Abstract
Cationic lipids are used for delivering nucleic acids (lipoplexes) into cells for both therapeutic and biological applications. A better understanding of the identified key-steps, including endocytosis, endosomal escape and nuclear delivery is required for further developments to improve their efficacy. Here, we developed a labelling protocol using aminated nanoparticles as markers for plasmid DNA to examine the intracellular route of lipoplexes in cell lines using transmission electron microscopy. Morphological changes of lipoplexes, membrane reorganizations and endosomal membrane ruptures were observed allowing the understanding of the lipoplex mechanism until the endosomal escape mediated by cationic lipids. The study carried out on two cationic lipids, bis(guanidinium)-tris(2-aminoethyl)amine-cholesterol (BGTC) and dioleyl succinyl paramomycin (DOSP), showed two pathways of endosomal escape that could explain their different transfection efficiencies. For BGTC, a partial or complete dissociation of DNA from cationic lipids occurred before endosomal escape while for DOSP, lipoplexes remained visible within ruptured vesicles suggesting a more direct pathway for DNA release and endosome escape. In addition, the formation of new multilamellar lipid assemblies was noted, which could result from the interaction between cationic lipids and cellular compounds. These results provide new insights into DNA transfer pathways and possible implications of cationic lipids in lipid metabolism.
INTRODUCTION
The delivery of nucleic acids into cells is increasingly of interest for therapeutic purposes, with a global relevance to any disease amenable to manipulation at the gene expression level. The development of non-viral vectors composed of plasmid DNA complexed to a number of molecules such as cationic lipids or polymers, so-called lipoplexes and polyplexes (1), respectively, is of particular importance for both in vitro and in vivo gene transfer strategies. Although their transfection efficiencies are less than those of viral systems, the potential risks associated with their use are lower than those associated with viral systems, encouraging efforts to improve non-viral transfection efficiencies [see (2–8) for recent reviews]. In vitro gene delivery using cationic lipids or cationic polymers has been studied extensively using various biochemical and photonic visualization techniques for delineating uptake, intracellular trafficking and transgene expression. More than one pathway of non-viral vector uptake has been identified, involving clathrin-dependent, clathrin-independent and cholesterol-dependent pathways. Fluorescence imaging studies have shown a rapid transport of polyplexes to the nucleus region (9,10). More recently, the dissociation of DNA from lipoplexes and from polyplexes as well as the kinetics and the location of DNA release from those complexes was analyzed using fluorescence resonance energy transfer (11,12). Labelled pDNA were localized in the cytoplasm and in the perinuclear space while polyplexes were observed to be associated with the nuclear membrane and within the nucleus revealing more precisely the intracellular unpacking of polyplexes and the distribution of released DNA (11). Interestingly, by coupling Cl− and pH-sensitive chromophores to polyamines, the efficacy of polyethylenimine or polyamine dendrimers containing titrable charges has been related to their extensive buffering capacity inducing endosomal Cl− accumulation and promoting the swelling and disruption of endosomes (13). Despite valuable information on the intracellular trafficking of DNA provided by fluorescent microscopy, structural details of processes related to DNA release, endosomal vesicle escape and transport to the nucleus remain less well established due to the lack of the resolution of available techniques.
To explore further the mechanisms of gene transfer regarding these crucial barriers, strategies providing a visualization of this process at a molecular scale are very much required. Transmission electron microscopy (TEM) is a very suitable imaging technique for observing cellular and subcellular structure at nanometer resolution. Since Felgner and colleagues (14) introduced the use of cationic lipids to improve the DNA transfer, TEM techniques including classic approaches (negative staining, metal-shadowing) or cryo-electron microscopy are currently used for structural characterization of synthetic vectors. However, a few attempts have been made by TEM to study their mechanisms of action (15–17). TEM analyses were carried out on ultrathin sections of plastic-embedded cells. While fixatives and stains preserve lipids, they are less efficient for DNA visualization. A major difficulty then concerns the identification of the transferred DNA and its location within the cell. The first steps of the internalization process could be described by direct observations because DNA complexes kept their genuine features (15,18). The TEM contribution was mainly focused on the study of lipoplexes and polyplexes interacting with cells and of their subsequent cellular entry providing evidence of a cell entry driven by endocytosis pathways. The complex size would play a role in the activation of endocytosis pathway. The clathrin-dependent pathway could be activated by small complexes while larger ones would involve preferentially macropinocytosis (19). If direct TEM observations allow the identification of clathrin vesicles due to the typical thickening of the vesicle membrane, it is more complicated for other endocytosis pathways since vesicles are featureless. Nevertheless, peculiar plasma membrane remodelling viewed by TEM can inform about the clathrin–independents pathways. For example, the deformations of plasma membrane triggered by PEI-ADN polyplexes during their engulfment are in favour of a mechanism involving adhesion molecules (syndecan) and their clustering into cholesterol-rich raft (20).
To study the intracellular traffic of transferred DNA, labelling strategies were developed. The detection of DNA was carried out using conventional immunolabelling technique performed on TEM sections. The labelling of oligonucleotides with digoxigenin revealed by immunocytochemical technique using horseradish peroxidase provides a positive signal for endosomes as well as for perinuclear space (21). Likewise biotinylated oligonucleotides were detected in cytoplasmic vesicles and in cytosol after immunolabelling (a gold bead-coupled second antibody) (22). Another approach relies on the direct labelling of DNA or transfection agents using gold particles as probe to study the intracellular fate of lipoplexes and polyplexes (16,23,24). When streptavidin–gold particles interacted with plasmid DNA chemically modified with biotin (16), gold particles were then encountered within endosomes and larger vesicles. No particle was detected into the cytosol and the nucleus suggesting that the labelling could prevent DNA from escaping the endosome. The transfection agent, PEI, was labelled with gold particles to study polyplexes trafficking (24). Then, dense structures larger than gold particles were observed at the plasma membrane, inside the cytoplasm and entering the nucleus. Although images of lipoplexes internalized into the cell showing their multilamellar organization have been obtained (18,25), unambiguous identification of the lipoplexes through the entirety of their intracellular routes is of crucial importance for a more in-depth investigation to guide the synthesis of future vectors. Investigations using labelled DNA or lipid allowed their detection within the cell, but the maturation of lipoplexes and/or endosomal escape process were not described (16,18,21–23). The main drawback to the study of non-viral vector trafficking at the nanometric scale is the absence of appropriate imaging probes that combine detection suitable for TEM observations and visualization of artefact-free lipoplex structures.
In the present study we used a 16-nm core-shell, silica-based nanoparticle (Np) containing a maghemite nanocrystal (easily recognizable by TEM) that possesses an aminated-modified surface (for its pDNA binding). These Nps enabled both tracking of labelled lipoplexes within the cell and also a description at the nanometre scale of the morphology of these lipoplexes. We characterized the interactions between Nps and pDNA using a co-sedimentation assay. As cationic lipids have been among the most efficient synthetic gene delivery reagents in vitro, we used guanidinium-cholesterol cationic lipid [bis(guanidinium)-tris(2-aminoethyl)amine-cholesterol (BGTC)] and dioleyl succinyl parmomycin (DOSP), respectively, to form lipoplexes (26–28). Lipoplexes labelled with Nps were visualized by cryo-TEM. These labelled lipoplexes were then incubated with adherent cells and the cellular traffic was studied on ultrafine plastic cell sections. The results showed morphological changes of the lipoplexes after cellular uptake, ruptured endosomal vesicles and newly-formed membrane assemblies, providing new insights into DNA transfer mediated by cationic lipids.
MATERIAL AND METHODS
Chemicals, plasmid and cationic liposomes
All chemicals including dextran sulphate (MW 500 000) were purchased from Sigma. Absolute ethanol (J.T. Baker) and ammonia (Carlo Erba) were used as received. Water was deionized (resistivity higher than 18 MΩ).
pCMV-Luc (6.3 kb) is a plasmid encoding the luciferase reporter gene under the control of the human cytomegalovirus immediate-early gene promoter (29). Plasmid DNA was purified from recombinant Escherichia coli using Endofree plasmid purification columns (Qiagen, Chatsworth, CA, USA). BGTC/DOPE and DOSP/DOPE liposomes were obtained as previously described (18,28,30).
Preparation and surface modification of core-shell, silica-based Nps
Silica-based Nps contained 7 nm diameter maghemite nanocrystals in a core-shell morphology, providing a high-contrast image. Maghemite Nps prepared by a coprecipitation method (31) were used as seeds in the reaction media for the growth of the controlled-size silica shell, according to the method derived from the Stöber process (32) and previously reported (33). For this study, 10–15 nm diameter γFe203@SiO2 Nps were generated. The particle surface was aminated with N-(trimethoxysilylpropyl)ethylenediamine using the procedure previously described (34). Surface-modified silica Nps were stored in 20 mM Tris–HCl buffer at pH 7.4 with a final concentration of 1 g/l.
Characterization of Np/pDNA interactions
A 30 µl aqueous solution containing 2.25 µg pCMV-Luc (pDNA) was gently mixed with an equal volume of Nps at the desired concentration. After a 30 min incubation time, the mix was centrifuged at 22 000g, for 30 min, at 20°C to pellet the total Nps content. To quantify pDNA that did not pellet with Nps, 15 µl aliquots of each supernatant were electrophoresed through a 1.2% agarose gel at 50 V/cm for 1 h. Gels stained using SYBR-green were scanned under UV illumination and pDNA was quantified using Image J software. As a control, pDNA alone was submitted to the same procedure and used as a reference for quantification measurement. The stability of Np/pDNA complexes regarding ionic strength was measured in a final volume of 100 µl at the desired NaCl concentration. The effect of pH on Np/pDNA complexes was measured using 15 mM sodium citrate (pH 4 and pH 5), 10 mM MES (pH 6), 5 mM HEPES (pH 7), 20 mM Tris–HCl (pH 8) and 5 mM sodium borohydrure (pH 9) buffers, respectively. The effect of the polyanion dextran sulphate on the stability of the Np/pDNA interaction was assessed after addition of dextran solution (at the desired final concentration) on the Np/pDNA complexes. According to the procedure described above, DNA recovered in the supernatant was submitted to gel electrophoresis and then quantified. All experiments were reproduced three times.
Labelling of lipoplexes
A total of 2.17 × 1012 Nps and 3 µg pDNA were mixed at a ratio Np/pDNA = 5 (mol/mol) in 300 mM NaCl. Following this, an equal volume of cationic liposome was added in pure water. The final lipid cationic charge ratio was equal to 4. For transfection assays, the formulation prepared in a 150 µl final volume was deposited in each well.
Cell culture transfection
H1299 human lung cancer cells were cultured at 37°C in 5% CO2/humidified atmosphere, in DMEM with glucose, l-glutamine and pyruvate, supplemented with 1% streptomycin/penicillin (GIBCO and Invitrogen Life Technologies, Carlsbad, CA) and 10% FCS (Eurobio, Courtaboeuf, France). At Day 1 before transfection, the cells were transferred onto 24-well culture plates, at 65 000 cells per well, resulting in ∼70–80% confluence 24 h later, prior to transfection.
Cells were transfected with labelled and unlabelled BGTC/DOPE–DNA and with DOSP/DOPE–DNA lipoplexes formulated at a cationic lipid/DOPE–DNA charge ratio of 4. Lipoplexes containing 3 µg luciferase plasmid were added to each well, i.e. 150 µl complex and 450 µl serum-free DMEM. After 3 h, the transfection medium was replaced by 1 ml fresh medium. Cells were cultured for an additional 21 h before gene expression was determined. Luciferase activity was measured using the Promega luciferase assay system (Madison, WI, USA), according to the procedure previously described (30). Transfection experiments were performed in triplicate. For TEM, cells were observed after a 3 h incubation period with lipoplexes.
Conventional TEM and cryo-TEM
For cryo-TEM, a 5 µl sample was deposited onto a holey carbon coated copper grid. The excess was blotted with a filter paper. Unstained samples were frozen into liquid ethane and the grids were mounted onto a Gatan 626 cryoholder, transferred into the microscope, and maintained at a temperature of ∼−175°C. Sample observations were performed with a FEI Tecnai F20 transmission electron microscope, operating at 200 kV. Low-dose images were recorded at a nominal magnification of 50 000× with a 2k × 2k USC1000 slow-scan CCD camera (Gatan, CA, USA).
For TEM, cells were processed for ultramicrotomy according to standard procedures. Briefly, after trypsin treatment, cells from eight wells were pooled, pelleted and fixed for 2 h in a mixture of 2.5% glutaraldehyde and 4% paraformaldehyde in 0.2 M cacodylate buffer (pH 7.4). Sample pellets were then post-fixed for 1 h at 4°C with 1% osmium tetroxide in the same buffer and were dehydrated with ethanol before embedding in Epon-Araldite. Thin sections (65 nm thickness) were stained successively with 5% uranyl acetate and 1% lead citrate. TEM observation was performed with a FEI tecnai F20 operated at 200 kV.
RESULTS
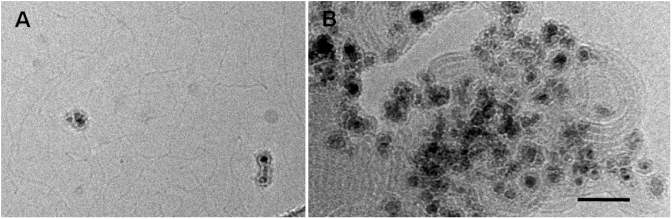
The present work aims to understand at a nanometre scale the gene transfer mechanism, from its cellular uptake to its delivery into the nucleus, using Np as the imaging probe. For this purpose, we labelled lipoplexes with Nps producing a high electron-scattering density easily observable by TEM (Figure 1). The Np with a 16.4 ± 4.8 nm mean diameter was a core-shell, silica-based Np containing 7 nm diameter maghemite nanocrystal, unambiguously recognizable by TEM (Supplementary Figure S1). Zeta potential measurements on these aminated-modified Nps showed that the Np surfaces exhibited a high density of positive charges brought by ammonium groups of the polysiloxane films in neutral and acidic media (Supplementary Figure S2). Its aminated-modified surface allowed the formation of Np/pDNA complexes mediated by electrostatic interactions (Figure 1A). To analyse the mechanism of action of lipoplexes, we focused our study on two cationic lipids, both known to be efficient DNA carriers. BGTC was made of a cholesterol hydrophobic moiety attached to guanidinium group and DOSP was composed of two aliphatic chains linked to an aminoglycoside head group. Thus, Np/pDNA complexes were mixed with either BGTC–DOPE or DOSP–DOPE liposomes to formulate labelled lipoplexes. Cryo-TEM image of labelled BGTC-lipoplexes revealed the presence of randomly-distributed Nps within lipid/pDNA assemblies (Figure 1B). This indicated that labelled lipoplexes kept their typical multilamellar organization and that their edges remained accessible to cell interaction. Thus, we expected that this labelling process should not modify the physicochemical properties of the lipoplexes, nor the transfection efficiency.
Figure 1.
Labelled lipoplexes with Nps. (A) Cryo images of pDNA interacting with Nps. (B) Labelled lipoplexes. Nps are incorporated into the lamellar assembly of pDNA/BGTC complexes. Note that the black dots within Nps correspond to small maghemite cores. Scale bar 50 nm.
Physico-chemical characterization of the interaction between pDNA and Np
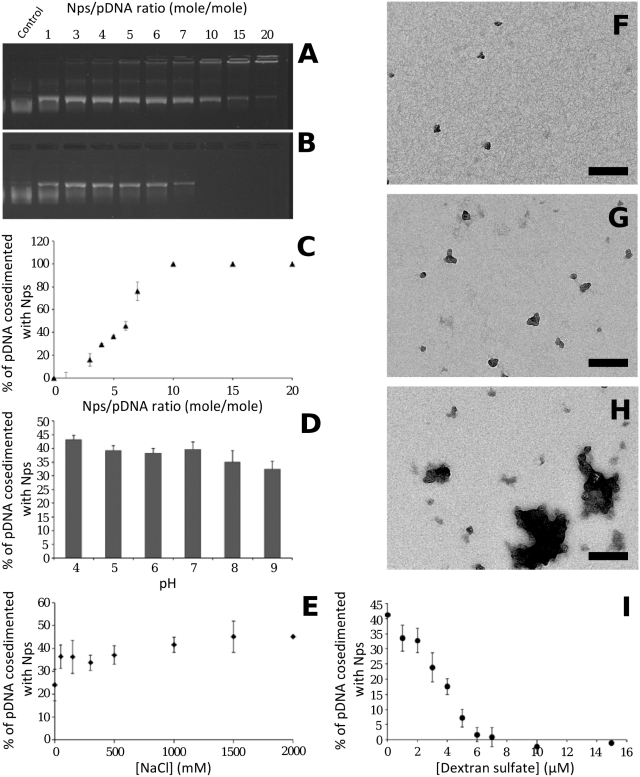
To adjust the Np/pDNA molar ratio (r) for lipoplex labelling, we first investigated the effect of increasing Nps on fixed DNA concentration (Figure 2). Figure 2A shows the results of agarose gel electrophoresis of pDNA, free and complexed with Nps. The mobility of pDNA was progressively restricted with increasing Np amount. DNA immobilization within the gel was due to Np/pDNA interactions. For quantification, Np/pDNA complexes were analysed after centrifugation (Figure 2B and C). At r = 1, 100% of DNA was present in the supernatant. For r ranging from 3 to 7, the amount of DNA increased gradually in the pellet. The formation of Np/DNA complexes was visualized by TEM (Figure 2F and G), showing few particles associated with DNA. At r = 10 and beyond, the total amount of pDNA was in the pellet. Large Np/pDNA complexes were formed as observed by TEM (Figure 2H). Our results were in good agreement with previous results on interactions between DNA and 26 nm aminated particles showing a similar cosedimentation profile (35). At a (w/w) ratio of 10, they showed that >90% of DNA was associated with particles. Note that in our conditions a (w/w) ratio of 10 corresponded to r = 17.8. It is interesting to note that, at high Np/pDNA ratios (>10), it has been reported that aminated silica particles enhanced transfection efficiency (35,36). To avoid this effect, we chose a lower Np/pDNA ratio. In subsequent experiments, we selected r = 5, for which no enhancing effect has been measured. We, then, studied the influence of pH on Np/pDNA complexes at r = 5. For a pH range between 4 and 9, the fraction of DNA present in the pellet varied between 42 and 32% (Figure 2D). At neutral and acidic pH no significant variation was observed suggesting that the complexes were stable under cellular pH conditions. In addition, the amount of pDNA in the supernatant remained similar from 0 to 2 M NaCl, indicating that Np/pDNA complexes were stable for these ionic strengths (Figure 2E). In addition, the complex stability was measured by competitive exchange with dextran sulphate (Figure 2I). As the concentration of dextran sulphate increased, DNA was recovered in supernatant. A 7 µM dextran sulphate was needed for a complete complex dissociation corresponding to a concentration range required for the dissociation of histone/DNA complexes or polyplexes (37,38). This result indicated that the release of DNA from Nps after exposure to polyanionic polymer was based on electrostatic interactions.
Figure 2.
Physicochemical characterization of Np/pDNA interactions. (A) Agarose gel electrophoresis of Np/pDNA complexes prepared at the ratio (r) given on top. (B and C) After centrifugation, gel electrophoresis of the supernatant (B), and pDNA cosedimented with Np plotted in (C). (D) Influence of pH on Np/pDNA complexes at r = 5. (E) Influence of NaCl on Nps/pDNA complexes at r = 5. (F–H) TEM images of Np/pDNA complexes at r = 1, r = 5 and r = 10, respectively. (I) Influence of polyanion (dextran sulphate; 500 000 kDa) on complexes stability. Scale bar 200 nm.
Transfection efficiency measurements of labelled and unlabelled lipoplexes
Labelled and unlabelled BGTC lipoplexes containing the reporter gene were incubated with a monolayer of H1299 pulmonary epithelial cells. Cell were incubated with lipoplexes for 3 h, then rinsed, and were cultured for an additional 21 h before gene expression determination. The measurements of luciferase activity revealed similar transgene expressions for r = 0, r = 5 and r = 10 (Table 1), indicating that at r = 5 the presence of Nps did not influence notably the transfection efficiency according to previous data (35,36).
Table 1.
Reporter gene expression obtained with labelled lipoplexes at different Nps/DNA ratios
| Lipoplexes | Luciferase activity |
|---|---|
| BGTC-based lipoplexes r = 0 | 2.24 ± 0.08 |
| BGTC-based lipoplexes r = 5 | 2.03 ± 0.13 |
| BGTC-based lipoplexes r = 10 | 1.83 ± 0.44 |
| DOSP-based lipoplexes r = 0 | 37.92 ± 5.12 |
| DOSP-based lipoplexes r = 5 | 26.72 ± 0.19 |
Expressed in nanograms of luciferase per milligram of protein. r corresponds to the Nps/DNA ratio.
This result suggested that the use of such small Nps should not interfere with the intracellular traffic of the lipoplexes. Interestingly, the particle size might be of importance regarding the DNA transfer, as it has been reported that the use of 230 nm silica Nps was able to enhance transfection efficiency (39). In our present work, the size of Nps is small with respect to the lipoplexes, such that it should not markedly change their morphologies or surface properties (Figure 1B). It is noteworthy that both labelled and unlabelled formulations with DOSP showed similar transfection efficiencies (Table 1).
In addition, the fate of Np/pDNA complexes incubated with cells for 3 h at r = 5 has been analyzed by TEM and the transgene expression has been measured after 24 h to evaluate the role of Nps as DNA carrier. DNA complexed with Nps did not lead to transgene expression. Moreover, TEM observations revealed that Nps were scarcely encountered within endosomes suggesting that few Np/DNA complexes could be internalized (Supplementary Figure S3). However we did not observe Nps into the cytosol indicating that they were not released from the endosome. The inability of endosomal escape could provide a possible explanation of the transfection inefficiency. These results validated the use of Nps as an imaging probe to study lipoplex trafficking.
EM observations of the intracellular fate of labelled BGTC lipoplexes
Plastic sections of cells observed by TEM allow the morphological characterization of BGTC lipoplexes interacting with cells and also after cellular uptake. As Np/pDNA complexes were stable under a broad range of saline and pH conditions and do not modify the transfection efficiency of parent BGTC–DOPE/DNA lipoplexes, lipoplex tracking was unambiguously provided by Np probes.
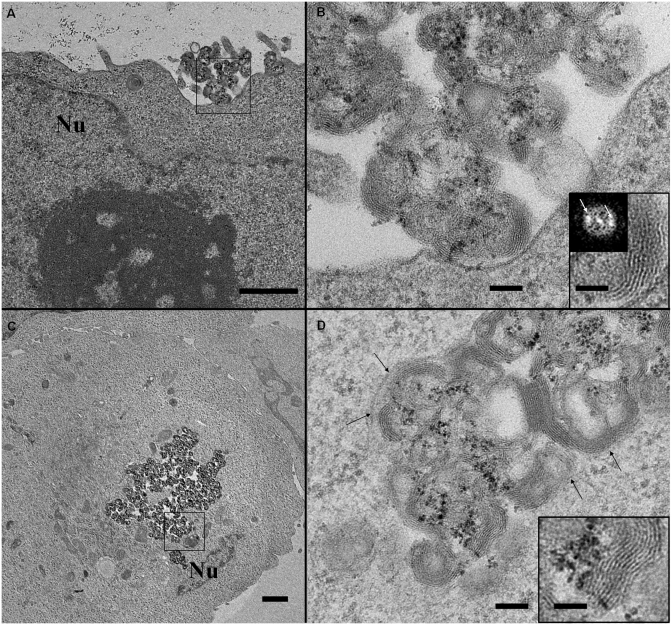
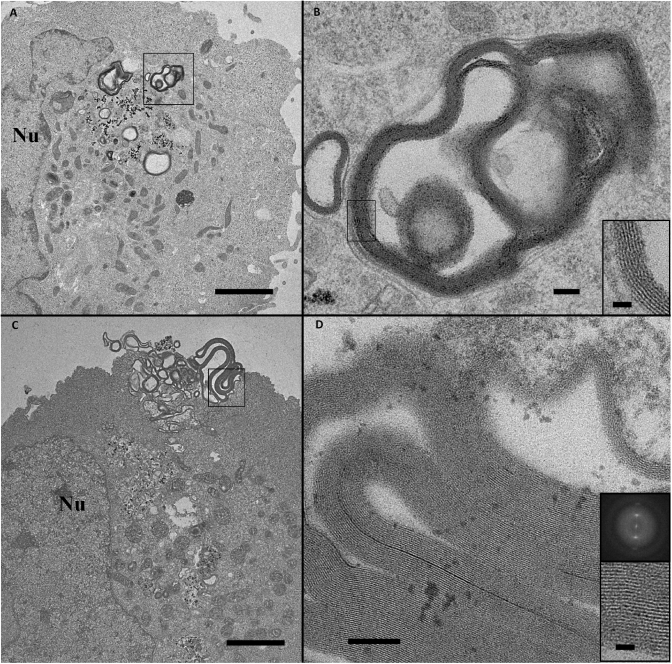
This analysis revealed morphological changes undergone by lipoplexes during their intracellular trafficking process. For the sake of clarity, we have classified these structural arrangements into three main parts, which may be related to typical steps of a gene delivery pathway. The first part relates to lipoplexes which kept their original morphology (Figure 3). Labelled lipoplexes were observed interacting with the plasma membrane (Figures 3A and B). They formed micron-sized clusters of roundish multilamellar structures containing Nps. The formation of these micron-sized clusters was due to the aggregation of lipoplexes induced by the culture medium use to grow the cells, as has already been described (40). Note that a precise determination of their size by dynamic light scattering measurement was not suitable due to the presence of large lipoplexes. The 6.5-nm repeat distance of lamellae (inset in Figure 3B) was similar to that visible on cryo-TEM image (Figure 1B), meaning that the repetitive motif corresponded to DNA strands complexed with cationic lipid bilayers. Lipoplexes whose morphologies resembled those present outside the cell, were also visible in the cytoplasm (Figure 3C and D). In addition to a similar morphology, the repeat distance between lipid membrane layers remained unchanged, i.e. 6.4–6.5 nm (inset in Figure 3D). Lipoplexes were contained in an endosomal vesicle (arrows in Figure 3D) indicating that their cellular entry had been achieved according to an endocytic process. This behaviour has been previously reported (18) and confirmed that their trafficking behaviour was not affected by labelling. Although the exact mechanism leading to large endosomes was not elucidated, the size of endosome containing the lipoplexes suggested a mechanism capable of large object entry. However a fusion of endocytotic vesicles as observed with clathrin-mediated internalization remained possible.
Figure 3.
TEM images of labelled BGTC lipoplexes at early stages of the DNA transfer route. (A, B) Labelled lipoplexes interacting with the plasma membrane. Large view (A) and an enlarged area (B) marked with black square in (A). Multilamellar structures (shown in inset in B) are typical of lipoplex assemblies with a 6.5-nm spacing provided by the distance between peaks (white arrows) on Fourier transform image. (C and D) Labelled lipoplexes with their genuine morphologies internalized into the cell via an endocytic process. (D) Enlarged view of marked area with black square in (C). Black arrows indicate the endosomal membrane. Inset: Enlargement of a 6.5-nm multilamellar assembly. Note that Nps (black densities) are clearly identified as well as the lipid layer structures (inset). Scale bars 1 µm (A and C), 100 nm (B and D) and 50 nm (inset). Nu: Nucleus.
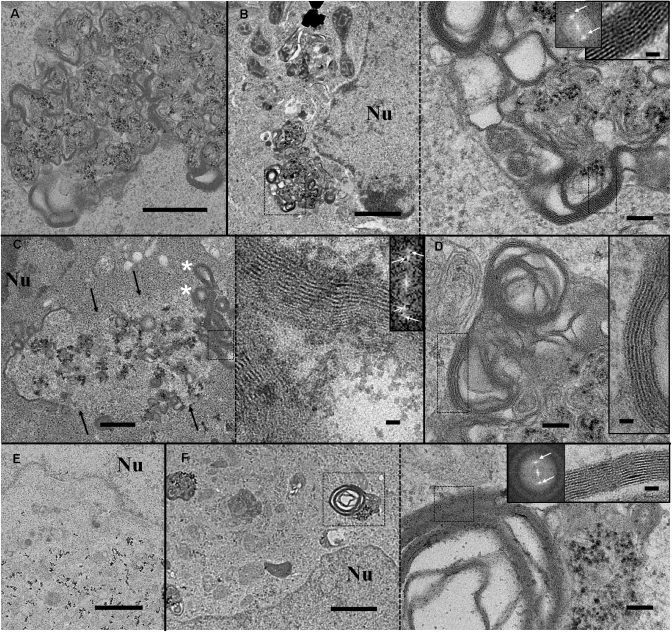
The second part concerns lipoplexes that underwent significant morphological changes. Typical views are presented in Figure 4. Lipoplexes looked different from genuine structures, as shown in Figure 3. Considerable lipid membrane reorganizations took place within the endocytic vesicles (Figure 4A–D). Roundish structures that were a landmark of genuine lipoplexes disappeared, suggesting that lipoplexes were destabilized. New, large, multilamellar structures with a 5.5 nm membrane to membrane repeat distance were visible at the endosomal vesicle edges (inset Figure 4B). Moreover, Nps were not associated with these multilayer structures, but rather remained mainly in the middle of the vesicle. These results showed that pDNA was not sandwiched within these new lipid structures and suggested that this important morphological rearrangement probably triggered the partial or complete dissociation of pDNA from cationic lipids. In addition, another aspect of morphological changes was encountered. The membrane of some endocytic vesicles containing lipoplexes had disappeared over a long distance revealing the loss of membrane integrity and a potential mixing of the endosomal compartment with the cytosol (Figure 4C). The endocytic compartment exhibited a brighter contrast than the cytosol, which delineated the ghost contour of the vesicle. At the periphery, multilamellar lipid structures were present (asterisks in Figure 4C). Inside the vesicle, Nps were visible, alone or associated with the few remaining lipoplexes. This membrane disruption provides a pertinent insight into endosomal membrane destabilization and pDNA endosomal escape. The presence of neo-formed multilamellar structures suggested that lipoplex reorganization occurred before the endosome rupture. However, we cannot exclude the possibility that it was concomitant with the release of pDNA from the lipoplexes. Unravelling this intriguing question would require further investigation. It is noteworthy that endocytic vesicles exhibiting morphological changes were observed close to the nucleus (Figure 4B, C and F), as was isolated Nps (Figure 4E). The transfer of pDNA from the endocytic vesicles to the nucleus may, then, be somewhat facilitated by this close vicinity. Despite close scrutiny, no Np was seen in the nucleus; hence a description of the ultimate step of nucleus entry was not achievable. It is possible that, during its transfer, pDNA dissociated from Nps.
Figure 4.
Gallery of labelled BGTC lipoplexes exhibiting various morphological modifications within endosomal vesicles that correspond to steps closely related to endosomal escape. (A) Typical morphological changes of lipoplexes show membrane reorganization and a loss of 6.5 nm multilamellar assemblies, suggesting DNA dissociation from cationic lipids. Scale bar 500 nm. (B) Reorganized lipoplexes close to the nucleus and an enlarged view corresponding to dashed black square (aside panel). New multilamellar structures are present at the vesicle boundaries. Insets: Enlarged image and its Fourier transform revealing a 5.5-nm membrane spacing. White arrows indicate peaks on Fourier transform image. Scale bars 1 µm, 100 nm and 20 nm. (C) Morphologies of modified lipoplexes after endosomal membrane disruption (black arrows). The dashed black square delineates the enlarged lipid assembly (aside panel) composed of two repeat distances. Inset: Fourier transform image showing two sets of bright peaks corresponding to a distance of 5.5 and 3.6 nm, respectively. Scale bars 500 nm and 20 nm. (D) Another example of lipid assembly with two membrane motifs clearly visible in enlarged area (inset). Scale bars 100 nm and 20 nm. (E) Nps in the vicinity of the nucleus. Scale bar 1 µm. (F) Low and high magnification view of vesicles containing Nps and composed of multilayered membranes with a 3.6-nm spacing motif (inset). Scale bars 1 µm, 100 nm and 20 nm. Nu: Nucleus.
Of interest, at the edge of the vesicle ghost (Figure 4C), multilamellar vesicles exhibited two lipid arrangements, with 5.5- and 3.6-nm spacing, respectively. The 5.5-nm lipid layers, similar to those in Figure 3B, were present in the outer part of the stack, facing the cytosol, while the 3.6-nm lipid layers were facing the luminal part of the vesicle. This peculiar lipid arrangement indicated that lipid–lipid interactions were modified during lipoplex trafficking. Indeed, this phenomenon was also observed, to a lesser extent, for other endocytic vesicles containing modified lipoplexes (Figure 4D). Although there is not yet a clear explanation of the driving forces underlying this mechanism, it could correspond to a peculiar rearrangement of the cationic lipid with cellular compounds, leading to a 3.6-nm packing. In accordance with this hypothesis, large and complex multilamellar vesicles exhibiting a 3.6-nm repeat distance were also observed in compartments containing Nps (Figure 4F).
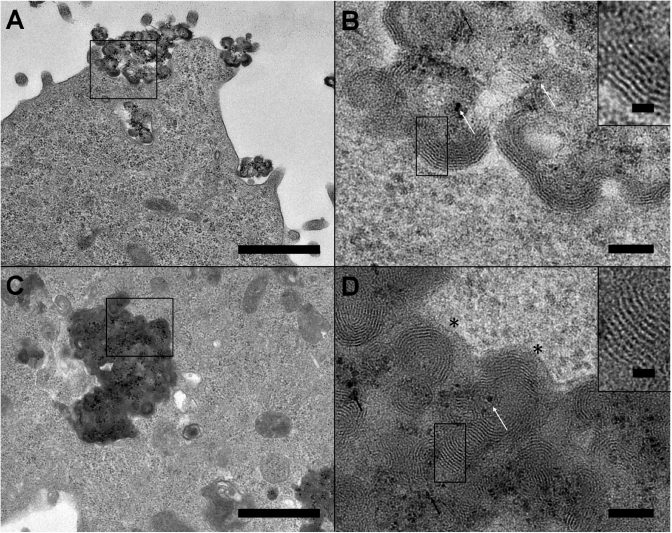
The third part reports on 3.6-nm spaced, large, multilamellar lipid structures devoid of Nps (Figure 5A and B). It is very tempting to propose that such lipid assemblies derived from those presented in Figure 4C and F, which would have gone through a further maturation process. It is interesting to note that these typical lipid structures, with the same regular lipid packing, were also observed outside the cell, suggesting that these lipid assemblies were rejected by the cell (Figure 5C and D). These data provided evidence that cells eliminated assemblies containing cationic lipids, and highlighted a mechanism involving the formation of pure multilamellar lipid assemblies with a typical membrane packing.
Figure 5.
Novel multilamellar lipid assemblies encountered inside (A and B) and outside (C and D) the cell, observed with BGTC cationic lipid. The enlarged views (B and D) correspond, respectively, to the marked areas (black squares). A 3.6-nm repeat motif is calculated from the peak distance on the enlarged image and its Fourier transform (inset). Scale bars 1 µm (A and C), 100 nm (B and D) and 20 nm (inset).
EM observations of the intracellular fate of labelled DOSP lipoplexes
Like BGTC lipoplexes, lipoplexes formulated with DOSP formed multilayered structures with DNA sandwiched in-between (28). Labelling DOSP lipoplexes with Nps did not modify their overall features, as observed in Figure 6 showing the early step of DOSP lipoplex entry. Several labelled lipoplexes (the enlarged view shows Nps marked with black arrows in Figure 6B) were observed in interaction with the plasma membrane (Figure 6A). As mentioned previously, typical features of lipoplexes exhibiting regular lamellar structures with a 6.5-nm repeat distance were also visible (Figure 6B and inset). Lipoplexes were observed intact internalized into the cell and were surrounded by an endosomal membrane (asterisk in Figure 6D) revealing an early intracellular pathway similar to that of BGTC lipoplexes.
Figure 6.
Labelled DOSP lipoplexes during cellular uptake and early steps of intracellular trafficking. (A and B) Labelled lipoplexes bound to the plasma membrane. Large view (A) and an enlarged area (B) marked with a black square in (A). Typical lipoplexes with a 6.5-nm lamellar spacing. Back dots (white arrows) correspond to Nps. C and D) Labelled lipoplexes surrounded by endosomal membrane after their internalization into the cell. (D) Enlarged view (corresponding to a black square drawn in C) shows typical 6.5 nm multilamellar arrangements (clearly visible in inset in D). Black arrows indicate Nps. Scale bars 1 µm (A and C), 100 nm (B and D) and 20 nm (inset).
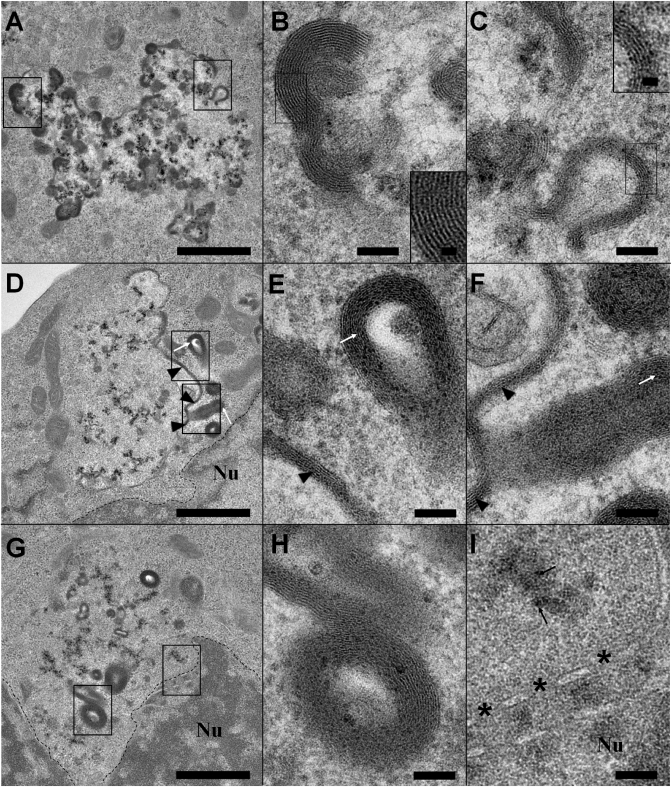
Figure 7 shows typical morphological changes of labelled DOSP lipoplexes at the level of the endocytic vesicle. Interestingly, vesicles appeared less dense than those in Figure 6C, and the endocytic membrane was missing over a long distance suggesting a release of material as observed for BGTC in Figure 4C. We did not, however, observe vesicles containing extensive membrane reorganization similar to that described in Figure 4A and B. These data suggested a more direct endosomal escape for DOSP than for BGTC. The endocytic vesicles contained Nps and lamellar structures located preferentially at their boundaries (Figure 7A, D and G). Two types of lamellar structures were observed, i.e. 6.5-nm repeat compact assemblies ascribed to lipoplexes (Figure 7B) and 5.5-nm multilamellar structures devoid of Nps, likely corresponding to newly-formed lipid stacks (Figure 7C–I). The 5.5-nm repeat assemblies had various shapes, i.e. long planar structures (Figure 7D and E), curved structures (Figure 7C) and elongated vesicles present at the vicinity of the endocytic vesicles (Figure 7E–H). The formation of the latter vesicles could arise from the budding of the planar structures as suggested by the curved shape (Figure 7C) and the protruding structure (Figure 7F). It is also important to note that these modifications occurred near the nuclear membrane dotted lines in Figure 7D and G), as noted with BGTC lipoplexes.
Figure 7.
Gallery of endosomes that contain labelled DOSP lipoplexes with deep morphological modifications providing snapshots on the endosomal escape process. (A–C) Partially disrupted endosome containing native labelled lipoplexes (B and inset) with a 6.5-nm repeat distance and new 5.5-nm repeat lipid structures devoid of Nps (C and inset). DNA dissociation from DOSP and its release from the endosome seem concomitant and probably occur in a more direct way than with BGTC, for which an intermediate step was observed. (D–F) Totally disrupted endosome containing only Nps in luminal volume. At its boundary, 5.5 nm multilamellar planar structures (arrow heads in E) and budding structures (white arrows in F) are present. (G–I) ‘Ghost’ endosome without visible membrane. The presence of Nps (black arrows) and multilamellar vesicles (H) allows its identification. Note its location close to the nucleus (I). Asterisks indicate nuclear pores. Scale bars: 1 µm (A, D and G), 100 nm (B, C, E, F, H and I) and 20 nm (insets).
DISCUSSION
Cationic lipid–DNA complexes have emerged as one of the major non-viral DNA delivery strategies over the last decade. Despite their wide use, a better understanding of the cellular and molecular mechanisms involved in lipoplex-mediated gene transfer is required to define the limiting barriers in DNA transfer and to optimize the vectors. As a general process, the cellular mechanisms can be divided into five main steps: (i) binding of the lipoplexes to the cell surface; (ii) entry of the lipoplexes into the cells mainly via endocytosis (a direct fusion with cell membrane cannot be excluded); (iii) DNA release into the cytosol; (iv) transport through the cytosol; and (v) entry of DNA into the nucleus and nuclear transcription.
To obtain more-detailed information on molecular mechanisms, we have studied, by TEM, the in vitro cellular uptake and intracellular fate of lipoplexes at the nanometre resolution scale. Lipoplexes were labelled with electron-dense Nps, allowing their clear identification all along their intracellular traffic. This pDNA labelling method relies on the stochastic mixture of Nps and pDNA, allowing the labelling of part or whole pDNA molecules. At r = 5, the labelled fraction of pDNA represents 40% of the total pDNA according to sedimentation assays, meaning that 60% remains label-free. However lipoplexes were formulated at a lipid/DNA charge ratio equal to 4 allowing the complexation of all pDNA molecules with cationic lipids. In these conditions, our cryoTEM and TEM observations revealed that all (>90%) lipoplexes contained Nps, meaning that the Np distribution was suitable to follow the intracellular route of almost all lipoplexes.
This study was performed with two cationic lipids, BGTC and DOSP. Both of these lipids have high transfection efficiencies; of the two the DOSP vector shows a better efficiency, up to 10-fold (Table 1 and Supplementary Table S1) (28), which can be explained by the intracellular trafficking mechanism. Both of the cationic lipids interact with the cell surface and are internalized through an endocytic process displaying common features of cellular uptake in the general framework of lipoplex-mediated gene transfer. However notable differences were observed for endosome escape. This step referring to DNA release from the endosome remains one of the steps for which very little structural information is available. It has been proposed that endosome escape would be triggered by the formation of a pore through the endosomal membrane, permitting DNA release (41). Our TEM snapshots indicate that membrane ruptures are larger than pores and also reveal lipoplex remodelling providing a more complete view of endosome escape mechanisms. For BGTC, two phases were observed; a deep reorganization of lipoplexes within the endosome with loss of the 6.5-nm characteristic repeat DNA–membrane distance (Figure 4A and B), and a more-advanced endosome change (Figure 4C) with large endosome rupture and the release of a large amount of material. For DOSP, lipoplex remodelling seems to occur in a more direct way, which would lead to better transfection efficiency. Indeed, endosomes contained highly-concentrated ‘native’ lipoplexes while showing a ruptured membrane, suggesting that DNA released from lipoplexes may concomitantly escape the endosome (Figure 7A and B). This ‘early’ DNA release could then prevent DNA degradation eventually occurring in endo-lysosomes (42–44). These data indicate different pathways of endosome escape that could also explain the difference in transfection efficiencies. Although we do not have a clear explanation, it is tempting to speculate that it is related to the nature of the cationic lipid. For a more complete understanding, further investigations on the respective role of the hydrophilic head group and the hydrophobic tail would be required.
Beyond endosome escape, the next step corresponds to DNA transport through the cytosol towards the nucleus. Our observations indicate that endosomal vesicles are located close to the nucleus. This accumulation near the nucleus, which was also reported for polymer–DNA complexes (45), suggests that DNA may reach the nucleus through either a passive diffusion or a short active transport. Concerning the passage of the nuclear barrier, our data show Nps close to, but not within, the nucleus. This absence deserves a few comments. One explanation could concern the size of Nps. They could be too large to pass through nuclear pore complexes despite their size is less than that of the pore which can be expanded to a size of ∼30 nm (46). Another explanation could be the dissociation of Nps from DNA molecules before entering the nucleus. Since interactions between DNA and Nps are based on electrostatic interactions, competition with polyanions or polycations such as histones could lead to the dissociation of Np/pDNA complexes (as shown in Figure 2I). Thus, Nps observed at the vicinity of the nucleus may correspond either to Np/pDNA complexes or to Nps released from the complexes suggesting that our labelling strategy based on mere electrostatic interactions is suitable to study the gene transfer process until the endosomal escape but seems less appropriate for the steps beyond. A third explanation could be related to the number of plasmid copies that enter nucleus. Quantification studies of pDNA copies in the nucleus showed that luciferase expression was detected with a minimum of 75 plasmids per nucleus after lipoplex and polyplex transfection (47) or even less (between 1 and 100 copies) after direct injection (48,49). Although estimation remains difficult, it is well accepted that this number is very low in comparison to the initial amount incubated with cells (47). In our labelling conditions, as about half of the pDNA is labelled, it is clear that we can expect few Nps per nucleus. In addition, EM observations using ultrafine cell sections provide a very limited view of the nucleus and, thus, reduce the chance of visualizing intranuclear Nps. As the probability of observing the phenomenon is very low, we cannot exclude the possibility that a few Nps could have passed the nuclear barrier. To summarize, our labelling strategy allows us to describe the intracellular fate of the lipoplexes and to track pDNA up to its endosomal release mediated by cationic lipids.
This study of lipoplex trafficking provides an overview after a 3-h incubation time. Since transgene expression is measured after 24 h, we have analyzed the aspect of lipoplexes after this period of time. Vesicles containing reorganized lipoplexes as well as those containing native lipoplexes were present although the latter were less frequently encountered. Lipoplex reorganizations were similar to those described after 3 h (data not shown). It is likely that lipoplexes remained attached onto the cell surface after rinsing, were progressively internalized and then underwent morphological changes leading to endosomal escape.
It was interesting to note that lipoplex remodelling was accompanied by the formation of new, multilamellar lipid structures Indeed, multilamellar assemblies with a 5.5-nm repeat distance were present in destabilized endosomes of DOSP lipoplex-treated cells. Although the mechanisms underlying endosomal escape remain ill-defined, evidence exists that specific compounds are required. The use of DOPE as a lipid helper is important for endosome release (50). In the same vein, it has been proposed that anionic lipids could be involved in the fusion process (41,51,52). Thus, it is possible that cellular compounds could interact with cationic lipid dissociated from DNA and then induce the formation of these new, multilamellar lipid structures. This lipid reorganization provides original insights into the fate of exogenous lipid (cationic and neutral lipid) introduced extensively into the cytoplasm. In the case of BGTC, the formation of multilayer vesicles with a 3.6-nm spacing was observed. These structures were present in large quantities in BGTC lipoplex-treated cells. They have been scarcely encountered in H1299 control cells, nor with DOSP lipoplex-treated cells (data not shown). This raises the question of why these structures are overexpressed by BGTC lipoplex-loaded H1299 cells. The H1299 cell line used in this study is derived from pulmonary type II epithelial cells which synthesize and secrete lamellar structures called lamellar bodies (LBs), specialized for the storage of pulmonary surfactant (53,54). LBs enriched in phospholipids and cholesterol are arranged in tightly-packed, concentric membrane sheets or lamellae with little or no inter-lamellar space. It has been shown that cholesterol enrichment of rat pulmonary type II epithelial cells increases LB production and that LBs accumulate extra cholesterol molecules, leading to an enhancement of cholesterol storage in LBs (55). In our conditions, a large amount of BGTC made of a cholesterol moiety as been introduced into the cytoplasm. It is likely that the intracellular BGTC has induced a similar process leading to the production of LB-like structures, unlike DOSP which is made of two aliphatic chains. The activation of such a process may well explain the secretion of LB-like structures, as observed in Figure 5, via a mechanism driven by the cellular machinery.
These data suggest that the nature of a cationic lipid can have an impact on lipid metabolism. This raises the question of whether the lipid could be involved in the process of DNA transfer. From our study, there is no evidence of a direct implication in this process. However, it has been shown that vector components can activate the cellular machinery to enhance gene expression. Indeed, the use of Pluronic polymer or incorporation of vinblastine into lipoplexes induces transcriptional activation of gene expression by activating the p53 and NF κB-signalling pathways (56,57). Thus, the role of the hydrophobic moiety of cationic lipids would deserve a more complete and systematic study.
CONCLUSION
DNA transfer using non viral vectors is a non-cellular process, meaning that this process is, to a lesser extent, not completely controlled by the cellular machinery but is, instead, more stochastically-mediated. This implies that the evolution of lipoplexes may be not synchronized and/or identical. Thus, it is not surprising that various aspects of lipoplex morphology were noted. The observation of labelled lipoplexes provided snapshots of morphological and structural modifications undergone by lipoplexes. For the first time, using Np probes, we were able to depict, at a nanometre resolution scale, the morphologies of lipoplexes, from their cellular uptake to pDNA release and endosome escape. We have identified evident landmarks of this intricate process, including lipid membrane reorganization and a potential mechanism for cationic lipid release, which clearly contribute to the understanding of the mechanism of action of lipoplexes. The development of this powerful approach for the investigation of specific intracellular pathways, and to identify parameters governing gene delivery, will guide the future synthesis of cationic vectors with optimized functions fulfilling the requirements of all steps involved in gene delivery.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
‘Association Française contre les myopathies’ (AFM); ‘Vaincre la Mucoviscidose’ (VLM); Conseil Régional d’Aquitaine (20071302007); VLM, PhD fellowship (to O.L.B.). Funding for open access charge: Centre National de la Recherche Scientifique.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors are very grateful to J. Lai Kee Him, S. Tan, A. Carpentier and Clothilde Gourden (IN-CELL-ART, Nantes, France) for their technical assistance. The authors are indebted to J.M. Lehn and P. Lehn for their stimulating discussions and constant supports of this work and for their pioneer works related to the synthesis of BGTC and DOSP cationic lipids.
REFERENCES
- 1.Felgner PL, Barenholz Y, Behr JP, Cheng SH, Cullis P, Huang L, Jessee JA, Seymour L, Szoka F, Thierry AR, et al. Nomenclature for synthetic gene delivery systems. Hum. Gene Ther. 1997;8:511–512. doi: 10.1089/hum.1997.8.5-511. [DOI] [PubMed] [Google Scholar]
- 2.Barteau B, Chèvre R, Letrou-Bonneval E, Labas R, Lambert O, Pitard B. Physicochemical parameters of non-viral vectors that govern transfection efficiency. Curr. Gene Ther. 2008;8:313–323. doi: 10.2174/156652308786070961. [DOI] [PubMed] [Google Scholar]
- 3.Khalil IA, Kogure K, Akita H, Harashima H. Uptake pathways and subsequent intracellular trafficking in nonviral gene delivery. Pharmacol. Rev. 2006;58:32–45. doi: 10.1124/pr.58.1.8. [DOI] [PubMed] [Google Scholar]
- 4.Hoekstra D, Rejman J, Wasungu L, Shi F, Zuhorn I. Gene delivery by cationic lipids: in and out of an endosome. Biochem. Soc. Trans. 2007;35:68–71. doi: 10.1042/BST0350068. [DOI] [PubMed] [Google Scholar]
- 5.Midoux P, Breuzard G, Gomez JP, Pichon C. Polymer-based gene delivery: a current review on the uptake and intracellular trafficking of polyplexes. Curr. Gene Ther. 2008;8:335–352. doi: 10.2174/156652308786071014. [DOI] [PubMed] [Google Scholar]
- 6.van der Aa MAEM, Mastrobattista E, Oosting RS, Hennink WE, Koning GA, Crommelin DJA. The nuclear pore complex: the gateway to successful nonviral gene delivery. Pharm. Res. 2006;23:447–459. doi: 10.1007/s11095-005-9445-4. [DOI] [PubMed] [Google Scholar]
- 7.Braun S. Muscular gene transfer using nonviral vectors. Curr. Gene Ther. 2008;8:391–405. doi: 10.2174/156652308786070998. [DOI] [PubMed] [Google Scholar]
- 8.Elouahabi A, Ruysschaert J. Formation and intracellular trafficking of lipoplexes and polyplexes. Mol. Ther. 2005;11:336–347. doi: 10.1016/j.ymthe.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 9.Godbey WT, Wu KK, Mikos AG. Tracking the intracellular path of poly(ethylenimine)/DNA complexes for gene delivery. Proc. Natl Acad. Sci. USA. 1999;96:5177–5181. doi: 10.1073/pnas.96.9.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suh J, Wirtz D, Hanes J. Efficient active transport of gene nanocarriers to the cell nucleus. Proc. Natl Acad. Sci. USA. 2003;100:3878–3882. doi: 10.1073/pnas.0636277100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen HH, Ho Y, Jiang X, Mao H, Wang T, Leong KW. Quantitative comparison of intracellular unpacking kinetics of polyplexes by a model constructed from quantum dot-FRET. Mol. Ther. 2008;16:324–332. doi: 10.1038/sj.mt.6300392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schneider S, Lenz D, Holzer M, Palme K, Süss R. Intracellular FRET analysis of lipid/DNA complexes using flow cytometry and fluorescence imaging techniques. J. Control Rel. 2010;145:289–296. doi: 10.1016/j.jconrel.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 13.Sonawane ND, Szoka FC, Verkman AS. Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes. J. Biol. Chem. 2003;278:44826–44831. doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- 14.Felgner PL, Gadek TR, Holm M, Roman R, Chan HW, Wenz M, Northrop JP, Ringold GM, Danielsen M. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl Acad. Sci. USA. 1987;84:7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Labat-Moleur F, Steffan AM, Brisson C, Perron H, Feugeas O, Furstenberger P, Oberling F, Brambilla E, Behr JP. An electron microscopy study into the mechanism of gene transfer with lipopolyamines. Gene Ther. 1996;3:1010–1017. [PubMed] [Google Scholar]
- 16.Zabner J, Fasbender AJ, Moninger T, Poellinger KA, Welsh MJ. Cellular and molecular barriers to gene transfer by a cationic lipid. J. Biol. Chem. 1995;270:18997–19007. doi: 10.1074/jbc.270.32.18997. [DOI] [PubMed] [Google Scholar]
- 17.Zhou X, Huang L. DNA transfection mediated by cationic liposomes containing lipopolylysine: characterization and mechanism of action. Biochim. Biophys. Acta. 1994;1189:195–203. doi: 10.1016/0005-2736(94)90066-3. [DOI] [PubMed] [Google Scholar]
- 18.Pitard B, Oudrhiri N, Vigneron JP, Hauchecorne M, Aguerre O, Toury R, Airiau M, Ramasawmy R, Scherman D, Crouzet J, et al. Structural characteristics of supramolecular assemblies formed by guanidinium-cholesterol reagents for gene transfection. Proc. Natl Acad. Sci. USA. 1999;96:2621–2626. doi: 10.1073/pnas.96.6.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonçalves C, Mennesson E, Fuchs R, Gorvel J, Midoux P, Pichon C. Macropinocytosis of polyplexes and recycling of plasmid via the clathrin-dependent pathway impair the transfection efficiency of human hepatocarcinoma cells. Mol. Ther. 2004;10:373–385. doi: 10.1016/j.ymthe.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 20.Kopatz I, Remy J, Behr J. A model for non-viral gene delivery: through syndecan adhesion molecules and powered by actin. J. Gene Med. 2004;6:769–776. doi: 10.1002/jgm.558. [DOI] [PubMed] [Google Scholar]
- 21.Lappalainen K, Miettinen R, Kellokoski J, Jääskeläinen I, Syrjänen S. Intracellular distribution of oligonucleotides delivered by cationic liposomes: light and electron microscopic study. J. Histochem. Cytochem. 1997;45:265–274. doi: 10.1177/002215549704500211. [DOI] [PubMed] [Google Scholar]
- 22.Akhtar S, Hughes MD, Khan A, Bibby M, Hussain M, Nawaz Q, Double J, Sayyed P. The delivery of antisense therapeutics. Adv. Drug Deliv. Rev. 2000;44:3–21. doi: 10.1016/s0169-409x(00)00080-6. [DOI] [PubMed] [Google Scholar]
- 23.Friend DS, Papahadjopoulos D, Debs RJ. Endocytosis and intracellular processing accompanying transfection mediated by cationic liposomes. Biochim. Biophys. Acta. 1996;1278:41–50. doi: 10.1016/0005-2736(95)00219-7. [DOI] [PubMed] [Google Scholar]
- 24.Thomas M, Klibanov AM. Conjugation to gold nanoparticles enhances polyethylenimine's transfer of plasmid DNA into mammalian cells. Proc. Natl Acad. Sci. USA. 2003;100:9138–9143. doi: 10.1073/pnas.1233634100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koynova R, Tarahovsky YS, Wang L, MacDonald RC. Lipoplex formulation of superior efficacy exhibits high surface activity and fusogenicity, and readily releases DNA. Biochim. Biophys. Acta. 2007;1768:375–386. doi: 10.1016/j.bbamem.2006.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vigneron JP, Oudrhiri N, Fauquet M, Vergely L, Bradley JC, Basseville M, Lehn P, Lehn JM. Guanidinium-cholesterol cationic lipids: efficient vectors for the transfection of eukaryotic cells. Proc. Natl Acad. Sci. USA. 1996;93:9682–9686. doi: 10.1073/pnas.93.18.9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pitard B, Oudrhiri N, Lambert O, Vivien E, Masson C, Wetzer B, Hauchecorne M, Scherman D, Rigaud JL, Vigneron JP, et al. Sterically stabilized BGTC-based lipoplexes: structural features and gene transfection into the mouse airways in vivo. J. Gene Med. 2001;3:478–487. doi: 10.1002/jgm.211. [DOI] [PubMed] [Google Scholar]
- 28.Desigaux L, Sainlos M, Lambert O, Chevre R, Letrou-Bonneval E, Vigneron J, Lehn P, Lehn J, Pitard B. Self-assembled lamellar complexes of siRNA with lipidic aminoglycoside derivatives promote efficient siRNA delivery and interference. Proc. Natl Acad. Sci. USA. 2007;104:16534–16539. doi: 10.1073/pnas.0707431104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pitard B, Pollard H, Agbulut O, Lambert O, Vilquin J, Cherel Y, Abadie J, Samuel J, Rigaud J, Menoret S, et al. A nonionic amphiphile agent promotes gene delivery in vivo to skeletal and cardiac muscles. Hum. Gene Ther. 2002;13:1767–1775. doi: 10.1089/104303402760293592. [DOI] [PubMed] [Google Scholar]
- 30.Letrou-Bonneval E, Chèvre R, Lambert O, Costet P, André C, Tellier C, Pitard B. Galactosylated multimodular lipoplexes for specific gene transfer into primary hepatocytes. J. Gene Med. 2008;10:1198–1209. doi: 10.1002/jgm.1212. [DOI] [PubMed] [Google Scholar]
- 31.Massart R. Preparation of aqueous magnetic liquids in alkaline and acidic media. IEEE Trans. Magn. 1981;17:1244–1245. [Google Scholar]
- 32.Stöber W, Fink A, Bohn E. Controlled growth of monodisperse silica spheres in the micron size range. J. Colloid Int. Sci. 1968;26:62–69. [Google Scholar]
- 33.Mornet S, Lambert O, Duguet E, Brisson A. The formation of supported lipid bilayers on silica nanoparticles revealed by cryoelectron microscopy. Nano Lett. 2005;5:281–285. doi: 10.1021/nl048153y. [DOI] [PubMed] [Google Scholar]
- 34.Trépout S, Mornet S, Benabdelhak H, Ducruix A, Brisson AR, Lambert O. Membrane protein selectively oriented on solid support and reconstituted into a lipid membrane. Langmuir. 2007;23:2647–2654. doi: 10.1021/la062227z. [DOI] [PubMed] [Google Scholar]
- 35.Kneuer C, Sameti M, Haltner EG, Schiestel T, Schirra H, Schmidt H, Lehr CM. Silica nanoparticles modified with aminosilanes as carriers for plasmid DNA. Int. J. Pharm. 2000;196:257–261. doi: 10.1016/s0378-5173(99)00435-4. [DOI] [PubMed] [Google Scholar]
- 36.Ravi Kumar MNV, Sameti M, Mohapatra SS, Kong X, Lockey RF, Bakowsky U, Lindenblatt G, Schmidt H, Lehr CM. Cationic silica nanoparticles as gene carriers: synthesis, characterization and transfection efficiency in vitro and in vivo. J. Nanosci. Nanotechnol. 2004;4:876–881. doi: 10.1166/jnn.2004.120. [DOI] [PubMed] [Google Scholar]
- 37.Paulson JR, Laemmli UK. The structure of histone-depleted metaphase chromosomes. Cell. 1977;12:817–828. doi: 10.1016/0092-8674(77)90280-x. [DOI] [PubMed] [Google Scholar]
- 38.Pichon C, Gonçalves C, Midoux P. Histidine-rich peptides and polymers for nucleic acids delivery. Adv. Drug Deliv. Rev. 2001;53:75–94. doi: 10.1016/s0169-409x(01)00221-6. [DOI] [PubMed] [Google Scholar]
- 39.Gemeinhart RA, Luo D, Saltzman WM. Cellular fate of a modular DNA delivery system mediated by silica nanoparticles. Biotechnol. Prog. 2005;21:532–537. doi: 10.1021/bp049648w. [DOI] [PubMed] [Google Scholar]
- 40.Turek J, Dubertret C, Jaslin G, Antonakis K, Scherman D, Pitard B. Formulations which increase the size of lipoplexes prevent serum-associated inhibition of transfection. J. Gene Med. 2000;2:32–40. doi: 10.1002/(SICI)1521-2254(200001/02)2:1<32::AID-JGM78>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 41.Xu Y, Szoka FC. Mechanism of DNA release from cationic liposome/DNA complexes used in cell transfection. Biochemistry. 1996;35:5616–5623. doi: 10.1021/bi9602019. [DOI] [PubMed] [Google Scholar]
- 42.Ross GF, Bruno MD, Uyeda M, Suzuki K, Nagao K, Whitsett JA, Korfhagen TR. Enhanced reporter gene expression in cells transfected in the presence of DMI-2, an acid nuclease inhibitor. Gene Ther. 1998;5:1244–1250. doi: 10.1038/sj.gt.3300721. [DOI] [PubMed] [Google Scholar]
- 43.Wattiaux R, Laurent N, Wattiaux-De Coninck S, Jadot M. Endosomes, lysosomes: their implication in gene transfer. Adv. Drug Deliv. Rev. 2000;41:201–208. doi: 10.1016/s0169-409x(99)00066-6. [DOI] [PubMed] [Google Scholar]
- 44.Lechardeur D, Verkman AS, Lukacs GL. Intracellular routing of plasmid DNA during non-viral gene transfer. Adv. Drug Deliv. Rev. 2005;57:755–767. doi: 10.1016/j.addr.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 45.Varga CM, Tedford NC, Thomas M, Klibanov AM, Griffith LG, Lauffenburger DA. Quantitative comparison of polyethylenimine formulations and adenoviral vectors in terms of intracellular gene delivery processes. Gene Ther. 2005;12:1023–1032. doi: 10.1038/sj.gt.3302495. [DOI] [PubMed] [Google Scholar]
- 46.Nigg EA. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 47.Cohen RN, van der Aa MAEM, Macaraeg N, Lee AP, Szoka FC. Quantification of plasmid DNA copies in the nucleus after lipoplex and polyplex transfection. J. Control. Release. 2009;135:166–174. doi: 10.1016/j.jconrel.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Capecchi MR. High efficiency transformation by direct microinjection of DNA into cultured mammalian cells. Cell. 1980;22:479–488. doi: 10.1016/0092-8674(80)90358-x. [DOI] [PubMed] [Google Scholar]
- 49.Ochiai H, Fujimuro M, Yokosawa H, Harashima H, Kamiya H. Transient activation of transgene expression by hydrodynamics-based injection may cause rapid decrease in plasmid DNA expression. Gene Ther. 2007;14:1152–1159. doi: 10.1038/sj.gt.3302970. [DOI] [PubMed] [Google Scholar]
- 50.Wrobel I, Collins D. Fusion of cationic liposomes with mammalian cells occurs after endocytosis. Biochim. Biophys. Acta. 1995;1235:296–304. doi: 10.1016/0005-2736(95)80017-a. [DOI] [PubMed] [Google Scholar]
- 51.Düzgüneş N, Goldstein JA, Friend DS, Felgner PL. Fusion of liposomes containing a novel cationic lipid, N-[2,3-(dioleyloxy)propyl]-N,N,N-trimethylammonium: induction by multivalent anions and asymmetric fusion with acidic phospholipid vesicles. Biochemistry. 1989;28:9179–9184. doi: 10.1021/bi00449a033. [DOI] [PubMed] [Google Scholar]
- 52.Hafez IM, Maurer N, Cullis PR. On the mechanism whereby cationic lipids promote intracellular delivery of polynucleic acids. Gene Ther. 2001;8:1188–1196. doi: 10.1038/sj.gt.3301506. [DOI] [PubMed] [Google Scholar]
- 53.Stratton CJ. The ultrastructure of multilamellar bodies and surfactant in the human lung. Cell Tissue Res. 1978;193:219–229. doi: 10.1007/BF00209036. [DOI] [PubMed] [Google Scholar]
- 54.Weaver TE, Na C, Stahlman M. Biogenesis of lamellar bodies, lysosome-related organelles involved in storage and secretion of pulmonary surfactant. Semin. Cell Dev. Biol. 2002;13:263–270. doi: 10.1016/s1084952102000551. [DOI] [PubMed] [Google Scholar]
- 55.Kolleck I, Guthmann F, Ladhoff A, Tandon NN, Schlame M, Rüstow B. Cellular cholesterol stimulates acute uptake of palmitate by redistribution of fatty acid translocase in type II pneumocytes. Biochemistry. 2002;41:6369–6375. doi: 10.1021/bi015980u. [DOI] [PubMed] [Google Scholar]
- 56.Yang Z, Zhu J, Sriadibhatla S, Gebhart C, Alakhov V, Kabanov A. Promoter- and strain-selective enhancement of gene expression in a mouse skeletal muscle by a polymer excipient Pluronic P85. J. Control. Release. 2005;108:496–512. doi: 10.1016/j.jconrel.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 57.Wang L, Dean DA, Macdonald RC. Effect of vinblastine on transfection: influence of cell types, cationic lipids and promoters. Curr. Drug Deliv. 2005;2:93–96. doi: 10.2174/1567201052772889. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.