Abstract
This report describes an integrated study on identification of potential markers for gastric cancer in patients’ cancer tissues and sera based on: (i) genome-scale transcriptomic analyses of 80 paired gastric cancer/reference tissues and (ii) computational prediction of blood-secretory proteins supported by experimental validation. Our findings show that: (i) 715 and 150 genes exhibit significantly differential expressions in all cancers and early-stage cancers versus reference tissues, respectively; and a substantial percentage of the alteration is found to be influenced by age and/or by gender; (ii) 21 co-expressed gene clusters have been identified, some of which are specific to certain subtypes or stages of the cancer; (iii) the top-ranked gene signatures give better than 94% classification accuracy between cancer and the reference tissues, some of which are gender-specific; and (iv) 136 of the differentially expressed genes were predicted to have their proteins secreted into blood, 81 of which were detected experimentally in the sera of 13 validation samples and 29 found to have differential abundances in the sera of cancer patients versus controls. Overall, the novel information obtained in this study has led to identification of promising diagnostic markers for gastric cancer and can benefit further analyses of the key (early) abnormalities during its development.
INTRODUCTION
Gastric cancer represents the second leading cause of cancer death worldwide, next only to lung cancer (1). In 2002, 934 000 new cases were reported worldwide. In the USA, ∼21 500 new cases of gastric cancer were diagnosed in 2008, with 10 800 deaths from the disease (2). The current 5-year survival rate of individuals diagnosed with gastric cancer is ∼24% (1), reflecting the reality that most cases are already in an advanced stage when diagnosed. As with other cancers, the challenge in early detection lies in the reality that the early symptoms tend to be relatively non-specific, and detection requires that invasive physical procedures, such as gastrointestinal endoscopy, be carried out on a regular basis, which may not be practical for general screening. The most ideal solution for early detection is to find reliable markers that can detect the cancer through simple blood tests.
Recent comparative transcriptomic studies have identified a number of gene markers of different types for gastric cancer, such as diagnostic markers [NF2 (3), NEK6 and INHBA (4)], prognostic markers [CDH17 (5), PDCD6 (6)], and gastric-cancer-associated genes [TSPAN1, Ki67 and CD34 (7)]. While exhibiting some predictive power, these gene markers were found highly inconsistent as identified by different studies (Supplementary Table S1), and none of them has reached the clinical trial stage. A few serum markers such as α-fetoprotein antigen (AFP), carcinoembryonic antigen (CEA), and CA19-9, identified through large-scale blood screening (8), have been used for gastric cancer detection. The detection sensitivities of these markers are, however, rather low, no more than 25% at the 90% specificity level (8), and hence they have not been widely used clinically for diagnostic purposes. Using immunoassay and proteomic techniques, a few new serum markers were recently proposed. including MUC1 and MUC5AC (8), pepsinogen C and pepsin A activation peptide (9), and Reprimo (10), although their true diagnostic power for gastric cancer, especially at an early stage, is yet to be thoroughly evaluated. More rigorous studies are in order on these proposed markers.
The lack of reliable serum markers for gastric cancer reflects the challenging nature of the problem, but also suggests the possibility that all the information derivable using the powerful omic techniques, in conjunction with computational approaches, may not have been fully utilized. For example, there have been only a few published large-scale studies attempting to link the information derivable from gene-expression profiles of cancer tissues to proteomic biomarker identification in patients’ sera. The general issue with the existing proteomic studies on serum marker identification is that the potential markers are probably of substantially lower abundance in comparison with other proteins in blood circulation, making their detection through large-scale screening extremely difficult. Our study indicates that transcriptomic analyses of cancer tissues can provide highly useful information in guiding proteomic studies in a search for protein markers in sera.
Herein a systematic study, aiming to ultimately identify serum markers for gastric cancer, is presented, where the following procedure was employed. Exon array chips were made on 80 pairs of gastric cancer and the adjacent noncancerous tissues from the same patients. Comparative analyses of gene expression data were performed to identify differentially expressed genes in cancer versus reference tissues. These genes were then subject to computational analyses to determine if their proteins may be secretory into blood circulation and thus potentially serve as serum markers. To rule out markers that are non-specific to gastric cancer, the predicted marker genes were compared with published microarray gene-expression data for other human diseases, including over 40 types of cancers. Then immunoassay and mass spectrometry (MS) analyses of serum samples from a subset of the 80 patients and healthy age- and gender-matched volunteers were used to verify the predicted markers.
MATERIALS AND METHODS
Sample collection
A total of 80 gastric cancer tissues and their adjacent noncancerous tissues were collected from 80 non-treated patients (4 of stage I, 7 of stage II, 54 of stage III and 15 of stage IV). To ensure the integrity of the mRNAs used for microarray experiments, all tissues were snap-frozen and stored in liquid nitrogen within 20 min after resection. In addition, blood samples were collected from the cancer patients before surgery, and control blood samples were collected from healthy age-/gender-matched volunteers involved in this study. All samples were collected at three affiliated hospitals of Jilin University College of Medicine and Jilin Provincial Cancer Hospital, Changchun, China. A written informed consent was obtained from all patients, which was approved by the Institutional Review Board (IRB) at the University of Georgia, Athens, GA, USA and by the Chinese IRB overseeing human subjects at Jilin University College of Medicine and Jilin Provincial Cancer Hospital. The histological subtype and pathologic stage of each tumor were determined according to the World Health Organization (WHO) and TNM classification system. Detailed patient information such as age, gender, and pathologic stage, is listed in Supplementary Table S2.
RNA preparation and microarray experiments
A total of 15 μg RNA was extracted from each tissue sample using Trizol (Invitrogen) followed by purification using the RNeasy Mini kit (QIAGEN) according to the manufacturer’s recommendation. Ratios of A260/A280 >1.9 and 28S/18S rRNA of 2 were used to ensure that the RNA samples were highly purified and not degraded.
The RNA samples were analyzed using the GeneChip Human Exon 1.0 ST (Affymetrix), following the protocol detailed in the Genechip Expression Analysis Technical Manual (P/N 900223). In brief, 1 μg of total RNA was used as template for synthesis of cDNA after rRNA reduction. Through reverse transcription in vitro, cRNA was obtained and used as the template for cDNA synthesis in the second cycle. Then cRNA was hydrolyzed by RNase H, and the sense strand DNA was digested by two endonucleases. Fragmented samples were labeled with DNA labeling reagent. The labeled samples were mixed with hybridization cocktail and hybridized to the microarray at 45°C, 60 rpm, and then incubated for 17 h. Next, the array was washed and stained on the GeneChip® Fluidics Station 450, using the appropriate fluidics script, before being inserted into the Affymetrix autoloader carousel and scanned using the GeneChip® Scanner 3000 with GeneChip® Operating Software (GCOS).
The raw exon array data (.CEL) collected on the 80 pairs of cancer and reference tissues is MIAME compliant and can be temporarily downloaded at http://csbl.bmb.uga.edu/∼juancui/Data/data.tgz.
Identification of differentially expressed genes in cancer versus reference tissues
We used the expression data of the 290 000 core probe sets supported by the full-length mRNAs from the Refseq database. The probe intensities were normalized using the quartile normalization by the MiDAS (Microarray Detection of Alternative Splicing) program (see Affymetrix white paper exon_alt_transcript_ analysis_ whitepaper.pdf), and then summarized to both exon- and gene-level expressions using the PLIER algorithm (11) (see ‘Guide to Probe Logarithmic Intensity Error (PLIER) Estimation’ at http://www.affymetrix.com/support/technical). All genes having very low expressions were then removed from our analysis since the signal-to-noise ratio is rather low among such genes; specifically, a gene was removed if its normalized average expression level in both the cancer and the reference groups was below 10. To detect genes with consistent differential expressions in cancer versus reference tissues, an analysis of variance (ANOVA) test and a paired Wilcoxon signed–rank test were performed, along with a third statistical test outlined as follows. For each gene, the number of pairs of cancer/reference tissues, Kexp, whose expression fold-change is larger than k (k is set to be 2) was counted; if the P-value for the observed Kexp was <0.05 (see details in Supplementary Procedure 1), the gene was considered to have differential expression in cancer versus reference tissues. All statistics are listed in Supplementary Table S3.
Bi-clustering analysis and pathway enrichment analysis
Our in-house bi-clustering QUBIC (QUalitative BI-Clustering) program (12) was used to identify statistically significant bi-clusters in the expression data. The basic idea of the algorithm is to find all subgroups of genes with similar expression patterns among some (to be identified) subsets of cancer tissues, and hence genes involved in each such pattern can possibly be used as signatures for cancer sub-typing or staging. Compared to existing algorithms, QUBIC can solve more general form of the bi-clustering problem in a computationally efficient manner (12).
The pathway enrichment analysis was first done using two popular programs, DAVID (13) and KOBAS (14). Additional curated pathway information from Human Pathway Interaction Database (15) was integrated to ensure a comprehensive coverage, and a P-value was calculated for each relevant pathway based on Fisher’s exact test on queried genes against the whole human genome.
Identification of gene signatures and prediction of blood-secreted proteins
A signature-selection procedure (see details in Supplementary Procedure 2) was used to systematically identify multi-gene combinations whose gene-expression levels can distinguish well between cancer and reference tissues. To ensure the robustness of the to-be-identified signatures, we randomly and repeatedly sampled the paired tissues and predicted the possible multi-gene signatures for each sample set; and then ranked the signatures derived from different sample sets using a consensus voting procedure (16).
Among the differentially expressed genes, we focused on those whose proteins are predicted to be secretory to blood circulation by our in-house program (17). To the best of our knowledge, this is the first program for prediction of blood secreted proteins. The basic idea of the algorithm is as follows. An extensive literature search led to a large collection of human proteins that are known to be blood secretory and detected by previous proteomic studies. A list of features shared by these blood secretory proteins was delineated, including their physical and chemical properties, amino acid sequence and motif, and structural features. Using these features, a support vector machine (SVM)-based classifier was trained to distinguish proteins that are blood secretory from those that are not. The trained classifier achieved ∼90% prediction sensitivity and 98% specificity based on a large test set consisting of 98 known blood-secretory proteins and 6601 non-blood-secretory proteins (17).
Public microarray data for gastric cancer as an independent validation set
Two public microarray datasets for gastric cancer from the GEO database were downloaded for comparative studies (Supplementary Procedure 3). The Kim dataset (18), with gene expression profiles of 50 gastric cancer patients in different cancer stages, was used to evaluate the effectiveness of the predictive markers through examining the consistency of their expression alteration across all samples. Given the ratio of the expression levels between the cancer tissues and the reference tissues, the data was normalized using the print-tip Lowess method in the marray package of the Bioconductor project (http://www.bioconductor.org) (19). The original study showed that the comparison of the log-transformed fold-changes between this dataset and the Affymetrix array is meaningful (18). The Xin dataset (20), with gene expression of 100 gastric cancer and 24 reference tissues, was first preprocessed using essentially the same procedure as described by Chen et al. (20). This data were used to test the robustness and effectiveness of the predicted markers through the same classification analysis applied on our dataset.
Specificity analysis of predicted gene markers through searches against public microarray data
To assess the specificity of the predicted gene markers to gastric cancer, a biomarker evaluation system (http://bioinfosrv1.bmb.uga.edu/DMarker/) was developed to check if the expression pattern of the predicted markers could also be caused by other human diseases. The basic idea is to collect all the microarray datasets for human diseases from the GEO (21), Oncomine (22) and SMD (23) databases after removing datasets with less than six samples in either cancer or the reference group, removing the genes with all their expression levels among the bottom 10% of the expression levels in each dataset, and genes with more than 50% values being missing within each dataset and 30% missing across all diseases. A marker is considered specific to gastric cancer only if no more than 30% of the patients and the control populations for any other disease dataset satisfy the expression cutoff for the marker, where 30% is determined empirically based on our studies.
Experimental validation of predicted serum markers
A combined approach of MS, antibody array and western blot analysis was used to validate the predicted serum markers. Briefly, for western blots, serum samples were incubated at 100°C for 5 min, separated using SDS–PAGE gels (Bio-Rad), and then transferred onto PVDF membranes. After blocking the non-specific binding sites, membranes were incubated overnight at 4°C with primary antibodies in 1.5% non-fat dry milk in TBST, followed by incubation with secondary antibodies for 2 h. The antibodies were from Abnova, Inc. (Taipei, Taiwan), Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) and Abcam, Inc. (Cambridge, MA). The enhanced chemiluminescence (ECL) reaction, using Western Lightning Chemiluminescence Reagent Plus (Perkin Elmer, USA), was then applied to the membranes, and the ECL membrane images were quantified using the ImageJ 1.34s software (http://rsb.info.nih.gov/ij/).
MS experiments were conducted on proteins extracted from gels (SDS–PAGE). The samples were digested with proteomics-grade Trypsin (Promega) and analyzed on an Agilent 1100 capillary LC (Palo Alto, CA) interfaced directly to an LTQ linear ion trap mass spectrometer (Thermo Electron, San Jose, CA). The peptide samples were loaded using positive N2 pressure on a PicoFrit 8 cm by 50 μm column (New Objective, Woburn, MA). Peptides were eluted from the column into the mass spectrometer during a 55 min linear gradient from 5 to 60% of total solution composed of mobile phase B at a flow rate of 200 nl min−1. The instrument was set to acquire MS/MS spectra on the nine most abundant precursor ions from each MS scan with a repeat count of three and repeat duration of 15 s. Data analysis was conducted using MASCOT (http://www.matrixscience.com), searching against the NCBI Human database and a reverse NCBI Human database. The assigned peptides were grouped into proteins using the ProteoIQ software (http://www.bioinquire.com). Only proteins with a false-discovery rate of <1% were considered statistically significant.
For the biotin-label-based array experiment, each sample (50 μl serum) was dialyzed, followed by a biotin-labeling step (Pierce, Rockford, IL, USA), where the primary amine was biotinylated. The biotin-labeled proteins were incubated with chips (RayBio®Biotin Label-Based Antibody Arrays, RayBiotech, Inc.) at room temperature for 2 h. After the incubation with HRP-streptavidin, the signals were visualized by chemiluminescence and then imaged by a Scan Array laser confocal slide scanner (PerkinElmer Life Science).
RESULTS
Differentially expressed genes in gastric cancer versus reference tissues
A total of 80 gastric cancer tissues and their adjacent reference tissues from the same 80 patients were collected (Supplementary Table S1). Microarray experiments were conducted on these tissues using the Affymetrix GeneChip Human Exon 1.0 ST Array, which covers 17 800 human genes. Using a set of criteria given in ‘Materials and Methods’ section, 2540 genes were found to exhibit differential expression patterns between the cancer and the reference tissues, of which 715 showed at least 2-fold changes in expression (detailed information of each gene is listed in Supplementary Table S3). The majority of these genes were up-regulated in cancer and only one-fifth was down-regulated (Supplementary Figure S1). In addition, 1276 genes were found to be differentially expressed in early-stage cancers (stages I–II), with 468 genes specific in early stage (i.e. having no substantial differences in the advanced cancers). Of the 1276 genes, 208 were consistently differentially expressed across all early-stage paired samples, 48 of which are gastrointestinal diseases-related (Supplementary Table S3).
Our observations about the altered gene-expressions in gastric cancer are largely consistent with the previous reports (Supplementary Table S1), such as the altered expression of TOP2A, CDK4 and CKS2 (24), CDH1 (25), GKN1, GKN2 and TFF1 (26,27). Some novel observations were also made. For example, a number of genes related to chromosomal amplifications, transcriptional regulation and signal transduction, such as CCNE1, POP4, RMP, UQCRFS1 and DKFZP762D096, were found to exhibit differential expressions in 55 of the 80 (∼68.7%) cancer samples used in our study. This percentage is significantly higher than that in a previous report (20). The observed up-regulation of two claudins, CLDN 7 and CLDN1, and down-regulation of DPT are found in most cancer tissues, regardless of their stage and subtype, potentially serving as gene markers for gastric cancer. We believe that the differences in some of our observations with the previous one may be due to different distributions of the patient populations in terms of age, gender and cancer subtype/stage used in different studies. In addition, a number of down-regulated genes were found by this study to be highly specific to gastric cancer. These include GIF, GNK1, GNK2, TFF1, GHL1, LIPF and ATP4A, providing a different type of marker with decreased abundance in cancer.
Enriched functional families and pathways
According to IPA (Ingenuity Pathway Analysis; http://www.ingenuity.com) annotation, 911 out of the 2540 differentially expressed genes are cancer-related, 219 related to antigen presence or immune responses, with 414 being gastric tissue specific. Among the 13 major IPA functional families, 9 and 10 families are found to be substantially enriched (P < 0.01) among the 2540 differentially expressed genes and the 911 cancer-related genes, respectively, when compared with the whole human genome (Supplementary Figure S2). Some interesting observations are made. For example, the level of enrichment in kinases, peptidases, enzymes, transporters and transmembrane receptors increases as cancer develops from early to advanced stages; transcription regulators are enriched only in early-stage cancers; and growth factors are more substantially enriched in early cancers than in advanced cancers. Among the 23 transcription factors that are consistently up- or down-regulated across at least 80% of all early-stage cancers in our study, signals possibly related to the early responses to cancer development were observed, such as the up-regulation of repressor E2F4/DP1, which may regulate cell growth and differentiation by the TGF-β signaling pathway, and the up-regulation of p14ARF, an upstream regulator of the TP53 pathway that inhibits the apoptosis.
Pathway enrichment analysis was conducted on the differentially expressed genes. Table 1 lists 13 such pathways, along with their statistical significance values. A few general cellular processes such as the cell cycle, DNA replication and cell communication were highly enriched by consistently up-regulated genes, while fatty acid metabolism, digestion and ion transport were enriched only by down-regulated genes. Most of these pathways start being up- or down-regulated in early-stage cancers and become increasingly more enriched as cancer advances. In addition to the pathways commonly associated with cancer development such as cell cycle and regulation, DNA damage and repair, cell growth, apoptosis and regulation, and estrogen receptor regulation pathways, a few gastric cancer-specific pathways were revealed. For example, a novel thyroid hormone-mediated gastric carcinogenic signaling pathway (28) is enriched with up-regulated genes (TTHY, PKM2, GRP78, FUMH, ALDOA and LDHA) in cancer tissues, most of which are in advanced stages.
Table 1.
Thirteen enriched pathways by differentially expressed genes
| Pathways | Number of genes |
P-value | |
|---|---|---|---|
| Stages I–II (specific) | All stages | ||
| Cell cycle | 22↑ (9↑) | 49↑ | 1.59E–21 |
| p53 signaling pathway | 10↑ (3↑) | 27↑ | 2.66E–12 |
| ECM-receptor interaction | 4↑ (–) | 31↑ | 8.18E–13 |
| Cell communication | 6↑ (–) | 34↑ | 4.70E–04 |
| Cell adhesion molecules (CAMs) | 4↑ (2↑) | 31↑ | 5.13E–04 |
| Role of BRCA1, BRCA2 and ATR in cancer susceptibility | 4↑ (–) | 10↑ | 2.90E–03 |
| E2F1 destruction pathway | 4↑ (–) | 6↑ | 8.00E–03 |
| Wnt signaling pathway | 4↑ (–) | 17↑ | 2.22E–02 |
| Focal adhesion | 4↑ (3↑) | 41↑ | 1.32E–09 |
| 3↓ (3↓) | 4↓ | 9.81E–02* | |
| Metabolism of xenobiotics by cytochrome P450 | 4↓ (–) | 16↓ | 7.21E–04* |
| Arginine and proline metabolism | 3↓ (–) | 3↓ | 1.16E–03* |
| Fatty acid metabolism | 3↓ (–) | 7↓ | 2.56E–03* |
| Insulin signaling pathway | 5↓ (–) | 7↓ | 9.37E–04* |
Upward arrow indicates for up-regulation and downward arrow indicates down-regulation. P-value is calculated for a pathway enriched in all stages except those marked with asterisks are for early stage only.
Effects of age and gender on gene expression data
The impact of age and gender on the expressions of the 2540 differentially expressed genes was assessed through multivariate analyses using ANOVA (11) and Cox Proportional Hazard Regression Model (29) (Supplementary Table S4). It was found that age significantly affects the expressions of 143 genes (P < 0.05), 113 of which further increase the differences in their expression levels between cancer and reference tissues, an observation that could have important implications to biomarker selection. For example, the average MUC1 expression level is substantially higher in cancer tissues of patients over 55 versus those under 55 (Supplementary Figure S3). Similar observations were made on a few other genes such as UBFD1, MDK and the other members of the Mucin family. The observation that age might impact on the expression of certain genes might correlate to some degree with the finding that poorly differentiated cancers are more aggressive and more common in young patients (30–32).
Possible gender-specific biases in our expression data were also examined, knowing that the male-to-female ratio of gastric cancer occurrences is about 2:1 (33). It was found that the expression levels of 59 genes, including WNT2, ARSE and KCNN2, are gender-dependent (Supplementary Table S4). An interesting observation was that the combination of age and gender has a more significant effect on the expression levels of 118 genes such as COL1A1, THY1 and CPS1. For genes like TIMP1 and ADH1A, older male patients have higher expression levels than younger female patients.
Among the differentially expressed genes specific to early-stage cancers, 28 and 9 genes are age- and gender-dependant, respectively; examples include P2RY6 and NSUN5. Other factors such as alcohol-/smoking-taking history were also evaluated using a multivariate analysis, and the detailed results are given in Supplementary Table S4.
Co-expressed gene clusters relevant to cancer classification
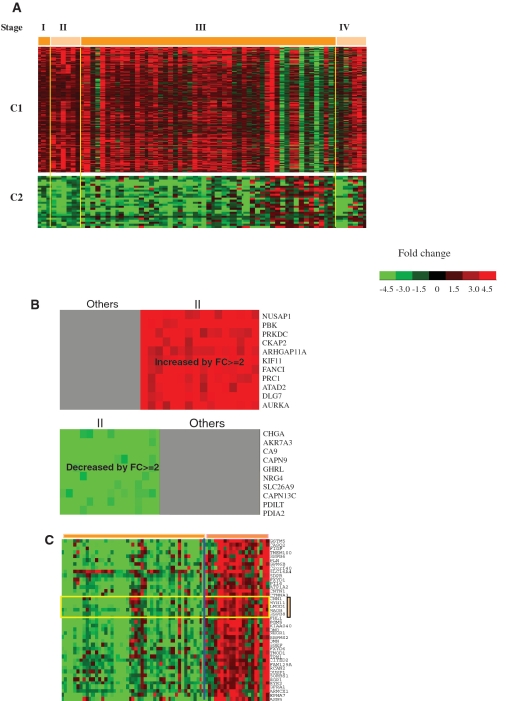
To discover novel associations of gene expression patterns with specific subtype and stage of gastric cancer, we have carried out a bi-clustering analysis of gene-expression data using an in-house program QUBIC (12). Twenty-one statistically significant bi-clusters have been identified which are cancer-, stage- or subtype-specific (Supplementary Table S5). Figure 1A shows two bi-clusters (C1 and C2) with the highest statistical significance among the 21, each representing a group of genes with correlated expressions across a majority of the 80 pairs of samples. Analyses of these two bi-clusters revealed that: (i) genes such as transcriptional regulators, growth factors and enzymes involved in cell-cycle regulation (STMN1 and CDCA8), transcription regulation (TCF19 and BRIP1), angiogenesis (IL8), chromosome integrity (TOP2A) and extracellular matrix remodeling (MMPs) were up-regulated starting at a very early stage of gastric cancer (in C1), while genes involved in metabolism are down-regulated (in C2); the coordinated expression patterns across genes in each of the two bi-clusters may suggest coordinated regulations across these genes; and (ii) most genes in C1 and C2 show discerning power between the cancer and the reference tissues, even at stage I.
Figure 1.
Identified bi-clusters across 80 samples over subsets of genes, where each row represents a gene and each column represent a pair of cancer/reference tissues. (A) C1 shows 244 genes that are consistently up-regulated in cancer versus reference tissues; C2 shows 95 genes, most of which are down-regulated. Note that the order of the tissue samples for different bi-clusters is not necessarily the same since the algorithm rearranges the order of tissue samples. (B) Two clusters specific to stage III, consisting of genes with at least 2-fold expression changes across most of stage III patients but not other stages. (C) A bi-cluster possibly subtype-specific, consisting of 42 genes. The six genes highlighted within orange bars are known to be subtype-associated in gastric cancer.
Some genes were found to exhibit distinct expression patterns specific to different cancer stages. For example, in Figure 1B, two gene clusters show at least 2-fold changes in gene expression across the majority of stage III cancer tissues versus the corresponding references, but not at other stages. This group of genes can serve as potential markers for measuring the progression of gastric cancer.
Another bi-cluster could provide useful information about subtypes. In this bi-cluster containing 42 genes (Figure 1C), 6, namely CNN1, MYH11, LMOD1, MAOB, HSPB8 and FHL1, have been previously reported to be differentially expressed between the intestinal and the diffuse subtypes of gastric cancer (18). Our analysis indicates that these 42 genes can better distinguish the two possible subtypes of gastric cancer (gene names listed in Supplementary Table S5) although further studies on a larger dataset are needed to confirm this suggestion. We noted that none of the 21 co-expression clusters was gender-specific.
Gene signatures for gastric cancer
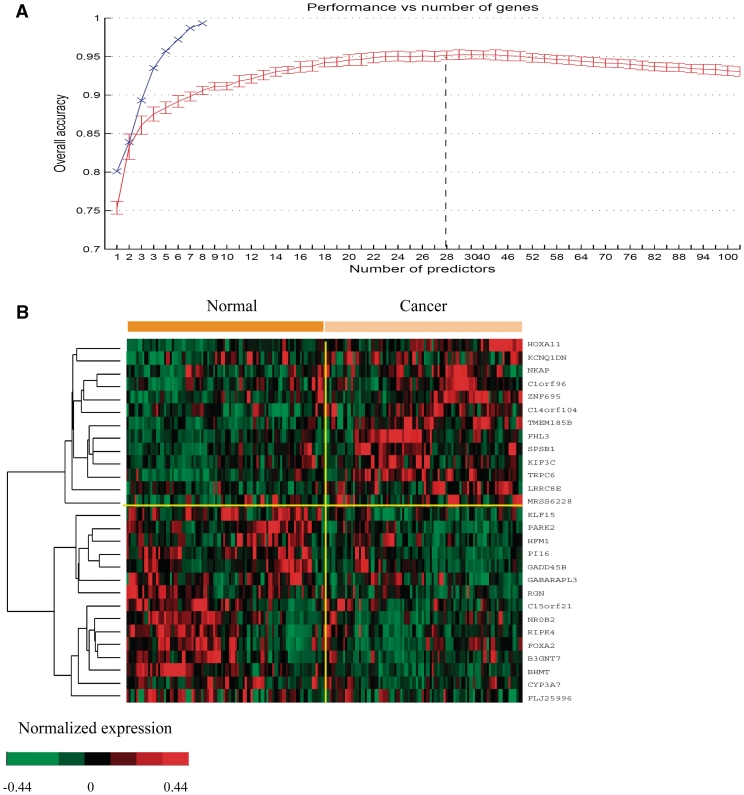
A number of genes were found to have distinguishing expression patterns between the cancer and the reference tissues, based on a classification analysis using RFE-SVM (see ‘Materials and Methods’ section). Figure 2A (red curve) summarizes the classification accuracies for the optimal k-gene combinations (markers) for k from 1 to 100, derived using our gene-signature identification program, where a 28-gene combination gives the best accuracy, having 95.9 and 97.9% agreement with the cancer and reference tissues, respectively (Figure 2B).
Figure 2.
(A) The red curve represents the overall classification accuracies of k-gene markers (k = 1,…, 100), which is the average of the best accuracies of 500 randomly selected subsets; the blue curve represents the best 5-fold cross validation accuracy of k-gene markers (k = 1, 2,…, 8), identified through an exhaustive search. (B) The heatmap for the best 28-gene marker, comprising of 13 up-regulated and 15 down-regulated genes. Among them, NKAP, TMEM185B, C14orf104 and C1orf96 are up-regulated, while KLF15, PI16 and GADD45B are down-regulated across >89% early-stage patients.
Our design of the RFE-SVM-based procedure took into consideration the classification accuracy, stability and reproducibility of the identified signatures, and hence the results are highly robust. A drawback of the method is that it may not necessarily find the best k-gene markers due to its heuristic nature, which allows us to search for large k-gene combinations efficiently. Thus, we have also carried out an exhaustive search for the best k-gene combinations by going through all k-gene combinations for small k values, specifically for all k ≤ 8, which guarantees to find the globally optimal combinations, using a linear SVM approach (34). This method, however, does not scale up to larger k values due to the intrinsic complexity of the exhaustive search. The performance of the identified k-gene markers was evaluated using 5-fold cross validation. In Figure 2A, the best accuracies of the so identified k-gene markers (k = 1, … , 8) are consistently better than those by RFE-SVM. These marker genes are found to be mostly associated with the regulation of cell growth and apoptosis such as cell cycle, ECM-receptor interaction, CDK regulation of DNA replication and the TNFR1 signaling pathway (Supplementary Table S7).
An interesting and unexpected observation was that some markers perform very well on some groups of patients, but not on others. For example, there is a discrepancy between the marker performance for patients of different gender and different ages, which is consistent with our previous observation that age and gender have considerable effects on expressions of some genes. To overcome this problem, separate marker searches were conducted for different genders. The detailed list of the markers so identified are given in Supplementary Table S7, including the top markers LIPG, INHBA and TTYH3 for women and WNT2, CD276 and MFAP2 for men.
A similar analysis on early-stage cancers was also carried out, and a number of promising markers specific to early-stage gastric cancer were identified for k ≤ 4 (Supplementary Table S7). Overall, we found that the best single-gene marker, namely HIST1H3F or CCL20, can have 94.4% classification agreement, 100% for cancer and 88.9% for reference tissues, respectively. This number improves to 97.3% when using the best four-gene combination (GAL3ST4-PPA1-HOXA13-HIST1H3F).
To examine the generality of the predicted gene markers, their classification accuracies were checked on previously published large microarray datasets for gastric cancer. On the Xin dataset (20), the success rates of our k-gene markers range from 81.7 to 100% when k goes from 1 to 7. When evaluated on the early-stage samples from the Kim dataset (18), our single-gene markers such as TFF3, CLDN4, MDK and MUC13 have consistent differential expression patterns across 80% (12 of 15) of the early-stage samples. Overall, these results indicate that our identified tissue markers are generally applicable. In addition, the specificity analysis enabled us to assess the specificity of each predicted marker, through which we obtained a few genes which are highly specific to the gastric cancer such as CKB and ATP4A.
Predicted serum markers for gastric cancer
Out of the 783 genes with at least 2-fold changes in expression levels across all cancer (or early-stage cancer) versus their reference tissues, a total of 136 genes were predicted to be blood-secretory by our program, respectively (17). A number of these proteins could potentially serve as serum markers for gastric cancer, which are grouped into three categories: (i) general markers for gastric cancer, (ii) markers specific to early-stage cancer and (iii) gender-specific markers. Table 2 lists 18 such proteins that we consider as promising serum markers for gastric cancer (details in Supplementary Table S8) based on their fold-change of expression in cancer tissue. Among them, MMP-1, Mucin-13 and cathepsin-B are effective gene discriminators between cancer and reference tissues, but they are not specific to gastric cancer according to our specificity analysis against public microarray data and the previous reports of their over-expression in other cancers such as breast, ovarian, lung and colon cancer (35). Endothelial lipase, gastric lipase, gastrin, gastric intrinsic factor, ghrelin and gastrokine 2 are, however, gastric tissue-specific, thus making them more promising serum markers for gastric cancer, particularly when used in conjunction with other markers.
Table 2.
Eighteen promising predictive markers for gastric cancer
| Serum marker | Stage efficiency |
Gender specificity |
|||
|---|---|---|---|---|---|
| General | Early | Female | Male | ||
| MMP1 | MMP-1, Matrix metalloproteinase 1 preproprotein | √ | |||
| MUC13 | Mucin 13 | √ | |||
| CTSB | Cathepsin B | √ | √ | ||
| GKN2 | Gastrokine 2 | √ | √ | ||
| GHRL | Appetite-regulating hormone (ghrelin) | √ | |||
| LIPF | Gastric triacylglycerol lipase (gastric lipase) | √ | √ | ||
| LIPG | Endothelial lipase | √ | √ | ||
| LIMK1 | LIM domain kinase 1 | √ | † | † | |
| GAST | Gastrin | √ | |||
| GIF | Gastric intrinsic factor | √ | |||
| AZGP1 | Zinc-alpha-2-glycoprotein | √ | √ | ||
| CLDN1 | Claudin-1 | √ | |||
| MDK | Midkine | √ | √ | ||
| TOP2A | Topo-IIA;DNA topoisomerase 2-alpha | √ | √ | ||
| CST1 | cystatin SN | √ | |||
| PDGFRB | PDGFR-β | √ | |||
| PGA4 | Pepsin A | √ | √ | ||
| COL10A1 | Collagen alpha-1(X) | √ | |||
Check mark denotes the condition that a gene shows good classification performance; dagger indicates that a gene has good classification accuracy but is gender-independent.
Experimental validation of the predicted markers
Western blot analysis was conducted to validate the 18 aforementioned proteins on serum samples from nine gastric cancer patients and five controls. Of the 18, 15 were successfully detected, among which 5 proteins, Mucin-13, gastric lipase, Topo IIA, gastrokine 2 and collagen alpha-1(X), show a detectable level of differential abundance between the sera of the cancer patients and the control group, with the first three showing substantial differences (Supplementary Figure S4A). This finding suggests that our prediction approach is sufficiently sensitive to predict potential serum markers.
The validation of the predicted markers was then expanded beyond those 18 proteins by conducting a large-scale screening on the serum samples using MS analyses and Biotin label-based antibody array experiments (36). Considering the sensitivity limitation of MS, the sera from the cancer patients and the controls was pooled, respectively, to enrich the potential markers. Excluding the native high abundance serum proteins (Supplementary Table S9), 81 proteins were identified with high confidence using MS. The Biotin array has 507 human proteins fixed on the array (details in Supplementary Table S10). Together, these two complementary techniques identified 67 out of the 136 computationally predicted potential serum markers, of which 24 proteins were found with differential abundance in sera samples from cancer patients versus the controls (Supplementary Table S10). The differential expressions of eight such proteins are shown in Supplementary Figure S4B, where it can be seen (top panels) that four proteins, EGFR (ErbB1), GRO (CXCL1), IL-1 alpha (IL1A) and osteoprotegerin (TNFRSF11B), are more highly expressed in cancer versus control sera, while the other four proteins (cf. bottom panels), including sFRP-4 (SFRP4), MMP-3 (MMP3), LIF and PARC (CCL18), consistently exhibit decreased abundance in the sera of cancer patients.
Among the other experimentally detected proteins, cathepsin B shows no significant differential abundance across the cancer versus control samples, a finding inconsistent with previous reports proposing it as a potential serum marker (37,38). MMP-1 and Topo-IIA have been previously suggested as cancer related in general (35), which is supported by our data. GKN2 and LIPF are gastric tissue-specific, and collagen alpha-1(X) and gastrin may be associated with other diseases or an immune response in general (details in Supplementary Table S8).
Overall, we have identified two types of potential serum markers: (i) proteins with substantially altered abundance in advanced cancer, including the proteins identified by antibody arrays, such as sFRP-4, MMP-3, LIF, PARC, EGF R, GRO, IL-1 alpha and osteoprotegerin, and three proteins identified by western blot, Mucin-13, gastric lipase and Topo-IIA. Among these potential markers, Mucin-13 functions in several signaling pathways that affect oncogenesis, motility, and cell morphology, and shows increased abundance in the sera of patients with advanced cancer; PARC (CCL18) also shows increased abundance in cancer sera, which is consistent with a previous report on childhood acute lymphoblastic leukemia (39); and (ii) proteins with moderately differential expression in early-stage cancer, like gastrokine 2, with decreased expression in cancer sera, which could be useful for detection of early-stage cancer since the abundance changes in half of the early-stage samples in our test, including one stage-I cancer. Clearly, further test on larger sample sets is needed.
DISCUSSION
Based on a transcriptomic analysis of 80 pairs of gastric cancer versus reference tissues, this study has identified a number of promising marker genes for gastric cancer in general, as well as for early-stage cancer. In addition, we have identified co-expressed gene clusters and enriched pathways. This information provides a basis for our further experimental study aimed to link genes with substantially altered gene expressions to possible mutations in the genomes of the cancer patients.
The exon array data collected through this study has provided wealth of information about splicing variants in gastric cancer. Based on these results, we have identified numerous abnormal splicing variants and abnormally expressed splicing variants in gastric cancer tissues, along with their associated pathways, particularly in early-stage gastric cancer. Our analysis of splicing variants (see details in Supplementary Procedure 4) and the analysis results will be reported elsewhere (manuscript in preparation).
Of the 136 predicted serum marker proteins in our study, 81 were identified in the serum samples by different experimental techniques, and 29 were found to have differential abundance in cancer versus control samples, which is highly encouraging. The discrepancy between the predicted markers and the validated ones is believed to be partially due to the fact that we made the serum marker prediction based on transcriptomic data (in conjunction with blood secretion prediction), rather than on proteomic data of cancer and reference tissues, the two of which are not necessarily always consistent for human proteins. Another possible reason could be due to the low abundance of the predicted marker proteins in sera. In addition, the MS and antibody arrays have detected proteins showing differential abundance in sera of cancer patients and the control group that were not predicted by our computational prediction, which can be attributed to: (i) some of the detected serum proteins with differential abundance may not be blood secretory proteins, instead they might have leaked into circulation; and (ii) the altered protein abundance in cancer versus reference tissues may not necessarily be reflected by the transcriptomic data as discussed above.
Clearly, the true effectiveness of the proposed markers for gastric cancer needs to be validated on larger sample sets, which we plan to do in the near future at the affiliated hospitals of Jilin University College of Medicine. One way to improve the efficacy of the proposed serum markers is to combine individual markers into multi-protein markers as was done for multi-gene markers. While detailed quantitative assessments of multi-protein markers are challenging due to the lack of accurate quantitative measures of these proteins in general, we have evaluated the detection accuracies based on the estimated protein abundance from the western blots. The preliminary results given in Supplementary Table S11 show that a k-protein marker (k ≤ 4) could give much improved detection accuracies than individual serum markers, which suggests a possible direction for further systematic analysis.
CONCLUSION
By integrating transcriptomic analyses, computational prediction and modeling, and proteomic analysis, we have generated a wealth of information on differentially expressed genes in gastric cancer versus reference tissues, their associated biological pathways, marker genes that can well distinguish cancer tissues from the reference tissues, and potential serum protein markers for gastric cancer. Substantial follow-up studies will be conducted to extend the analysis and clarify in greater detail the information generated by this study in revealing information on carcinogenesis and cancer progression. Limited clinical tests will be conducted to assess the effectiveness of the predictive markers based on the 80 pairs of samples investigated. The novel data generated in this study should provide highly useful to other researchers, possibly leading to vastly improved methods for gastric cancer detection and treatment.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This study was supported in part by National Science Foundation (DBI-0354771, ITR-IIS-0407204, CCF-0621700, DBI-0542119), the National Institutes of Health (1R01GM075331), a ‘Distinguished Scholar’ grant from the Georgia Cancer Coalition, and seed funding from the University of Georgia. Funding for open access charge: Research grant (Y.X.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors are grateful to all the individuals who participated in this study. In particular, they thank Drs Xiaoyan Lv, Yang Gao and Hongyu zhang of the Jilin University College of Medicine, for their great efforts in sample collection. They also gratefully acknowledge Dr Guojun Li of Computational and Systems Biology Laboratory and Ms Judy Gray at the University of Georgia for helpful discussions and technical assistance. Their appreciation is also extended to Dr Yan Wang of Jilin University for his help in comparing our predicted markers with microarray data from other human diseases. Last but not the least, they thank all the 80 anonymous patients for voluntarily contributing their tissue and serum samples to support this cancer study.
REFERENCES
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J. Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Lochhead P, El-Omar EM. Gastric cancer. Br. Med. Bull. 2008;85:87–100. doi: 10.1093/bmb/ldn007. [DOI] [PubMed] [Google Scholar]
- 3.Taddei A, Castiglione F, Degl'Innocenti DR, Buccoliero AM, Garbini F, Tommasi C, Freschi G, Bechi P, Messerini L, Taddei GL. NF2 expression levels of gastrointestinal stromal tumors: a quantitative real-time PCR study. Tumori. 2008;94:551–555. doi: 10.1177/030089160809400417. [DOI] [PubMed] [Google Scholar]
- 4.Takeno A, Takemasa I, Doki Y, Yamasaki M, Miyata H, Takiguchi S, Fujiwara Y, Matsubara K, Monden M. Integrative approach for differentially overexpressed genes in gastric cancer by combining large-scale gene expression profiling and network analysis. Br. J. Cancer. 2008;99:1307–1315. doi: 10.1038/sj.bjc.6604682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee HJ, Nam KT, Park HS, Kim MA, Lafleur BJ, Aburatani H, Yang HK, Kim WH, Goldenring JR. Gene expression profiling of metaplastic lineages identifies CDH17 as a prognostic marker in early stage gastric cancer. Gastroenterology. 2010 doi: 10.1053/j.gastro.2010.04.008. 2010, 139, 213–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamada Y, Arao T, Gotoda T, Taniguchi H, Oda I, Shirao K, Shimada Y, Hamaguchi T, Kato K, Hamano T, et al. Identification of prognostic biomarkers in gastric cancer using endoscopic biopsy samples. Cancer Sci. 2008;99:2193–2199. doi: 10.1111/j.1349-7006.2008.00935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L, Li X, Wang GL, Wang Y, Zhu YY, Zhu J. Clinicopathological significance of overexpression of TSPAN1, Ki67 and CD34 in gastric carcinoma. Tumori. 2008;94:531–538. doi: 10.1177/030089160809400415. [DOI] [PubMed] [Google Scholar]
- 8.Xu Y, Zhang L, Hu G. Potential application of alternatively glycosylated serum MUC1 and MUC5AC in gastric cancer diagnosis. Biologicals. 2009;37:18–25. doi: 10.1016/j.biologicals.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 9.Kon OL, Yip TT, Ho MF, Chan WH, Wong WK, Tan SY, Ng WH, Kam SY, Eng A, Ho P, et al. The distinctive gastric fluid proteome in gastric cancer reveals a multi-biomarker diagnostic profile. BMC Med. Genomics. 2008;1:54. doi: 10.1186/1755-8794-1-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernal C, Aguayo F, Villarroel C, Vargas M, Diaz I, Ossandon FJ, Santibanez E, Palma M, Aravena E, Barrientos C, et al. Reprimo as a potential biomarker for early detection in gastric cancer. Clin. Cancer Res. 2008;14:6264–6269. doi: 10.1158/1078-0432.CCR-07-4522. [DOI] [PubMed] [Google Scholar]
- 11.Affymetrix. Alternative Transcript Analysis Methods for Exon Arrays. 2005 Version 1.1 11 October. Affymetrix Santa Clara. [Google Scholar]
- 12.Li G, Ma Q, Tang H, Paterson AH, Xu Y. QUBIC: a qualitative biclustering algorithm for analyses of gene expression data. Nucleic Acids Res. 2009;37:e101. doi: 10.1093/nar/gkp491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 14.Wu J, Mao X, Cai T, Luo J, Wei L. KOBAS server: a web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 2006;34:W720–W724. doi: 10.1093/nar/gkl167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, Buetow KH. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang ZQ, Han LY, Lin HH, Cui J, Jia J, Low BC, Li BW, Chen YZ. Derivation of stable microarray cancer-differentiating signatures using consensus scoring of multiple random sampling and gene-ranking consistency evaluation. Cancer Res. 2007;67:9996–10003. doi: 10.1158/0008-5472.CAN-07-1601. [DOI] [PubMed] [Google Scholar]
- 17.Cui J, Liu Q, Puett D, Xu Y. Computational prediction of human proteins that can be secreted into the bloodstream. Bioinformatics. 2008;24:2370–2375. doi: 10.1093/bioinformatics/btn418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim SY, Kim JH, Lee HS, Noh SM, Song KS, Cho JS, Jeong HY, Kim WH, Yeom YI, Kim NS, et al. Meta- and gene set analysis of stomach cancer gene expression data. Mol. Cells. 2007;24:200–209. [PubMed] [Google Scholar]
- 19.Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X, Leung SY, Yuen ST, Chu KM, Ji J, Li R, Chan AS, Law S, Troyanskaya OG, Wong J, et al. Variation in gene expression patterns in human gastric cancers. Mol. Biol. Cell. 2003;14:3208–3215. doi: 10.1091/mbc.E02-12-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrett T, Suzek TO, Troup DB, Wilhite SE, Ngau WC, Ledoux P, Rudnev D, Lash AE, Fujibuchi W, Edgar R. NCBI GEO: mining millions of expression profiles – database and tools. Nucleic Acids Res. 2005;33:D562–D566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherlock G, Hernandez-Boussard T, Kasarskis A, Binkley G, Matese JC, Dwight SS, Kaloper M, Weng S, Jin H, Ball CA, et al. The Stanford Microarray Database. Nucleic Acids Res. 2001;29:152–155. doi: 10.1093/nar/29.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El-Rifai W, Frierson HF, Jr, Harper JC, Powell SM, Knuutila S. Expression profiling of gastric adenocarcinoma using cDNA array. Int. J. Cancer. 2001;92:832–838. doi: 10.1002/ijc.1264. [DOI] [PubMed] [Google Scholar]
- 25.Becker KF, Atkinson MJ, Reich U, Becker I, Nekarda H, Siewert JR, Hofler H. E-cadherin gene mutations provide clues to diffuse type gastric carcinomas. Cancer Res. 1994;54:3845–3852. [PubMed] [Google Scholar]
- 26.Hippo Y, Taniguchi H, Tsutsumi S, Machida N, Chong JM, Fukayama M, Kodama T, Aburatani H. Global gene expression analysis of gastric cancer by oligonucleotide microarrays. Cancer Res. 2002;62:233–240. [PubMed] [Google Scholar]
- 27.Moss SF, Lee JW, Sabo E, Rubin AK, Rommel J, Westley BR, May FE, Gao J, Meitner PA, Tavares R, et al. Decreased expression of gastrokine 1 and the trefoil factor interacting protein TFIZ1/GKN2 in gastric cancer: influence of tumor histology and relationship to prognosis. Clin. Cancer Res. 2008;14:4161–4167. doi: 10.1158/1078-0432.CCR-07-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu R, Li Z, Bai S, Zhang H, Tang M, Lei Y, Chen L, Liang S, Zhao YL, Wei Y, et al. Mechanism of cancer cell adaptation to metabolic stress: proteomics identification of a novel thyroid hormone-mediated gastric carcinogenic signaling pathway. Mol. Cell Proteomics. 2009;8:70–85. doi: 10.1074/mcp.M800195-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Peduzzi P, Concato J, Feinstein AR, Holford TR. Importance of events per independent variable in proportional hazards regression analysis. II. Accuracy and precision of regression estimates. J. Clin. Epidemiol. 1995;48:1503–1510. doi: 10.1016/0895-4356(95)00048-8. [DOI] [PubMed] [Google Scholar]
- 30.Hirahashi M, Yao T, Matsumoto T, Nishiyama K, Oya M, Iida M, Tsuneyoshi M. Intramucosal gastric adenocarcinoma of poorly differentiated type in the young is characterized by Helicobacter pylori infection and antral lymphoid hyperplasia. Mod. Pathol. 2007;20:29–34. doi: 10.1038/modpathol.3800714. [DOI] [PubMed] [Google Scholar]
- 31.Gajdos C, Tartter PI, Bleiweiss IJ, Bodian C, Brower ST. Stage 0 to stage III breast cancer in young women. J. Am. Coll. Surg. 2000;190:523–529. doi: 10.1016/s1072-7515(00)00257-x. [DOI] [PubMed] [Google Scholar]
- 32.Mintzer D, Glassburn J, Mason BA, Sataloff D. Breast cancer in the very young patient: a multidisciplinary case presentation. Oncologist. 2002;7:547–554. doi: 10.1634/theoncologist.7-6-547. [DOI] [PubMed] [Google Scholar]
- 33.Chandanos E, Lagergren J. Oestrogen and the enigmatic male predominance of gastric cancer. Eur. J. Cancer. 2008;44:2397–2403. doi: 10.1016/j.ejca.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 34.Vapnik V. The Nature of Statistical Learning Theory. New York: Springer-Verlag; 1995. [Google Scholar]
- 35.Poola I, DeWitty RL, Marshalleck JJ, Bhatnagar R, Abraham J, Leffall LD. Identification of MMP-1 as a putative breast cancer predictive marker by global gene expression analysis. Nat. Med. 2005;11:481–483. doi: 10.1038/nm1243. [DOI] [PubMed] [Google Scholar]
- 36.Lin Y, Huang R, Chen LP, Lisoukov H, Lu ZH, Li S, Wang CC, Huang RP. Profiling of cytokine expression by biotin-labeled-based protein arrays. Proteomics. 2003;3:1750–1757. doi: 10.1002/pmic.200300530. [DOI] [PubMed] [Google Scholar]
- 37.Ebert MP, Kruger S, Fogeron ML, Lamer S, Chen J, Pross M, Schulz HU, Lage H, Heim S, Roessner A, et al. Overexpression of cathepsin B in gastric cancer identified by proteome analysis. Proteomics. 2005;5:1693–1704. doi: 10.1002/pmic.200401030. [DOI] [PubMed] [Google Scholar]
- 38.Poon TC, Sung JJ, Chow SM, Ng EK, Yu AC, Chu ES, Hui AM, Leung WK. Diagnosis of gastric cancer by serum proteomic fingerprinting. Gastroenterology. 2006;130:1858–1864. doi: 10.1053/j.gastro.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 39.Struyf S, Schutyser E, Gouwy M, Gijsbers K, Proost P, Benoit Y, Opdenakker G, Van Damme J, Laureys G. PARC/CCL18 is a plasma CC chemokine with increased levels in childhood acute lymphoblastic leukemia. Am. J. Pathol. 2003;163:2065–2075. doi: 10.1016/S0002-9440(10)63564-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.