Abstract
The polypurine/polypyrimidine (pPu/pPy) tract of the human vascular endothelial growth factor (VEGF) gene is proposed to be structurally dynamic and to have potential to adopt non-B DNA structures. In the present study, we further provide evidence for the existence of the G-quadruplex structure within this tract both in vitro and in vivo using the dimethyl sulfate (DMS) footprinting technique and nucleolin as a structural probe specifically recognizing G-quadruplex structures. We observed that the overall reactivity of the guanine residues within this tract toward DMS was significantly reduced compared with other guanine residues of the flanking regions in both in vitro and in vivo footprinting experiments. We also demonstrated that nucleolin, which is known to bind to G-quadruplex structures, is able to bind specifically to the G-rich sequence of this region in negatively supercoiled DNA. Our chromatin immunoprecipitation analysis further revealed binding of nucleolin to the promoter region of the VEGF gene in vivo. Taken together, our results are in agreement with our hypothesis that secondary DNA structures, such as G-quadruplexes, can be formed in supercoiled duplex DNA and DNA in chromatin in vivo under physiological conditions similar to those formed in single-stranded DNA templates.
INTRODUCTION
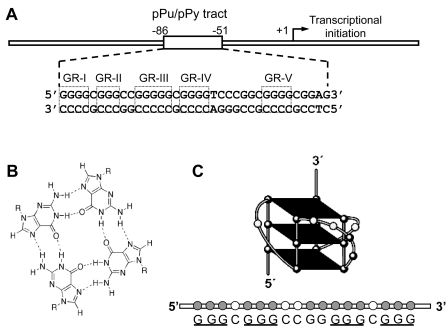
The switch to an angiogenic phenotype in cancer cells is often mediated by increased expression of vascular endothelial growth factor (VEGF), which is a pluripotent cytokine and angiogenic growth factor (1–3). VEGF consists of two identical subunits and it binds to VEGF receptors on the surfaces of endothelial cells (4). The interaction of VEGF and its cognate receptors stimulates the proliferation, migration, survival and permeability of endothelial cells, promoting the formation of new blood vessels (1–3). VEGF expression, which is frequently elevated in many types of cancer (1–3), is mainly regulated at the transcriptional level (5) and its expression is induced by a variety of factors, including hypoxia, pH, activated oncogenes, inactivated tumor suppressor genes and growth factors (6–9). The molecular basis of VEGF gene expression has been extensively studied by characterizing the cis-acting elements and transcription factors involved in constitutive VEGF expression in human cancer cells (10). This study revealed that the proximal 36-bp region (–85 to −50 relative to transcription initiation site) is essential for basal or inducible VEGF promoter activity in several human cancer cell lines (Figure 1A). These cis-regulatory elements contain at least three Sp1 binding sites, which consist of a polypurine/polypyrimidine (pPu/pPy) sequence. The pPu/pPy tracts are overly represented in the proximal promoter region of many TATA-less mammalian genes, including EGF-R, c-Myc, VEGF, HIF-1a, c-Myb, malic enzyme, I-R, AR, c-Src, c-Ki-Ras, TGFb and PDGF A-chain (11–13). These sequences have been proposed to be very dynamic in their conformation, easily adopting non-B-DNA conformations, such as melted DNA, hairpin structures, slipped helices or others, under physiological conditions, when there is negative supercoiling (14–18). In particular, G-rich sequences of these tracts have been reported to form parallel or antiparallel G-quadruplex structures consisting of two or more G-tetrads in the presence of monovalent cations such as Na+ and K+, as shown in Figure 1B (19). Significantly, the polypurine (guanine) tract of the VEGF promoter contains at least four runs of three or more contiguous guanines separated by one or more bases, corresponding to a general motif capable of forming an intramolecular G-quadruplex: VEGF: 5′-G4 (C) G3 (CC) G5 (C) G4 (TCCCGGC) G4 (CGG)-3′ (18,20). Using the electrophoretic mobility shift assay (EMSA), dimethyl sulfate (DMS) footprinting, the DNA polymerase stop assay, circular dichroism (CD) spectroscopy and molecular modeling (20), our previous studies demonstrated that the G-rich strand in this tract within the VEGF proximal promoter is indeed capable of forming intramolecular parallel G-quadruplex structures in vitro in the presence of K+ (Figure 1C) which can be further stabilized by various G-quadruplex interactive agents (20). We also demonstrated the dynamic nature of this tract in the VEGF promoter region using in vitro footprinting experiments with DNase I and S1 nuclease (18). Furthermore, the same study revealed that the identical changes in footprinting patterns by both nucleases in the presence of KCl were significantly enhanced by the presence of the G-quadruplex-interactive agent telomestatin, raising the possibility that telomestatin interacts with G-quadruplex structure(s) formed by the G-rich sequence within this tract (18). We also observed that selected G-quadruplex-interactive agents could inhibit the messenger RNA (mRNA) expression of these genes in various human cancer cells, including kidney cancer cells, suggesting that the transcription of these genes can be controlled by ligand-mediated G-quadruplex stabilization (20). Taken together, our previous studies provide critical support for the idea that G-quadruplex structures may play structural roles in vivo and therefore provide insight into novel methodologies for rational drug design.
Figure 1.
(A) Schematic representation of the human VEGF gene promoter region containing the polypurine/polypyrimidine tract proximally located at the transcription initiation site. Five G-repeats within this tract are indicated in open boxes (GR-1-GR-V). (B) Schematic representation of the hydrogen-bonded G-tetrad. (C) Schematic illustration of a major parallel G-quadruplex structure formed by the VEGF G-rich single strand in the presence of K+. The guanine residues involved in the formation of tetrads are underlined.
In the present study, in order to obtain more direct evidence for the existence of the G-quadruplex structures in the pPu/pPy region of the VEGF proximal promoter, we employed both in vitro and in vivo DMS footprinting techniques and utilized a nucleolin protein to detect these structures. DMS footprinting is known to be particularly useful for probing G-quadruplexes in vitro, because these structures require N7 of guanine and there is protection of the guanine residues involved in G-quadruplex formation from N7 methylation by DMS (21). Nucleolin is an abundant nucleolar protein that has been implicated in remodeling the chromatin structure, rDNA transcription, rRNA maturation, ribosome assembly and nucleo-cytoplasmic transport (22–23). Nucleolin is also reported to be a G-quadruplex binding protein that binds tightly and specifically to both the four-stranded and the two-stranded G-quadruplex DNA with a nanomolar dissociation constant for binding (24). Recently, we also found that nucleolin binds in vitro to the c-Myc G-quadruplex structure with high affinity and to other known quadruplex structures (25). Overall, the results of our study described here strongly support the presence of G-quadruplex structures in the promoter region of the VEGF gene both in vitro and in vivo.
MATERIALS AND METHODS
Cell culture
The A-498 renal cancer cell line was obtained from ATCC and the cells were cultured in RPMI-1640 media (GIBCO BRL) supplemented with 10% heat-inactivated fetal bovine serum in a humidified atmosphere containing 5% CO2 at 37°C.
Plasmid DNA
For in vitro footprinting of the VEGF promoter region, we used the supercoiled form of the luciferase reporter plasmid pGL3-VEGFP, which was constructed by ligating an 837-bp VEGF promoter region (–787 to +50 relative to the transcription initiation site) into the KpnI and NheI sites of pGL3-basic basic vector (Promega, Madison, WI, USA), as described previously (10).
DMS Footprinting of the VEGF G-rich single strand
The 32P-labeled VEGF G-rich single strand d(CCCCGGGGCGGGCCGGGGGCGGGGTCCC) was denatured by heating at 90°C for 5 min and then cooled slowly to room temperature for 2–3 h in 20 mM Tris–HCl buffer with 100 mM KCl. The annealed DNA (20 µl) was treated with 1% DMS for 2 min to methylate DNA and modification reactions were stopped by adding 80 µl stop buffer containing 0.3 M NaOAC (pH 6.0) and 3 μg of calf thymus DNA, ethanol precipitated twice, and treated with piperidine (10%). After hot piperidine treatment, the cleaved products were resolved on a 16% denaturing polyacrylamide gel as described previously (20).
Electrophoretic mobility shift assay
To determine the binding of nucleolin to the VEGF G-quadruplex, the VEGF G-rich single strand d(CCCCGGGGCGGGCCGGGGGCGGGGTCCC) was 5′-end-labeled with 32P, denatured by heating at 90°C for 5 min, then slow-cooled to room temperature with 100 mM KCl to allow the formation of G-quadruplex structures in the DNA oligomer. This folded DNA oligomer was incubated with purified recombinant maltose-binding protein (MBP)–nucleolin fusion proteins (MBP-nucleolin) to form protein–DNA complexes, and then subjected to a 5% or 8% native polyacrylamide gel electrophoresis (PAGE) to separate the nucleolin–G-quadruplex complex from unbound DNA. This MBP-tagged nucleolin carries human nucleolin residues 284–709 (pNuc-1,2,3,4-RGG9), including all four RNA-binding domains (RBDs) and the C-terminal domain, and is fused at the N terminus to Escherichia coli maltose-binding protein as described previously (25). In control experiments, the corresponding VEGF C-rich single strand d(GGGACCCCGCCCCCGGCCCGCCCCGGGG), the mutant VEGF G-rich single strand d(CCCCGGGGCGGGCCGGCGGCGGGGTCCC), and the VEGF oligomer duplex were also used.
Chromatin immunoprecipitation assays
The occupancy of the VEGF promoter by nucleolin and RNA polymerase II in vivo was determined by the chromatin immunoprecipitation (ChIP) assay as previously described (26). In brief, A-498 cells were grown to 80–90% confluence in 15-cm culture plates. After cross-linking with 1% formaldehyde in serum-free medium for 10 min, phosphate-glycine buffer was added to a final concentration of 0.125 M and cells were washed twice with ice-cold phosphate-buffered saline (PBS). The chromatin lysate was sonicated on ice to an average DNA length of 400 bp. Chromatin was pre-cleared with blocked Sepharose A, and immunoprecipitation was performed with mouse monoclonal anti-nucleolin, anti-Sp1 and anti-RNA polymerase II antibodies and 8 μg of mouse IgG as the negative control. All antibodies and IgG were obtained from Santa Cruz Biotechnology. Following an initial 5-min denaturation at 95°C, the polymerase chain reaction (PCR) amplification involved 40 cycles of 94°C for 60 s, 55°C for 30 s, 72°C for 30 s using 32P-5′-end labeled primers designed to the proximal promoter region (–242 to +48) of VEGF [forward d(TCGCTCGCCATTGGATCTCGAGGA)3 and reverse d(GCCCGACACACTGGCCGAAGCGACGA)] and the proximal promoter region (–273 to +71) of HIF-1α [forward d(TCGCTCGCCATTGGATCTCGAGGA) and reverse d(GCCCGACACACTGGCCGAAGCGACGA)]. PCR was also performed using primers designed to a nonspecific promoter region 1 kb upstream (−1079 to −874) of the transcription start site of VEGF [forward d(CCTCAGTTCCCTGGCAACATCTG) and reverse d(GAAGAATTTGGCACCAAGTTTGT)]. All PCR amplification reactions were then analyzed on a 6% nondenaturing PAGE and visualized with a Phosphorimager (Molecular Dynamics).
Polymerase chain reaction
Template complementary DNA (cDNA) was prepared from total RNA extracted from A-498 cells using an RNeasy mini kit (Qiagen, Valencia, CA, USA), according to the manufacturer’s protocol. VEGF and actin cDNAs were detected by PCR using forward and backward primers specific for each gene. Primer sequences for the VEGF gene were designed against a common region to all isoforms of the VEGF mRNA: forward primer, d(TGCATTGGAGCCTTGCCTTG) (nucleotides 1054–1073 from NM 003376.4); reverse primer, d(CGGCTCACCGCCTCGGCTTG) (nucleotides 1664–1683 from NM 003376.4). In parallel, the amplification of β-actin cDNA was done as an internal standard according to the manufacturer’s protocol (Ambion). All of the reactions involved an initial denaturation at 95°C for 3 min, followed by 40 cycles for VEGF or 30 cycles for β-actin at 94°C for 30 s, 55°C for 30 s and 72°C for 40 s on a GeneAmp PCR system 9600 (Perkin-Elmer).
In vitro footprinting of the VEGF promoter region with DMS
A supercoiled form of the plasmid pGL3-VEGFP was incubated in the presence of 100 mM KCl at 37°C for 1 h and then treated with DMS (0.2%) for 2 min as previously described (27). DNA was precipitated with ethanol and resuspended in double-distilled water after vacuum drying. To map DMS reactive sites on the plasmid DNA, linear amplification by PCR was performed using Thermo Sequenase Cycle Sequencing kit (USB) with a 32P-labeled gene-specific primer 1V d(CCCAGCGCCACGACCTCCGAGCTACC) spanning +23 and +49 of the promoter region to amplify the top strand of the plasmid DNA as previously described (20). PCR was carried out using cycling conditions consisting of an initial 4-min denaturation step at 94°C, 1 min at 60°C and 1 min at 72°C, for a total of 40 cycles. Sequencing ladders (A, G, T and C) as size markers were generated by the dideoxy method with a 32P-labeled gene-specific primer 1V and the plasmid pGL3-VEGFP.
In vivo genomic footprinting with DMS
A498 cells were incubated for 48 h with and without TMPyP4 at the IC50 concentration (50 µM). For DMS footprinting, cells were washed with PBS and then incubated with PBS containing 2% DMS for 2 min. Treated cells were lysed and incubated overnight with DNA lysis buffer (50.0 mM Tris–HCl, pH 8.5, 50 mM NaCl, 25 mM EDTA, pH 8.0, 0.5% SDS and 300 µg/ml of proteinase K), and genomic DNA was purified by phenol–chloroform extraction and ethanol precipitation. The DNA was cleaved with piperidine and the cleavage sites were identified by LM-PCR as described below (28).
Ligation-mediated PCR
The isolated genomic DNA, after piperidine treatment, was analyzed by ligation-mediated (LM)-PCR to map the cleavage sites on the (+) strand of the VEGF promoter region (28). The first-strand synthesis was accomplished with a gene specific primer 1V d(CCCAGCGCCACGACCTCCGAGCTACC) spanning +23 and +49 of the promoter region annealed on genomic DNA and Vent DNA polymerase (New England Biolabs) using a thermal cycle of 10 min at 95°C, 30 min at 60°C and 10 min at 76°C as previously described (28). The reaction mixtures were extracted once by phenol–chloroform and precipitated with ethanol to purify DNA. The purified DNA was then ligated with 100 pmol of unidirectional linker L11 d(GAATTCAGATC) and L26 d(GCGGTGACCCCGGGAGATCTGAATTC) in 20 µl of ligation solution [250 mM Tris–HCl (pH 7.7)/10 mM MgCl2/20 mM dithiothreitol/3 mM ATP/0.005% bovine serum albumin/20 U of T4 DNA ligase (Promega)]. After incubation for 12–16 h at 17°C, samples were extracted once by phenol–chloroform and precipitated with ethanol to purify and concentrate DNA. The purified ligated DNAs were amplified by PCR using a nested primer 2V d(CCCGGCTGCCCCAAGCCTCCGCGA) spanning +1 and +23 and the linker oligonucleotide (L26). PCR was carried out using cycling conditions consisting of an initial 4-min denaturation step at 94°C, 1 min at 60°C and 1 min at 72°C, for a total of 40 cycles. Labeling of the (+) strand was performed with 32P-end-labeled nested primer 2V. Subsequently, the samples were phenol–chloroform-extracted, ethanol-precipitated, resuspended in sequencing loading buffer and separated in 10% polyacrylamide, 7 M urea sequencing gels. Sequencing ladders (A, G, T and C) as size markers were generated by the dideoxy sequencing method with a gene specific primer 1V and the plasmid pGL3-VEGFP.
RESULTS
Formation of G-quadruplex structures by the G-rich strand of the pPu/pPy tract of the VEGF promoter
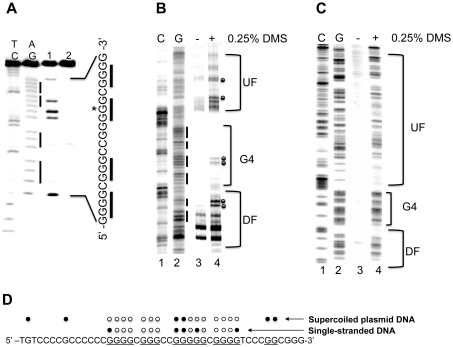
DMS footprinting is useful for fine mapping the presence of G-quadruplex structures within the promoter region (20,21). The formation of G-quadruplex structures requires N7 of guanine, which is subsequently protected from N7 methylation by DMS (20,21). Consistent with our previous studies (20), DMS footprinting revealed that a unique G-quadruplex structure is formed in the G-rich single strand of the pPu/pPy tract of the VEGF promoter region. This tract has the capacity to form a unique intramolecular G-quadruplex structure in the presence of K+, which requires four guanine blocks (GR-I to GR-IV), consisting of 12 total guanines (Figure 1A). DMS cleavage of the VEGF G-rich single strand showed that three stacked G-tetrads formed by four G-stretches were protected from methylation by DMS (except one guanine residue indicated by an asterisk), whereas two guanine residues within the central loop showed hypersensitivity to DMS (Figure 2A, lane 1). We further examined G-quadruplex formation in the promoter DNA duplex of the VEGF gene (787 to +50) using in vitro DMS footprinting experiments with a supercoiled form of plasmid pGL3-VEGFP. As shown in Figure 2B (lane 4), under supercoiled conditions DMS shows dramatically reduced reactivity within the four 5′-end runs of guanines (bracket ‘G4′) on the G-rich strand of the pPu/pPy tract of the VEGF promoter region, where G-quadruplex structures are proposed to be present. In contrast, the flanking regions (Brackets ‘UF’ and ‘DF’) of the proposed G-quadruplex-forming sequence show enhanced reactivity to DMS (Figure 2B, lane 4), presumably because of the presence of locally unwound structures at the junctions between normal duplex regions and stable secondary structures. Interestingly, a moderate reactivity of DMS toward some of the guanine residues from GR-III was found, suggesting a loop region of the G-quadruplex structures as observed in studies using a single-stranded G-rich DNA (Figure 2A, lane 1). The reactivity of DMS toward the guanine residues within the complete G-rich region is normal in the linearized form of plasmid pGL3-VEGFP (Figure 2C, lane 4), showing that negative supercoiling is required to drive the local unwinding of the pPu/pPy tract, allowing a specific G-rich region to form a G-quadruplex. The results of DMS footprinting for the VEGF G-rich single strand and the G-rich region within a supercoiled plasmid pGL3-VEGFP in the presence of 100 mM KCl are summarized in Figure 2D.
Figure 2.
(A) DMS footprinting of the VEGF G-rich single strand in the presence of 100 mM KCl. AG and TC lanes represent purine and pyrimidine- specific Maxam–Gilbert sequencing reactions, respectively. Lanes 1 and 2 correspond to the G-rich strand DMS treated and untreated, respectively. (B and C) Autoradiogram showing DMS modification sites on the G-rich strand of a supercoiled (B) or linearized (C) form of pGL3-VEGF plasmid. Lanes 1 and 2 represent respective cytosine and guanine-specific dideoxysequencing reactions using the same primer as for the extension reactions. Lanes 3 and 4 correspond to the pGL3-VEGF plasmids that are untreated and treated with DMS (0.25%), respectively. Each band reflects extension reaction products of a gene specific primer designed to anneal to the G-rich strand of the plasmid DNA to map the DMS modification sites on the same strand. The bracket ‘G4’ represents a proposed G-quadruplex-forming region and brackets ‘UF’ and ‘DF’ represent the up- and downstream flanking regions of the G-quadruplex-forming sequence. (D) Summary of the results from both DMS footprinting experiments. The DMS-protected guanine residues within the G-rich sequences of the VEGF promoter are indicated by open circles, and closed circles indicate the guanine residues methylated by DMS. Data shown are representative of at least two experiments.
In vivo DMS footprinting
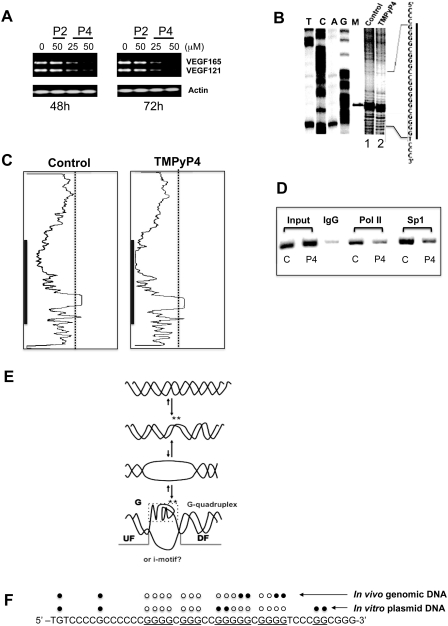
The DMS-based genomic footprinting analysis of the VEGF promoter was carried out to investigate the potential link between alterations in DNA conformation and changes in gene expression using A498 cells as a model cell line. We first examined the effects of G-quadruplex-interactive agents on the expression of the VEGF gene in A498 renal carcinoma cells using TMPyP4 and TMPyP2, a positional isomer of TMPyP4 that has low affinity for G-quadruplexes (29). The IC50 values for both TMPyP2 and TMPyP4 in A-498 cell lines were ∼50 µM when their antiproliferative activities were assessed with the MTT assay. The treatment of A498 cells with 25 and 50 µM TMPyP4 resulted in a significant repression of VEGF gene expression after 48 h, while TMPyP2 did not cause a decrease in VEGF gene expression (Figure 3A). This result suggests that the G-quadruplex-binding activity of TMPyP4 is responsible for reducing the transcription of the VEGF gene in the presence of TMPyP4. Next, in vivo DMS footprinting via LM-PCR of the G-rich strand of the pPu/pPy tract of the VEGF promoter was carried out to determine the presence of G-quadruplex structures within this region. As shown in Figure 3B, genomic DMS footprinting analysis of the VEGF promoter revealed that the three guanine repeats at the 3′ side of the G-rich strand of the VEGF promoter were strongly protected, while remarkable DMS hypersensitivity sites in vivo were found at guanine residues located at the third guanine repeat (GR-III) (Figure 3B, lane 1). This unusual hypersensitivity is believed to be the result of enhanced reactivity of DMS toward the guanine residues located in the loop region of the G-quadruplex structures formed within the pPu/pPy tract of the VEGF promoter region. A similar observation was made in in vitro DMS footprinting experiments using a supercoiled plasmid DNA as shown in Figure 2B (lane 4). It is worthwhile to note that G-quadruplex DNA structures formed at this region in vivo might differ from those formed in vitro because two hypersensitive guanine residues in the GR-III from the in vivo protection experiment do not correspond to the loop guanines presented in our VEGF G-quadruplex model deducted from in vitro experiments. Densitometric scans (Figure 3C) of the autoradiogram in Figure 3B revealed that DMS protection was only slightly enhanced within the same region of the G-rich strand following TMPyP4 treatment (Figure 3B, lane 2). This result suggests the presence of a permanently folded DNA structure into a G-quadruplex in vivo at the pPu/pPy tract of the VEGF promoter even in the absence of G-quadruplex-interactive agents, such as TMPyP4. Next, we performed ChIP assays to examine whether the recruitment of Sp1 and RNA polymerase II to the VEGF gene promoter is attenuated by TMPyP4 consistent with reduced transcription of this gene in the presence of TMPyP4 (Figure 3A). The treatment of A498 cells with 50 µM TMPyP4 for 48 h resulted in decreased recruitment of Sp1 and RNA polymerase II to the VEGF promoter DNA as shown in Figure 3D. This result suggests that the interaction of TMPyP4 with G-quadruplexes in the VEGF promoter would prevent the binding of Sp1 and RNA polymerase II to the VEGF promoter in vivo, resulting in repression of the VEGF gene. These results also support our hypothesis that enhanced protection of guanine residues from methylation by DMS is not simply due to the increased association of nuclear proteins, but to the formation of G-quadruplex structures as well. The results from both footprinting experiments are summarized in Figure 3E and F.
Figure 3.
(A) RT-PCR analysis of VEGF expression in A498 cells treated with DMS vehicle (control), 50 µM TMPyP2 (P2) or 25 and 50 µM TMPyP4 (P4) for 48 and 72 h. (B) In vivo DMS footprinting analysis of the VEGF promoter region in A498 cells treated for 24 h with DMS vehicle (lane 1) or 50 µM TMPyP4 (lane 2). Lanes A, G, T and C represent the products of sequencing reactions with the same template as a size marker. Lane M represents a 109-bp-sized marker. (C) Densitometric scans of the autoradiogram in (B). The gray bars indicate the guanine repeats, which are involved in G-quadruplex formation. (D) ChIP analysis to determine the effect of TMPyP4 on recruitment of RNA polymerase II and Sp1 to the VEGF promoter region containing the polypurine/polypyrimidine tract in A498 renal carcinoma cells treated with either DMSO control or 25 µM TMPyP4 for 48 h. Recruitment of RNA polymerase II (Pol II) and Sp1 to the VEGF proximal promoter was assessed using primers specific to the VEGF promoter. One percent of total input DNA was used as a loading control (input), and isotype-matched IgG was used as an internal control for the immunoprecipitation (IgG). (E) Proposed equilibrating forms of the pPu/pPy tract of the VEGF promoter in genomes. The asterisks indicate the guanine residues within the G-quadruplex-forming region that show hyperreactivity toward DMS, because they may be located within the locally unwound region or loop region of the G-quadruplex structures. (F) Summary of the results from both in vitro and in vivo DMS footprinting experiments. The DMS-protected guanine residues within the G-rich sequences of the VEGF promoter are indicated by open circles, and closed circles indicate the guanine residues either methylated or hypermethylated by DMS. Data shown are representative of at least two experiments.
Use of the G-quadruplex-specific recognition protein nucleolin to verify the presence of G-quadruplex structures in the promoter region of the VEGF gene in vitro and in vivo
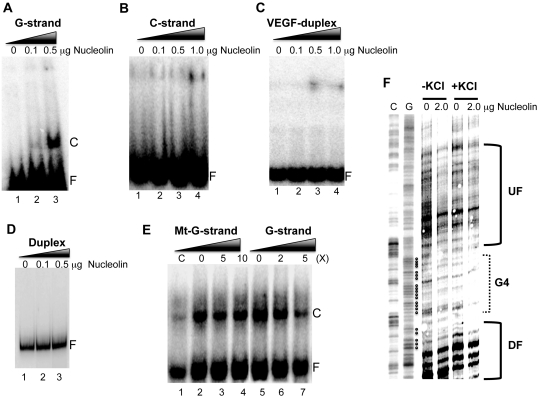
In this study, we utilized nucleolin as a probe for G-quadruplex structures to identify their presence in the promoter region of the VEGF gene both in vitro and in vivo based on the assumption that nucleolin could bind to the G-quadruplex structures formed by this tract. We first confirmed the binding of nucleolin to the intramolecular G-quadruplex structure formed by the G-rich strand of the VEGF proximal promoter region. To determine the binding of nucleolin to the VEGF G-quadruplex, a 5′-end 32P-labeled DNA oligomer, spanning the G-quadruplex-forming region of the G-rich sequence, was folded into G-quadruplex structures as described in the ‘Materials and Methods’ section. This folded DNA oligomer was incubated with nucleolin to form protein–DNA complexes, and then subjected to a native PAGE to separate the nucleolin–G-quadruplex complex from unbound DNA by the difference in the electrophoretic mobility. As shown in Figure 4A, the VEGF G-quadruplex forms a complex with nucleolin, while a complementary C-rich strand (Figure 4B), VEGF duplex oligomer (Figure 4C), and PCR-amplified 202 bp duplex DNA containing the proximal promoter region of the VEGF gene (Figure 4D), fail to form a complex with nucleolin, indicating a possible interaction of the VEGF-G-quadruplex with nucleolin. The results of competition experiments shown in Figure 4E also confirmed that nucleolin was able to specifically bind to the VEGF G-quadruplex. A 5-fold excess of the VEGF G-rich strand prevented the binding of nucleolin, while a 10-fold excess of the mutant VEGF-G-rich strand had no effect.
Figure 4.
(A) Nucleolin binding to the intramolecular G-quadruplex structure formed by the G-rich strand of the VEGF proximal promoter region. The VEGF G-quadruplex incubated with various concentrations of nucleolin to determine the formation of the complex with nucleolin. (B–D) The complementary VEGF C-rich strand (B), VEGF oligomer duplex (C) and PCR-amplified duplex DNA representing the proximal promoter region of the VEGF gene (D) were incubated with various concentrations of nucleolin to determine the formation of complex with nucleolin. (E) Competition experiments to measure the specificity of nucleolin binding to the VEGF G-quadruplex. Two different amounts of competitors were added to the binding reaction with 1 μg nucleolin. Lanes 2–4 are competition experiments with mutant G-strand (Mt-G-strand) competitors at 0×, 5× and 10×, respectively, and lanes 5–7 are competition experiments with the VEGF-G-strand at 0×, 2× and 5×, respectively. The ‘C’ and ‘F’ in panels A–E represent the complex and free form of DNA with nucleolin, respectively. (F) Nucleolin binding to the pPu/pPy tract of the VEGF proximal promoter region in supercoiled plasmid DNA. DNase I footprinting was used to determine the binding of nucleolin to the promoter region of the VEGF gene with the plasmid pGL3-VEGFP, which contains the VEGF promoter region from –787 to +50. The bracket ‘G4’ represents a proposed G-quadruplex-forming region and brackets ‘UF’ and ‘DF’ represent up- and downstream flanking regions of the G-quadruplex-forming sequence. Data shown are representative of at least two experiments.
In our previous study, it was demonstrated that the transition of B-form DNA to G-quadruplexes occurs at the G-rich strand of the pPu/pPy tract of the VEGF promoter region under negative supercoiling stress (18). This suggests that negative supercoiling could generate binding sites for nucleolin on this tract. Thus, DNase I footprinting was used to determine the binding of nucleolin to the promoter region of the VEGF gene using the plasmid pGL3-VEGFP, which contains the VEGF promoter region from −787 to +50. In this experiment, nucleolin was incubated with pGL3-VEGFP in the absence or presence of 50 mM KCl at 37°C for 1 h and then probed with DNase I as described previously. DNase I cleavage sites in the plasmids incubated with nucleolin were then mapped using linear amplification by PCR with 32P-labeled gene-specific primers as described in ‘Materials and Methods’ section. As shown in Figure 4F, the DNase I protected region located at approximately −53 to −123 bp includes the predicted G-quadruplex-forming region. This result suggests that nucleolin interacts with G-quadruplex structure(s) formed by the G-rich sequence within the pPu/pPy region of the VEGF gene.
Nucleolin can bind to the VEGF proximal promoter in cells
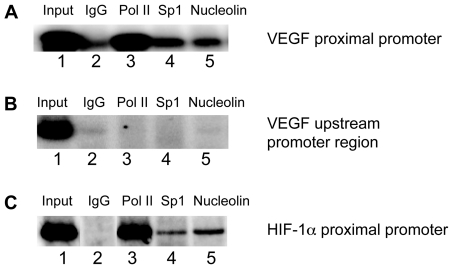
Next, we carried out a ChIP assay to determine whether nucleolin is associated with the proximal promoter region of the VEGF gene in A498 renal carcinoma cells. As shown in Figure 5A, we observed the PCR amplification product of the proximal promoter region (–242 to +48) after immunoprecipitation of the cross-linked chromatin with the anti-nucleolin antibody (lane 5) in addition to the anti-RNA polymerase II (lane 3) and anti-Sp1 antibody (lane 4), while immunoprecipitates with purified mouse IgG as a negative control resulted in no PCR amplified products (lane 2). These results support the idea that nucleolin as well as RNA polymerase II bind to the VEGF promoter spanning the nucleotides from −242 to +48, suggesting the presence of specific G-quadruplex structures within the proximal promoter region of this gene in vivo. To further determine the specificity of nucleolin binding to the G-quadruplex-forming region, we also amplified the 5′ upstream promoter region (–1079 to −874) of the VEGF gene, not containing any putative G-quadruplex-forming regions, and no PCR products indicative of the upstream region of the VEGF promoter were found (Figure 5B). As a positive control, we also amplified the proximal promoter region (–273 to +71) of the HIF-1α gene, which contains a putative G-quadruplex-forming region. As shown in Figure 5C, PCR products indicative of this region were observed in the input sample (lane 1), RNA polymerase, Sp1 and nucleolin-specific immunoprecipitations.
Figure 5.
(A) ChIP analysis to determine the binding of nucleolin to the VEGF promoter region containing the polypurine/polypyrimidine tract in A498 renal carcinoma cells. Recruitment of RNA Polymerase II (Pol II) (lanes 3), Sp1 (lanes 4) and nucleolin (lanes 5) to the VEGF proximal promoter was assessed using primers specific to the VEGF promoter (–248 to +48). One percent of total input DNA was used as a loading control (lane 1) and isotype-matched IgG was used as an internal control for the immunoprecipitation (lane 2). (B) PCR amplification of immunoprecipitated DNAs using primers specific to the 5′ upstream promoter region (–1079 to –874) of the VEGF gene as a negative control. (C) PCR amplification of immunoprecipitated DNAs using primers specific to the proximal promoter region (−273 to +71) of the HIF−1α gene as a positive control. Data shown are representative of at least two experiments.
DISCUSSION
The pPu/pPy tract (−85 to −50 relative to transcription initiation site) within the proximal promoter region of the VEGF gene is essential for basal or inducible expression of this gene in several human cancer cell lines (5, 10). The G-rich strand of this tract contains four runs of at least three contiguous guanines separated by one or more bases, conforming to a general motif capable of forming an intramolecular G-quadruplex (18,20). Our previous studies demonstrated that this G-rich sequence is able to form an intramolecular propeller-type parallel-stranded G-quadruplex structure in vitro (20). Our other study also revealed a significant change in the digestion patterns within this region with both DNase I and S1 nuclease after the addition of KCl and telomestatin (18), suggesting the possible interaction of the G-quadruplex-interactive agent telomestatin with a G-quadruplex-forming region within the VEGF gene in supercoiled DNA.
In this report, we have further examined the possible presence of G-quadruplex structures both in vitro and in vivo in the pPu/pPy tract within the proximal promoter region of the VEGF gene using DMS and nucleolin as chemical and protein probes, respectively. DMS footprinting has been widely used for fine mapping of the presence of G-quadruplex structures within G-rich single-stranded DNA since the formation of G-quadruplex structures requires the N7 of guanine, which is protected from N7 methylation by DMS (19,21). Our recent study also demonstrated that DMS footprinting is useful for probing G-quadruplexes within the duplex DNA in vitro (27). As demonstrated in this study, four consecutive guanine tracts at the 5′ side of the G-rich strand within the pPu/pPy tract of the VEGF promoter, which are predicted to be involved in the formation of the G-tetrad, show diminished reactivity to DMS under supercoiled conditions. By contrast, the immediate flanking regions of the proposed G-quadruplex-forming sequence show enhanced reactivity to DMS, presumably because of the presence of locally unwound structures at the junctions between normal duplex regions and stable secondary structures. Interestingly, guanine residues in a predicted central loop region showed hypersensitivity to DMS. Overall, our findings in the present study are in line with our hypothesis that G-quadruplex structures could be formed by the G-rich strand of the pPu/pPy tract within the VEGF promoter region and these structures might participate in the regulation of this gene’s transcription.
In vivo DMS-LMPCR footprinting of the G-rich strand of the pPu/pPy tract of the VEGF promoter also provided interesting information regarding the possibility of the presence of G-quadruplex structures within this region. As described in Figure 3B, most of the protected guanine residues correspond to those that are known to be involved in the formation of G-quadruplexes in vitro. This is consistent with the results of in vitro DMS footprinting of the G-rich strand in both a single-stranded and double-stranded form in supercoiled DNA. Furthermore, the presence of DMS-hypersensitive guanines within the loop region of the predicted G-quadruplex structures on the G-rich strand would reflect the presence of a stable G-quadruplex within the VEGF promoter region. However, two DMS-hypersensitive guanine residues within the GR-III do not correspond to the loop guanines presented in our VEGF G-quadruplex model deducted from in vitro DMA protection experiments. This implies the possibility that G-quadruplex DNA structures actually forming in this region in vivo might differ from those observed in in vitro experiments. It is very difficult to experimentally determine the G-quadruplex structures that actually exist in vivo. However, the possibility of the existence of different forms of G-quadruplexes in vivo cannot be completely excluded because the internal nuclear environment could differ from those in vitro with respect to ionic composition and concentration, pH, protein composition and concentration, polyamine concentration, etc. Interestingly, DMS protection was only slightly enhanced within the same region of the G-rich strand following TMPyP4 treatment, although the occupancy of both Sp1 and RNA polymerase of the proximal promoter region of the VEGF gene sharply decreased after a 48-h incubation in the presence of 50 µM TMPyP4. We speculate that TMPyP4 might not significantly facilitate the formation of G-quadruplexes in this region, but would interact with the already-existing G-quadruplexes. The G-quadruplexes complexed with TMPyP4 could be more efficient than G-quadruplex structures alone in preventing the transcription components, such as Sp1 and RNA polymerase II, from binding to the VEGF promoter region, thus resulting in reduced expression of the VEGF gene. The result of ChIP assays also supports the notion that enhanced protection of the guanine residues from methylation by DMS in both untreated and TMPyP4-treated cells is due to the formation of G-quadruplex structures rather than the increased association of nuclear proteins.
In several previous studies, the abundant nucleolar protein, nucleolin, has been shown to bind both intra- and intermolecular G-quadruplex DNA with very high affinity (24,25), suggesting that G-quadruplex structures are binding targets of nucleolin in vivo. Based on these observations, we utilized a nucleolin protein as a structural probe to identify G-quadruplex structures formed within the duplex DNA both in vitro and in vivo in this study. In support of the pPu/pPy tract of the VEGF promoter region having similar G-quadruplex structures both in vivo and in vitro, DNase I footprinting of the plasmid pGL3-VEGFP revealed the binding of nucleolin to the G-quadruplex forming region of the VEGF promoter. A ChIP assay further confirmed the in vivo binding of nucleolin to the VEGF promoter region containing the pPu/pPy tract. The results presented here support the use of nucleolin for both in vitro and in vivo structural probing to identify the formation of G-quadruplex structures within the G-rich sequences that affect transcription of many mammalian genes in vivo. The results with both in vitro and in vivo DMS footprinting experiments show that predictions of intracellular G-quadruplex structures, especially structures involving local interactions, can be made on the basis of in vitro reactivity of DMA toward the guanine residues involved in the formation of these structures.
Several lines of experimental evidence from previous studies support a potential formation of G-quadruplex structures in the promoter region of the VEGF gene (18,20). Our findings from this study further provide strong evidence that the G-rich region within pPu/pPy tract of the VEGF promoter region could adopt stable G-quadruplex structures, which could be probed with DMS both in vitro and in vivo. In conjunction with DMS footprinting, protein probes such as nucleolin, which specifically recognize G-quadruplex structures, could be used to investigate whether G-quadruplex structures are formed in vitro and in vivo from putative G-quadruplex forming sequences. Our results also support the idea that nucleolin can function as a transcriptional regulator of mammalian genes (25), whose promoter regions are able to form G-quadruplex structures under physiological conditions. Thus, ongoing studies are in progress in our laboratory to define a possible role for nucleolin in regulating transcription of the VEGF gene. Although it is not yet known whether these noncanonical DNA structures can serve as a substrate for these transcription factors in an in vivo system, their specific interaction with non-B-DNA structures in an in vitro system suggests that alternative DNA conformations may naturally exist and may also play a crucial role in the regulation of many genes containing highly dynamic conformations in their promoter regions. Such alternative structures, although speculative at this point, may be of relevance for the new therapeutic approach that proposes the use of small molecule drugs as transcriptional regulators.
FUNDING
The National Institutes of Health (CA109069); Arizona Biomedical Research Commission (ADCRC#8020). Funding for open access charge: The National Institutes of Health (CA109069).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Dr. Allison Hays for critical reading of the final version of the article and figures. We also thank Dr. Keping Xie for providing pGL3-V789 (pGL3-VEGFP) for this study.
REFERENCES
- 1.Martiny-Baron G, Marme D. VEGF-mediated tumour angiogenesis: a new target for cancer therapy. Curr. Opin. Biotechnol. 1995;6:675–680. doi: 10.1016/0958-1669(95)80111-1. [DOI] [PubMed] [Google Scholar]
- 2.Folkman J. Role of angiogenesis in tumor growth and metastasis. Semin. Oncol. 2002;29:15–18. doi: 10.1053/sonc.2002.37263. [DOI] [PubMed] [Google Scholar]
- 3.Jain RK. Tumor angiogenesis and accessibility: role of vascular endothelial growth factor. Semin. Oncol. 2002;29:3–9. doi: 10.1053/sonc.2002.37265. [DOI] [PubMed] [Google Scholar]
- 4.Goodsell DS. The molecular perspective: VEGF and angiogenesis. Stem Cells. 2003;21:118–119. doi: 10.1634/stemcells.21-1-118. [DOI] [PubMed] [Google Scholar]
- 5.Jośko J, Mazurek M. Transcription factors having impact on vascular endothelial growth factor (VEGF) gene expression in angiogenesis. Med. Sci. Monit. 2004;10:RA89–RA98. [PubMed] [Google Scholar]
- 6.Brieger J, Weidt EJ, Schirmacher P, Storkel S, Huber C, Decker HJ. Inverse regulation of vascular endothelial growth factor and VHL tumor suppressor gene in sporadic renal cell carcinomas is correlated with vascular growth: an in vivo study on 29 tumors. J. Mol. Med. 1999;77:505–510. doi: 10.1007/s001099900022. [DOI] [PubMed] [Google Scholar]
- 7.Laughner E, Taghavi P, Chiles K, Mahon PC, Semenza GL. HER2 (neu) signaling increases the rate of hypoxia-inducible factor 1alpha (HIF-1α) synthesis: novel mechanism for HIF-1-mediated vascular endothelial growth factor expression. Mol. Cell. Biol. 2001;21:3995–4004. doi: 10.1128/MCB.21.12.3995-4004.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H, Ye D, Xie X, Chen B, Lu W. VEGF, VEGFRs expressions and activated STATs in ovarian epithelial carcinoma. Gynecol. Oncol. 2004;94:630–635. doi: 10.1016/j.ygyno.2004.05.056. [DOI] [PubMed] [Google Scholar]
- 9.Schafer G, Cramer T, Suske G, Kemmner W, Wiedenmann B, Hocker M. Oxidative stress regulates vascular endothelial growth factor-A gene transcription through Sp1- and Sp3-dependent activation of two proximal GC-rich promoter elements. J. Biol. Chem. 2003;278:8190–8198. doi: 10.1074/jbc.M211999200. [DOI] [PubMed] [Google Scholar]
- 10.Shi Q, Le X, Abbruzzese JL, Peng Z, Qian CN, Tang H, Xiong Q, Wang B, Li XC, Xie K. Constitutive Sp1 activity is essential for differential constitutive expression of vascular endothelial growth factor in human pancreatic adenocarcinoma. Cancer Res. 2001;61:4143–4154. [PubMed] [Google Scholar]
- 11.Beckman JS, Weber JL. Survey of human and rat micro-satellites. Genomics. 1992;12:627–631. doi: 10.1016/0888-7543(92)90285-z. [DOI] [PubMed] [Google Scholar]
- 12.Behe MJ. An overabundance of long oligopurine tracts occurs in the genome of simple and complex eukaryotes. Nucleic Acids Res. 1995;23:689–695. doi: 10.1093/nar/23.4.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brahmachari SK, Sarkar PS, Raghavan S, Narayan M, Maiti AK. Polypurine/polypyrimidine sequences as cis-acting transcriptional regulators. Gene. 1997;190:17–26. doi: 10.1016/s0378-1119(97)00034-6. [DOI] [PubMed] [Google Scholar]
- 14.Brahmachari SK, Ramesh N, Shouche YS, Mishra RK, Bagga R, Meera G. Unusual DNA structures: sequence requirements and role in transcriptional control. In: Sarma RH, Sarma MH, editors. Structure & Methods, Volume 2: DNA Protein Complexes & Proteins. Schenectady, NY: Adenine Press; 1990. pp. 33–49. [Google Scholar]
- 15.Evans T, Efstratiadis A. Sequence-dependent S1 nuclease hypersensitivity of a heteronomous DNA duplex. J. Biol. Chem. 1986;261:14771–14780. [PubMed] [Google Scholar]
- 16.McCarthy JG, Heywood SM. A long polypyrimidine/polypurine tract induces an altered DNA conformation on the 3′ coding region of the adjacent myosin heavy chain gene. Nucleic Acids Res. 1987;15:8069–8085. doi: 10.1093/nar/15.19.8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z, Lin XH, Qiu QQ, Deuel TF. Modulation of transcription of the platelet-derived growth factor A-chain gene by a promoter region sensitive to S1 nuclease. J. Biol. Chem. 1992;267:17022–17031. [PubMed] [Google Scholar]
- 18.Sun D, Guo K, Rusche JJ, Hurley LH. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005;33:6070–6080. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simonsson T. G-quadruplex DNA structures–variations on a theme. Biol. Chem. 2001;382:621–628. doi: 10.1515/BC.2001.073. [DOI] [PubMed] [Google Scholar]
- 20.Sun D, Liu WJ, Guo K, Rusche JJ, Ebbinghaus S, Gokhale V, Hurley LH. The proximal promoter region of the human vascular endothelial growth factor gene has a G-quadruplex structure that can be targeted by G-quadruplex-interactive agents. Mol. Cancer Ther. 2008;7:880–889. doi: 10.1158/1535-7163.MCT-07-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dabrowiak JC, Goodisman J, Ward B. Quantitative DNA footprinting, Methods Mol. Biol. 1997;90:23–42. doi: 10.1385/0-89603-447-X:23. [DOI] [PubMed] [Google Scholar]
- 22.Ginisty H, Sicard H, Roger B, Bouvet P. Structure and functions of nucleolin. J. Cell. Sci. 1999;112:761–772. doi: 10.1242/jcs.112.6.761. [DOI] [PubMed] [Google Scholar]
- 23.Mongelard F, Bouvet P. Nucleolin: a multiFACeTed protein. Trends Cell Biol. 2007;17:80–86. doi: 10.1016/j.tcb.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 24.Hanakahi LA, Sun H, Maizels N. High affinity interactions of nucleolin with G-G-paired rDNA. J. Biol. Chem. 1999;274:15908–15912. doi: 10.1074/jbc.274.22.15908. [DOI] [PubMed] [Google Scholar]
- 25.González V, Guo K, Hurley LH, Sun D. Identification and characterization of nucleolin as a c-myc G-quadruplex-binding protein. J. Biol. Chem. 2009;284:23622–23635. doi: 10.1074/jbc.M109.018028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niu G, Wright KL, Huang M, Song L, Haura E, Turkson J, Zhang S, Wang T, Sinibaldi D, Coppola D, et al. Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis. Oncogene. 2002;21:2000–2008. doi: 10.1038/sj.onc.1205260. [DOI] [PubMed] [Google Scholar]
- 27.Sun D, Hurley LH. The importance of negative superhelicity in inducing the formation of G-quadruplex and i-motif structures in the c-Myc promoter: implications for drug targeting and control of gene expression. J. Med. Chem. 2009;52:2863–2874. doi: 10.1021/jm900055s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drouin R, Therrien JP, Angers M, Ouellet S. In vivo DNA analysis. Methods Mol. Biol. 2001;148:175–219. doi: 10.1385/1-59259-208-2:175. [DOI] [PubMed] [Google Scholar]
- 29.Grand CL, Han H, Muñoz RM, Weitman S, Von Hoff DD, Hurley LH, Bearss DJ. The cationic porphyrin TMPyP4 down-regulates c-MYC and human telomerase reverse transcriptase expression and inhibits tumor growth in vivo. Mol. Cancer Ther. 2002;1:565–73. [PubMed] [Google Scholar]