Abstract
The HMG-domain transcription factor Sox10 is expressed throughout oligodendrocyte development and is an important component of the transcriptional regulatory network in these myelin-forming CNS glia. Of the known Sox10 regulatory regions, only the evolutionary conserved U2 enhancer in the distal 5′-flank of the Sox10 gene exhibits oligodendroglial activity. We found that U2 was active in oligodendrocyte precursors, but not in mature oligodendrocytes. U2 activity also did not mediate the initial Sox10 induction after specification arguing that Sox10 expression during oligodendroglial development depends on the activity of multiple regulatory regions. The oligodendroglial bHLH transcription factor Olig2, but not the closely related Olig1 efficiently activated the U2 enhancer. Olig2 bound U2 directly at several sites including a highly conserved one in the U2 core. Inactivation of this site abolished the oligodendroglial activity of U2 in vivo. In contrast to Olig2, the homeodomain transcription factor Nkx6.2 repressed U2 activity. Repression may involve recruitment of Nkx6.2 to U2 and inactivation of Olig2 and other activators by protein–protein interactions. Considering the selective expression of Nkx6.2 at the time of specification and in differentiated oligodendrocytes, Nkx6.2 may be involved in limiting U2 activity to the precursor stage during oligodendrocyte development.
INTRODUCTION
Development of a differentiated cell from a pluripotent precursor is regulated by a complex network of transcription factors that interact in many ways and influence each other’s expression and activity. In the central nervous system (CNS), many different neuronal and glial cell types have to be generated from a common pool of precursor cells. Oligodendrocytes represent one of these glial cell types. In the white matter, they form myelin sheaths around axons and thereby allow rapid saltatory conduction along the axon.
In recent years, much has been learnt about the origin of oligodendrocytes and the stages of their development during vertebrate and particularly during mammalian embryogenesis (1). In the mouse spinal cord, oligodendrocyte precursors (OLPs) are predominantly derived from neuroepithelial cells in a well-defined region of the ventral part of the ventricular zone (2). From this pMN domain, specified OLPs colonize the spinal cord parenchyma, accumulate preferentially at the end of embryogenesis in the marginal zone as the future white matter where they start to undergo terminal differentiation and express myelin genes before eventually providing myelin sheaths to axons from several neurons.
Several transcription factors have been identified that are expressed in developing and mature oligodendrocytes (3,4). These include bHLH, homeodomain, zinc finger and high-mobility group (HMG) domain proteins. They are required for various aspects of oligodendrocyte development including specification, identity, survival, lineage progression and terminal differentiation. The bHLH protein Olig2 and the HMG domain transcription factor Sox10 are particularly important. Olig2 is already expressed in neuroepithelial cells before specification and continues to be present throughout oligodendroglial development. OLPs are absent in the spinal cord of Olig2-deficient mice, pointing to the essential role of Olig2 during the early phases of oligodendroglial development (5,6). Deletion of the closely related and co-expressed Olig1, in contrast, rather causes a terminal differentiation defect arguing that Olig1 and Olig2 function predominantly during different phases of oligodendroglial development (7,8). One of the genes that is induced immediately after oligodendroglial specification is Sox10 (5,6). Once induced, Sox10 remains present in oligodendroglia (9,10) where it first ensures OLP survival together with its close relative Sox9 (11) and later drives terminal differentiation as a direct activator of myelin gene expression (10).
Although much has been learnt about the function of single transcription factors, only few data exist on the interplay of these factors and their cross-regulations within the oligodendroglial transcriptional network. Recently, we have identified an enhancer ∼36 kb upstream of the mouse Sox10 gene that is capable of driving transgene expression in oligodendroglia (12). This enhancer was also active in several other glial cell types of the peripheral nervous system (PNS), including Schwann cells and satellite glia, but not in enteric glia. Here, we focused on the oligodendroglial activity of this enhancer to determine how Sox10 expression is regulated in the oligodendrocyte lineage and how it is influenced by other components of the oligodendroglial transcriptional network.
MATERIALS AND METHODS
Transgenic animals
Sox10lacZ/+ mice in which Sox10 coding sequences were replaced on one allele by lacZ marker sequences, and mice carrying the lacZ transgene under control of the hsp68 minimal promoter and the mouse U2 enhancer (U2-lacZ) have been described before (12,13).
U2mut-lacZ resembled the previously described U2-lacZ transgene except for the presence of an inactivating mutation in the Olig2 binding site bHLH4 (Figure 4A). After separation from the vector backbone and purification, the U2mut-lacZ transgene was microinjected as a SpeI/KpnI fragment into the male pronucleus of fertilized FVB oocytes. Transgenic mice were generated from injected oocytes according to standard techniques. Founder mice and transgenic offspring were identified and genotyped by PCR analysis of DNA prepared from tail biopsies using the U2-specific forward primer 5′-GGCACAGAAAGGTCTCTTTG-3′ and the lacZ-specific reverse primer 5′-AGTAGCTGTCAGCGTCTGGT-3′ as described previously (12).
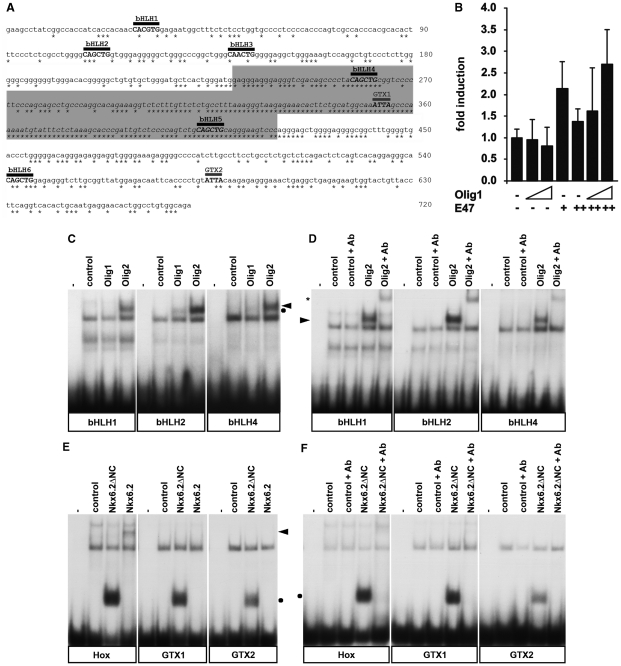
Figure 4.
The Sox10 U2 enhancer contains binding sites for Olig2 and Nkx6.2. (A) Sequence of mouse U2. Asterisks below the sequence indicate positions that are fully conserved among mouse, human and chicken. The 190 bp core region (gray box) is particularly enriched for conserved positions. Putative binding sites are marked by a bar above the sequence. Potential Olig2 binding sites are designated as bHLH1–bHLH6, potential Nkx6.2 binding sites as GTX1 and GTX2. Numbers on the right indicate the exact nucleotide position. (B) Transient transfections were performed in 33B oligodendroglioma with the U2 luc reporter and pCMV-based expression plasmids for Olig1 and E47 as indicated. Increasing amounts of expression plasmid were used as indicated by the triangles (Olig1) and + versus ++ (E47). Luciferase activities in extracts from transfected cells were determined in three experiments each performed in duplicates. The luciferase activity obtained for U2-luc in the absence of ectopic transcription factor was arbitrarily set to 1. Fold inductions in the presence of transcription factors were calculated and are presented as mean ± SD. (C and D) EMSA was performed with radiolabeled double-stranded oligonucleotides encompassing the potential Olig2 binding sites bHLH1, bHLH2 and bHLH4 (for sequence, see Figure 5A). Oligonucleotides were incubated in the absence (−) or presence of extracts from 293 cells that were either untransfected (control) or transfected with expression plasmids for myc-tagged versions of Olig1 or Olig2. Antibodies against the tag (+Ab) were added as indicated. The Olig2-containing complex is marked by an arrowhead, the Olig1-containing complex by a dot and the supershifted complexes by asterisks. All other complexes are unspecific as they were also obtained with the control extracts. (E and F) EMSA was carried out with radiolabeled double-stranded oligonucleotides encompassing GTX1, GTX2 (for sequence, see Figure 5A) and the known high-affinity Nkx6.2 binding sequence HoxA5/A6 (Hox). Oligonucleotides were incubated in the absence (−) or presence of extracts from 293 cells that were either untransfected (control) or transfected with expression plasmids for T7-tagged versions of full-length Nkx6.2 (fl) or amino- and carboxyterminally truncated Nkx6.2 (ΔNC). Antibodies against the tag (+Ab) were added as indicated. The complex containing full-length Nkx6.2 is marked by an arrowhead, the complex containing the ΔNC version by a dot.
Tissue preparation, histological staining, immunohistochemistry and documentation
Embryos from 11.5 days post-coitum (dpc) to 16.5 dpc, newborn (P1) to 14-day-old (P14) pups and adult mice were sacrificed and underwent fixation in 1 or 4% paraformaldehyde depending on their further use. After fixation, whole embryos and tissues were immediately stained for β-galactosidase activity or were cryoprotected in sucrose and frozen at −80°C in tissue freezing medium (Leica, Bensheim, Germany) in preparation for cryotome sectioning.
Detection of β-galactosidase activity was performed after fixation in 1% paraformaldehyde on whole mount embryos (11.5 dpc) or 20 µm transverse cryosections by incubation in 1% X-gal for several hours at 37°C.
For immunohistochemistry, 10 µm cryotome sections from the forelimb level of genotyped, age-matched mouse embryos and pups were incubated with the following primary antibodies: anti-β-galactosidase goat antiserum (1:500 dilution, Biotrend), anti-Olig2 rabbit antiserum (1:1000 dilution, Chemicon), anti-Olig1 rabbit antiserum (1:5000 dilution, gift of C. Stiles, Dana-Farber Cancer Institute, Boston, MA, USA) anti-PDGFR-α rabbit antiserum (1:80 dilution, NeoMarkers), anti-MBP rabbit antiserum (1:200 dilution, NeoMarkers), anti-Sox10 guinea pig antiserum [1:1000 dilution (14)], anti-Nkx6.2 guinea pig antiserum (1:1000 dilution, gift of J. Ericson, Karolinska Institutet, Stockholm, Sweden), anti-Nkx2.2 mouse monoclonal (1:400 dilution, Developmental Studies Hybridoma Bank, Iowa City, IA, USA) and anti-NeuN mouse monoclonal (1:600 dilution, Chemicon). Secondary antibodies conjugated to Cy2 and Cy3 immunofluorescent dyes (Dianova) were used for detection. In case of Nkx2.2 antibodies, signal intensity was amplified using the TSA™-Plus Cy3 System (Perkin Elmer, Boston, MA, USA).
Samples were analyzed and documented either with a Leica inverted microscope (DMIRB) equipped with a cooled MicroMax CCD camera (Princeton Instruments, Trenton, NJ, USA) or with a Leica MZFLIII stereomicroscope equipped with an Axiocam (Zeiss, Oberkochem, Germany).
Plasmids
All expression plasmids were derived from pCMV5 and thus carried coding sequences under control of the cytomegalovirus immediate early promoter. The eukaryotic pCMV5 expression plasmid for myc-tagged Olig2 has been described previously (15). Using a similar strategy, coding sequences for full-length Olig1 and different Olig2 fragments (amino acids 2–180, amino acids 181–323) were cloned as BamHI/BglII fragments behind a myc-tag into the BglII site of pCMV5-myc. Untagged versions of Olig1 (accession number NM_021770.3) and Olig2 (accession number NM_016967.2) (16) were inserted into pCMV5 as KpnI/EcoRI and SmaI fragments, respectively. The full-length Nkx6.2 open reading frame (accession number NM_183248.2) was amplified by PCR and placed into pCMV5 as T7-tagged and untagged version using BamHI and HindIII sites. Nkx6.2 fragments coding for amino acids 1–133, amino acids 134–216 and amino acids 134–277 were inserted as T7-tagged versions only. The E47 coding sequence (accession number BC018260.1, gift of Jonas Muhr, Karolinska Institutet, Stockholm, Sweden) was placed between BglII and HindIII sites of pCMV5. Coding sequences for Nkx2.2 and Nkx6.1 were transferred into pCMV5 from corresponding pcDNA3 and pCAGG expression plasmids (gifts of L. Sussel, Columbia University, New York, NY, USA and J. Ericson, Karolinska Institutet, Stockholm, Sweden). For reporter gene assays the U2-luc plasmid was used. It contained the luciferase reporter gene under control of the 0.67 kb U2 region (12) upstream of the β-globin minimal promoter. Transcription factor binding sites within U2 were mutated using the QuickChange XL Site-directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA).
Cell culture, transient transfection, extract preparation, EMSA and co-immunoprecipitation
HEK293 cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% fetal calf serum (FCS) and transfected by the polyethylenimine (PEI) technique using 10 µg pCMV5-based expression plasmid per 100-mm plate. Cells were harvested 48 h post-transfection for extract preparation (17) and ectopic expression of the respective transcription factor was verified by western blotting using anti-myc tag (1:10 000 dilution, Cell Signaling Technology) or anti-T7 tag (1:10 000 dilution, Novagen) mouse monoclonals. With these extracts, electrophoretic mobility shift analyses (EMSA) were performed as described (18) using 32P-labeled 25-bp double-stranded oligonucleotides containing putative Olig2 or Nkx6.2 binding sites (for sequences, see Figure 5A). In supershift experiments, antibodies were used in a 1:40 dilution and pre-incubated for 30 min with the protein extracts before addition of 32P-labeled oligonucleotide probes.
Figure 5.
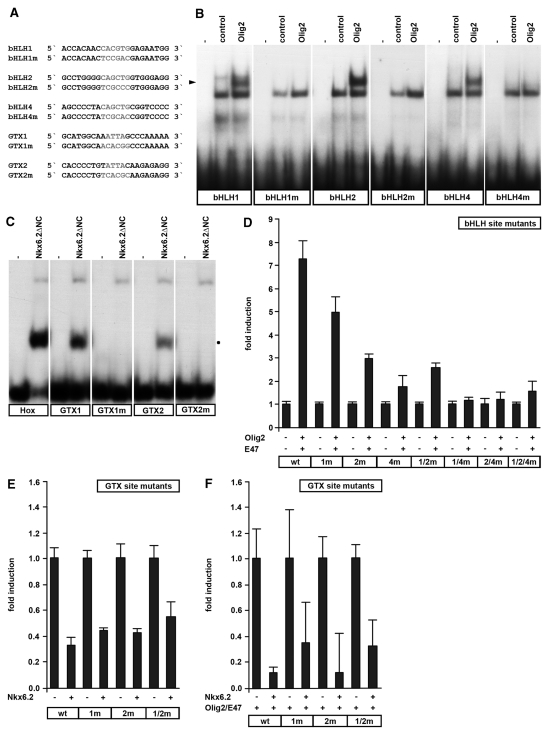
Activation of the U2 enhancer requires binding sites for Olig2, whereas repression by Nkx6.2 is independent of binding sites. (A) Sequence of oligonucleotides with wild-type and mutant binding sites for Olig2 and Nkx6.2. (B and C) EMSA was performed with wild-type and mutant binding sites for Olig2 (B) and Nkx6.2 (C). Oligonucleotides were incubated in the absence (−) or presence of extracts from 293 cells which were either untransfected (control) or transfected with expression plasmids for tagged versions of Olig2 or Nkx6.2 ΔNC. The position of the Olig2-specific complex is highlighted by an arrowhead, the position of the Nkx6.2-specific complex by a dot. (D–F) Transient transfections were performed in 33B oligodendroglioma with different versions of the U2 luc reporter and expression plasmids for Olig2, E47 (D), Nkx6.2 (E) or Olig2, E47 and Nkx6.2 (F) as indicated (+). The U2 luc reporter had either wild-type sequences (wt) or carried one or multiple mutations in the Olig2 binding sites (bHLH1m, bHLH2m, bHLH4m, bHLH1/2m, bHLH1/4m, bHLH2/4m, bHLH1/2/4m) (D) or the Nkx6.2 (GTX1m, GTX2m, GTX1/2m) binding sites (E and F). Luciferase activities in extracts from transfected cells were determined in three experiments each performed in duplicates. The luciferase activity obtained for each reporter in the absence of ectopic transcription factor was arbitrarily set to 1. Fold inductions in the presence of transcription factors were calculated and are presented as mean ± SD. Statistically significant (P ≤ 0.001) reporter gene activation by Olig2 and E47 (D) was obtained for the wt, 1m, 2m and 1/2m U2 luc reporters as determined by Student’s t-test. Nkx6.2-dependent repression was statistically significant (P ≤ 0.001) for all reporters in E and F.
Coimmunoprecipitations were carried out with HEK293 protein extracts diluted 1:5 in TEN buffer (100 µM EDTA, 100 mM NaCl, 0.5% NP-40, 20 mM Tris, pH 7.4, 10 µg/ml aprotinin, 10 µg/ml leupeptin, 2 mM dithiothreitol). Tagged proteins were precipitated overnight at 4°C using monoclonal antibodies directed against the tag and protein G sepharose beads. After separation from the supernatant and three consecutive washing steps with TEN buffer, precipitated proteins were eluted from protein G sepharose beads and detected by western blot analysis.
Luciferase assay
Rat 33B oligodendroglioma cells were maintained in DMEM containing 10% FCS and transfected with SuperFect® Transfection Reagent (Qiagen, Hilden, Germany) on 24-well tissue culture plates using varying amounts (25–500 ng, usually 100 ng) of pCMV5-based expression vectors and 500 ng of luciferase reporter plasmid. Cells were harvested 48 h post-transfection. Luciferase activity was determined in the presence of luciferin substrate by detection of chemiluminescence.
Chromatin immunoprecipitation
Chromatin immunoprecipitation assays were performed as described (19) on 33B cells transfected with pCMV5-myc-Olig2 or pCMV-T7-Nkx6.2, and on dissected, trypsinized spinal cord of 18.5 dpc-old mouse embryos. Proteins were cross-linked to DNA in 1% formaldehyde. After addition of glycine, chromatin was prepared and sheared to an avarage fragment length of 300–600 bp using a Sonoplus HD2070 homogenisator (Bandelin, Berlin, Germany). Immunoprecipitations were performed overnight at 4°C using monoclonal α-myc-tag or α-T7-tag IgG (for 33B cells) or polyclonal α-Olig2 antibodies (for spinal cords) as well as control preimmune serum and protein G sepharose beads. DNA was purified from input and precipitated chromatin after cross-link reversal and subjected to PCR. The following primer pairs were used for detection in 33 cycles of standard PCR using an annealing temperature of 57°C: 5′-CGACAGCCCGTACAGCTGC-3′ and 5′-GTGACTGAGAGTCTGAGAGC-3′ for rat U2, 5′-CGACAGCCCCTACAGCTGC-3′ and 5′-GTGACTGAGAGTCTGAGAGC-3′ for mouse U2, 5′-CTAGGCAAGCACTCTGTCAA-3′ and 5′-GCCTCAGCATGGTTCTAGTT-3′ for the rat genomic control region and 5′-GCAGACAGCAGTTGGAGACC-3′ and 5′-CCTGTGCCCAGACTCTTGTT-3′ for mouse U5 (12).
RESULTS
Expression of the Sox10 U2 enhancer in the developing spinal cord occurs predominantly in OLPs
Among the previously identified Sox10 enhancers, U2 was the only one active in oligodendroglial cells (12). In the original study, experiments were performed on spinal cords of 16.5 dpc-old embryos. At this stage, oligodendrocyte specification is already over and most oligodendroglial cells are in the OLP stage. It is thus not clear from the published data whether U2 fully recapitulates the expression pattern of Sox10 which is turned on immediately after the specification event in OLPs and remains expressed in mature oligodendrocytes (10).
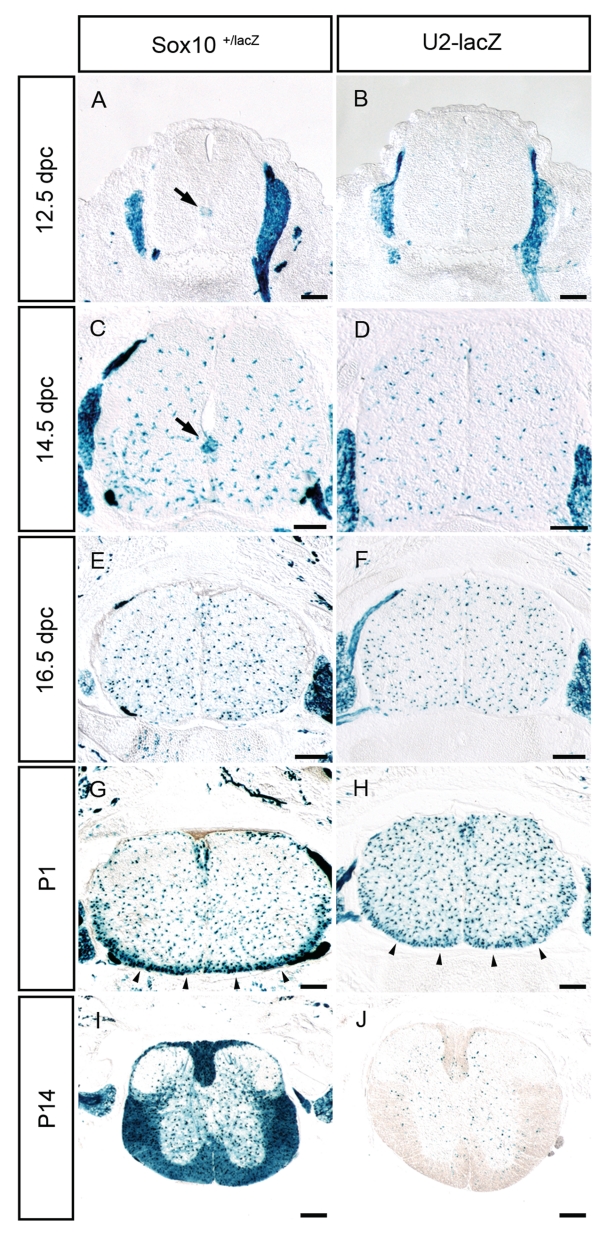
We, therefore, compared the U2-lacZ expression pattern in the spinal cord at various days of embryonic and post-natal development with lacZ expression from the Sox10lacZ allele that faithfully mimicks Sox10 expression (13). Whereas OLPs were already visible at 12.5 dpc by X-gal staining near the pMN domain in spinal cords of Sox10+/lacZ embryos (Figure 1A, arrow), age-matched spinal cords of U2-lacZ transgenic embryos exhibited no X-gal staining within the spinal cord (Figure 1B). In contrast, X-gal staining was comparable in both genotypes outside the spinal cord in dorsal root ganglia and along peripheral nerves (Figure 1A and B). At 12.75 dpc X-gal staining started to become detectable in U2-lacZ transgenic embryos but only in Sox10-positive cells that had migrated some distance from the ventricular zone (Figure 2A and A′). At 14.5 dpc, a high number of lacZ-positive cells was detectable in U2-lacZ and Sox10+/lacZ embryos (Figure 1C and D). In both genotypes, OLPs were stained throughout the spinal cord parenchyma. In Sox10+/lacZ embryos, the origin of OLPs was still marked by a particular high density of X-gal stained cells near the pMN domain (Figure 1C, arrow). This staining was again missing in the U2-lacZ embryos (Figure 1D). Compared to lacZ expression in Sox10+/lacZ embryos, the U2-lacZ transgene is thus turned on in the oligodendrocyte lineage with delay.
Figure 1.
The U2-lacZ transgene is expressed in embryonic and early post-natal spinal cord. LacZ expression was detected colorimetrically using X-gal substrate on transverse sections from the forelimb level of Sox10lacZ/+ (A, C, E, G and I) and age-matched U2-lacZ (B, D, F, H and J) mice at 12.5 dpc (A and B), 14.5 dpc (C and D), 16.5 dpc (E and F), P1 (G and H) and P14 (I and J). The pMN domain as the main origin for spinal cord OLPs is highlighted in A and C by an arrow. Arrowheads in G and H point to the marginal zone. No lacZ staining was detected in wild-type littermates under identical conditions. Size bars correspond to 200 µm.
Figure 2.
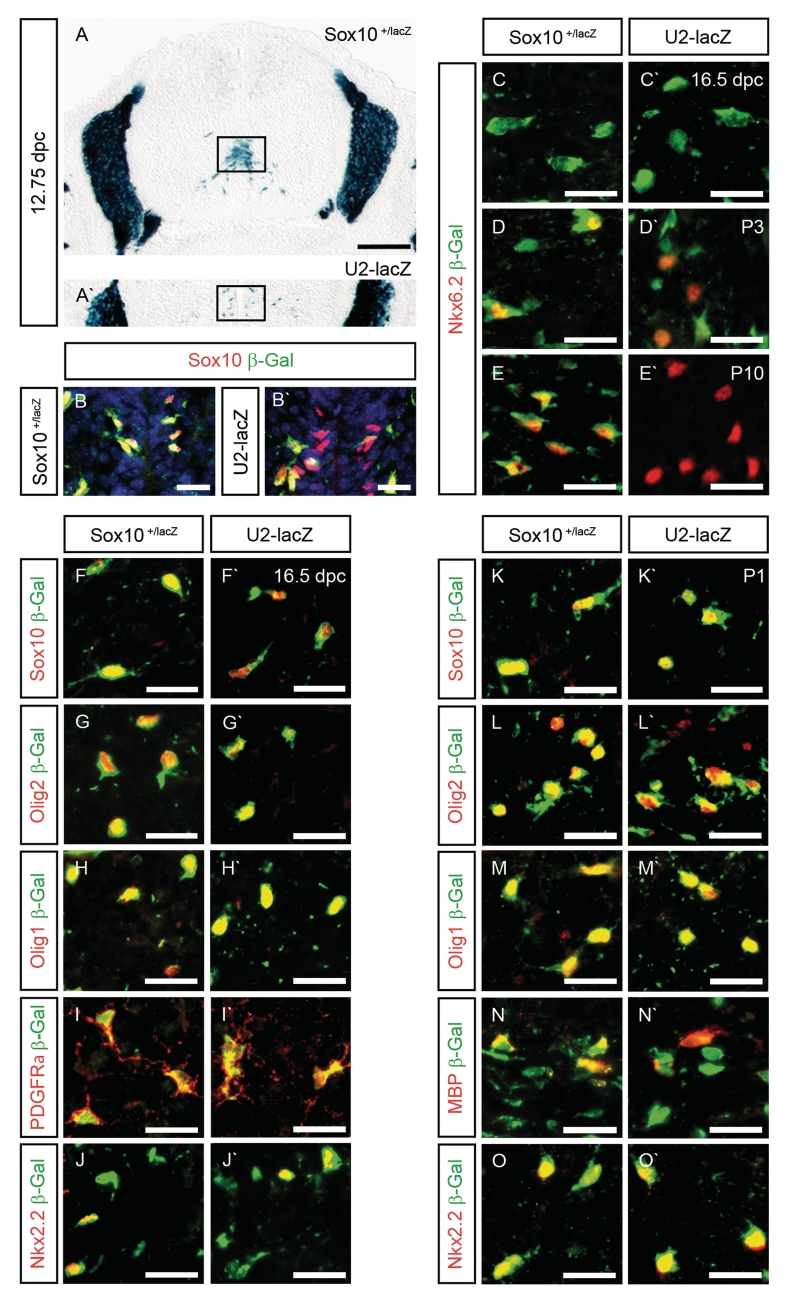
Expression of the U2-lacZ transgene is restricted to OLPs in the embryonic and early post-natal spinal cord. (A and A′) The location of the pMN domain (boxed area) was determined on transverse sections from the forelimb level of Sox10lacZ/+ (A) and U2-lacZ (A′) embryos at 12.75 dpc by X-gal staining. (B–O and B′–O′) Co-immunohistochemistry was performed on transverse spinal cord sections (forelimb level) of Sox10lacZ/+ (B–O) and age-matched U2-lacZ (B′–O′) mice at 12.75 dpc (B and B′), 16.5 dpc (C, C′, F, F′,G, G′,H, H′, I, I′, J and J′), P1 (K, K′,L, L′, M, M′, N, N′, O and O′), P3 (D and D′) and P10 (E and E′) using antibodies directed against β-galactosidase (in green) in combination with antibodies directed against Sox10 (B, B′, F, F′, K and K′), Nkx6.2 (C, C′, D, D′, E, E′), Olig2 (G, G′, L and L′), Olig1 (H, H′, M and M′), PDGFRα (I, I′), Nkx2.2 (J, J′, O and O′) and MBP (N, N′) as oligodendroglial markers (all in red). Nuclei in B and B′ were counterstained with Dapi (blue). Pictures were taken from the following regions: pMN domain (B and B′), mantle zone (F, F′, G, G′, H, H′, I and I′) and ventral marginal zone (C, C′, D, D′, E, E′, J, J′, K, K′, L, L′, M, M′, N, N′, O and O′). Size bars correspond to 200 µm (A and A′) and 25 µm (B–O and B′–O′).
At 16.5 dpc and P1, the staining pattern of spinal cords from Sox10+/lacZ and U2-lacZ mice were very similar with OLPs being detected throughout the spinal cord (Figure 1E–H). At P1, OLP density was higher in the ventral marginal zone as the future white matter region where terminal differentiation of oligodendrocytes starts at this time (Figure 1G and H, arrowheads). Closer inspection revealed that the difference between staining of the marginal zone and the remainder of the parenchyma was more pronounced in Sox10+/lacZ than in U2-lacZ mice (Figure 1G and H). There are two reasons for the more intense X-gal staining in the marginal zone. On the one hand, the density of oligodendroglial cells is higher. On the other, oligodendroglia in this region start terminal differentiation and upregulate Sox10 during the process (10). As we did not find any significant difference in overall oligodendroglial numbers in the marginal zone of Sox10+/lacZ and U2-lacZ spinal cords (data not shown), activity of the U2 enhancer is probably not upregulated during terminal differentiation.
At later post-natal stages, U2-lacZ activity is drastically downregulated compared to Sox10lacZ activity. Whereas spinal cords of P14-old Sox10+/lacZ mice exhibited intense X-gal staining throughout the spinal cord with particularly high levels in the white matter, there were only a few remaining X-gal positive cells in age-matched spinal cords of U2-lacZ mice (Figure 1I and J). As most oligodendroglia have converted from OLPs to myelinating oligodendrocytes at this time, we conclude that U2 activity is turned off during terminal differentiation. U2 thus contributes to Sox10 expression in the oligodendrocyte lineage during a particular time window only.
So far, our conclusions are based on the X-gal staining pattern. To confirm that the stained cells indeed represent oligodendroglia, we additionally performed co-immunohistochemistry with antibodies directed against β-galactosidase (Figure 2). This confirmed the absence of U2-lacZ transgene expression at 12.5 dpc (data not shown). At 12.75 dpc, the first β-galactosidase-positive cells became detectable in U2-lacZ spinal cords in the vicinity of the pMN domain (Figure 2B′). These cells were farther away from the pMN domain than β-galactosidase-positive cells in age-matched Sox10+/lacZ spinal cords (Figure 2B) in agreement with the X-gal staining pattern (Figure 2A and A′). They were fewer and represented only a fraction of the Sox10-positive cells, whereas β-galactosidase labeled all Sox10-positive cells in Sox10+/lacZ spinal cords (Figure 2B and B′). As the Sox10-positive cells without β-galactosidase in the U2-lacZ spinal cords were the ones closer to the pMN domain, we conclude that β-galactosidase expression is delayed in newly specified OLPs relative to Sox10 expression.
At 14.5 dpc and 16.5 dpc, the β-galactosidase staining pattern was very similar in U2-lacZ and Sox10+/lacZ spinal cords (Figure 2F–J and F′–J′ and data not shown). In both genotypes, β-galactosidase-positive cells in the spinal cord were also stained with antibodies against Sox10, Olig2 and Olig1 as markers for cells of the oligodendrocyte lineage (Figure 2F, F′, G, G′, H and H′ and data not shown). There was also a near complete overlap with PDGFRα, a marker for OLPs (Figure 2I and I′ and data not shown). This was expected, as virtually all oligodendroglia are OLPs at various stages of their development at this time. A fraction of the β-galactosidase-positive cells in the marginal zone also expressed Nkx2.2 at 16.5 dpc (Figure 2J and J′). These cells represent late-stage OLPs (11,20).
At P1, β-galactosidase-positive cells still expressed Sox10, Olig2 and Olig1 (Figure 2K, K′, L, L′, M and M′). More than 90% of all β-galactosidase-positive cells were also labeled by antibodies directed against Nkx2.2 (Figure 2O and O′). In the marginal zone of spinal cords from Sox10+/lacZ mice, there were now β-galactosidase-positive cells that also expressed MBP as a marker for terminally differentiating and differentiated oligodendrocytes (Figure 2N). These cells were also found in U2-lacZ mice (Figure 2N′). Nkx6.2 is expressed in the oligodendrocyte lineage during specification and after terminal differentiation (21,22). From late embryogenesis onwards, Nkx6.2 thus selectively marks mature oligodendrocytes (20). In agreement, there was no Nkx6.2 expression at 16.5 dpc in β-galactosidase-positive cells of Sox10+/lacZ and U2-lacZ spinal cords (Figure 2C and C′). At P3, Nkx6.2 was found in a fraction of β-galactosidase-positive cells in the marginal zone (Figure 2D and D′). In Sox10+/lacZ spinal cords, the double-positive cells corresponded to cells with high β-galactosidase expression levels in agreement with β-galactosidase upregulation during oligodendroglial differentiation (Figure 2D). In U2-lacZ spinal cords, in contrast, β-galactosidase expression was lower in Nkx6.2-positive cells than in Nkx6.2-negative oligodendroglia (Figure 2D′). By P10, β-galactosidase had disappeared from Nkx6.2-positive oligodendrocytes in U2-lacZ spinal cords altogether (Figure 2E′), whereas >98% of Nkx6.2-positive cells expressed β-galactosidase in Sox10+/lacZ spinal cords (Figure 2E). Taken together, these results argue that the U2 enhancer may still be active for a short time at the onset of terminal differentiation before being turned off. Alternatively, β-galactosidase protein persists for some time after its expression has already ceased.
The Sox10 U2 enhancer is activated by Olig2 and repressed by Nkx6.2 in cell culture
Having determined the temporal expression pattern of the U2 enhancer, we searched for activating transcription factors. Olig2 was an interesting candidate as this class B bHLH protein is not only essential for oligodendrocyte specification, but continues to be expressed in OLPs and even in terminally differentiating oligodendrocytes (5,6). More importantly, chicken neural tube electroporation studies had placed Olig2 genetically upstream of Sox10 (6,23).
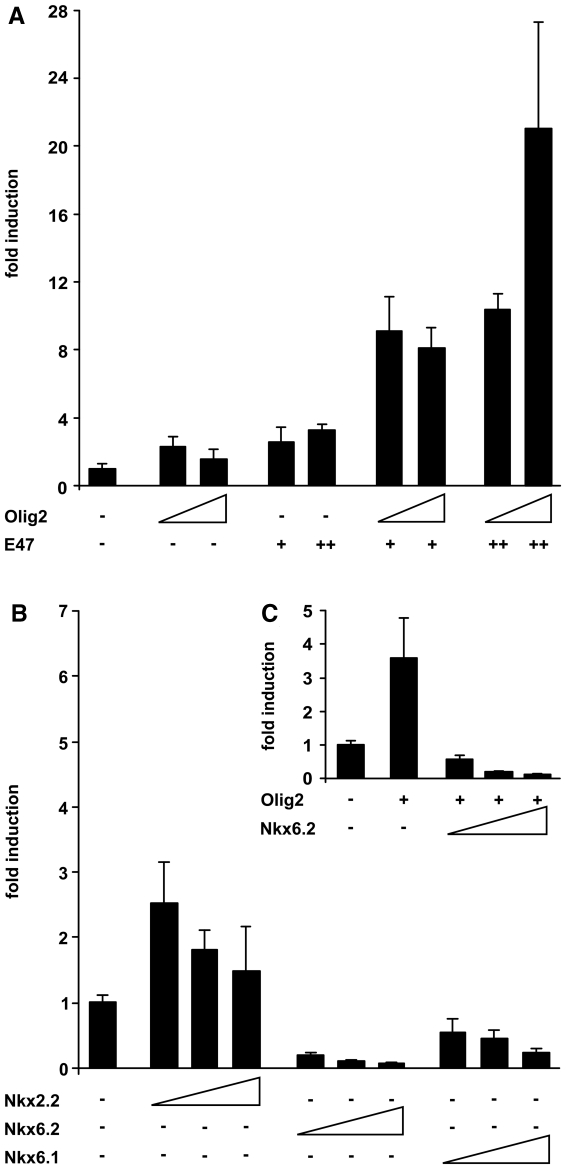
To analyze whether Olig2 is able to influence U2 activity, we performed transient transfections with a U2-luc reporter in 33B oligodendroglioma cells which endogenously express Sox10 and Olig2 (data not shown). Whereas co-transfection of the U2-luc reporter with increasing amounts of an empty expression plasmid did not change luciferase activities, a small but reproducible increase in luciferase activity was observed after co-transfection of Olig2 (Figure 3A). Similar slight increases of luciferase reporter gene activity were obtained in co-transfection with the Class A bHLH protein E47. As Olig2 and E47 have been shown to physically interact and as class A and Class B bHLH proteins often function as heterodimers in vivo (24,25), we also analyzed the effect of combined Olig2 and E47 on U2 enhancer activity. Intriguingly, we now obtained a robust induction of U2 activity that ranged from 8- to 20-fold depending on the amounts of co-transfected expression plasmids (Figure 3A).
Figure 3.
U2 activity in transiently transfected oligodendroglioma cells is modulated by the presence of Olig2 and Nkx6.2. Transient transfections were performed in 33B cells with a luciferase reporter under control of the Sox10 U2 enhancer and the β-globin minimal promoter (U2-luc). Empty pCMV expression plasmids (−) or expression plasmids for Olig2 (A and C), E47 (A), Nkx6.2 (B and C), Nkx6.1 (B), Nkx2.2 (B) and various combinations thereof (A and C) were co-transfected as indicated below the bars. Increasing amounts of expression plasmid were used as indicated by the triangles (Olig2, Nkx6.2, Nkx6.1 and Nkx2.2) and + versus ++ (E47). Luciferase activities in extracts from transfected cells were determined in three experiments each performed in duplicates. The luciferase activity obtained for U2-luc in the absence of ectopic transcription factor was arbitrarily set to 1. Fold inductions in the presence of transcription factors were calculated and are presented as mean ± SD. High statistical significance (P ≤ 0.001) was obtained for activation rates obtained by Olig2 and E47 and for repression rates obtained by Nkx6.2 or Nkx6.1 as determined by Student’s t-test.
Co-transfection of Nkx2.2 with the U2-luc reporter resulted in a mild increase in U2 activity (Figure 3B). There was furthermore no additive or synergistic increase of U2 activity when Nkx2.2 was combined with Olig2 and E47 (data not shown). While this leaves open the possibility that Nkx2.2 contributes to U2 activity in late stage OLPs, Olig2 clearly appears to be the more potent activator in combination with its E47 heterodimerization partner. This fits well with the established in vivo relationships between Sox10, Nkx2.2 and Olig2 (26).
When co-transfected with the U2-luc reporter in 33B cells, Nkx6.2 decreased U2 activity in a concentration-dependent manner (Figure 3B). The closely related Nkx6.1 acted similarly, but was less effective (Figure 3B). Interestingly, Nkx6.2 did not only decrease basal U2 activity, which is likely dependent on the endogenous Olig2 in 33B cells. It also counteracted the increased U2 activity obtained after co-transfection of Olig2 (Figure 3C). It therefore appears plausible that Nkx6.2 may inhibit U2 activity during those phases of oligodendrocyte development where it is expressed, i.e. in differentiated oligodendrocytes and around the time of oligodendroglial specification (20–22). Nkx6.1 may support Nkx6.2 in its repressive function at the time of oligodendroglial specification when both factors are co-expressed.
The Sox10 U2 enhancer contains binding sites for Olig2 and Nkx6.2
To understand the mechanism by which Olig2 and Nkx6.2 may influence the activity of the U2 enhancer, we searched its sequence for potential binding sites. Within the region present in our U2 constructs, six sites were identified with similarity to a bHLH consensus binding site (Figure 4A). These sites were designated bHLH1–bHLH6. Of these sites, only bHLH4 and bHLH5 were present in the U2 core that exhibits strong evolutionary conservation between mammals and birds. bHLH1, bHLH2 and bHLH3, in contrast, were located in the upstream flank. This is still well conserved among mammals, but not between mammals and birds. bHLH6 was in the 3′-flank. Sequence conservation of the sites corresponded to overall sequence conservation of their environment. Only bHLH4 and bHLH5 exhibited full conservation in mammals and birds. Sites bHLH2 and bHLH6 were conserved in all mammals, whereas bHLH1 was only identical in mice and humans and bHLH3 exhibited conservation in rodents only.
All sites were tested by EMSA for their Olig2-binding ability. However, only bHLH1, bHLH2 and bHLH4 yielded specific complexes with extracts containing myc-tagged Olig2 (Figure 4C and data not shown). The presence of Olig2 within the complex was confirmed by supershift with myc-tag antibodies (Figure 4D). Interestingly, all three sites bound Olig2 more strongly than the closely related Olig1 (Figure 4C). In the presence of similar amounts of protein, bHLH2 and bHLH4 yielded only faint complexes with Olig1. With bHLH1, no Olig1-specific complex was observed. In agreement with the poorer binding ability, Olig1 induced the U2-luc reporter only marginally even in combination with E47 (Figure 4B).
Within the U2 enhancer we also detected two potential binding sites for Nkx6.2 (Figure 4A). Site GTX1 was present in the U2 core and highly conserved, GTX2 in contrast was in the less conserved 3′-flank. In EMSA, none of the two sites bound significant amounts of full-length Nkx6.2 (Figure 4E). However, even the HoxA5/A6 positive control oligonucleotide with its high-affinity binding site for Nkx6.2 (27) bound very little full-length Nkx6.2. Therefore, we switched to a shortened version of Nkx6.2 that consisted of amino acids 134–216 and essentially corresponded to the homeodomain (Figure 6A). With this shortened Nkx6.2 protein, we obtained protein–DNA complexes both for GTX1 and GTX2 (Figure 4E). Compared to the HoxA5/A6 positive control, less complex was formed with GTX1 and even less with GTX2 suggesting that the three sites bind Nkx6.2 with different avidity. Addition of Nkx6.2-specific antibodies interfered with complex formation thus confirming the presence of the shortened Nkx6.2 version in the respective complexes (Figure 4F).
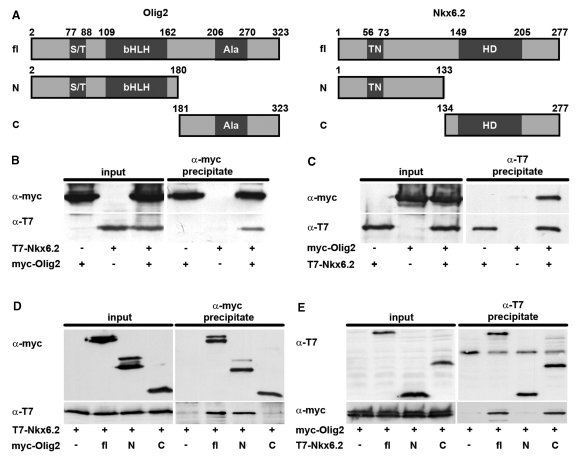
Figure 6.
Olig2 and Nkx6.2 interact via their DNA-binding domains. (A) Schematic representation of the domain structure of Olig2 and Nkx6.2 (fl) and the N-terminal (N) and C-terminal (C) parts of each protein used in co-immunoprecipitations. Numbers indicate the amino acid positions that mark end or beginning of the proteins and its domains. (B and C) Co-immunoprecipitations were performed on extracts containing full-length versions of myc-tagged Olig2, T7-tagged Nkx6.2 or both with antibodies directed against the myc-tag (α-myc) (B) and the T7-tag (α-T7) (C) as indicated above the panels. (D) Additionally, co-immunoprecipitations were carried out on extracts containing the myc-tagged full-length Olig2 (fl), its N-terminal (N) or C-terminal (C) parts in the presence of T7-tagged Nkx6.2 and antibodies directed against the myc-tag. (E) In the reverse experiment, T7-tagged full-length Nkx6.2 (fl), as well as its N-terminal (N) or C-terminal (C) parts were tested for their ability to precipitate full-length myc-tagged Olig2 in co-immunoprecipitations with antibodies directed against the T7-tag. Proteins in the input (1/10 in B, C and 1/20 in D, E) and in the precipitate were visualized by western blotting using the two tag-specific antibodies as indicated on the left of each panel in B–E.
Only Olig2-dependent activation, but not Nkx6.2-dependent repression requires binding sites in U2
To determine whether the binding sites mediate the effects of Olig2 and Nkx6.2 on the U2 enhancer, we mutated each site (Figure 5A) and confirmed by EMSA that the mutations completely abolished binding of Olig2 and Nkx6.2, respectively (Figure 5B and C). Then, we checked how binding site mutations affect the Olig2-dependent activation of the U2-luc reporter in transiently transfected 33B cells. In these experiments, we co-transfected E47 to boost induction rates (Figure 5D). Qualitatively similar results were obtained in transfections with Olig2 alone (data not shown). Olig2-dependent induction rates for a U2-luc reporter with mutant bHLH1 site were only 69% as high as induction rates for the wild-type construct (Figure 5D). Mutation of bHLH2 reduced activation rates to 41%, mutation of bHLH4 even to 25% of wild-type levels. It thus appears that all identified Olig2 binding sites contribute to the induction, however, to different extents. This conclusion was also confirmed when U2-luc reporters were analyzed that contained multiple binding site mutations. Mutation of both bHLH1 and bHLH2 reduced induction rates to similar levels as mutation of bHLH2 alone. In contrast, all U2-luc reporters in which binding site mutations were combined with a bHLH4 mutation had lost their Olig2 responsiveness completely. Among the three binding sites, bHLH4 therefore appears to be the most important one. Interestingly, this is the site within the conserved core of the U2 enhancer.
Analogous studies were also performed with the U2-luc reporter to map the repressive effect of Nkx6.2 to the identified binding sites. However, Nkx6.2 similarly repressed wild-type U2-luc and mutant versions in which either GTX1 or GTX2 were mutated (Figure 5E). Even a U2-luc reporter in which both Nkx6.2 binding sites were simultaneously mutated was still strongly repressed by Nkx6.2 (Figure 5E). Similar results were obtained in transfections in the presence or absence of co-transfected Olig2 (compare Figure 5E to F). We conclude that the potential Nkx6.2 binding sites are dispensable for Nkx6.2-dependent repression.
There are many ways in which Nkx6.2 may exert its repressive effect. Interaction with activating transcription factors such as Olig2 and ensuing inactivation is one possibility. To investigate this possibility, we performed co-immunoprecipitation experiments on extracts of 293 cells in which tagged versions of full-length Nkx6.2 and Olig2 were expressed after transient transfection (Figure 6A). When precipitations were carried out with antibodies against the tag of Olig2, Nkx6.2 was efficiently co-precipitated if present in the extract (Figure 6B). Under reciprocal conditions, Olig2 was similarly co-precipitated with antibodies directed against the Nkx6.2-specific tag (Figure 6C). Nkx6.2 could thus exert part of its repressive effect by interacting and functionally interfering with Olig2.
Using truncated versions of both Nkx6.2 and Olig2, we also determined the interacting regions in both proteins. Olig2 was divided into two fragments, an N-terminal part that corresponds to amino acids 2–180 and a C-terminal part that encompasses amino acids 181–323. Of these two protein fragments, only the N-terminal part precipitated Nkx6.2 (Figure 6D). This fragment contains the DNA-binding bHLH domain.
Nkx6.2 was also divided into an N-terminal region (corresponding to amino acids 1–133) and a C-terminal part (corresponding to amino acids 134–277) (Figure 6A). The C-terminal fragment of Nkx6.2 interacted as efficiently with Olig2 as the full-length protein, whereas Olig2 could not be detected in the precipitate of the N-terminal fragment (Figure 6E). Considering that the C-terminal part of Nkx6.2 contains the homeodomain, our results suggest that the DNA-binding domains of both proteins interact.
Olig2 binding is essential for activity of the U2 enhancer in OLPs in vivo
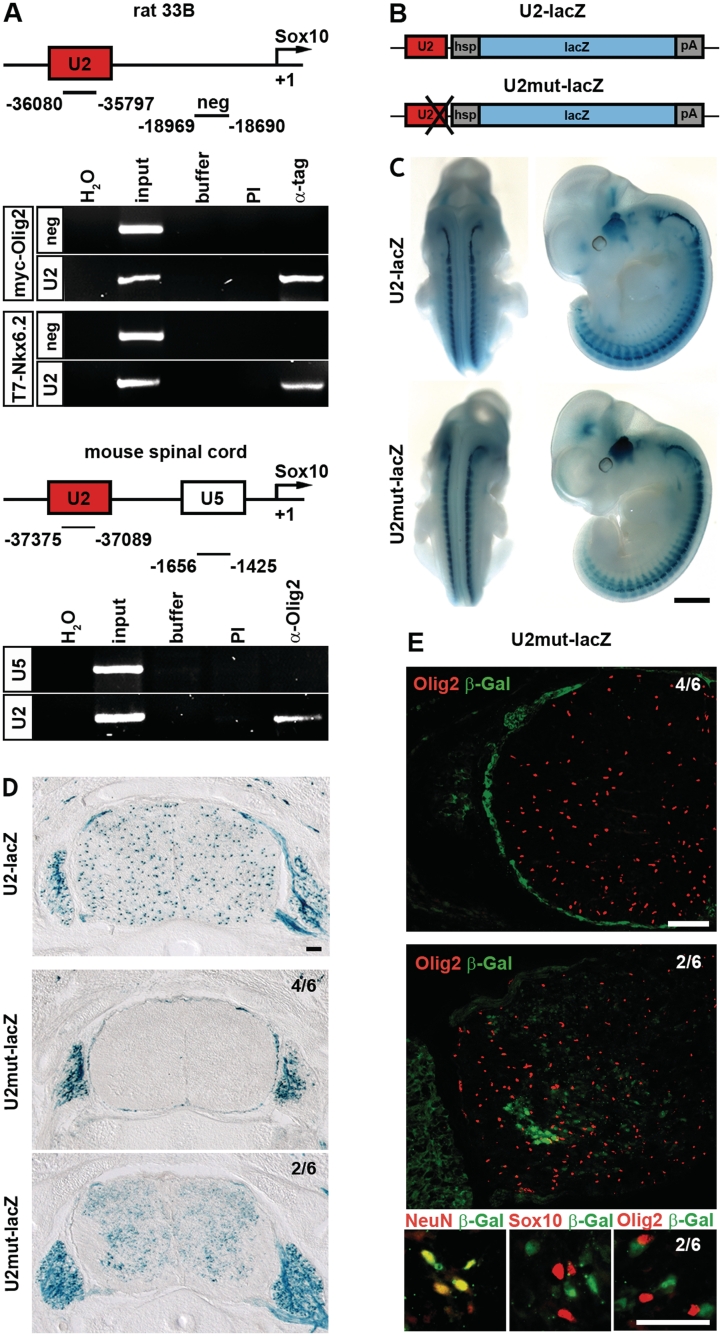
As tissue culture and in vitro experiments pointed to Olig2 as a direct activator of U2, we set out to confirm this in vivo. First, we studied if Olig2 can bind to U2 in its native chromatin context (Figure 7A). When 33B cells were transfected with myc-tagged Olig2 and chromatin immunoprecipitations were carried out with antibodies directed against the myc tag, U2 was specifically recovered in the precipitate, whereas a size-matched control region located between U2 and the transcriptional start of the Sox10 gene was not. Control IgG, furthermore, failed to precipitate the U2 fragment.
Figure 7.
Olig2 binding is essential for U2 activity in vivo. (A) Chromatin immunoprecipitation was performed on 33B cells (upper panels) transfected with myc-tagged Olig2 (myc-Olig2) or T7-tagged Nkx6.2 (T7-Nkx6.2), and on spinal cords from four 18.5 dpc-old wild-type mice (lower panels) in the absence (buffer) and presence of antibodies (PI, preimmune serum; α-tag, anti-myc or anti-T7 tag antibodies; α-Olig2, anti-Olig2 antibodies). PCR was applied on the immunoprecipitate to detect U2, a non-conserved region between U2 and the transcriptional start of Sox10 (neg) and U5. These regions of the Sox10 locus were also amplified from 1/10 of the material used for immunoprecipitation (input). (H2O), water control. (B) Schematic representation of the transgenic constructs consisting of U2 in wild-type (U2) or mutant (U2mut) version, the hsp68 minimal promoter (hsp), the lacZ marker gene (lacZ) and a SV40 polyA signal (pA). (C and D) LacZ activity was detected colorimetrically using X-gal substrate on whole embryos at 11.5 dpc (C) and on transverse sections (forelimb level) at 16.5 dpc (D) in embryos carrying the U2-lacZ transgene in wild-type or mutant version. The whole mount stainings at 11.5 dpc (C) were documented from the side and from the back. All transgenic lines carrying the U2mut-lacZ transgene looked similar at this stage. Transverse sections at 16.5 dpc (D) exhibited some differences. In most U2mut-lacZ transgenic lines (4/6), X-gal staining had completely disappeard from the spinal cord. A minority (2/4) showed a residual weak and diffuse X-gal staining. (E and F) Co-immunohistochemistry was performed at 16.5 dpc on transverse spinal cord sections of the two types of U2mut-lacZ transgenic embryos (4/6 and 2/6) using antibodies directed against β-galactosidase (in green) in combination with antibodies directed against the oligodendroglial markers Sox10 and Olig2 and the neuronal marker NeuN (all in red) as indicated. Size bars correspond to 1 mm in C and 100 µm in D and E.
While this ascertains that Olig2 can bind to the U2 enhancer in its native genomic context, it does not prove that this is really the case in OLPs of the developing spinal cord. To address this latter question, we performed additional chromatin immunoprecipitations on spinal cord from 18.5-dpc old embryos as OLPs are very prominent in this tissue during late embryogenesis. Using anti-Olig2 antibodies, the U2 enhancer was again precipitated in significant amounts (Figure 6A). This contrasts with U5, an enhancer without activity in oligodendroglia (12). Neither fragment was precipitated under identical conditions by preimmune serum (Figure 7A). All available evidence thus suggests that Olig2 is bound to the U2 enhancer in OLPs.
Using anti-T7-tag antibodies, we were also able to immunoprecipitate U2 from chromatin of 33B cells transfected with T7-tagged Nkx6.2 (Figure 7A). Considering that we found no evidence for a role of the potential Nkx6.2 binding sites in U2 repression, we have to conclude that Nkx6.2 binding to U2 is either irrelevant for repression or, if relevant, that recruitment to U2 does not require direct binding to DNA, but is instead mediated by interacting proteins such as Olig2.
Because bHLH4 had emerged as the most important Olig2-binding site in vitro, we generated a transgenic construct in which a lacZ reporter gene was driven by the hsp68 minimal promoter in combination with a U2 version in which the bHLH4 site had been mutated (U2mut-lacZ, Figure 7B). Apart from the bHLH4 mutation, this transgenic construct corresponded exactly to the original U2-lacZ transgene (Figure7B) (12). Transgenic lines were generated with the mutant construct and activity of the mutant enhancer was determined by X-gal staining in six transgenic lines. U2 had previously been shown to be active throughout the PNS of mouse embryos at 11.5 dpc. This activity remained unaffected by the introduced mutation. Cranial ganglia, dorsal root ganglia and peripheral nerves were similarly stained in U2mut-lacZ and U2-lacZ transgenic embryos at this early time and staining patterns were indistinguishable (Figure 7C). This agrees well with the fact that Olig2 is restricted in occurrence and function to the CNS. Differences became only detectable at later time points such as 14.5 (data not shown) and 16.5 dpc, when OLPs were present (Figure 7D). These differences did not concern the PNS expression which was still comparable between U2-lacZ and U2mut-lacZ embryos. Within the spinal cord, however, alterations were obvious. Whereas spinal cords of U2-lacZ embryos had a speckled appearance because of the transgene expression in OLPs, spinal cords of U2mut-lacZ embryos either lacked X-gal staining (progeny from 4 out of 6 analyzed founders) or exhibited a weaker and more diffuse X-gal staining (progeny from 2 out of 6 analyzed founders). The weak diffuse staining was furthermore specifically excluded from the marginal zone which is rich in oligodendroglia (Figure 7D) arguing that oligodendroglial expression of the U2 enhancer was also abolished in these embryos by the introduced mutation.
Additional immunohistochemical studies confirmed the complete absence of β-galactosidase-positive cells in the embryonic spinal cord of most embryos (Figure 7E, 4/6). OLPs were, however, present in normal numbers as indicated by the presence of Olig2 and Sox10-positive cells (Figure 7E and data not shown). In those embryos where diffuse staining was observed for the U2mut-lacZ transgene, β-galactosidase was found to co-localize predominantly with NeuN-positive neurons rather than with Sox10-positive or Olig2-positive OLPs (Figure 7F, 2/6). We therefore conclude that the Olig2 binding site in the conserved core of the U2 enhancer is absolutely essential for activity in OLPs. In its absence, the U2 activity in the CNS is either completely lost or reduced and redirected to neurons.
DISCUSSION
Recently, we have identified the evolutionary conserved U2 element in the distal 5′-flanking region of the Sox10 gene and characterized it as an enhancer with broad activity in the developing PNS and much more restricted oligodendroglial activity in the CNS (12). Here, we focused on the CNS-specific activity of the U2 enhancer to understand how Sox10 expression is regulated in oligodendroglia. By following expression of the transgenic reporter in U2-lacZ mice, we show that activity of the U2 enhancer is restricted to the precursor stage in the oligodendrocyte lineage. As oligodendroglia underwent terminal differentiation and expressed myelin genes, U2 was turned off. There was furthermore a significant delay between the onset of Sox10 expression in newly specified OLPs and the onset of U2 activity. These results argue that U2 is only partly responsible for the Sox10 expression pattern in the oligodendrocyte lineage and that there must be other not yet identified regulatory regions that drive expression in newly specified oligodendrocytes immediately after their specification as well as in differentiating and mature oligodendrocytes. The seemingly simple, continuous expression of Sox10 in the oligodendrocyte lineage thus results from the combined activity of at least two regulatory regions. This is somewhat reminiscent of the regulation of the MBP gene which codes for a major component of the myelin in both PNS and CNS. Correspondingly, MBP is expressed in Schwann cells of the PNS and oligodendrocytes of the CNS. Additionally expression levels need to be regulated over time in both cell types. In oligodendrocytes, there is first a phase of relatively low expression followed by maximal expression during the phase of active myelination before downregulation occurs again in mature oligodendrocytes to adjust for levels that are adequate for myelin maintenance. In this case as well, the expression pattern is the result of many different enhancers that are spread over large distances of the MBP genomic locus (28,29).
Activity of the mammalian U2 enhancer has also been analyzed in zebrafish and found to be present in MBP-expressing oligodendrocytes (30). Oligodendroglial U2 activity thus differs slightly between the two species and appears extended in zebrafish compared to mouse. Mechanistically, it would be interesting to determine the cause for this difference. The physiological relevance is less clear, however, as the U2 enhancer is not conserved in its sequence in the zebrafish genome.
Another important finding of our study is the identification of Olig2 as a main activator of the U2 enhancer. Olig2 is restricted in its expression to the CNS. Thus, it can only be involved in regulating U2 activity in oligodendroglia and not in Schwann cells and satellite glia of the PNS. In the spinal cord, Olig2 is first expressed ventrally in a defined domain of the ventricular zone where it is successively required for the specification of motor neurons and the specification of OLPs (5,6). Its further expression remains closely associated with the oligodendrocyte lineage arguing that Olig2 continues to function in these cells. Electroporations in the neural tube of developing chicken embryos have furthermore suggested that Olig2 is genetically upstream of Sox10 (6,23,26). By showing that Olig2 binds to the U2 enhancer and activates it both in vitro and in vivo, we here show for the first time that the influence of Olig2 on Sox10 expression is direct. Our in vitro data furthermore suggest that Olig2 performs this function primarily as a heterodimer with ubiquitous Class A bHLH proteins such as E47. This fits with the observation that Olig2 and E47 can be co-immunoprecipitated and jointly bind to a bHLH site in vitro (24,25). We furthermore provide evidence both in vitro as well as in vivo that Olig2 activates the U2 enhancer through several sites of which bHLH4 is the predominant one. This site is part of the conserved U2 core and strongly conserved in mammalian and avian species suggesting that U2 activity in oligodendroglia is under Olig2 control in a wide variety of species.
It also needs to be emphasized that our study provides overwhelming evidence for an activating role of Olig2. This was by no means expected as several previous studies had come to the conclusion that Olig2 primarily functions as a transcriptional repressor during dorsoventral patterning of the early spinal cord (16,31,32). Among the few studies that had shown an activating role for Olig proteins is the work by Li and colleagues, who had found MBP expression in zebrafish to be activated by Olig1 in cooperation with Sox10 (33). In mouse, both Olig1 and Olig2 synergistically activate MBP expression (15,33). These species-specific differences notwithstanding, it can be concluded that Olig proteins function both as repressors and activators, probably depending on the transcription factors with which they cooperate (34). Sox10 as one of these transcription factors is both a target gene and an interactor of Olig2, pointing to the intricate relationships within the oligodendroglial transcriptional network.
As Olig2 is more active in OLPs, whereas the related Olig1 performs its main function in differentiating oligodendrocytes (5–8), Olig2 is physiologically more relevant for U2 activation. Interestingly, we also found that Olig2 binds better than Olig1 in EMSA and is a much better U2 activator in transient transfection assays. We thus provide molecular evidence that the two closely related Olig proteins are not completely identical in their function and possess at least partly different properties.
While our data show that oligodendroglial expression is under control of Olig2, they do not reveal how Olig2 causes the initial induction of Sox10 in newly specified OLPs as this initial induction does not depend on U2. Again we would argue that Olig2 function is context dependent and that the initial induction requires a transcriptional partner which is unable to function on U2.
The loss of a required partner protein may also explain the decline of U2 activity once OLPs turn into myelinating oligodendrocytes. However, other mechanisms also contribute. It is known for instance that mature oligodendrocytes have lower levels of Olig2 than OLPs. The transient increase of nuclear Olig1 during the active phase of terminal differentiation probably has very little influence on U2 activity as Olig1 is not an efficient U2 activator. In mature oligodendrocytes, Olig1 is furthermore mainly cytoplasmic and thus not active as a transcription factor (8). The reduced amounts of Olig2 will be distributed among several regulatory regions and protein complexes. As we have shown in this study, one of the complexes that can form contains Olig2 and Nkx6.2. Because of the restricted expression pattern of Nkx6.2 in the oligodendrocyte lineage (20), this complex can form in mature oligodendrocytes but not in OLPs. Considering further the inhibitory influence of Nkx6.2 on U2 activity, it seems plausible that Nkx6.2 is involved in the downregulation of U2 activity in oligodendrocytes through inactivation of Olig2. The same mechanism may also explain why U2 is not immediately induced in newly specified OLPs. At that time, Nkx6.2 is about to be down-regulated, but still present (21,22). Nkx6.1 exhibits the same expression pattern during this early time, and also represses U2 activity. Their joint presence may prevent U2 from being activated and require an independent, Nkx6.2 insensitive enhancer for Sox10 induction in this earliest phase of oligodendroglial development. U2 would then become active after disappearance of Nkx6.2 and Nkx6.1.
In contrast to Nkx6.2 and Nkx6.1, Nkx2.2 did not repress U2 activity. This fits with the observation that the U2-lacZ transgene is co-expressed with Nkx2.2 in late stage OLPs. It also indicates that the two groups of Nkx proteins perform at least partially different roles during oligodendrocyte development.
Analysis in transgenic mice indicates that the Olig2 binding site is absolutely essential for U2 activity in OLPs, but dispensable for U2 activity in PNS glia. It will be interesting to define in future experiments the cis-acting elements within U2 for PNS expression and to see how they are arranged relative to the Olig2 binding site and to other binding sites that are additionally required for oligodendroglial expression. The presence of such additional sites is expected as enhancer activity is usually dependent on the combined activity of several transcription factors. The fact that U2 activity does not completely mirror Olig2 occurrence further corroborates the need for additional factors. Very often, cooperating transcription factors are bound in close vicinity to each other on the enhancer that they jointly activate. Inspection of the sequence around the identified Olig2 site, however, did not point to obvious candidates for such interacting transcription factors. Their future identification and the characterization of their interplay with Olig2 will further deepen our insight into the transcriptional network that drives oligodendrocyte development.
FUNDING
All experimental work was funded by the Deutsche Forschungsgemeinschaft (We1326/9 to M.W.). Funding for open access charge: Deutsche Forschungsgemeinschaft (We1326/9 to M.W.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENT
We thank J. Ericson, J. Muhr, C. Stiles and L. Sussell for providing cDNAs and antibodies.
REFERENCES
- 1.Richardson WD, Kessaris N, Pringle N. Oligodendrocyte wars. Nat. Rev. Neurosci. 2006;7:11–18. doi: 10.1038/nrn1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rowitch DH. Glial specification in the vertebrate neural tube. Nat. Rev. Neurosci. 2004;5:409–419. doi: 10.1038/nrn1389. [DOI] [PubMed] [Google Scholar]
- 3.Wegner M. A matter of identity: transcriptional control in oligodendrocytes. J. Mol. Neurosci. 2008;35:3–12. doi: 10.1007/s12031-007-9008-8. [DOI] [PubMed] [Google Scholar]
- 4.Li H, He Y, Richardson WD, Casaccia P. Two-tier transcriptional control of oligodendrocyte differentiation. Curr. Opin. Neurobiol. 2009;19:479–485. doi: 10.1016/j.conb.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, Rowitch DH. Common developmental requirement for olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- 6.Zhou Q, Anderson DJ. The bHLH transcription factors olig2 and olig1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- 7.Xin M, Yue T, Ma Z, Wu FF, Gow A, Lu QR. Myelinogenesis and axonal recognition by oligodendrocytes in brain are uncoupled in Olig1-null mice. J. Neurosci. 2005;25:1354–1365. doi: 10.1523/JNEUROSCI.3034-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arnett HA, Fancy SP, Alberta JA, Zhao C, Plant SR, Kaing S, Raine CS, Rowitch DH, Franklin RJ, Stiles CD. bHLH transcription factor Olig1 is required to repair demyelinated lesions in the CNS. Science. 2004;306:2111–2115. doi: 10.1126/science.1103709. [DOI] [PubMed] [Google Scholar]
- 9.Kuhlbrodt K, Herbarth B, Sock E, Hermans-Borgmeyer I, Wegner M. Sox10, a novel transcriptional modulator in glial cells. J. Neurosci. 1998;18:237–250. doi: 10.1523/JNEUROSCI.18-01-00237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stolt CC, Rehberg S, Ader M, Lommes P, Riethmacher D, Schachner M, Bartsch U, Wegner M. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev. 2002;16:165–170. doi: 10.1101/gad.215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finzsch M, Stolt CC, Lommes P, Wegner M. Sox9 and Sox10 influence survival and migration of oligodendrocyte precursors in the spinal cord by regulating PDGF receptor {alpha} expression. Development. 2008;135:637–646. doi: 10.1242/dev.010454. [DOI] [PubMed] [Google Scholar]
- 12.Werner T, Hammer A, Wahlbuhl M, Bösl MR, Wegner M. Multiple conserved regulatory elements with overlapping functions determine Sox10 expression in mouse embryogenesis. Nucleic Acids Res. 2007;35:6526–6538. doi: 10.1093/nar/gkm727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Britsch S, Goerich DE, Riethmacher D, Peirano RI, Rossner M, Nave KA, Birchmeier C, Wegner M. The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev. 2001;15:66–78. doi: 10.1101/gad.186601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maka M, Stolt CC, Wegner M. Identification of Sox8 as a modifier gene in a mouse model of Hirschsprung disease reveals underlying molecular defect. Dev. Biol. 2005;277:155–169. doi: 10.1016/j.ydbio.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 15.Wißmüller S, Kosian T, Wolf M, Finzsch M, Wegner M. The High-mobility-group domain of Sox proteins interacts with the DNA-binding domains of many transcription factors. Nucleic Acids Res. 2006;34:1735–1744. doi: 10.1093/nar/gkl105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun T, Dong H, Wu L, Kane M, Rowitch DH, Stiles CD. Cross-repressive interaction of Olig2 and Nkx2.2 transcription factors in developing neural tube associated with formation of a specific physical complex. J. Neurosci. 2003;23:9547–9556. doi: 10.1523/JNEUROSCI.23-29-09547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schlierf B, Ludwig A, Klenovsek K, Wegner M. Cooperative binding of Sox10 to DNA: requirements and consequences. Nucleic Acids Res. 2002;30:5509–5516. doi: 10.1093/nar/gkf690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stolt CC, Schlierf A, Lommes P, Hillgärtner S, Werner T, Kosian T, Sock E, Kessaris N, Richardson WD, Lefebvre V, et al. SoxD proteins influence multiple stages of oligodendrocyte development and modulate SoxE protein function. Dev. Cell. 2006;11:697–710. doi: 10.1016/j.devcel.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 19.Schlierf B, Werner T, Glaser G, Wegner M. Expression of Connexin47 in oligodendrocytes is regulated by the Sox10 transcription factor. J. Mol. Biol. 2006;361:11–21. doi: 10.1016/j.jmb.2006.05.072. [DOI] [PubMed] [Google Scholar]
- 20.Cai J, Zhu Q, Zheng K, Li H, Qi Y, Cao Q, Qiu M. Co-localization of Nkx6.2 and Nkx2.2 homeodomain proteins in differentiated myelinating oligodendrocytes. Glia. 2010;58:458–468. doi: 10.1002/glia.20937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai J, Qi Y, Hu X, Tan M, Liu Z, Zhang J, Li Q, Sander M, Qiu M. Generation of oligodendrocyte precursor cells from mouse dorsal spinal cord independent of Nkx6 regulation and Shh signaling. Neuron. 2005;45:41–53. doi: 10.1016/j.neuron.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 22.Vallstedt A, Klos JM, Ericson J. Multiple dorsoventral origins of oligodendrocyte generation in the spinal cord and hindbrain. Neuron. 2005;45:55–67. doi: 10.1016/j.neuron.2004.12.026. [DOI] [PubMed] [Google Scholar]
- 23.Sun T, Echelard Y, Lu R, Yuk DI, Kaing S, Stiles CD, Rowitch DH. Olig bHLH proteins interact with homeodomain proteins to regulate cell fate acquisition in progenitors of the ventral neural tube. Curr. Biol. 2001;11:1413–1420. doi: 10.1016/s0960-9822(01)00441-9. [DOI] [PubMed] [Google Scholar]
- 24.Lee S-K, Lee B, Ruiz EC, Pfaff SL. Olig2 and Ngn2 function in opposition to modulate gene expression in motor neuron progenitor cells. Genes Dev. 2005;19:282–294. doi: 10.1101/gad.1257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samanta J, Kessler JA. Interactions between ID and OLIG proteins mediate the inhibitory effects of BMP4 on oligodendroglial differentiation. Development. 2004;131:4131–4142. doi: 10.1242/dev.01273. [DOI] [PubMed] [Google Scholar]
- 26.Liu Z, Hu X, Cai J, Liu B, Peng X, Wegner M, Qiu M. Induction of oligodendrocyte differentiation by Olig2 and Sox10: evidence for reciprocal interactions and dosage-dependent mechanisms. Dev. Biol. 2007;302:683–693. doi: 10.1016/j.ydbio.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 27.Awatramani R, Scherer S, Grinspan J, Collarini E, Skoff R, O'Hagan D, Garbern J, Kamholz J. Evidence that the homeodomain protein Gtx is involved in the regulation of oligodendrocyte myelination. J. Neurosci. 1997;17:6657–6668. doi: 10.1523/JNEUROSCI.17-17-06657.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forghani R, Garofalo L, Foran DR, Farhadi HF, Lepage P, Hudson TJ, Tretjakoff I, Valera P, Peterson A. A distal upstream enhancer from the myelin basic protein gene regulates expression in myelin-forming schwann cells. J. Neurosci. 2001;21:3780–3787. doi: 10.1523/JNEUROSCI.21-11-03780.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farhadi HF, Lepage P, Forghani R, Friedman HC, Orfali W, Jasmin L, Miller W, Hudson TJ, Peterson AC. A combinatorial network of evolutionarily conserved myelin basic protein regulatory sequences confers distinct glial-specific phenotypes. J. Neurosci. 2003;23:10214–10223. doi: 10.1523/JNEUROSCI.23-32-10214.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Antonellis A, Huynh JL, Lee-Lin S-Q, Vinton RM, Renaud G, Loftus SK, Elliot G, Wolfsberg TG, Green ED, McCallion AS, et al. Identification of neural crest and glial enhancers at the mouse Sox10 locus through transgenesis in zebrafish. PLoS Genet. 2008;4:e1000174. doi: 10.1371/journal.pgen.1000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu QR, Yuk D, Alberta JA, Zhu Z, Pawlitzky I, Chan J, McMahon AP, Stiles CD, Rowitch DH. Sonic hedgehog-regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron. 2000;25:317–329. doi: 10.1016/s0896-6273(00)80897-1. [DOI] [PubMed] [Google Scholar]
- 32.Zhou Q, Wang S, Anderson DJ. Identification of a novel family of oligodendrocyte lineage-specific basic helix-loop-helix transcription factors. Neuron. 2000;25:331–343. doi: 10.1016/s0896-6273(00)80898-3. [DOI] [PubMed] [Google Scholar]
- 33.Li H, Lu Y, Smith HK, Richardson WD. Olig1 and Sox10 interact synergistically to drive myelin basic protein transcription in oligodendrocytes. J. Neurosci. 2007;27:14375–14382. doi: 10.1523/JNEUROSCI.4456-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gokhan S, Marin-Husstege M, Yung SY, Fontanez D, Casaccia-Bonnefil P, Mehler MF. Combinatorial profiles of oligodendrocyte-selective classes of transcriptional regulators differentially modulate myelin basic protein gene expression. J. Neurosci. 2005;25:8311–8321. doi: 10.1523/JNEUROSCI.1850-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]