
| Water | 0 | 0 |
| RMSD of | ||
| Bond lengths (Å) | 0.009 | 0.01 |
| Bond angles (°) | 1.257 | 1.343 |
| Average B factor (Å2) | 77.7 | 86.9 |
| Ramachandran plot | ||
| Most favored (%) | 85.7 | 83.3 |
| Allowed region (%) | 14.3 | 16.6 |
| Disallowed region (%) | 0.0 | 0.1 |
The numbers in parentheses are for the last shell. Rwork = Σ|Fo – Fc|/ΣFo for reflections of work set. Rfree = Σ|Fo – Fc|/ΣFo for reflections of test set (5% of total reflections).
Site-directed mutagenesis
Site-directed mutagenesis of hTYW5 was performed with the plasmid pTYW5, according to the manufacturer’s instructions (Stratagene). Introduced mutations were confirmed by DNA sequencing. The mutant proteins were overexpressed in E. coli cells in the same manner as the wild-type protein, purified by Ni affinity chromatography, and dialyzed against Tris–HCl, pH 8.0, containing 300 mM NaCl and 1 mM DTT.
Gel filtration chromatography analysis
The purified hTYW5 and ΔC TYW5 proteins were loaded onto a HiLoad 16/60 Superdex 200 column (GE Healthcare). The column, which was equilibrated in 20 mM Tris–HCl buffer (pH 8.0), containing 300 mM NaCl and 1 mM DTT, was run at 0.5 ml/min in the same buffer as that used for crystallization.
Isolation and purification of yeast tRNAPhe
The Saccharomyces cerevisiae BY4742 strain (Mat α; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0) lacking TYW4 (YOL141w::kanmx4) was obtained from EUROSCARF. The ΔTYW4 strain was cultivated in YPD (2% peptone, 1% yeast extract and 2% glucose). Total RNA was extracted from the cultured cells. tRNAPhe containing the yW-72 intermediate was isolated and purified by reciprocal circulating chromatography (24). The DNA probe complementary to the yeast tRNAPhe used in this study is 5′-tgcgaattctgtggatcgaacacaggacct-3′.
In vitro activity assays
Hydroxylation of yW-72 was performed basically as described (17). The reaction mixture (50 μl) contained 40 mM Tris–HCl (pH 7.6), 10 mM KCl, 1 mM DTT, 3 mM MgCl2, 4 mM ascorbic acid, 1.5 mM Fe(II)SO4, 4 mM 2-OG, 30 pmol of tRNAPhe with yW-72 and 2.8 μM recombinant hTYW5p variant. The reactions were incubated for 1 h at 37°C. The tRNAPhe was recovered by ISOGEN (Wako), precipitated by ethanol and then subjected to RNaseA digestion. The RNA fragments were analyzed by capillary LC nano ESI/MS.
Gel shift assay
The wild-type and mutant TYW5 proteins were mixed with the annealed yeast tRNAPhe containing yW-72, at ratios of 1:1.2 and 2:1.2, respectively. Each mixture was incubated at 37°C for 15 min, and then electrophoresed in a 5% acrylamide gel (20 mA) in TBE buffer, for 1 h at room temperature. The gel was stained with ethidium bromide and CBB to detect both tRNA and proteins.
RESULTS AND DISCUSSION
Overall structure
The crystal structure of hTYW5 was determined by the multiwavelength anomalous diffraction (MAD) method, using a selenomethionine-derivatized crystal (Figure 2). We also solved the structure of the cofactor-bound form, by soaking the crystal in buffer containing 2-OG and NiCl2, the latter of which acts as an inhibitor, instead of Fe(II) ion. The final models of the apo and cofactor-bound forms were refined at 2.5 and 2.8 Å resolutions, to free R-factors of 28.1 and 29.9%, respectively (Table 1 and 2). The data collection and refinement statistics are summarized in Table 1 and 2. The root mean square (RMS) deviation between the apo and cofactor-bound forms is 0.31 Å, indicating that they are almost identical.
Figure 2.
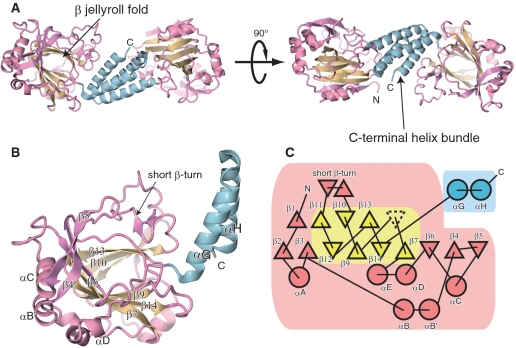
Overall structure of hTYW5. (A) and (B) Ribbon representations of the (A) hTYW5 dimer and (B) protomer. The N-terminal catalytic domain, the β-jellyroll fold and the C-terminal helix bundle are colored pink, cyan and yellow, respectively. (C) A secondary structure topology diagram of human TYW5. The α-helices and β-strands are represented by circles and triangles, respectively. The N-terminal catalytic domain, the β-jellyroll fold and the C-terminal helix bundle are colored as in Figures 2A and B. The jellyroll cores of the other 2-OG dependent oxygeneses generally comprise eight β-strands, whereas hTYW5 has only seven strands. The missing β-strand (β8) is represented by the dashed-line triangle.
The crystal contains two hTYW5 dimers per asymmetric unit. The overall structure of the hTYW5 protomer consists of the N-terminal catalytic domain (residues 1–270) and the C-terminal helical domain (residues 271–311) (Figure 2). In the apo and cofactor-bound structures, one of the dimer molecules contains partially disordered regions (residues 1–3, 63–66, 71, 92 and 312–315 for chain A). The relatively high R-free factors for both structures can be ascribed to this structural disorder.
The central core of the catalytic domain contains a β-strand jellyroll fold, which is homologous to the structures of the other Fe(II)/2-OG oxygenases and JmjC domains (13,25). While the structures of the Fe(II)/2-OG dependent oxygenases typically have a jellyroll fold with eight β-strands, the jellyroll fold of hTYW5 has only seven β strands (β7 and β9–β14), which are surrounded by additional α-helices (αA, αB, αB′, αC, αD, αE), β-strands (β1–β6), and several loops (Figure 2C). In the C-terminal domain, the helices αG and αH form extensive interactions with those of another protomer, thereby creating an antiparallel four-helix bundle. The physiological relevance of this dimer formation is discussed below.
Structural comparison with FIH
The amino-acid sequence of hTYW5 shares considerable sequence similarity (26% identity) with human FIH, which catalyzes the hydroxylation of the Asn803 residue of Hif1α (Supplementary Figure S1). FIH belongs to the JmjC domain-containing protein family, which catalyzes the hydroxylation using Fe(II) ion and 2-OG as cofactors (26,27). As expected from the amino-acid sequence similarity, hTYW5 has a similar tertiary structure to FIH, and shares the core structure with the other JmjC-domain-containing proteins. A structural similarity search with the DALI program (28) also indicated that the structure of hTYW5 is highly similar to that of FIH (r.m.s.d. = 1.98 Å for 210 Cα atoms) (Figure 3).
Figure 3.
Structural comparison of hTYW5 and FIH. (A) The overall structure of hTYW5. One protomer of the dimer is colored as in Figure 2, while the other protomer is grey. The directions of the catalytic pockets are indicated by arrows. A close-up view of the C-terminal helices (αG and αH) of hTYW5, which form the anti-parallel four-helix bundle, is shown below. (B) The overall structure of human FIH. The crystal structure of FIH (PDB ID: 1mze) is shown in a ribbon representation. The N-terminal catalytic domain, the C-terminal helix bundle and the β-jellyroll fold are colored purple, orange and green, respectively. The directions of the catalytic pockets are indicated by arrows. (C) and (D) Structural comparison of the active sites of (C) hTYW5 and (D) FIH. The crystal structures of the cofactor-bound forms of hTYW5 and FIH complexed with substrate peptide (PDB ID: 1h2k) are shown in ribbon representations. The long loop between β4 and β5 of FIH is colored orange. The TYW5-specific short, basic β-turn, and the long insertions between β6 and αD and between β14 and αG, which adopt different structures from those in FIH, are highlighted in blue and green, respectively. The substrate peptide of FIH is shown in pink.
However, several structural differences exist between hTYW5 and FIH. First, although both hTYW5 and FIH have α-helical C-terminal domains that form the dimerization interface, the arrangements of these helices are completely different (Figure 3A and B). The four α helices of the C-terminal domains of FIH adopt a rhomboid-shaped arrangement, whereas those of hTYW5 form an anti-parallel four-helix bundle. As a consequence, the relative orientations of the catalytic domains of the dimers are also different. The two entrances of the active sites of the FIH dimer are oriented in opposite directions, whereas those of the hTYW5 dimer are oriented in nearly perpendicular directions. Secondly, the structures around the entrance of the active site are different (Figure 3C and D). In the FIH-peptide complex structure, the substrate peptide is sandwiched between the β sheet (β7 and β8) and the long loops (loops between β4 and β5 and between β10 and β11). In contrast, in hTYW5, these loops are replaced by a short basic loop and a short basic β-turn, respectively. These structural differences may reflect the substrate differences between hTYW5 and FIH (i.e. nucleotide and peptide, respectively). Moreover, in hTYW5, the long loop insertions between β6 and αD and between β14 and αG occupy the entrance of the catalytic pocket. Interestingly, in FIH, the corresponding entrance site is empty, and is used as the binding site for the substrate peptide (Figure 3C and D). Therefore, this loop insertion in hTYW5 occupies a similar position to the substrate peptide in the FIH complex, thus allowing the substrate binding site to have a smaller, deeper pocket suitable for accommodating the yW-72 base. It is also interesting to note that a similar substrate switching mechanism exists between TYW4 and protein phosphatase methyltransferase 1 (PPM1): TYW4 methylates the acp side chain of the yW base precursor (Figure 1), while PPM1 catalyzes the methylation of the peptide substrate (29,30). In TYW4, a β-hairpin in the C-terminal Kelch-repeat domain is inserted into the entrance of the catalytic pocket of the N-terminal domain, thus switching the peptide substrate (in the case of PPM1) to a nucleobase substrate by reducing the space in the catalytic pocket (12). Such a substrate switching mechanism may be a general strategy in the evolution of RNA modification enzymes from peptide modification enzymes.
The active site structure of hTYW5
The active site of the typical Fe(II)/2-OG oxygenases is located in the center of the β jellyroll fold, and consists of a highly conserved His-X-(Asp/Glu)-Xn-His signature motif. These conserved residues provide three chemical groups to chelate the Fe(II) ion, while two additional chelating groups are supplied by the C-1 carboxylate and C-2 ketone groups of 2-OG.
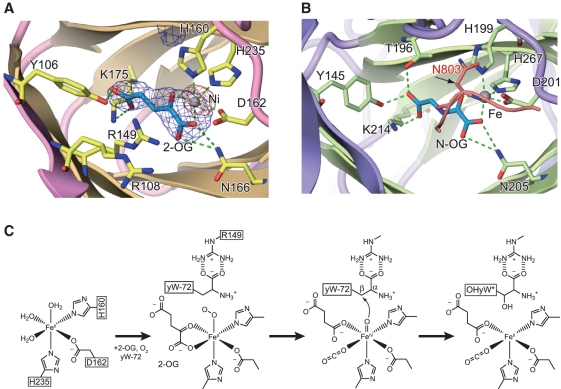
In the crystal structure of the cofactor-bound TYW5, we observed a strong peak in the anomalous difference Fourier map, calculated from the dataset collected at the peak wavelength of the Ni K-shell absorption edge. Therefore, we assigned this peak to the Ni(II) ion bound to the catalytic site (Figure 4A). As in the typical Fe(II)/2-OG oxygenases, the Ni(II) ion is coordinated by the signature motif of His160, Asp162 (on the loop connecting strands β7 and β9) and His235 (on β13), and mimics the physiological cofactor, Fe(II) ion (Figure 4A). Furthermore, in this cofactor-bound form, we clearly observed an electron density peak corresponding to the 2-OG cofactor (Figure 4A). The bound 2-OG cofactor is recognized by the conserved residues among TYW5 and FIH in the catalytic pocket, where the ε-amino group of Lys175 and the hydroxyl group of Tyr106 hydrogen bond to the C-5 carboxyl moiety of 2-OG, and the side chain amino group of Asn166 hydrogen bonds to the C-1 carboxyl moiety of 2-OG (Figure 4A). The side-chain conformations of these residues in hTYW5, as well as the 2-OG recognition manner, are also similar to those in FIH.
Figure 4.
The active site and catalytic mechanism of TYW5. (A) An unbiased Fo – Fc omit map around 2-OG (contoured at 3.0σ) and an anomalous Fourier map around the nickel ion (contoured at 20.0σ) are shown in blue and red, respectively. The omit map was calculated after removing the 2-OG cofactor from the model. (B) The active site of FIH complexed with the substrate peptide. The same color code as in Figure 3B is used. The substrate peptide of FIH is colored red. FIH catalyzes the hydroxylation of the β-carbon of Asn803, which is encircled with the β carbon indicated by an arrow. (C) Proposed catalytic mechanism by hTYW5.
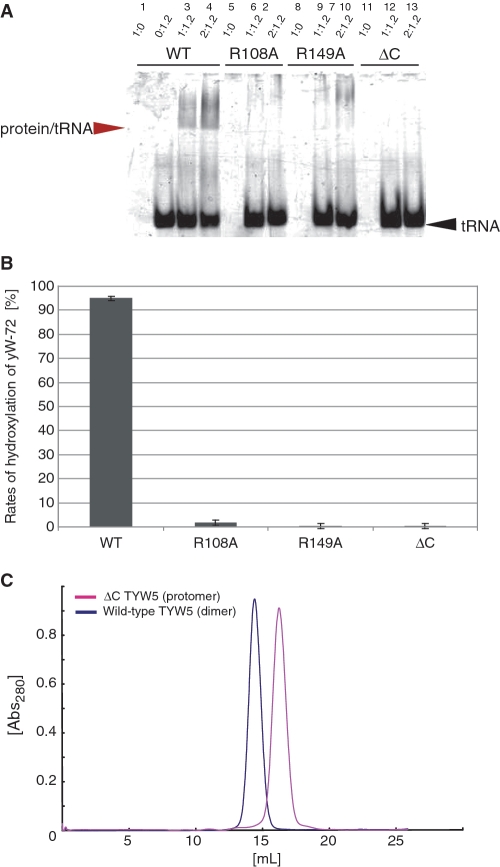
In contrast, some residues are specifically conserved among the TYW5 enzymes. In particular, Arg108 and Arg149, residing near the active site, are strictly conserved (Supplementary Figure S1). These Arg residues correspond to Asp148 and Leu188 in FIH, respectively. To elucidate the function of these Arg residues, we substituted them with Ala residues (R108A and R149A) and measured their hydroxylation activities (Figure 5B). The results showed that both mutations abolished the ability to form OHyW* from yW-72-containing tRNAPhe, indicating that these residues are essential for the enzymatic activity. To further clarify the role of these Arg residues, we performed a tRNA gel shift analysis with the mutants. Both mutants lacked the ability to bind tRNA (Figure 5A), suggesting that these Arg residues are involved in the tRNA recognition. Given that the binding mode of the acp side chain of the yW-72 base to TYW5 is the same as that of the target Asn residue in the substrate peptide to FIH, we can predict that these Arg residues are located close to the carboxyl group of the acp side chain. Therefore, these residues are likely to be involved in the recognition of the substrate yW-72 base moiety of tRNA. This hypothesis is in good agreement with the previous observation that hTYW5 has almost no hydroxylation activity for the fully modified yW base (17), in which the carboxyl group of the acp side chain is methylated and thus can no longer form a salt bridge with the conserved Arg residues.
Figure 5.
Mutant analysis. (A) Gel-shift assay. The wild-type hTYW5 and mutant hTYW5 (R108A, R149A and ΔC hTYW5) proteins were mixed with tRNAPhe containing the yW-72 modification at molar ratios of 1:1.2 and 2:1.2, respectively, and then analyzed. The gel was stained with ethidium bromide. The tRNA band and the protein/tRNA complex band are indicated by black and red arrows, respectively. (B) LC/MS analysis. LC/MS fragment analyses of RNaseA-digested tRNAPhe with yW-72, treated with recombinant wild-type TYW5 and mutant hTYW5 (R108A, R149A and ΔC hTYW5) in the presence of 2-OG and Fe2+. (C) Gel filtration chromatography of wild-type hTYW5 and ΔC hTYW5. The absorbances at 280 nm of wild-type hTYW5 and ΔC hTYW5 are shown as blue and pink lines, respectively. The molecular mass of the hTYW5 monomer is 36.5 kDa. Wild-type hTYW5 was eluted at the size expected for a dimer, whereas ΔC hTYW5 was eluted at the size expected for a monomer.
Based on the present crystal structure and functional analysis, as well as the proposed catalytic mechanism for other Fe(II)/2-OG oxygenases (25), we propose the following hydroxylation mechanism by TYW5 (Figure 4C). At first, TYW5 binds Fe(II) ion, 2-OG, molecular oxygen, and the yW-72 residue at position 37 of the tRNAPhe precursor. In this stage, the carboxyl group of yW-72 is recognized by the guanidinium group of the conserved Arg149. Next, the highly reactive oxoferryl species is generated from Fe(II), 2-OG and O2, by converting 2-OG into succinate and carbon dioxide. Finally, the reactive oxygen of the oxoferryl species attacks the β carbon of the acp side chain of yW-72, to form OHyW*.
Structural comparison with other JmjC-domain-containing proteins and Fe(II)/2-OG oxygenases
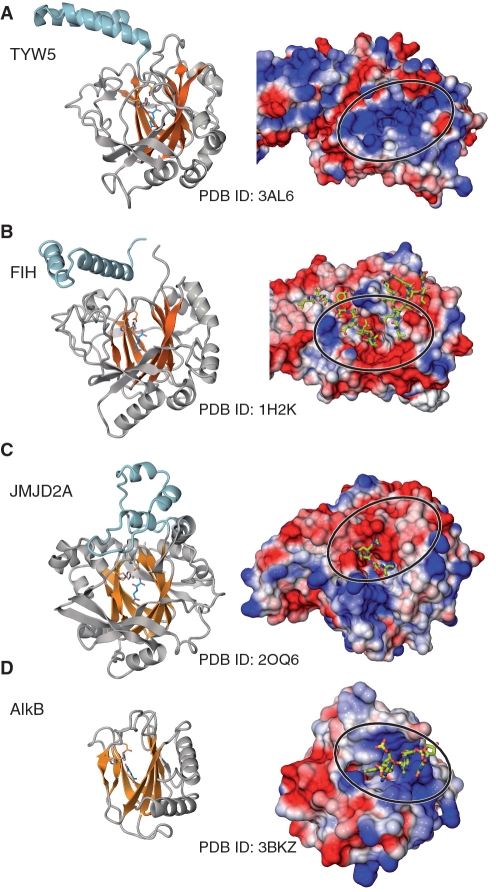
The most prevalent JmjC-domain-containing proteins are histone demethylases, which catalyze the hydroxylation of the methyl group of the target methyl-lysine residues in histones, and the unstable hydroxymethyl lysine thus generated is then spontaneously converted into formaldehyde and lysine. The crystal structure of JMJD2A, which is involved in the demethylation of di- and tri-methylated H3K9 and H3K36, was reported (31,32). As expected from the sequence similarity, hTYW5 also has a similar β-jellyroll core structure to that of JMJD2A (Figure 6A and C) (r.m.s.d. = 2.87 Å for 185 Cα atoms). The structural arrangements of the catalytic residues that ligate the Fe(II) and 2-OG cofactors are also well conserved between hTYW5 and JMJD2A. However, the structural similarity to JMJD2A is limited to the core structure, and the surrounding structural elements around the β-jellyroll fold are different. JMJD2A also has an additional α-helical C-terminal domain, but the arrangement of the α helices is completely different from those of both hTYW5 and FIH (Figure 6A, B and C). This is consistent with the fact that this C-terminal domain of JMJD2A is not involved in the dimer formation. Furthermore, the resultant molecular surface and its electrostatic potential around the catalytic pocket are completely different; the catalytic pocket of hTYW5 is positively charged, whereas that of JMJD2A is negatively charged (Figure 6A and C). This is reasonable, because JMJD2A recognizes the positively charged histone tail, whereas hTYW5 binds the negatively charged tRNA.
Figure 6.
Structural comparison of (A) TYW5 with other JmjC-domain-containing proteins, (B) FIH, (C) JMJD2A and Fe(II)/2-OG dependent oxygenase and (D) AlkB. In each panel, the structure is shown in both ribbon and surface models. In the ribbon models in the left column, the core β sheets of the β jellyroll fold are colored orange. The structures were aligned using the SSM algorithm implemented in COOT (32). In the surface models in the right column, the molecular surfaces of the positively charged regions are colored blue and those of the negatively charged regions are colored red, with the intensity of the color proportional to the local potential calculated by the program APBS (35). The bound substrates are shown in stick models. The surfaces around the catalytic pocket are indicated by circles.
AlkB, another member of the Fe(II)/2-OG oxygenases, recognizes the polynucleotide chain. AlkB catalyzes oxidative dealkylation, to repair alkylated DNA (33). AlkB and TYW5 share functional similarity, in that both enzymes recognize a nucleotide base and hydroxylate its exocyclic alkyl carbon atom. However, AlkB is not classified as a JmjC-domain-containing protein, and its amino-acid sequence is highly divergent from that of TYW5. As a result, the structure of AlkB (34) is also completely different from that of TYW5, and only the arrangement of the secondary structural elements of the β-jellyroll fold is conserved (Figure 6A and D) (r.m.s.d. = 2.43 Å for 118 Cα atoms). However, interestingly, the surface of AlkB has a positively charged patch for binding to the phosphate backbone of the polynucleotide chain, which is reminiscent of the positively charged patch around the catalytic pocket in hTYW5 (Figure 6D). A putative tRNA recognition mechanism by this positively charged patch in TYW5 is discussed below.
Dimer formation by the C-terminal helix bundle domain and possible tRNA recognition mechanism
The C-terminal domain, containing two anti-parallel α-helices, forms extensive interactions with that of another protomer, through the core hydrophobic interactions augmented by the surrounding hydrogen-bonding interactions (Figure 3A). The resultant area buried by the interaction is 1365 Å2 per protomer. To determine whether the dimer observed in the crystal is also formed in solution, we subjected the wild-type (wt) hTYW5 and the C-terminal helix bundle deletion mutant (ΔC hTYW5) to gel filtration chromatography. The results revealed that wt hTYW5 also forms a dimer in solution, whereas the ΔC hTYW5 mutant behaves as a monomer, indicating the importance of the C-terminal helix bundle for dimer formation (Figure 5C). Furthermore, we analyzed the tRNA-binding ability and the hydroxylation activity of the ΔC hTYW5 mutant, by a gel shift assay and an LC/MS analysis (Figure 5A and B), respectively. The ΔC hTYW5 mutant showed drastically decreased tRNA binding and hydroxylation activities, as compared to those of the wt hTYW5. Therefore, these results suggest that the dimer formation mediated by the C-terminal domain is essential for the tRNA recognition and subsequent RNA modification by hTYW5.
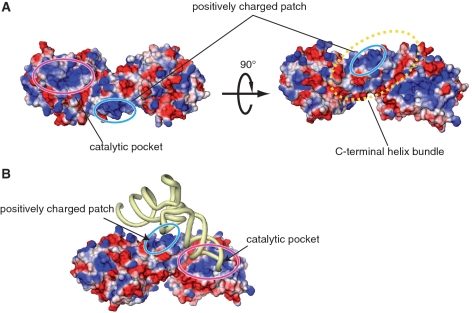
The surface electrostatic potential of hTYW5 revealed the existence of a large, positively charged patch around the catalytic pocket (Figure 7A), formed by highly conserved, basic residues. In contrast, the catalytic pockets of the other Jmj-C-containing proteins that interact with polypeptides are not positively charged (Figure 6A, B and C). Therefore, the positively charged catalytic pocket of hTYW5 may be involved in the recognition of the tRNA anticodon stem loop, to orient nucleotide 37 correctly within the catalytic site. In addition, a large, positively charged patch exists on the C-terminal helix bundle, which is created by the conserved basic residues (Lys274, Lys281 and Arg284) of the αG helices of both protomers upon dimer formation (Figure 7A). We hypothesize that this positively charged patch on the C-terminal helix bundle is involved in tRNA binding. Based on the present structure and functional analyses, we created a docking model of the TYW5–tRNAPhe complex, in which the D arm is captured by the positively charged patch (Figure 7B), and the anticodon loop is directed into the positively charged catalytic pocket. Further clarification of the mechanism of tRNAPhe recognition by hTYW5 must await the structure determination of the TYW5–tRNAPhe complex.
Figure 7.
The proposed tRNAPhe recognition mechanism by TYW5. (A) The electrostatic surface potential of hTYW5. The colors of the molecular surfaces of the positively charged regions are based on the local electrostatic potential, calculated by the program APBS (35). (B) Docking model of TYW5 and tRNAPhe. The tRNA molecule is colored yellow. In (A) and (B), the catalytic pocket, the C-terminal helix bundle and the positively charged patch are indicated by circles.
ACCESSION NUMBERS
The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes: 3AL5, 3AL6).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grant for the National Project on Protein Structural and Functional Analyses from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) (to O.N.); grants from MEXT (to O.N. and T.S.); Kurata Memorial Hitachi Science and Technology Foundation grants (to O.N.). Funding for open access charge: Grant for the National Project on Protein Structural and Functional Analyses from the Ministry of Education, Culture, Sports, Science and Technology (MEXT, to O.N.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank the beam-line staffs at BL41XU of SPring-8 (Hyogo, Japan) and NW12A and NE3A of PF-AR (Tsukuba, Japan) for technical help during data collection and Arisa Inagaki for technical assistance.
REFERENCES
- 1.Czerwoniec A, Dunin-Horkawicz S, Purta E, Kaminska KH, Kasprzak JM, Bujnicki JM, Grosjean H, Rother K. MODOMICS: a database of RNA modification pathways. 2008 update. Nucleic Acids Res. 2009;37:D118–D121. doi: 10.1093/nar/gkn710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruce AG, Uhlenbeck OC. Enzymatic replacement of the anticodon of yeast phenylalanine transfer ribonucleic acid. Biochemistry. 1982;21:855–861. doi: 10.1021/bi00534a007. [DOI] [PubMed] [Google Scholar]
- 3.Thiebe R, Poralla K. Origin of the nucleoside Y in yeast tRNAPhe. FEBS Lett. 1973;38:27–28. doi: 10.1016/0014-5793(73)80504-6. [DOI] [PubMed] [Google Scholar]
- 4.Carlson BA, Kwon SY, Chamorro M, Oroszlan S, Hatfield DL, Lee BJ. Transfer RNA modification status influences retroviral ribosomal frameshifting. Virology. 1999;255:2–8. doi: 10.1006/viro.1998.9569. [DOI] [PubMed] [Google Scholar]
- 5.Waas WF, Druzina Z, Hanan M, Schimmel P. Role of a tRNA base modification and its precursors in frameshifting in eukaryotes. J. Biol. Chem. 2007;282:26026–26034. doi: 10.1074/jbc.M703391200. [DOI] [PubMed] [Google Scholar]
- 6.Konevega AL, Soboleva NG, Makhno VI, Semenkov YP, Wintermeyer W, Rodnina MV, Katunin VI. Purine bases at position 37 of tRNA stabilize codon-anticodon interaction in the ribosomal A site by stacking and Mg2+-dependent interactions. RNA. 2004;10:90–101. doi: 10.1261/rna.5142404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Crecy-Lagard V, Brochier-Armanet C, Urbonavicius J, Fernandez B, Phillips G, Lyons B, Noma A, Alvarez S, Droogmans L, Armengaud J, et al. Biosynthesis of wyosine derivatives in tRNA: an ancient and highly diverse pathway in Archaea. Mol. Biol. Evol. 27:2062–2077. doi: 10.1093/molbev/msq096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goto-Ito S, Ito T, Kuratani M, Bessho Y, Yokoyama S. Tertiary structure checkpoint at anticodon loop modification in tRNA functional maturation. Nat. Struct. Mol. Biol. 2009;16:1109–1115. doi: 10.1038/nsmb.1653. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki Y, Noma A, Suzuki T, Senda M, Senda T, Ishitani R, Nureki O. Crystal structure of the radical SAM enzyme catalyzing tricyclic modified base formation in tRNA. J. Mol. Biol. 2007;372:1204–1214. doi: 10.1016/j.jmb.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 10.Goto-Ito S, Ishii R, Ito T, Shibata R, Fusatomi E, Sekine SI, Bessho Y, Yokoyama S. Structure of an archaeal TYW1, the enzyme catalyzing the second step of wye-base biosynthesis. Acta Crystallogr. D. 2007;63:1059–1068. doi: 10.1107/S0907444907040668. [DOI] [PubMed] [Google Scholar]
- 11.Umitsu M, Nishimasu H, Noma A, Suzuki T, Ishitani R, Nureki O. Structural basis of AdoMet-dependent aminocarboxypropyl transfer reaction catalyzed by tRNA-wybutosine synthesizing enzyme, TYW2. Proc. Natl Acad. Sci. USA. 2009;106:15616–15621. doi: 10.1073/pnas.0905270106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki Y, Noma A, Suzuki T, Ishitani R, Nureki O. Structural basis of tRNA modification with CO2 fixation and methylation by wybutosine synthesizing enzyme TYW4. Nucleic Acids Res. 2009;37:2910–2925. doi: 10.1093/nar/gkp158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 14.Clissold PM, Ponting CP. JmjC: cupin metalloenzyme-like domains in jumonji, hairless and phospholipase A2beta. Trends Biochem. Sci. 2001;26:7–9. doi: 10.1016/s0968-0004(00)01700-x. [DOI] [PubMed] [Google Scholar]
- 15.Lando D, Peet DJ, Gorman JJ, Whelan DA, Whitelaw ML, Bruick RK. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002;16:1466–1471. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clifton IJ, McDonough MA, Ehrismann D, Kershaw NJ, Granatino N, Schofield CJ. Structural studies on 2-oxoglutarate oxygenases and related double-stranded beta-helix fold proteins. J. Inorg. Biochem. 2006;100:644–669. doi: 10.1016/j.jinorgbio.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 17.Noma A, Ishitani R, Kato M, Nagao A, Nureki O, Suzuki T. Expanding role of the Jumonji C domain as an RNA hydroxylase. J. Biol. Chem. 2010 doi: 10.1074/jbc.M110.156398. In press, doi10.1074/jbc.M110.156398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iyer LM, Abhiman S, de Souza RF, Aravind L. Origin and evolution of peptide-modifying dioxygenases and identification of the wybutosine hydroxylase/hydroperoxidase. Nucleic Acids Res. 2010;38:5261–5279. doi: 10.1093/nar/gkq265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneider TR, Sheldrick M. Substructure solution with SHELXD. Acta Crystallogr. D. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 20.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol. Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 21.Terwilliger TC, Berendzen J. Bayesian correlated MAD phasing. Acta Crystallogr. D. 1997;53:571–579. doi: 10.1107/S0907444997005398. [DOI] [PubMed] [Google Scholar]
- 22.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 23.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 24.Miyauchi K, Ohara T, Suzuki T. Automated parallel isolation of multiple species of non-coding RNAs by the reciprocal circulating chromatography method. Nucleic Acids Res. 2007;35:e24. doi: 10.1093/nar/gkl1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hausinger RP. FeII/alpha-ketoglutarate-dependent hydroxylases and related enzymes. Crit. Rev. Biochem. Mol. Biol. 2004;39:21–68. doi: 10.1080/10409230490440541. [DOI] [PubMed] [Google Scholar]
- 26.Dann CE, 3rd, Bruick RK, Deisenhofer J. Structure of factor-inhibiting hypoxia-inducible factor 1: An asparaginyl hydroxylase involved in the hypoxic response pathway. Proc. Natl Acad. Sci. USA. 2002;99:15351–15356. doi: 10.1073/pnas.202614999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elkins JM, Hewitson KS, McNeill LA, Seibel JF, Schlemminger I, Pugh CW, Ratcliffe PJ, Schofield CJ. Structure of factor-inhibiting hypoxia-inducible factor (HIF) reveals mechanism of oxidative modification of HIF-1 alpha. J. Biol. Chem. 2003;278:1802–1806. doi: 10.1074/jbc.C200644200. [DOI] [PubMed] [Google Scholar]
- 28.Holm L, Sander C. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 29.De Baere I, Derua R, Janssens V, Van Hoof C, Waelkens E, Merlevede W, Goris J. Purification of porcine brain protein phosphatase 2A leucine carboxyl methyltransferase and cloning of the human homologue. Biochemistry. 1999;38:16539–16547. doi: 10.1021/bi991646a. [DOI] [PubMed] [Google Scholar]
- 30.Kalhor HR, Luk K, Ramos A, Zobel-Thropp P, Clarke S. Protein phosphatase methyltransferase 1 (Ppm1p) is the sole activity responsible for modification of the major forms of protein phosphatase 2A in yeast. Archives Biochem. Biophys. 2001;395:239–245. doi: 10.1006/abbi.2001.2558. [DOI] [PubMed] [Google Scholar]
- 31.Chen ZZ, Zang JY, Whetstine J, Hong X, Davrazou F, Kutateladze TG, Simpson M, Mao QL, Pan CH, Dai SD, et al. Structural insights into histone demethylation by JMJD2 family members. Cell. 2006;125:691–702. doi: 10.1016/j.cell.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 32.Ng SS, Kavanagh KL, McDonough MA, Butler D, Pilka ES, Lienard BMR, Bray JE, Savitsky P, Gileadi O, von Delft F, et al. Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity. Nature. 2007;448:87–91. doi: 10.1038/nature05971. [DOI] [PubMed] [Google Scholar]
- 33.Mishina Y, He C. Oxidative dealkylation DNA repair mediated by the mononuclear non-heme iron AlkB proteins. J. Inorg. Biochem. 2006;100:670–678. doi: 10.1016/j.jinorgbio.2005.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang CG, Yi CQ, Duguid EM, Sullivan CT, Jian X, Rice PA, He C. Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA. Nature. 2008;452:U961–U964. doi: 10.1038/nature06889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.