Abstract
In many bacterial species, the multi-enzyme RNA degradosome assembly makes key contributions to RNA metabolism. Powering the turnover of RNA and the processing of structural precursors, the RNA degradosome has differential activities on a spectrum of transcripts and contributes to gene regulation at a global level. Here, we report the isolation and characterization of an RNA degradosome assembly from the α-proteobacterium Caulobacter crescentus, which is a model organism for studying morphological development and cell-cycle progression. The principal components of the C. crescentus degradosome are the endoribonuclease RNase E, the exoribonuclease polynucleotide phosphorylase (PNPase), a DEAD-box RNA helicase and the Krebs cycle enzyme aconitase. PNPase and aconitase associate with specific segments in the C-terminal domain of RNase E that are predicted to have structural propensity. These recognition ‘microdomains’ punctuate structurally an extensive region that is otherwise predicted to be natively disordered. Finally, we observe that the abundance of RNase E varies through the cell cycle, with maxima at morphological differentiation and cell division. This variation may contribute to the program of gene expression during cell division.
INTRODUCTION
RNA turnover is an essential activity in all organisms and contributes to the regulation of gene expression in both specific and general ways (1,2). For many bacterial species, the early stages of transcript degradation and RNA processing are mediated largely by the endoribonuclease, RNase E (3). In Escherichia coli, RNase E not only contributes to the degradation of the majority of mRNAs, but also functions in the maturation of precursors of structured RNA, such as 5S and 16S ribosomal RNA, tRNA and the RNase P ribozyme (4). RNase E is also involved in the gene silencing mediated by small non-coding RNAs and the RNA chaperone Hfq (5–7). The enzyme is well conserved in the proteobacteria phylum, and homologues are found in plastids of certain plants (4,8,9). Considering the many roles of RNase E, it is perhaps not surprising that the enzyme is essential for the survival of E. coli (10,11).
In E. coli and related γ-proteobacteria, RNase E organizes a multi-enzyme assembly, referred to as the RNA degradosome (3). The C-terminal half of E. coli RNase E forms the scaffolding for the degradosome assembly. This region is ∼530 residues in length, but has remarkably low sequence complexity, consistent with predictions that it lacks globular compactness, and is predominantly unstructured (12). However, the segment is punctuated by short regions of predicted structural propensity which correspond to the binding sites for RNA, the cytoplasmic membrane and two of the principal degradosome partner proteins, namely the glycolytic enzyme enolase, and the 3′→5′ exoribonuclease polynucleotide phosphorylase (PNPase) (4,12,13). The remaining principal degradosome component, the DEAD-box RNA helicase B (RhlB), has been shown to interact with a region of RNase E predicted to have low structural complexity, C-terminal to an RNA-binding site with predicted propensity to form secondary structure (12). The question of why enolase and RNase E associate remains unanswered, but the interaction is likely to be functionally important judging from its conservation (14,15) and requirement for response to phosphosugar stress (7). The finding that metabolic enzymes are associated with ribonucleases even in bacterial species that lack an RNase E homologue further indicates that the interaction has functional consequence (16). In addition to the canonical components, the degradosome is also associated with other proteins in sub-stoichiometric amounts, including the RNA chaperone Hfq, ribosomal proteins, polyphosphate kinase and protein chaperones (4,7,17,18).
Degradosomes and related assemblies are likely to be almost ubiquitous amongst diverse bacterial lineages (19–21). In eukaryotes and archaea, an analogous multi-enzyme complex, the exosome, is assembled on a core that resembles bacterial PNPase (22). In the case of RNase E-mediated degradosomes, the portion of the enzyme that organizes the different assemblies shows marked sequence divergence amongst family members. From the handful of degradosome assemblies characterized thus far, it is apparent that these complexes are diverse in composition (4,19,23,24). The emerging picture of the degradosome is a dynamic, multi-modular assembly with compositional variation, conformational flexibility and phylogenetic diversity.
In this study, we have identified an RNase E-mediated degradosome assembly in the Gram-negative α-proteobacterium Caulobacter crescentus. This organism has proven to be an excellent experimental system for studying prokaryotic cell cycle regulation and morphological development (25). The cell-cycle states of C. crescentus are morphologically distinct, with cells in the pre-DNA synthesis gap (G1) having a single polar flagellum and pili, and cells in DNA synthesis phase (S) having a polar stalk but no flagellum or pili. Cell division is asymmetric from the sedentary stalked form, producing daughter cells in the morphologically distinct free-swimming form. These morphological differences permit the isolation of populations in defined stages of the cell cycle via a facile density centrifugation step (26).
Using co-immunopurification we identify the components of the C. crescentus degradosome assembly as RNase E, a DEAD-box helicase, PNPase and most surprisingly aconitase. This association of a Krebs cycle enzyme with the C. crescentus RNA degradosome highlights the recurrent evolution of physical associations between the enzymes of RNA degradation and central metabolism. We show that the RNase E level varies in a cell-cycle-dependent manner, with maxima at the G1-to-S transition and at the point of cell division. Finally, we comment on how the regulation of RNase E abundance may contribute to post-transcriptional gene regulation during cell division.
MATERIALS AND METHODS
Caulobacter crescentus cell cultures and growth
For all immunopurification and synchronization experiments, the C. crescentus strain NA1000, a derivative of CB15 that lacks holdfast and can be synchronized, was used (26). Unless mentioned otherwise, NA1000 was grown at 30°C in either peptone-yeast extract (PYE) (27) or M2G minimal medium (28).
Co-immunopurification of C. crescentus degradosome complex
Anti-RNase E antibodies were raised in rabbits against the peptide (CRDDSGDEDDTPIRSRR) by Cambridge Research Biochemicals. The N-terminal C was added for peptide purification. The co-immunopurification protocol is a modification of the procedure developed for the affinity purification of FLAG-tagged E. coli RNase E complexes (7). C. crescentus cells were grown asynchronously in 1 l of PYE to A600 of ∼0.4, harvested by centrifugation and resuspended in 30 ml of lysis buffer (20 mM Na-phosphate pH 8.0). Cells were flash-frozen in liquid nitrogen and stored at −80°C until required. Cell pellets were thawed on ice and supplemented with 5 mM EDTA and protease inhibitor tablets (Roche) prior to lysis by passage three times through a high-pressure homogenizer (emulsiflex). The cell suspension was centrifuged (30 min, 30 000g) and the supernatant was incubated with activated or blank protein A sepharose as required (2 h, 4°C). The protein A sepharose was collected and washed in a mini-chromatography column (Bio-Rad) three times with lysis buffer supplemented with 100 mM NaCl. To elute proteins bound to anti-RNase E, the protein A-sepharose was incubated with 30 μl of the anti-RNase E antigenic peptide diluted to 1 mg/ml in lysis buffer (30 min, 4°C) and eluate was collected by centrifugation. For analysis by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) the entire volume of eluate was loaded into a single well of a 4–12% polyacrylamide NuPage gel (Invitrogen).
Cell-cycle synchronization
To synchronize a population of C. crescentus NA1000, swarmer cells were isolated by Ludox density centrifugation and resuspended in fresh media (26). Typically, a 50-ml synchronous culture was generated and progression through the cell division cycle was monitored by taking 1-ml samples at 20-min intervals and western blotting for the presence of the flagellar protein FliF. Cells were examined by optical microscopy.
Western blotting
Whole-cell extracts or immune-precipitated fractions were separated on SDS-PAGE gels and proteins electro-transferred onto poly(vinylidene fluoride) (PVDF) membranes. The following primary antibody dilutions were used: affinity-purified C. crescentus RNase E antibody, 1:500; E. coli PNPase crude antiserum, 1:1000; E. coli aconitase crude antiserum, 1:1000; C. crescentus FliF crude antiserum, 1:5000. For all western blots secondary goat anti-rabbit IgG horseradish peroxidase conjugate was used (Sigma) at a dilution of 1:5000, and blots were visualized on photographic film following incubation in enhanced chemiluminescence substrate (GE Healthcare). The photographic films were subsequently digitized, and the intensities of bands were measured with Scion Image software (Scion Corp.).
9S RNA processing assay
A reaction master mix containing 0.3 μM 9S RNA, 25 mM Tris–Cl pH 8, 10 mM MgCl2, 25 mM NaCl, 25 mM KCl, 1 mM DTT, 0.5 U/μl of RNaseOUT™ (Invitrogen) was prepared. 9S RNA substrate was preheated at 50°C for 2 min then cooled to room temperature prior to addition to the reaction buffer. All reactions were initiated with the addition of 0.5 μM E. coli RNase E catalytic domain (kindly provided by Kasia Bandrya) or C. crescentus RNase E catalytic domain. Reactions were incubated at 37°C, and 5 μl aliquots were withdrawn at time points 0, 15 min and quenched by 5 μl 2 × proteinase K buffer (200 mM Tris–HCl, pH 8, 25 mM EDTA, 300 mM NaCl, 2% w/v sodium dodecylsulphate), supplemented with 0.5 mg/ml Tritirachium album Proteinase K (Sigma–Aldrich) and incubated for 20 min at 50°C to ensure all enzymes were digested. Ten microlitres of 2× RNA loading dye (Fermentas) was added to all reactions and denatured at 90°C for 5 min before being analysed on an 8% denaturing polyacrylamide gel. The RNA substrates/products were visualized by SYBR gold stain. All experiments were carried out in an RNase-free environment.
Nuclear magnetic resonance data collection and structure analysis
The nuclear magnetic resonance (NMR) sample was prepared containing 1 mM EKPRRGWWRR, 50 mM sodium phosphate (pH 7.5), 20 µM 3,3,3-trimethylsilylpropionate (TSP) and 10% D2O, to a final volume of 550 µl in a 5 mm Ultra-Imperial grade NMR tube (Wilmad). Spectra for sequential assignment and structure determination were recorded at 298 K and 278 K at a 1H frequency of 500.13 MHz on a Bruker DRX spectrometer using standard procedures (29). Few NOE cross peaks were observed in homonuclear NOESY spectra acquired at 298 K, but numerous resonances were detected in experiments collected at 278 K. Interestingly, only 1HN sites showed significant changes in chemical shift between 278 and 298 K. The appearance of NOE cross peaks at 278 K is therefore probably the result of the increased solvent viscosity producing a longer overall rotational correlation time, rather than changes in the dynamics of the peptide.
Sequential assignments were obtained at 278 K for residues Lys-2 to Arg-10 via standard methods (30), and then transferred to TOCSY spectra collected at 298 K. Homonuclear TOCSY (τmix 60 ms) and NOESY experiments (τmix 350 ms) were collected with 256 and 1024 pairs of complex points and acquisition times of 51 ms and 102 ms in the indirect and direct 1H dimensions, respectively. All spectra were processed and interpreted using the CcpNmr Azara and Analysis packages (31). A total of 201 unambiguously assigned inter-proton distance restraints were harvested from the 278 K NOESY spectrum as input for structure determination. All structure calculations were carried out using the default settings of ARIA version 1.2 (32). Nine rounds of ARIA calculations were performed, with 20 structures calculated for the first eight iterations and 50 structures in the last of 50 structures determined in the final round. The 20 lowest energy models were selected for refinement in explicit solvent, yielding an ensemble with no distance restraint violations >0.3 Å and acceptable covalent geometry statistics (Table 1). Structure images were generated using PyMOL, version 1.1 (33) (http://pymol.org).
Table 1.
Statistics for the EKPRRGWWRR peptide solution structure
| NMR distance constraints | ||
| Intra-residue | 107 | |
| Sequential | 59 | |
| Medium range | 35 | |
| Long range | 0 | |
| Total | 201 | |
| Structure statistics | ||
| Distance constraint violations (Å) | 0.0249 ± 0.0038 | |
| Idealized bond length violations (Å) | 0.0046 ± 0.0003 | |
| Idealized bond angle violations (°) | 0.62 ± 0.05 | |
| Idealized improper angle violations (°) | 1.46 ± 0.20 | |
| Final energy EL-J (kJ mol−1) | –81.8 ± 3.5 | |
| Assessment of backbone quality according to the Ramachandran plot | ||
| Most favoured region | 50.0% | |
| Additionally allowed region | 37.5% | |
| Average pair-wise root mean square deviations | ||
| Backbone (Å) | Heavy atoms (Å) | |
| Residues 1–10 | 1.57 ± 0.63 | 2.63 ± 0.66 |
| Residues 5–10 | 0.44 ± 0.22 | 1.45 ± 0.36 |
RESULTS
Identification of a putative degradosome complex in C. crescentus
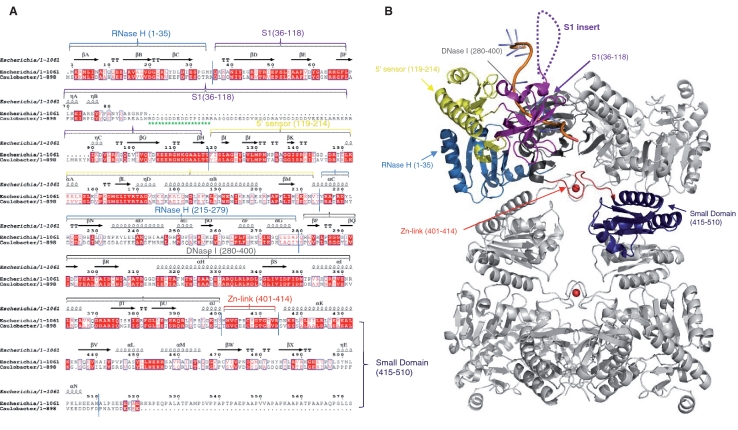
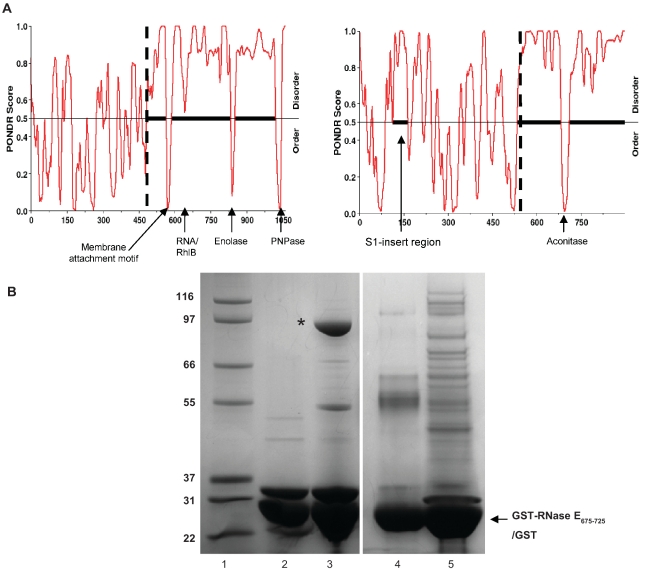
A rabbit polyclonal antibody was raised against a 16-mer peptide (RDDSGDDEDDTPIRSRR) corresponding to a segment of the S1 subdomain of the C. crescentus RNase E catalytic domain. The antigenic peptide is part of a poorly conserved insert in α-proteobacteria that is absent in RNase E of other bacterial classes (Figure 1A), although it does occur in plant homologues (34). We suspected that the S1 insert region would provide a suitable antibody binding epitope as this moiety is highly charged and, according to the PONDR disorder prediction algorithm (35,36), is intrinsically unstructured (Figure 4A). The crystal structure of the E. coli RNase E catalytic domain (37) suggests that the S1 subdomain should be exposed and accessible (Figure 1B).
Figure 1.
RNase E of α-proteobacteria have a distinguishing S1 domain insert not found in RNase E of other bacterial classes. (A) Structure based sequence alignment of the E. coli and C. crescentus RNase E catalytic domains. The secondary structural elements of E. coli RNase E are shown on the lines above the sequence alignment using the PDB file 2BX2. The arrows indicate β-sheet, the coils indicate α-helices, TT indicates β turns and η indicates 310 helices. Red letters indicate homology and blue boxes show similarity. The red highlights indicate identity across the sequences. The green stars represent the antigenic peptide used in this study. Structural sub-domains of RNase E are coloured (RNase H: light blue; S1: purple; 5′ sensor: yellow; DNase I: dark grey; zinc link: red; small domain: dark blue). Alignments were prepared using CLUSTALW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html) and ESPript (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). (B) E. coli RNase E NTD tetramer in complex with 13-mer RNA (pale orange). Structural sub-domains are highlighted for one protomer, coloured as in Figure 1A. The position of the S1 insert absent in E. coli RNase E catalytic domain is represented by the dashed loops.
Figure 4.
Identification of an aconitase binding region in C. crescentus RNase E. (A) Disorder prediction based on RNase E protein sequence from E. coli (left) and C. crescentus (right). N-terminal catalytic domains and C-terminal scaffold domains are separated by a vertical dashed line and extended regions of disorder are represented with a solid horizontal black line. C-terminal microdomains and the C. crescentus S1 insert are indicated with arrows. (B) Pull downs from cell extracts using immobilised C. crescentus micro-domain (RNase E675–725). Lane 1, molecular weight marker. Lane 2, input GST-RNase E675–725. Lane 3, the material eluted from GST–microdomain fusion, the asterisk indicates aconitase. Lane 4, input GST. Lane 5, material eluted from pulldown with GST alone. The gel is Coomassie stained.
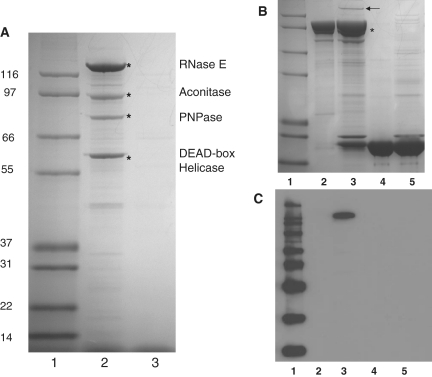
Caulobacter crescentus RNase E was isolated directly from cell lysates using the anti-RNase E antibody immobilized on protein A-sepharose. Following washing, proteins forming stable complexes with RNase E were eluted and analysed by denaturing gel electrophoresis (Figure 2A). Major protein bands of ∼125 kDa and 60 kDa were reproducibly present in pull-down experiments. Mass spectrometer analyses on combined LysC and trypsin digested samples (PNAC, University of Cambridge) revealed that the band at 125 kDa is the full-length RNase E (16 unique proteolytic peptides identified, protein identification code CC1877). Like its E. coli homologue (38), C. crescentus RNase E runs at a higher than expected position on a denaturing gel, with an apparent mass of 125 kDa by SDS-PAGE, but a calculated molecular mass of 100 kDa. The anomalous migration of RNase E from both species is probably due to their unusual sequence composition. The mass spectrometry data also shows that the band at 60 kDa is a predicted DEAD-box RNA helicase (CC1847, 17 unique peptides identified). Of the five E. coli DEAD-box helicases, the closest homologue of C. crescentus CC1847 helicase is RhlE, and not the canonical degradosome helicase, RhlB. However, it has been shown that in E coli RhlE can associate with RNase E and substitute functionally for RhlB in RNA degradation (39).
Figure 2.
Identification of an RNA degradosome complex in C. crescentus. (A) SDS-PAGE analysis of immunoprecipitate from cell lysate of C. crescentus using RNase E antibody (lane 2). Cell lysates were applied to protein–A sepharose with bound antibody raised against C. crescentus RNase E and eluted with the peptide epitope. Degradosome components identified by MALDI fingerprinting are marked with an asterisk and correspond to RNase E, aconitase, polynucleotide phosphorylase and a DEAD-box RNA helicase. Lane 1 shows molecular weight standard (kDa). Lane 3 is a control in which C. crescentus lysates were eluted from protein–A sepharose in the absence of the anti-RNase E antibody. The gel is Coomassie stained. (B) Identification of PNPase partners by pull down. Cell lysates were applied to a glutathione matrix with bound fusion protein of PNPase and glutathionine-S-transferase (GST) and eluted with glutathione. Lane 1, molecular weight marker. Lane 2, GST–PNPase input. Lane 3, material eluted from glutathione-sepharose pre-incubated with GST–PNPase and C. crescentus cell lysate. Lane 4, GST alone input. Lane 5, control: elution from glutathione sepharose following incubation with GST and C. crescentus cell lysate. The asterisk indicates the position of GST-PNPase, and RNase E is marked with an arrow. The gel is Coomassie stained. (C) Western blot of same gel, probed with antibody against C. crescentus RNase E.
Several other protein bands were observed in the immunoprecipitates; however, western blot analysis with the anti-RNase E antibody indicated that many of these correspond to RNase E fragments and are likely to arise from proteolysis (data not shown). As found in the purification of the E. coli homologue (38), it was essential to have protease inhibitors in the purification buffers to obtain full-length C. crescentus RNase E. Particularly, the presence of 5 mM EDTA in the immunoprecipitation buffers significantly reduced proteolysis of RNase E. Using these conditions, we were able to identify two further co-purifying proteins by mass spectrometry. A band at 80 kDa was found to be polynucleotide phosphorylase (PNPase; CC0034, 11 proteolytic peptides identified), and most surprisingly, a band of ∼90 kDa corresponded to aconitase A (CC3667, 12 proteolytic peptides identified). These data suggest that the composition of the degradosome complex in C. crescentus shows great similarity to the canonical E. coli complex, with the exception of an exchange of the glycolytic enzyme enolase for the Krebs cycle enzyme aconitase. Several other possible substoichiometric components of the degradosome complex were also noted in the pull-down experiments, and one of these was identified as the 27-kDa small ribosomal subunit protein S3 (CC1254, 6 proteolytic peptides identified). This protein is also associated with RNase E in E. coli (17).
Following the identification of PNPase in the co-immunopurification of RNase E, an additional pull-down experiment was performed using N-terminally GST-tagged PNPase as bait. After incubation with C. crescentus cell lysate and purification by glutathione sepharose chromatography, the major proteins eluted from the resin by glutathione-containing buffer were the input GST-PNPase and a protein of ∼125 kDa (Figure 2B). A western blot using anti-RNase E antibody confirmed that this 125-kDa protein was indeed RNase E (Figure 2C). The apparent low level of RNase E retrieved in this experiment is presumably due to competition between GST-PNPase and endogenous PNPase in the cell lysate for binding to RNase E.
A conserved PNPase recognition motif in α-proteobacterial RNase E
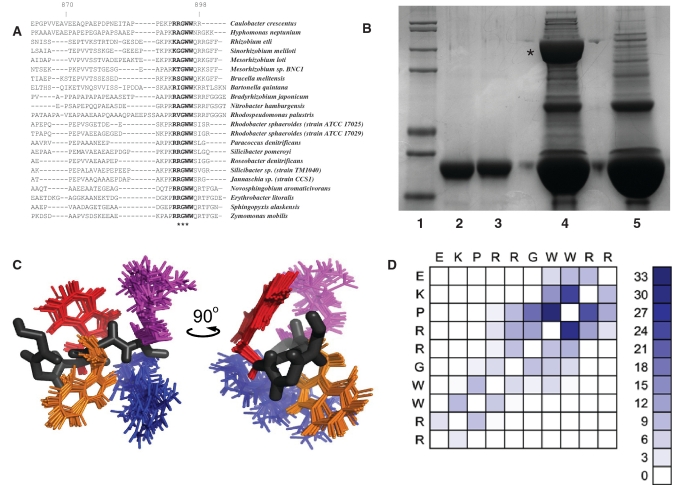
On comparing the C. crescentus RNase E protein sequence with α-proteobacterial homologues, it became apparent that the C-terminal scaffold domain shows great diversity even within the same bacterial class. However, there is a strikingly conserved sequence motif at the very C-terminus of the protein, with glycine–tryptophan–tryptophan being absolutely conserved (Figure 3A). An N-terminal GST fusion of the 14 C-terminal residues of RNase E containing the GWW motif (TAPPEKPRRGWWRR) was generated and used as bait in a pull-down experiment with C. crescentus cell lysate. The GST-fusion peptide interacted predominantly with a single protein of ∼80 kDa, and MALDI mass spectrometry analysis identified 17 unique peptides from this protein corresponding to C. crescentus PNPase (CC0034) (Figure 3B).
Figure 3.
A PNPase interaction motif in C. crescentus RNase E. (A) Sequence alignment of multiple α-proteobacterial RNase E sequences (C-terminal end only). The absolutely conserved GWW motif is highlighted with asterisks. (B) Pull-down experiment with GST-GWW peptide. The GWW bait sequence is from the C-terminus of C. crescentus RNase E (tappekprrGWWrr). Lane 1, molecular weight marker. Lane 2, GST–GWW input. Lane 3, GST only input. Lane 4, elution from glutathione sepharose following incubation with GST-GWW and C. crescentus cell lysate. Lane 5, elution from glutathione sepharose following incubation with GST and C. crescentus cell lysate. Co-purifying PNPase is marked with an asterisk. (C) Overlay of the 20 best NMR structural models of the 10-mer peptide (EKPRRGWWRR), showing the sidechains of residues WWRR only. A single peptide backbone trace is shown in black with WWRR coloured orange, red, magenta and blue, respectively. (D) NOE residue interaction matrix, highlighting inter-residue contacts clustering about the GWW motif.
The PNPase recognition motif has propensity to form a structured microdomain
Proton NMR spectra were collected from a synthetic peptide containing the GWW motif: EKPRRGWWRR. The backbone amide chemical shift dispersion observed between Lys-2 and Trp-8 (1.33 p.p.m.) is consistent with the peptide possessing a degree of residual structure. A total of 201 unambiguously assigned inter-proton distance restraints were harvested from a NOESY spectrum collected at the 278 K as input for structure determination. Lowest energy models were selected for refinement in explicit solvent, yielding an ensemble with no distance restraint violations >0.3 Å and acceptable covalent geometry statistics (Table 1). Consistent with the distribution of medium range NOEs, superposition of the backbone traces of the final ensemble indicates that the first four residues of the peptide are poorly defined. However, between residues Arg-5 and Arg-10 the structures overlay closely, yielding coordinate position root mean square deviations (RMSDs) of 0.44 Å for backbone atoms and 1.45 Å for all heavy atoms (Table 1). Over this region, the peptide adopts a conformation that positions the side-chains of Trp-7 and Trp-8 and of Lys-9 and Lys-10 on opposite sides of the backbone (Figure 3C). It remains to be seen whether this conformation is retained when the motif docks into its recognition site on the surface of PNPase, or if structural rearrangements occur.
Identification of the aconitase binding region in RNase E
The protein disorder prediction software PONDR has previously been used to identify natively unstructured regions within the E. coli RNase E protein (12). In this analysis, the C-terminal domain was predicted to be intrinsically disordered, with the exception of four short segments that have greater structural propensity (Figure 4A, left). These ‘micro-domain’ segments were all subsequently shown to mediate molecular interactions between RNase E and cognate partners (13–15,40,41). PONDR disorder analysis of the C. crescentus RNase E protein shows that the well-conserved N-terminal catalytic domain is predicted to be structurally ordered, with the exception of the S1 insert. The C-terminal domain of C. crescentus RNase E is predicted to be mostly intrinsically disordered and to contain only one structural micro-domain, compared with four in the E. coli homologue (Figure 4A, right). This single micro-domain comprises residues 675–725 (RNase E675–725), and shares little sequence identity with any of the known microdomains of E. coli RNase E, and as such may reflect a novel interaction mode between RNase E and a degradosome partner protein in C. crescentus. To explore the potential interaction with the micro-domain, an N-terminal GST fusion of RNase E675–725 was used as bait in a pull-down experiment after incubation with C. crescentus cell lysate. The fusion construct was able to interact with one predominant protein of ∼90 kDa in the pull down, and MALDI mass spectrometry analysis identified 12 unique peptides from the 90-kDa band corresponding to aconitase A (CC3667) (Figure 4B).
The catalytic domain of C. crescentus RNase E displays processing activity comparable to the E. coli homologue
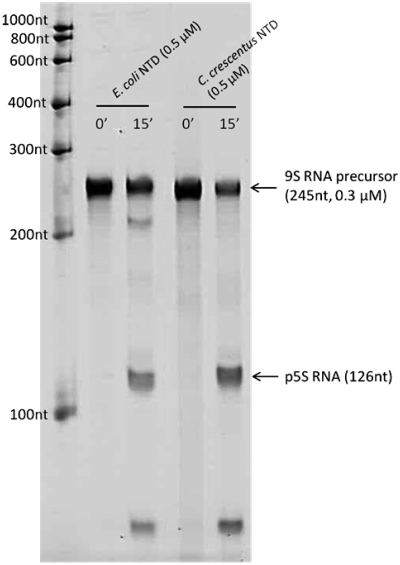
To test whether RNase E of C. crescentus displays ribonuclease activity similar to the E. coli homologue, a construct of the N-terminal catalytic domain of C. crescentus RNase E (NTD, residues 1–575) was generated, similar to a previously studied catalytic domain construct of E. coli RNase E (37,42). The activity of C. crescentus RNase E NTD was compared in vitro to that of E. coli RNase E NTD using an assay for the processing of E. coli 9S RNA, which is a precursor of 5S ribosomal RNA (43,44). Processing of 9S RNA by E. coli RNase E releases the p5S precursor (126 nt), containing three extra residues at each end flanking the sequence of the mature 5S RNA (120 nt). Following a 15-min reaction using the same enzyme:substrate ratio (1.0:0.6), p5S products were generated by both E. coli and C. crescentus NTD constructs (Figure 5). These data show that E. coli and C. crescentus RNase E catalytic domains have comparable activities and specificity for the 9S substrate and are likely to use similar mechanisms for recognition of complex, folded substrate and for catalysis.
Figure 5.
9S ribosomal RNA precursor processing by C. crescentus and E. coli RNase E catalytic domain. Digestion time points (0, 15 min) were analysed by denaturing polyacrylamide gel and detected with SYBR Gold stain. The p5S product, as seen to be released by both RNase E enzymes, is a precursor of the mature 120 nt 5S rRNA. The length of RNA species were determined by comparing to RiboRuler™ low-range RNA marker (Fermentas), in the leftmost lane.
Cell-cycle variation of RNase E
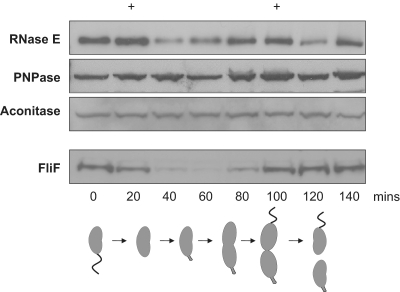
The abundance of degradosome components during the cell division cycle was monitored in a synchronous C. crescentus culture by western blotting. A pattern of variation in the abundance of RNase E was apparent, with two distinct maxima: first at the point of differentiation between swarmer and stalked cell forms (∼30 min), and also prior to cell division (∼100 min). Strikingly, following the differentiation between swarmer and stalked forms the apparent level of RNase E in the cell drops to approximately one-third of the maximum amount, before returning to near-peak levels at the point of cell division (Figure 6). The same synchronous cell cultures were probed with an antibody against the flagellar protein FliF, and the expected loss of signal for FliF following the differentiation from swarmer to stalked cells, before peaking again towards the end of the cell division cycle, confirmed that the cultures were successfully synchronized.
Figure 6.
Cell-cycle variation of RNase E. (Top) Representative western blots of RNase E, PNPase, aconitase and FliF protein abundance during the cell division cycle. The two maxima of RNase E abundance are marked at the top with +. (Bottom) Cartoon representation of the stages of C. crescentus cell division.
Antibodies raised against E. coli PNPase and E. coli aconitase both successfully cross-reacted with the homologous C. crescentus proteins. The abundance of PNPase and aconitase in C. crescentus was monitored during the cell division cycle and, despite minor fluctuations, both proteins showed a near-constant level of abundance, with both maintaining at least 65% of the maximal level. It is estimated that only a small fraction (5–10%) of the total cellular amount of PNPase is associated with the E. coli degradosome at any one time (45) and, as such, a constant total protein concentration would not directly reflect stoichiometric variations of this protein within the degradosome complex. Antibodies against E. coli DEAD box helicases (RhlB, RlhE, CsdA and SrmB) were used to probe western blots of C. crescentus cell lysate, but unfortunately none of these successfully cross-reacted with C. crescentus DEAD-box helicase CC1847.
DISCUSSION
Using an antibody raised against a short peptide from RNase E, corresponding to an exposed insert in the S1 RNA-binding domain, we have been able to co-immunopurify a degradosome-like complex in C. crescentus and to identify its principal protein components. As the antigenic peptide is predicted to be exposed and not to mediate protein–protein interactions that underpin the degradosome, the immunoprecipitations were anticipated to maintain the integrity of the assembly.
The C. crescentus degradosome, like its E. coli counterpart, includes RNase E, a DEAD-box RNA helicase (CC1847) and PNPase. The protein sequence of CC1847 shares greater sequence similarity with E. coli RhlE (39% identity) rather than with the canonical E. coli degradosome helicase, RhlB (32% identity). A second predicted DEAD-box protein encoded in the C. crescentus genome (CC0835) shows even greater sequence identity to RhlE (45% identity) than CC1847. The conservation of helicase–ribonuclease interactions emphasizes its functional importance (21). This is also supported by the recent finding that a DEAD-box helicase is a partner of the RNase Y endoribonuclease of Bacillus subtilis (20), which is not evolutionarily related to RNase E.
While the E. coli degradosome contains the glycolytic enzyme enolase, the C. crescentus degradosome carries aconitase from the tricarboxylic acid cycle. The presence of aconitase in the C. crescentus degradosome was an unexpected and intriguing discovery. Aconitase is an iron-dependent enzyme, and it has been noted that in the absence of iron, eukaryotic aconitase and the homologous iron-responsive element binding proteins (IRE-BP) can bind to IRE RNA elements as part of regulatory response (46). It remains to be tested whether C. crescentus aconitase plays an analogous metal-responsive role, but its physical linkage with the major RNA degradative machinery may promote the turnover of targeted transcripts in a metal-dependent fashion.
The C. crescentus RNA degradosome appears to differ from the degradosome of the closely related α-proteobacterium Rhodobacter capsulatus, which is comprised of RNase E, two DEAD-box RNA helicases and the Rho transcription termination factor (23). It is interesting to note that the R. capsulatus degradosome lacks enolase, and contains only sub-stoichiometric amounts of PNPase. On the other hand, the Bacillus subtilis degradosome-like assembly recruits both enolase and phosphofructokinase (16). The apparent compositional variation of degradosomes and degradosome-like assemblies in evolution perhaps reflects the capacity of the natively unstructured segment of the scaffolding ribonuclease for modular interactions that meet specialized regulatory requirements in different organisms (19).
The C-terminal domain of RNase E shows marked sequence diversity across bacterial classes, and we have also noted that the sequence varies greatly even amongst species of the same α-proteobacterial class. Surprisingly, amongst the diversity of the C-terminal domain, we found a strikingly conserved GWW motif at the very C-terminus of the protein. We have shown that this conserved sequence motif is involved with the interaction with PNPase in C. crescentus. Presumably, given the sequence conservation of the motif, this region will play a similar role in PNPase recognition in all members of the α-proteobacteria. Our NMR data indicate that this motif displays a persistent conformation in isolation, which might dock into an exposed binding site in PNPase. Docking of this motif into an exposed binding site in PNPase may either stabilize the secondary structure or induce a conformational adjustment in this region; this awaits experimental elucidation.
One of the proteins identified in our co-immunoprecipitation experiments with RNase E is the 27-kDa small ribosomal subunit protein S3 (CC1254). The presence of S3 was not unexpected, as several ribosomal proteins have previously been found as sub-stoichiometric components of the E. coli degradosome (47). More recently, five ribosomal proteins (L2, L3, L4, S3 and S4) were shown to co-purify with the E. coli degradosome, although it is thought that only L4 is able to interact directly with RNase E, and this interaction is proposed to regulate RNase E activity (48). Interestingly, in the co-immunopurification experiments described here, S3 was almost undetectable in the absence of EDTA in the binding buffer, whereas the relative level of the other degradosome components remained constant.
The crystal structure of the catalytic N-terminal domain of E. coli RNase E has been solved in the presence of short RNA substrates, and more recently in an RNA-free apo form (37,49). In all of these structures, RNase E forms a homo-tetramer, with a marked conformational switch between RNA-free and RNA-bound forms, which is predicted to be important for substrate recognition. The catalytic domain of RNase E is well conserved at the protein sequence level: the key determinants of the fold and active site are common between the E. coli and C. crescentus RNase E, so we expect the enzymes to have very similar structure, mechanism and behaviour. Indeed, the recombinant C. crescentus RNase E NTD has comparable activity and specificity for processing precursors of 5S ribosomal RNA. We also observe that, in common with its E. coli homologue, the recombinant C. crescentus NTD has 5′-end sensitivity for certain substrates (V. Chan et al., unpublished data).
By comparing the intensity of western blots on samples of C. crescentus where the cell number had been calculated by serial dilution and plating, against the intensities from western blots against known concentrations of recombinant RNase E, we estimate that each C. crescentus cell contains 1000–3000 molecules of RNase E protomers (data not shown). This would correspond to 250–750 RNase E tetramers, and likely a similar number of degradosome machines. Similar estimations have been made for the number of RNase E in E. coli (50).
Recent evidence indicates that transcription and translation are co-compartmentalized in bacteria (51). Intriguingly, RNase E is localized to active sites of transcription in C. crescentus. The functional reason for this is not yet clear, but RNase E may be involved in RNA degradation following translation. The limited number of RNase E tetramers may mean that there are few degradosomes at each transcription locus, or that the machines are highly processive.
Caulobacter crescentus has been developed as a model system to study complex processes such as progression through the cell cycle or morphological change. These phenomena require complex orchestration of many processes, and, in the case of C. crescentus, involve cell wall synthesis, pili and flagella assembly/disassembly, cell curvature, and stalk genesis (52). The coordination of such tasks is likely to require networks of communication between different processes. Our finding that RNase E abundance varies during the cell cycle and maximizes at the G1→ S transition is of particular interest with regard to the potential contributions of post-transcriptional regulation in programs of cell-cycle progression and morphogenesis. The coordinated activity of an exo-ribonuclease, RNase R, has previously been described in C. crescentus, where the regulatory RNA SsrA (tmRNA) is degraded following the G1–S transition in an RNase R-dependent manner (53) and the tmRNA may be compartmentalized (54). Further studies will clarify whether variation of the level of RNase E contributes in an active way to the unfolding of events required for cell-cycle progression and morphogenesis.
FUNDING
Funding for open access charge: The Wellcome Trust.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank AJ Carpousis, Zbyszek Pietras, Kasia Bandyra, Lucy Shapiro, Harley McAdams and Tom Taverner for helpful comments and suggestions. We thank AJ Carpousis for generously providing antibodies against E. coli enolase and polynucleotide phosphorylase. We thank Philip Aldridge and Urs Jenal for providing C. crescentus strains and FliF antibody, and for practical advice on synchronisation experiments. We also thank Jeff Green for providing the anti-aconitase antibody, Len Packman for mass spectrometry analyses and Mark Carrington for generous use of the fluorescence microscope.
REFERENCES
- 1.Grunberg-Manago M. Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annu. Rev. Genet. 1999;33:193–227. doi: 10.1146/annurev.genet.33.1.193. [DOI] [PubMed] [Google Scholar]
- 2.Wilusz CJ, Wilusz J. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet. 2004;20:491–497. doi: 10.1016/j.tig.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 3.Carpousis AJ. The RNA degradosome of Escherichia coli: an mRNA degrading machine assembled on RNase E. Annu. Rev. Microbiol. 2007;61:71–87. doi: 10.1146/annurev.micro.61.080706.093440. [DOI] [PubMed] [Google Scholar]
- 4.Carpousis AJ, Luisi BF, McDowall KJ. Endonucleolytic initiation of mRNA decay in Escherichia coli. Prog. Mol. Biol. Transl. Sci. 2009;85:91–135. doi: 10.1016/S0079-6603(08)00803-9. [DOI] [PubMed] [Google Scholar]
- 5.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfeiffer V, Papenfort K, Lucchini S, Hinton JCD, Vogel J. Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat. Struct. Mol. Biol. 2009;16:840–846. doi: 10.1038/nsmb.1631. [DOI] [PubMed] [Google Scholar]
- 7.Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bollenbach TJ, Schuster G, Stern DB. Cooperation of endo- and exoribonucleases in chloroplast mRNA turnover. Prog. Nucleic Acid Res. Mol. Biol. 2004;78:305–337. doi: 10.1016/S0079-6603(04)78008-3. [DOI] [PubMed] [Google Scholar]
- 9.Mudd EA, Sullivan S, Gisby MF, Mironov A, Kwon CS, Chung WI, Day A. A 125 kDa RNase E/G-like protein is present in plastids and is essential for chloroplast development and autotrophic growth in Arabidopsis. J. Exp. Bot. 2008;59:2597–2610. doi: 10.1093/jxb/ern126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jain C, Deana A, Belasco JG. Consequences of RNase E scarcity in Escherichia coli. Mol. Microbiol. 2002;43:1053–1064. doi: 10.1046/j.1365-2958.2002.02808.x. [DOI] [PubMed] [Google Scholar]
- 11.Takada A, Nagai K, Wachi M. A decreased level of FtsZ is responsible for inviability of RNase E-deficient cells. Genes Cells. 2005;10:733–741. doi: 10.1111/j.1365-2443.2005.00872.x. [DOI] [PubMed] [Google Scholar]
- 12.Callaghan AJ, Aurriko JP, Grossmann JG, Kühnel K, Poljak L, Ilag P, Robinson C, Chandran V, Carpousis AJ, Symmons MF, et al. Studies of the RNA degradosome-organising domain of the Escherichia coli RNase E. J. Mol. Biol. 2004;340:965–979. doi: 10.1016/j.jmb.2004.05.046. [DOI] [PubMed] [Google Scholar]
- 13.Khemici V, Poljak L, Luisi BF, Carpousis AJ. The RNase E of Escherichia coli is a membrane-binding protein. Mol. Microbiol. 2008;70:799–813. doi: 10.1111/j.1365-2958.2008.06454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandran V, Luisi BF. Recognition of enolase in the Escherichia coli RNA degradosome. J. Mol. Biol. 2006;358:8–15. doi: 10.1016/j.jmb.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 15.Nurmohamed S, McKay D, Robinson CV, Luisi BF. Molecular recognition between Escherichia coli enolase and ribonulcease E. Acta Cryst., D. 2010;66:1036–1040. doi: 10.1107/S0907444910030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Commichau FM, Rother FM, Herzberg C, Wagner E, Hellwig D, Lehnik-Habrink M, Hammer E, Volker U, Stulke J. Novel activities of glycolytic enzymes in Bacillus subtilis: interactions with essential proteins involved in mRNA processing. Mol. Cell. Proteomics. 2009;8:1350–1360. doi: 10.1074/mcp.M800546-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Butland G, Peregrin-Alvarez JM, Li J, Yang W, Yang X, Canadien V, Starostine A, Richards D, Beattie B, Krogan N, et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 18.Hu P, Janga1 SC, Babu M, Javier Díaz-Mejía1 J, Butland G, Yang W, Pogoutse O, Guo X, Phanse S, Wong P, et al. Global functional atlas of Escherichia coli encompassing previously uncharacterised proteins. PLoS Biol. 2009;7:e1000096. doi: 10.1371/journal.pbio.1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marcaida MJ, DePristo MA, Chandran V, Carpousis AJ, Luisi BF. The RNA degradosome: life in the fast lane of adaptive molecular evolution. Trends Biochem. Sci. 2006;31:359–365. doi: 10.1016/j.tibs.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 20.Lehnik-Habrink M, Pförtner H, Rempeters L, Pietack N, Herzberg C, Stülke J. The RNA degradosome in Bacillus subtilis: identification of CshA as the major RNA helicase in the multiprotein complex. Mol. Microbiol. 2010 doi: 10.1111/j.1365-2958.2010.07264.x. [DOI] [PubMed] [Google Scholar]
- 21.Ait-Bara S, Carpousis AJ. Characterization of the RNA degradosome of Pseudoalteromonas haloplanktis: conservation of the RNase E-RhlB interaction in the γ-proteobacteria. J. Bacteriol. 2010;192:5413–5423. doi: 10.1128/JB.00592-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evguenieva-Hackenberg E, Klug G. RNA degradation in archaea and Gram-negative bacteria different from Escherichia coli. Prog. Mol. Biol. Transl. Sci. 2009;85:275–317. doi: 10.1016/S0079-6603(08)00807-6. [DOI] [PubMed] [Google Scholar]
- 23.Jäger S, Fuhrmann O, Heck C, Hebermehl M, Schiltz E, Rauhut R, Klug G. An mRNA degrading complex in Rhodobacter capsulatus. Nucleic Acids Res. 2001;29:4581–4288. doi: 10.1093/nar/29.22.4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erce MA, Low JK, March PE, Wilkins MR, Takayama KM. Identification and functional analysis of RNase E of Vibrio angustum S14 and two-hybrid analysis of its interaction partners. Biochim. Biophys. Acta. 2009;1794:1107–1114. doi: 10.1016/j.bbapap.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 25.Shen X, Collier J, Dill D, Shapiro L, Horowitz M, McAdams HH. Architecture and inherent robustness of a bacterial cell-cycle control system. Proc. Natl Acad. Sci. USA. 2008;105:11340–11345. doi: 10.1073/pnas.0805258105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evinger M, Agabian N. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 1977;132:294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poindexter JS. Biological properties and classification of the Caulobacter group. Bacteriol. Rev. 1964;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson RC, Ely B. Isolation of spontaneously derived mutants of Caulobacter crescentus. Genetics. 1977;86:25–32. doi: 10.1093/genetics/86.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palmer AG, Cavanagh J, Rance M, Skelton NJ, Fairbrother WJ. Protein NMR Spectroscopy: Principles and Practice. 2nd edn. London: Academic Press; 2006. [Google Scholar]
- 30.Wűthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley Interscience; 1986. [Google Scholar]
- 31.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 32.Linge JP, Williams MA, Spronk CAEM, Bonvin AMJJ, Nilges M. Refinement of protein structures in explicit solvent. Proteins. 2003;50:496–506. doi: 10.1002/prot.10299. [DOI] [PubMed] [Google Scholar]
- 33.DeLano W. Schrödinger: LLC; 2010. The PyMOL Molecular Graphics System, Version 1.1. [Google Scholar]
- 34.Schein A, Sheffy-Levin S, Glaser F, Schuster G. The RNase E/G-type endoribonuclease of higher plants is located in the chloroplast and cleaves RNA similarly to the E. coli enzyme. RNA. 2008;14:1057–1068. doi: 10.1261/rna.907608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng K, Radivojac P, Vucetic S, Dunker AK, Obradovic Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinformatics. 2006;7:208. doi: 10.1186/1471-2105-7-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xue B, Dunbrack RL, Williams RW, Dunker AK, Uversky VN. PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta – Proteins Proteomics. 2010;1804:996–1010. doi: 10.1016/j.bbapap.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Callaghan AJ, Marcaida MJ, Stead JA, McDowall KJ, Scott WG, Luisi BF. Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nature. 2005;437:1187–1191. doi: 10.1038/nature04084. [DOI] [PubMed] [Google Scholar]
- 38.Worrall JAR, Gorna M, Crump N, Phillips L, Tuck A, Price A, Bavro V, Luisi BF. Reconstitution and analysis of the multienzyme Escherichia coli RNA degradosome. J. Mol. Biol. 2008;382:870–883. doi: 10.1016/j.jmb.2008.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khemici V, Toesca I, Poljak L, Vanzo NF, Carpousis AJ. The RNase E of Escherichia coli has at least two binding sites for DEAD-box RNA helicases: functional replacement of RhlB by RhlE. Mol. Microbiol. 2004;54:1422–1430. doi: 10.1111/j.1365-2958.2004.04361.x. [DOI] [PubMed] [Google Scholar]
- 40.Nurmohamed S, Vaidialingam B, Callaghan AJ, Luisi BF. Crystal structure of Escherichia coli polynucleotide phosphorylase core bound to RNase E, RNA and manganese: implications for catalytic mechanism and RNA degradosome assembly. J. Mol. Biol. 2009;389:18–33. doi: 10.1016/j.jmb.2009.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chandran V, Poljak L, Vanzo N, Leroy A, Núñez Miguel R, Fernandez-Recio J, Parkinson J, Burns C, Carpousis AJ, Luisi BF. Recognition and cooperation between the ATP-dependent RNA helicase RhlB and ribonuclease RNase E. J. Mol. Biol. 2007;367:113–132. doi: 10.1016/j.jmb.2006.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Callaghan AJ, Grossmann JG, Redko YU, Ilag LL, Moncrieffe MC, Symmons MF, Robinson CV, McDowall KJ, Luisi BF. Quaternary structure and catalytic activity of the Escherichia coli ribonuclease E amino-terminal catalytic domain. Biochemistry. 2003;42:13848–13855. doi: 10.1021/bi0351099. [DOI] [PubMed] [Google Scholar]
- 43.Carpousis AJ, Leroy A, Vanzo N, Khemici V. Escherichia coli RNA degradosome. Methods Enzymol. 2001;342:333–345. doi: 10.1016/s0076-6879(01)42556-0. [DOI] [PubMed] [Google Scholar]
- 44.Cormack RS, Mackie GA. Structural requirements for the processing of Escherichia coli 5S ribosomal RNA by RNase E in vitro. J. Mol. Biol. 1992;228:1078–1090. doi: 10.1016/0022-2836(92)90316-c. [DOI] [PubMed] [Google Scholar]
- 45.Liou GG, Jane WN, Cohen SN, Lin NS, Lin-Chao S. RNA degradosomes exist in vivo in Escherichia coli as multicomponent complexes associated with the cytoplasmic membrane via the N-terminal region of RNase E. Proc. Natl Acad. Sci. USA. 2001;98:63–68. doi: 10.1073/pnas.011535498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Constable A, Quick S, Gray NK, Hentze MW. Modulation of the RNA-binding activity of a regulatory protein by iron in vitro: switching between enzymatic and genetic function? Proc. Natl Acad. Sci. USA. 1992;89:4554–4558. doi: 10.1073/pnas.89.10.4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Besarab DA, Kaberdin VR, Wei CL, Liou GG, Lin-Chao S. RNA components of Escherichia coli degradosome: evidence for rRNA decay. Proc. Natl Acad. Sci. USA. 1998;95:3157–3161. doi: 10.1073/pnas.95.6.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh D, Chang SJ, Lin PH, Averina OV, Kaberdin VR, Lin-Chao S. Regulation of ribonuclease E activity by the L4 ribosomal protein of Escherichia coli. Proc. Natl Acad. Sci. USA. 2009;106:864–869. doi: 10.1073/pnas.0810205106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koslover DJ, Callaghan AJ, Marcaida MJ, Garman EF, Martick M, Scott WG, Luisi BF. The crystal structure of Escherichia coli RNase E apoprotein and a mechanism for RNA degradation. Structure. 2008;16:1238–1244. doi: 10.1016/j.str.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kido M, Yamanaka K, Mitani T, Niki H, Ogura T, Hiraga S. RNase E polypeptides lacking a carboxyl-terminal half suppress a mukB mutation in Escherichia coli. J. Bacteriol. 2006;178:3917–3925. doi: 10.1128/jb.178.13.3917-3925.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Llopis PM, Jackson AF, Sliusarenko O, Surovtsev I, Heinritz J, Emonet T, Jacobs-Wagner C. Spatial organization of the flow of genetic information in bacteria. Nature. 2010;466:77–81. doi: 10.1038/nature09152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ebersbach G, Jacobs-Wagner C. Exploration into the spatial and temporal mechanisms of bacterial polarity. Trends Microbiol. 2007;15:101–108. doi: 10.1016/j.tim.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 53.Hong SJ, Tran QA, Keiler KC. Cell cycle-regulated degradation of tmRNA is controlled by RNase E and SmpB. Mol. Microbiol. 2005;57:565–575. doi: 10.1111/j.1365-2958.2005.04709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Russell JH, Keiler KC. Subcellular localization of a bacterial regulatory RNA. Proc. Natl Acad. Sci. USA. 2009;106:16405–16409. doi: 10.1073/pnas.0904904106. [DOI] [PMC free article] [PubMed] [Google Scholar]