Abstract
RIG-I recognizes molecular patterns in viral RNA to regulate the induction of type I interferons. The C-terminal domain (CTD) of RIG-I exhibits high affinity for 5′ triphosphate (ppp) dsRNA as well as blunt-ended dsRNA. Structures of RIG-I CTD bound to 5′-ppp dsRNA showed that RIG-I recognizes the termini of dsRNA and interacts with the ppp through electrostatic interactions. However, the structural basis for the recognition of non-phosphorylated dsRNA by RIG-I is not fully understood. Here, we show that RIG-I CTD binds blunt-ended dsRNA in a different orientation compared to 5′ ppp dsRNA and interacts with both strands of the dsRNA. Overlapping sets of residues are involved in the recognition of blunt-ended dsRNA and 5′ ppp dsRNA. Mutations at the RNA-binding surface affect RNA binding and signaling by RIG-I. These results provide the mechanistic basis for how RIG-I recognizes different RNA ligands.
INTRODUCTION
The RIG-I like receptors (RLRs) are cytosolic pattern recognition receptors of viral RNA that mediate the innate anti-viral immune responses (1–5). The RLR family has three proteins, RIG-I, MDA5 and LGP2 (1,3,6). This family of proteins contains a DExD/H-Box RNA helicase domain and a C-terminal RNA-binding domain. Both RIG-I and MDA5 also contain two caspase activation and recruitment domains (CARDs) at their N-termini (7). In contrast, LGP2 lacks the CARDs and associated signaling capability. It regulates RIG-I and MDA5 signaling (8–11). RIG-I and MDA5 sense overlapping sets of viruses (12–14). The essential roles of RIG-I and MDA5 in antiviral immunity were confirmed in gene knock-out mice (12,15).
Since RNA can fold into complicated three-dimensional structures and contain various modifications, especially at their termini, it is not clear what is the exact structural feature of the viral RNA recognized by the RLRs and how RNA binding activates RLR signaling. RNA transcripts containing a 5′ triphosphate (ppp) potently trigger the activation of RIG-I upon transfection into cells (16,17). Recent studies using chemically synthesized dsRNA or hairpin RNA containing 5′ ppp further demonstrated that base pairing near the 5′ ppp is necessary for RIG-I activation (18–20). In addition, the genomic RNA of influenza A and Sendai viruses were identified as physiological agonist of RIG-I (20,21). It was suggested that the panhandle structure of the influenza A virus genomic RNA is likely recognized by RIG-I (20,22). Moreover, blunt-ended dsRNA without 5′ ppp can also activate RIG-I signaling in transfected cells (11,23). To make ligand recognition even more complex, the digestion of the dsRNA mimetic poly (I:C) by RNase III to fragments of <1 kb transforms poly (I:C) from an activator of MDA5 to an activator of RIG-I (24). Consistent with this, short RNA generated by RNase L digestion of cellular RNA induced interferon (IFN) production via RIG-I and MDA5 (25). These results indicated that the 5′ ppp is not absolutely needed for the activation of RIG-I by RNA. Clearly, how RIG-I recognizes such a wide range of ligands need to be better defined.
The CTD of RIG-I is the RNA-binding domain and in vitro binding studies using purified RLR CTDs and RNAs with defined structures have been widely used to elucidate the structural features of RNAs recognized by the RLRs (11,26–30). The RIG-I CTD binds 5′ ppp dsRNA, blunt-ended dsRNA and 5′ ppp ssRNA, with significantly higher affinity for 5′ ppp dsRNA (30). The crystal structure of LGP2 CTD bound to an 8-bp blunt-ended dsRNA provided the first evidence that the termini of dsRNA are recognized by the RLRs (11). The crystal structures of RIG-I CTD bound to a 14-bp GC-rich and a 12-bp AU-rich 5′ ppp dsRNA confirmed that RIG-I CTD also recognizes the termini of 5′ ppp dsRNA (30). A similar mode of RNA binding was observed in the structure of RIG-I CTD bound to a different 12-bp dsRNA (31). These structures showed that the 5′ ppp of the dsRNA is recognized by a cluster of positively charged residues including Lys858, Lys861, Lys888 and His847 (30,31). In contrast, both MDA5 and LGP2 CTDs bind blunt-ended dsRNA with no preference for 5′ ppp dsRNA (11,27). Consistent with this, structures of MDA5 and LGP2 CTDs showed that these two proteins lack the ppp-binding site observed in RIG-I CTD. Comparisons of the structures of RIG-I CTD bound to 5′ ppp dsRNA and LGP2 CTD bound to blunt-ended dsRNA showed that the dsRNA were in dramatically different orientations (30). These observations led us to hypothesize that RIG-I CTD will bind blunt-ended dsRNA and 5′ ppp dsRNA in different ways (30).
To understand the promiscuous RNA binding by RIG-I, we determined the crystal structure of RIG-I CTD bound to a 14-bp blunt-ended dsRNA at 2.4-Å resolution and compared the structure of RIG-I CTD bound to 5′ ppp dsRNA of the same sequence. These two structures revealed that distinct but overlapping sets of residues are involved in the binding of blunt-ended dsRNA and 5′ ppp dsRNA. The orientation of blunt-ended dsRNA relative to RIG-I CTD is dramatically different from that of the 5′ ppp dsRNA. Mutagenesis of key residues at the RNA-binding surface affected RNA binding in vitro and RIG-I signaling in vivo.
MATERIALS AND METHODS
Purification, crystallization and structural determination
Human RIG-I CTD was expressed and purified as described (32). The 14-bp blunt-ended dsRNA was chemically synthesized by IDT (Coralville, IA, USA). The dsRNA was mixed with excess RIG-I CTD and the 2:1 (RIG-I CTD:dsRNA) complexes were purified by gel filtration chromatography on a Superdex75 (1.6 × 60) column (GE Healthcare). The complex was concentrated to ∼20 mg/ml and crystallized in 27% polyethylene glycol 500 monomethyl ether (PEG MME 500) in 0.1 M HEPES buffer at pH 7.5. The crystals were flash frozen in liquid nitrogen. Diffraction data were collected using a Rigaku RAXIS IV++ detector and processed with HKL2000 (32). The complex crystallizes in space group P212121 with cell dimensions: a = 36.99 Å, b = 70.30 Å, c = 125.54 Å. The crystallographic asymmetric unit (ASU) contains one 2:1 (RIG-I CTD:dsRNA) complex. Structures of RIG-I CTD in the complex was determined by molecular replacement with MOLREP (33) using the structure of RIG-I CTD bound to a 14-bp 5′ ppp dsRNA as search model (PDB code: 3LRN). The structural model was rebuilt using O (34). After several rounds of refinement with CNS (35), electron density for the dsRNA became obvious. A 14-bp dsRNA was manually docked into the electron density map and rebuilt with O. The structure was refined using CNS. Statistics of data collection and structural refinement are shown in Table 1. The structural figures were generated with PyMol (http:///www.pymol.org).
Table 1.
Statistics of crystallographic analysis
| Diffraction data | |
| Space group | P212121 |
| Unit cell | a = 36.99 Å, b = 70.30 Å, c = 125.54 Å |
| Asymmetric unit | one 2:1 RIG-I CTD:dsRNA complex |
| Resolution (Å) | 50.0–2.40 (2.49–2.40)a |
| Unique reflections | 13 384 |
| Redundancy | 6.0 (5.8) |
| Completeness | 99.1% (98.0%) |
| <I/σI> | 29.5 (5.3) |
| Rsym(%) | 8.6 (44.8) |
| Refinement | |
| Resolution (Å) | 50–2.4 Å |
| Reflections (F > 0) (total/test set) | 12 860/1329 |
| Protein atoms | 2098 |
| RNA atoms | 596 |
| Zinc ions | 2 |
| Solvent atoms | 133 |
| Rcryst/Rfree (%) | 21.7/27.9 |
| R.m.s.d. bond length | 0.006 Å |
| R.m.s.d. bond angle | 1.32° |
aValues in the parentheses are for the highest resolution shell; 10% of reflections are used in the test set for Rfree calculation.
RNA-binding studies
Different forms of RNA were generated either by chemical synthesis or by in vitro transcription using T7 RNA polymerase. The in vitro transcribed RNA were purified by gel filtration chromatography and analyzed by denaturing polyacrylamide gel electrophoresis. Sequences of all the RNA and DNA oligos used in this study are shown in Supplementary Table S1. Double-stranded RNA was generated by heating the ssRNA at 95°C for 5 min and cooling gradually to room temperature. In the binding studies by gel filtration chromatography, the RNA samples (at ∼100 µM) were mixed with equal volume of RIG-I CTD (at ∼200 µM) and 100 µl of samples were injected over a Superdex200 (10/300 GL) column (GE Healthcare) eluted with a buffer containing 20 mM Tris and 150 mM NaCl at pH 7.5.
In the binding studies by electrophoretic mobility shift assays (EMSAs), each type of RNA was mixed with RIG-I CTD at a molar ratio 1:3 at a final concentration of ∼1.6:5 µM. The mixtures were resolved on 12% polyacrylamide gel and stained with ethidium bromide. Mutants of RIG-I CTD were generated using the Quikchange mutagenesis kit (Stratagene). The mutant proteins were expressed and purified the same way as the wild-type protein. RNA-binding studies for the mutants were conducted by EMSA and gel filtration chromatography.
IFN-β luciferase reporter gene assays
IFN-β luciferase reporter gene assays were used to analyze the activities of three different forms of RNA in stimulating RIG-I signaling in HEK 293T cells according to protocols described previously (30). Briefly, the cells were transfected with plasmid of RIG-I along with IFN-β luciferase reporter plasmid and Renilla luciferase control plasmid. After 24 h, three different forms of RNA (Supplementary Table S1) at 5, 10 and 50 nM concentrations were transfected into the cells to stimulate RIG-I dependent signaling. The ratios of firefly luciferase reporter over Renilla luciferase control were determined. Signaling of RIG-I mutants were analyzed under similar conditions in cells stimulated with these three forms of RNA at a final concentration of 50 nM.
RESULTS
RIG-I CTD binds blunt-ended dsRNA
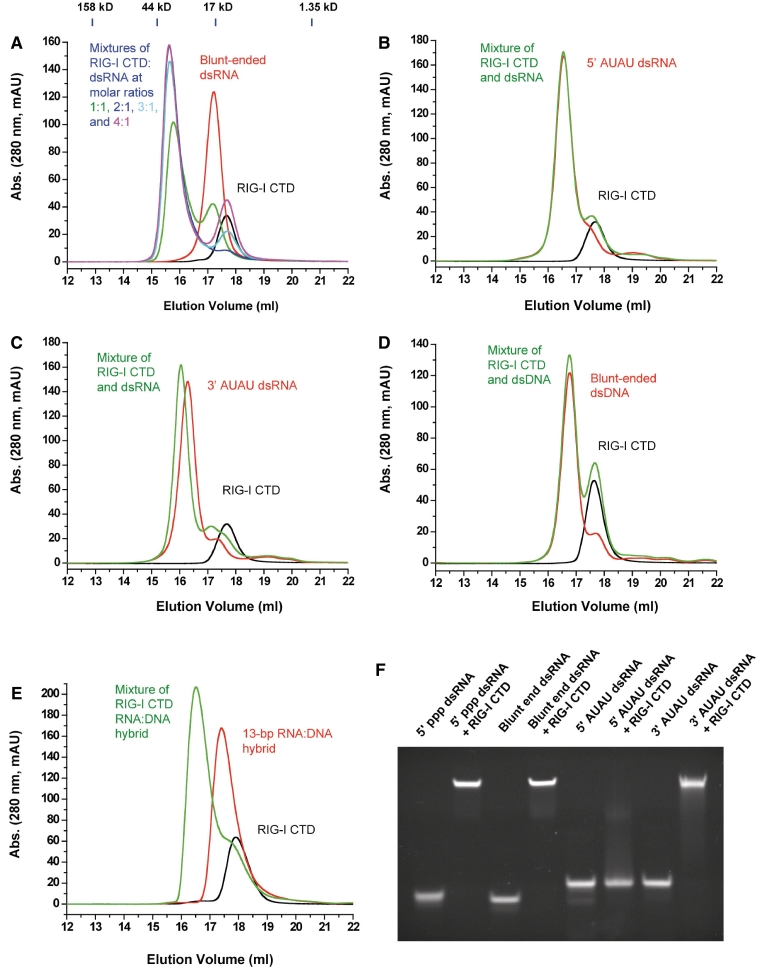
We synthesized a 14-bp blunt-ended dsRNA, two 14-bp dsRNA with either 5′ or 3′ overhangs, a 14-bp dsDNA and a 13-bp RNA:DNA hybrid (Supplementary Table S1) and studied their binding interactions with RIG-I CTD. Gel filtration chromatography showed that RIG-I CTD binds blunt-ended dsRNA and forms a stable complex (Figure 1A). Based on the gel filtration chromatograms of the RIG-I CTD:dsRNA mixtures at different molar ratios (Figure 1A), the stoichiometry between RIG-I CTD and the 14-bp blunt-ended dsRNA is estimated to be 2:1. Binding studies by EMSA also showed that RIG-I CTD binds blunt-ended dsRNA as well as 5′ ppp dsRNA (Figure 1F). However, the non-phosphorylated 14-bp dsRNA with 5′ overhangs failed to bind RIG-I CTD (Figure 1B and F). Although the dsRNA with 3′ overhangs still binds RIG-I CTD by EMSA (Figure 1F), gel filtration chromatography showed it binds RIG-I CTD with reduced affinity and does not form the stoichiometric 2:1 complex (Figure 1C). In addition, gel filtration chromatography showed that RIG-I CTD does not bind a 14-bp dsDNA with the same sequence as the 14-bp blunt-ended dsRNA (Figure 1D). Furthermore, a 13-bp blunt-ended RNA:DNA hybrid also binds RIG-I CTD (Figure 1E). The stoichiometry between RIG-I CTD and the RNA:DNA hybrid is estimated to be 1:1.
Figure 1.
RIG-I CTD binds blunt-ended dsRNA without 5′ ppp. (A) Binding studies of RIG-I CTD with the 14-bp blunt-ended dsRNA by gel filtration chromatography. RIG-I CTD, the 14-bp blunt-ended dsRNA and mixtures of RIG-I CTD:dsRNA at molar ratios 1:1 to 4:1 were injected over a superdex200 column. Elution profile of RIG-I CTD is in black and the dsRNA in red. Elution profiles of mixtures of RIG-I CTD and the 14-bp dsRNA at molar ratios 1:1, 2:1, 3:1 and 4:1 are in green, blue, cyan and purple, respectively. The molecular masses of four protein standards and their elution positions are shown above the chromatograms. (B) Binding studies of RIG-I CTD with a 14-bp dsRNA containing 5′ AUAU overhangs. A 2:1 mixture of RIG-I CTD and the dsRNA was analyzed by gel filtration chromatography. (C) Binding studies of RIG-I CTD with a 14-bp dsRNA containing 3′AUAU overhangs. The molar ratio between RIG-I CTD and the dsRNA is 2:1 in the sample. (D) Binding studies of RIG-I CTD with a 14-bp blunt-ended dsDNA with the same sequence as the 14-bp blunt-ended dsRNA. The molar ratio between RIG-I CTD and the dsDNA is 2:1. (E) Binding studies of RIG-I CTD with a 13-bp RNA:DNA hybrid. The molar ratio between RIG-I CTD and the RNA:DNA hybrid is 2:1. (F) Binding studies of RIG-I CTD with the 14-bp 5′ ppp dsRNA, 14-bp blunt-ended dsRNA and 14-bp dsRNA with either 5′ or 3′ overhangs by EMSA.
Overall structure of RIG-I CTD bound to blunt-ended dsRNA
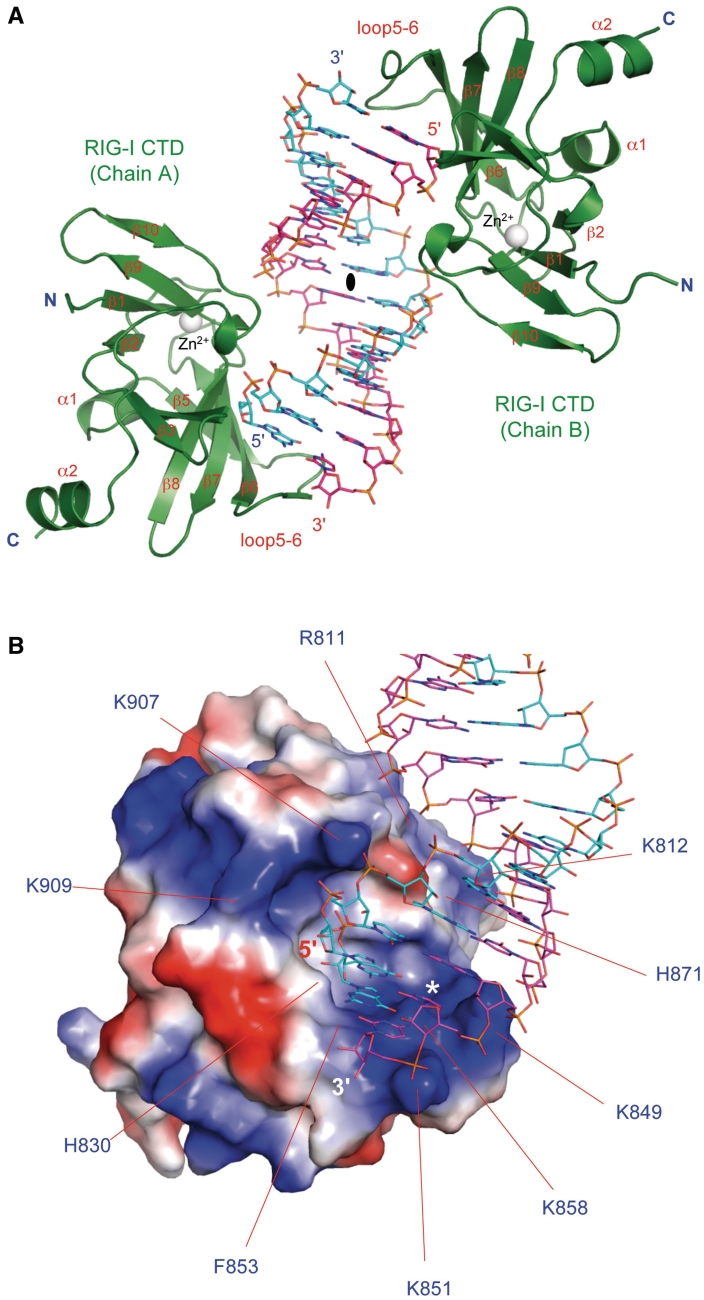
To elucidate the structural basis of blunt-ended dsRNA recognition by RIG-I, we determined the crystal structure of RIG-I CTD bound to a 14-bp blunt-ended dsRNA (Figure 2A and Supplementary Figure S1). The structure showed that one RIG-I CTD bound to each terminus of the blunt-ended dsRNA, forming a complex with pseudo 2-fold symmetry (Figure 2A). Structures of the two RIG-I CTDs in the complex are essentially the same (r.m.s.d. 0.6 Å) and the two RIG-I CTDs bind the dsRNA in similar ways. Discussions of the structure will be based on the interface between RIG-I CTD chain A and the dsRNA due to better-defined electron density throughout this interface.
Figure 2.
Crystal structure of human RIG-I CTD bound to a 14-bp blunt-ended dsRNA. (A) Structure of RIG-I CTD bound to the dsRNA. RIG-I CTDs are shown by the green ribbons. The dsRNA is shown by the sticks representation. Carbon atoms of the two RNA strands are colored cyan and pink, respectively. The zinc ion bound to RIG-I CTD is shown by the gray sphere. The complex exhibits pseudo 2-fold non-crystallographic symmetry. The pseudo 2-fold axis is shown by the black oval. (B) Surface electrostatics of RIG-I CTD. Positively charged surface is colored blue and negatively charged surface red. The blunt-ended dsRNA bound to RIG-I CTD is shown by the stick models. Key residues mediating blunt-ended dsRNA recognition are labeled. Location of the ppp-binding site for 5′ ppp dsRNA is indicated by the white asterisk.
The overall structure of the RIG-I CTD:dsRNA complex looks like two hands holding a stack of chips (Figure 2A). The exposed base pairs at the termini of the dsRNA stack on the loop connecting strands β5 and β6 (loop 5–6), interacting with RIG-I CTD through hydrophobic interactions (Figure 3A). The thumb, corresponding to the loop connecting strand β10 and the C-terminal helix, reaches into the major groove of the dsRNA and interacts with the RNA through electrostatic interactions (Figure 2A). The structure of RIG-I CTD shows shape and charge complementarity with the first-turn of the blunt-ended dsRNA (Figure 2B). The average shape correlation statistics (Sc) shows the two interfaces have an average Sc of 0.67, a better match compared to the interface between RIG-I CTD and 5′ ppp dsRNA (Sc = 0.62); an Sc value of 1.0 indicates a perfect fit between two interacting surfaces (36).
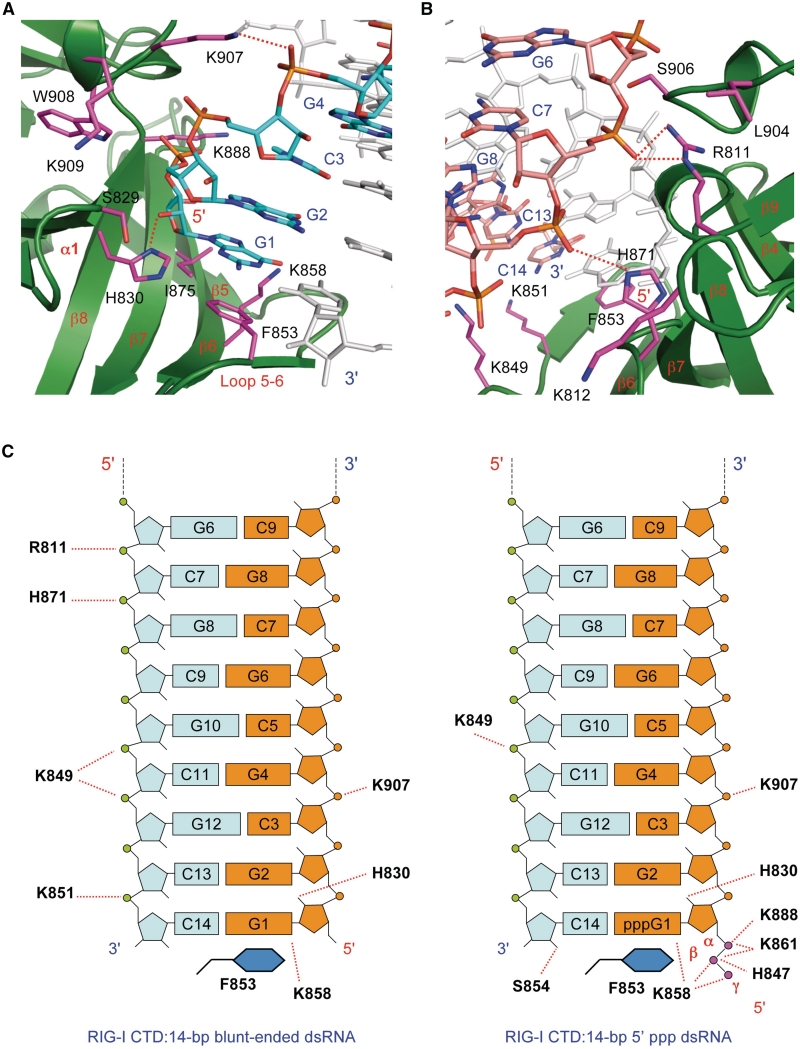
Figure 3.
Structural basis of blunt-ended dsRNA recognition by RIG-I CTD. (A) Interactions between the 5′ 4 nt of the 14-bp blunt-ended dsRNA and RIG-I CTD. The dsRNA is shown by the stick models. Carbon atoms of the RNA strand interacting with RIG-I CTD are colored cyan. The complementary strand is colored light gray. RIG-I CTD is shown by the green ribbons. Key residues involved in RNA binding are shown by the magenta stick models. (B) Interactions between complementary strand and RIG-I CTD. The complementary strand is shown by the stick models with carbon atoms colored pink. (C) Schematic representations of the interactions between RIG-I CTD and the 14-bp blunt-ended dsRNA (left) and a 14-bp 5′ ppp dsRNA (right). Nucleotides in the two dsRNA are labeled the same way for comparison. Interactions between the dsRNA and one of the two RIG-I CTD molecules are shown.
The 5′ 4 nt of the dsRNA make major contributions to RNA binding, contributing to ∼770 Å2 of buried surface area (Figure 3A). However, unlike similar dsRNA with 5′ ppp, the complementary strand also makes significant contribution to binding (buried surface area of ∼540 Å2, Figure 3B and C). The total buried surface area at the blunt-ended dsRNA:RIG-I CTD interface (∼1310 Å2) is slightly larger than that at the 5′ ppp dsRNA: RIG-I CTD interface (∼1200 Å2).
Interactions between RIG-I CTD and the blunt-ended dsRNA
The interaction between the blunt-ended dsRNA and RIG-I CTD contains a mixture of electrostatic, direct and solvent mediated hydrogen bonding, hydrophobic and van der Waals interactions (Supplementary Table S2). The first 4 nt at the 5′ end of the dsRNA interact with RIG-I CTD in a similar manner as dsRNA containing 5′ ppp (Figure 3A) (30). The 2′-hydroxyl of nucleotide G1 forms a hydrogen bond with the side chain of His830 (Figure 3A). This is the only direct contact between the ribose and the protein. This hydroxyl group also forms two solvent-mediated hydrogen bonds with backbone amine and carbonyl groups of Tyr831 and Ile887. This ribose also has van der Waals contacts with two hydrophobic residues Val886 and Ile875. The second nucleotide (G2) interacts with RIG-I CTD through three solvent-mediated hydrogen bonds with residues Ser906, Trp908 and Glu890. G2 also has van der Waals contacts with Lys888 and His830. The third nucleotide C3 interacts with RIG-I CTD mainly through solvent mediated hydrogen bonds between its phosphodiester and the backbone amines of Trp908 and Lys909. The side chain amine of Lys907 interacts with the phosphodiester of G4 through electrostatic interactions (Figure 3A and C). Although similar interactions were also observed in the complex between 5′ ppp dsRNA and RIG-I CTD, the 5′ ppp makes additional electrostatic interactions with RIG-I CTD (Figure 3C).
Several nucleotides from the complementary strand also make significant contributions to blunt-ended dsRNA binding (Figure 3B and C). Key contributions likely come from positively charged Arg811 and His871 that interact with the phosphodiester of C7 and G8 through electrostatic interactions (Figure 3B and C). Notably, these interactions are not observed in the structures of 5′ ppp dsRNA bound to RIG-I CTD (Figure 3C). The side chain of Lys849 is within 4 Å of the phosphodiester of C11 and G12, and might interact with the RNA through additional electrostatic interactions (Figure 3B and C). Moreover, the side chain amine of Lys851 is 3.7 Å away from the phosphate of C14 and interacts with the RNA through electrostatic interactions (Figure 3B and C). Apart from these electrostatic interactions, three solvent-mediated hydrogen bonds are observed between nucleotides C5 and G6 and residues Leu904, Arg811 and Ser906.
The exposed GC base pair at the termini of the dsRNA interacts with RIG-I CTD primarily through hydrophobic interactions by stacking its guanine ring on the phenyl side chain of Phe853 (Figure 3A). The guanine base also forms a hydrogen bond with the side chain of Lys858 (Figure 3A). A purine base at the 5′-terminus of the dsRNA should stack more effectively on the phenyl ring of Phe853 than a pyrimidine base.
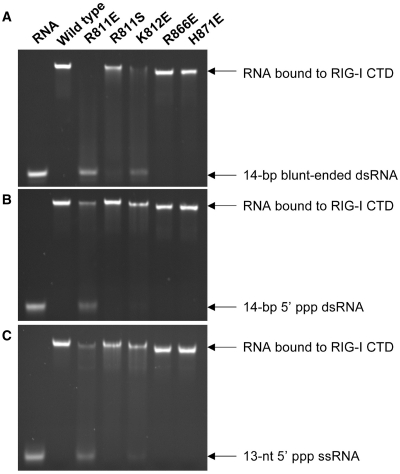
The structure of RIG-I CTD bound to the blunt-ended dsRNA indicates that residues Arg811, Lys812 and His871 may play critical roles in the specific binding of blunt-ended dsRNA. To test this prediction, we purified RIG-I CTD mutants R811E, R811S, K812E, K812S, H871E and conducted RNA-binding studies (Figure 4 and Supplementary Figure S2). Substitution of Arg811 with glutamate almost abolished blunt-ended dsRNA binding and significantly reduced binding to both 5′-ppp dsRNA and ssRNA (Figure 4). Replacement of Arg811 by a hydrophilic serine reduced the binding of blunt-ended dsRNA, but did not affect binding to triphosphorylated dsRNA or ssRNA (Figure 4). Substitution of the nearby Lys812 by glutamate dramatically reduced blunt-ended dsRNA binding, but only slightly reduced binding to triphosphorylated dsRNA or ssRNA (Figure 4). Replacement of Lys812 by a neutral serine residue did not affect RNA binding (Supplementary Figure S3). Surprisingly, the replacement of His871 by glutamate did not affect the binding to all three forms of RNA (Figure 4), suggesting that the His871 makes only minimal contribution to RNA binding. As expected, the replacement of Arg866, a residue not at the RNA-binding surface, by glutamate did not affect RNA binding (Figure 4). The slightly slower migration of the R866E and H871E complex is likely due to the changes of the surface electrostatics of the mutant proteins. In addition, our previous studies showed that mutation K907E disrupted blunt-ended dsRNA as well as 5′-ppp dsRNA binding (30). Together, these results suggest that the electrostatic interactions between Arg811 and Lys907 and the dsRNA are essential for blunt-ended dsRNA binding. Furthermore, previous studies showed that substitution of Phe853 by serine also reduced dsRNA binding (30), suggesting the hydrophobic interaction between Phe853 and the exposed base pair is also needed for effective RNA binding.
Figure 4.
Mutations at the blunt-ended dsRNA-binding surface affect RNA binding by RIG-I CTD. (A) Binding studies of wild-type and mutants of RIG-I CTD with the 14-bp blunt-ended dsRNA by EMSA. (B) Binding studies of RIG-I CTD mutants with a 14-bp 5′ ppp dsRNA. (C) Binding studies of RIG-I CTD mutants with a 13-nt 5′ ppp ssRNA.
Blunt-ended dsRNA and 5′-ppp dsRNA bind RIG-I CTD in different orientations
The structure presented above and the structures of 5′ ppp dsRNA bound to RIG-I CTD (30) revealed that RIG-I CTD binds triphosphorylated and non-phosphorylated dsRNA in dramatically different ways (Figure 5A). The blunt-ended dsRNA needs to swing towards RIG-I CTD by ∼15° relative to 5′-ppp dsRNA to increase its contacts with the protein (Figure 5A). Surprisingly, the interactions between the 5′ 4 nt of the blunt-ended dsRNA and RIG-I CTD are similar to those observed in the structures of 5′-ppp dsRNA bound to RIG-I CTD (30). No rotation along the helical axis of the dsRNA is necessary for RIG-I CTD to interact with these two different forms of dsRNA. It is obvious that the specific interactions between the ppp and RIG-I CTD determine how 5′-ppp dsRNA binds RIG-I CTD, while the interactions between the phosphodiester backbones of the dsRNA and the protein determine how the blunt-ended dsRNA binds RIG-I CTD. Superposition of RIG-I CTDs in the two complexes showed that the structures of RIG-I CTDs are almost the same (r.m.s.d. 0.5 Å, Figure 5A), indicating only minor conformational adjustment of RIG-I CTD is needed to bind these two forms of RNA. These structures show that RIG-I CTD is a versatile RNA-binding module that uses similar binding surfaces to recognize different forms of RNA.
Figure 5.
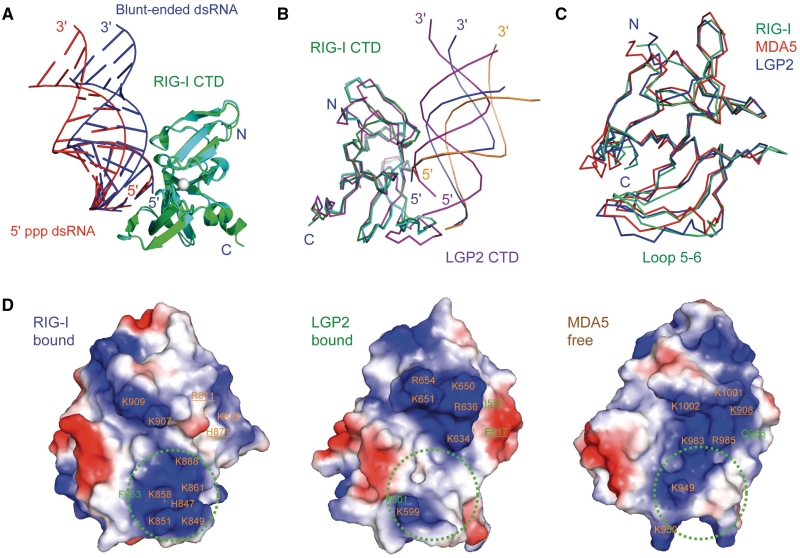
RIG-I and LGP2 CTDs bind blunt-ended dsRNA and 5′ ppp dsRNA differently. (A) Superposition of RIG-I CTD bound to the 14-bp blunt-ended dsRNA and a 14-bp 5′ ppp dsRNA with the same sequence. RIG-I CTDs bound to the blunt-ended dsRNA (blue) and the 5′ ppp dsRNA (red) are shown by the green and cyan ribbons, respectively. (B) Superposition of RIG-CTD bound to the blunt-ended dsRNA and the 5′ ppp dsRNA and LGP2 CTD bound to blunt-ended dsRNA. The blunt-ended dsRNA bound to RIG-I CTD is shown by the blue ribbons. The 5′ ppp dsRNA bound to RIG-I CTD is shown by the orange ribbons. A 14-bp dsRNA (magenta ribbons) is superimposed on the 8-bp dsRNA in the LGP2 CTD:dsRNA complex structure to facilitate comparisons. (C) Superposition of RIG-I (ligand bound), MDA5 (ligand free) and LGP2 (ligand bound) CTD structures. The RNA-binding surfaces of the proteins face the reader. (D) Electrostatics of the RNA-binding surfaces of the RLR CTDs. Positively charged surfaces are colored blue and negatively charged surfaces are red. The ppp-binding site of RIG-I CTD and corresponding regions of LGP2 and MDA5 are indicated by the green circles. Key residues involved in RNA-binding are labeled. Residues of RIG-I CTD interacting with the complementary strand of the dsRNA and corresponding residues in MDA5 and LGP2 CTDs are underlined.
Comparison of the structures of RIG-I and LGP2 CTDs bound to blunt-ended dsRNA showed that the dsRNA bind the two CTDs in significantly different orientations as well (Figure 5B). The interaction between LGP2 CTD and the dsRNA is more extensive (buried surface area of ∼1540 Å2) and shows slightly higher shape complementarity (Sc = 0.70) (11). To bind the LGP2 CTD, the dsRNA needs to rotate along its helical axis and swing towards the protein by ∼15° relative to the position of the blunt-ended dsRNA bound to RIG-I CTD (Figure 5B). Even though the overall structures of RIG-I and LGP2 CTDs bound to dsRNA are similar (r.m.s.d. 1.35 Å), the conformations of loop5–6 that contact the exposed base pairs are quite different (Figure 5B and C). The surface electrostatics of RIG-I and LGP2 CTDs are also dramatically different (Figure 5D). Critical residues Arg811 and His871 involved in blunt-ended dsRNA binding by RIG-I CTD are replaced by hydrophobic Ile557 and negatively charged Glu617 in LGP2 CTD (Figure 5D). These structural differences resulted in the dramatically different mode of blunt-ended dsRNA binding by RIG-I and LGP2 CTDs.
The overall structures of RIG-I, LGP2 and MDA5 CTDs are highly conserved (r.m.s.d. 1.5 Å, Figure 5C). The largest structural differences occur at the N-terminal or C-terminal regions, which are quite flexible (Figure 5C). The loops (loop5–6) connecting strands β5 and β6 also show high degree of flexibility (Figure 5C). The conformation of this loop may determine how the RLRs recognize the exposed base pair at the termini of the dsRNA and likely influence the orientation of the RNA bound to the RLRs. RNA-binding studies by NMR spectroscopy showed that MDA5 CTD uses a similar binding surface as RIG-I and LGP2 to bind blunt-ended dsRNA (27).
Mutations at the RNA-binding surface affect RIG-I signaling
The activities of three different forms of RNA in stimulating RIG-I signaling were examined by IFN-β luciferase reporter gene assays. These studies show that both blunt-ended dsRNA and 5′ ppp dsRNA were capable of activating RIG-I-dependent reporter activities (Figure 6A). However, when the RNAs were used at 5 nM concentrations, 5′ ppp dsRNA shows higher activity than blunt-ended dsRNA or 5′ ppp ssRNA (Figure 6A). Together with the results from our previous studies (30), we demonstrated that blunt-ended dsRNA can stimulate RIG-I signaling in cells.
Figure 6.
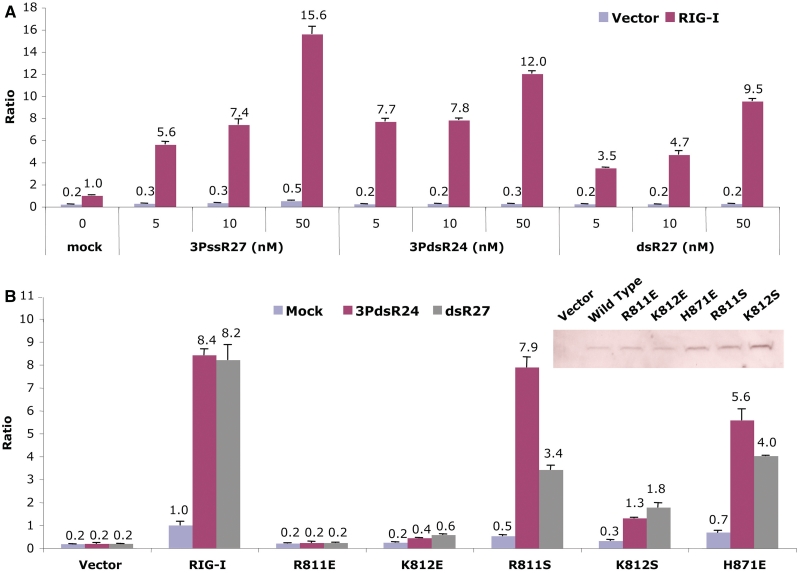
Mutations at the RNA-binding surface affect RIG-I signaling. (A) Effects of structures of the RNA ligands on the activation of IFN-β luciferase production. The assay shows the activities of a 27-nt 5′ ppp ssRNA (3PssR27), a 24-bp 5′ ppp dsRNA (3PdsR24) and a 27-bp blunt-ended dsRNA (dsR27) in stimulating RIG-I signaling in HEK 293T cells. The error bars correspond to the standard deviations of signals from three independent transfections. (B) Effects of mutations at the RNA-binding surface on IFN-β luciferase production in cells transfected with a 27-bp blunt-ended dsRNA and a 24-bp dsRNA with 5′ ppp. Expression of the wild-type and mutants of RIG-I in the transfected cells were confirmed by western blot (inset).
To test how mutations of key residues at the RNA-binding surface affect RIG-I signaling, we generated five mutants in the context of full-length RIG-I and conducted IFN-β luciferase assays. Expressions of wild-type and mutants of RIG-I in the transfected cells were confirmed by western blot (Figure 6B, inset). Substitution of Arg811 with glutamate abolished RIG-I signaling for both blunt-ended dsRNA and 5′ ppp dsRNA (Figure 6B). Although the replacement of Lys812 by glutamate does not disrupt 5′ ppp dsRNA binding by RIG-I CTD, this mutation abolished signaling for both blunt-ended dsRNA and 5′ ppp dsRNA (Figure 6B). However, substitution of His871 with glutamate only reduced RIG-I signaling to 5′ ppp dsRNA by ∼30%, but reduced signaling by blunt-ended dsRNA by ∼50% (Figure 6B). Consistent with results from RNA-binding studies by RIG-I CTD mutants, replacement of the positively charged Arg811 with serine only slightly reduced RIG-I signaling by 5′ ppp dsRNA, but reduced the signaling by blunt-ended dsRNA by ∼60% (Figure 6B). In contrast, a serine substitution of the nearby Lys812 reduced signaling by blunt-ended dsRNA and 5′ ppp dsRNA by ∼80% (Figure 6B). These results demonstrated that the two positively charged residues, Arg811 and Lys812, play critical roles in the recognition of blunt-ended dsRNA by RIG-I. In addition, previous studies demonstrated that substitutions of any of the three positively charged residues, Lys888, Lys907 or Lys909, by glutamate also abolished RIG-I activation by both blunt-ended dsRNA and 5′ ppp dsRNA (30). Moreover, hydrophobic interactions between the exposed base pair and RIG-I CTD are also important for the function of RIG-I; substitution of Phe853 by serine severely decreased RIG-I signaling in response to all the three forms of RNA (30).
DISCUSSION
To elucidate the structural features of viral RNA recognized by RIG-I, we expressed human RIG-I CTD, conducted RNA-binding studies, and in this and previous studies, determined the structures of RIG-I CTD bound to two different forms of RNA. These studies demonstrated that RIG-I CTD binds triphosphorylated dsRNA or ssRNA as well as blunt-ended dsRNA (30). The crystal structures of RIG-I CTD bound to a 14-bp blunt-ended dsRNA and 5′ ppp dsRNA revealed that distinct but overlapping sets of residues contact the RNA (30). Mutations of key residues at the RNA-binding surface differentially affect RNA binding and signaling by RIG-I. These results demonstrated that RIG-I CTD is a versatile RNA-binding module capable of recognizing various RNA structures. The crystal structures of RIG-I, LGP2 and MDA5 CTDs in isolation (27–29) and the structures of RIG-I and LGP2 CTDs bound to RNA (11,30) allowed us to compare the RNA-binding properties of the RLRs.
A key question about the functions of the RLRs is what structural features of RNAs are recognized by these proteins. Biochemical, biophysical and cell-based approaches have been employed to elucidate the structures of RIG-I agonists. Several different techniques such as surface plasmon resonance (SPR), gel filtration chromatography and EMSA have been used to study the RNA-binding properties of RIG-I. These studies clearly showed that RIG-I CTD binds 5′ ppp ssRNA or dsRNA and blunt-ended dsRNA, but does not associate with dsDNA (11,30). Non-phosphorylated dsRNA with 5′ overhangs failed to bind RIG-I CTD, while dsRNA with 3′ overhangs retained some binding. In contrast, triphosphorylated dsRNA with either 3′ or 5′ overhangs still bind RIG-I CTD (30). Although the RNA:DNA hybrid still binds RIG-I CTD, it forms a 1:1 instead of a 1:2 complex with RIG-I CTD, suggesting that RIG-I CTD only recognizes one terminus of the hybrid with the RNA at the 5′-end. It is likely that RIG-I CTD does not bind the terminus of the hybrid with the DNA at the 5′-end. The affinity of 5′ ppp dsRNA (∼0.3 nM) for RIG-I CTD is higher than that of 5′ ppp ssRNA (∼5 nM) or blunt-ended dsRNA (∼8 nM) (30). The affinities of RIG-I CTD for blunt-ended dsRNA and 5′ ppp dsRNA determined by isothermal titration calorimetry (ITC) are significantly lower than those determined by SPR (31). This is likely due to the techniques used and the salt concentrations in the samples. These studies demonstrated that blunt-ended dsRNA and triphosphorylated dsRNA or ssRNA are all ligands for RIG-I. Consistent with these results, all three forms of RNA activate RIG-I signaling in cells (20,37).
The structures of RIG-I CTD bound to blunt-ended dsRNA and 5′ ppp dsRNA provided insight into how RIG-I recognizes different forms of RNA. The two structures revealed that overlapping sets of residues are involved in the recognition of these two forms of RNA (30). The first 4 nt at the 5′ end of triphosphorylated dsRNA make major contributions to 5′ ppp dsRNA binding. The ppp interacts with a cluster of positively-charged residues including Lys851, Lys858, Lys861, His847 and Lys888 through multiple electrostatic interactions (30). The phosphodiester backbone makes additional interactions with the protein through a combination of electrostatic, hydrogen bonding and van der Waals interactions. Based on the structure of RIG-I CTD bound to 5′ ppp dsRNA, removal of the ppp should dramatically reduce dsRNA binding (30). However, RIG-I CTD can bind blunt-ended dsRNA without ppp with nanomolar affinity (30). This is likely achieved through additional electrostatic interactions between the complementary strand and Arg811 and His871 in RIG-I CTD. Consistent with this observation, replacement of Arg811 with glutamate had severe effects on blunt-ended dsRNA binding, but had a more modest effect on the binding of triphosphorylated dsRNA or ssRNA binding. Moreover, substitution of R811 with serine only reduced the signaling of blunt-ended dsRNA.
The 2′-hydroxyl of the ribose makes limited contributions to dsRNA binding. Only the 2′-hydroxyl of the 5′-terminal nucleotide is involved in direct and solvent mediated hydrogen bonds with RIG-I CTD. Modification of this hydroxyl by methylation reduced the activity of 5′ ppp dsRNA in stimulating RIG-I signaling (31). It is likely the steric hindrance caused by this modification reduced RNA binding. However, it is not clear why methylation of the 2′-hydroxyl of the second nucleotide also reduced RIG-I signaling (31), since this ribose makes no direct contact with RIG-I CTD.
RIG-I CTD in complex with the blunt-ended dsRNA crystallized in a different crystal form compared to the complex with corresponding 5′ ppp dsRNA. The blunt-ended dsRNA complex did not crystallize under the conditions of the 5′ ppp dsRNA complex, and the 5′ ppp dsRNA complex did not crystallize under the conditions for the blunt-ended dsRNA complex. The crystal packings for the two complexes are significantly different. This suggested the two forms of dsRNA bind RIG-I CTD in different orientations in solution. Although the ppp can be flexible in solution, it has to adopt a specific conformation to bind RIG-I effectively, so it is unlikely that 5′ ppp dsRNA will bind RIG-I CTD in a similar orientation as the blunt-ended dsRNA. This orientation may reduce the specific interactions between the ppp and RIG-I CTD.
Crystal structures of RIG-I and LGP2 CTDs bound to dsRNA showed that the two proteins use similar binding surfaces to interact with RNA (Figure 5B). Results from NMR titration of MDA5 CTD with an 8-bp blunt-ended dsRNA suggested that similar binding surface is involved in RNA binding by MDA5 (27,38). Like RIG-I, both MDA5 and LGP2 CTD bind to the termini of the blunt-ended dsRNA (11,27). Some major differences in RNA binding by the three RLRs should be emphasized, however. Although LGP2 CTD binds dsRNA with 3′ overhangs, it does not bind dsRNA with 5′ overhangs (11). MDA5 CTD only binds blunt-ended dsRNA but not dsRNA with 5′ or 3′ overhangs (27). The RNA-binding surfaces of the three proteins exhibit dramatically different electrostatic potential (Figure 5D) that should contribute to different modes of protein:RNA interactions. The cluster of positively charged residues around Lys861 and Lys858 involved in ppp recognition is only observed at the RNA-binding surface of RIG-I (Figure 5D), explaining why both LGP2 and MDA5 CTDs have no preference for RNA ligands with 5′ ppp (27,30). The differences in surface electrostatics also explain why RIG-I and LGP2 CTDs bind dsRNA in different ways. The differences should contribute to the function of these three innate immune receptors in the detection of viral infections.
ACCESSION NUMBERS
The atomic coordinates and structure factors of RIG-I CTD bound to the 14-bp blunt-ended dsRNA have been deposited with the RCSB Protein Data Bank under the accession codes: 3OG8.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of Allergy and Infectious Diseases (Grant AI087741 to P.L. and Grant AI073335 to C.K.); the Robert Welch Foundation (Grant A-1687 to P.L.). Funding for open access charge: National Institute of Health (grant number AI87741 to P.L.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 2.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 3.Yoneyama M, Fujita T. RNA recognition and signal transduction by RIG-I-like receptors. Immunol. Rev. 2009;227:54–65. doi: 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]
- 4.Pichlmair A, Reis e Sousa C. Innate recognition of viruses. Immunity. 2007;27:370–383. doi: 10.1016/j.immuni.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 5.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 6.Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M, Jr, Akira S, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 7.Yoneyama M, Fujita T. Function of RIG-I-like receptors in antiviral innate immunity. J. Biol. Chem. 2007;282:15315–15318. doi: 10.1074/jbc.R700007200. [DOI] [PubMed] [Google Scholar]
- 8.Rothenfusser S, Goutagny N, DiPerna G, Gong M, Monks BG, Schoenemeyer A, Yamamoto M, Akira S, Fitzgerald KA. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J. Immunol. 2005;175:5260–5268. doi: 10.4049/jimmunol.175.8.5260. [DOI] [PubMed] [Google Scholar]
- 9.Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, Tsujimura T, Fujita T, Akira S, Takeuchi O. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc. Natl Acad. Sci. USA. 2010;107:1512–1517. doi: 10.1073/pnas.0912986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Venkataraman T, Valdes M, Elsby R, Kakuta S, Caceres G, Saijo S, Iwakura Y, Barber GN. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J. Immunol. 2007;178:6444–6455. doi: 10.4049/jimmunol.178.10.6444. [DOI] [PubMed] [Google Scholar]
- 11.Li X, Ranjith-Kumar CT, Brooks MT, Bharmaiah S, Herr AB, Kao C, Li P. The RIG-I like receptor LGP2 recognizes the termini of double-stranded RNA. J. Biol. Chem. 2009;284:13881–13891. doi: 10.1074/jbc.M900818200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 13.Takeuchi O, Akira S. Innate immunity to virus infection. Immunol. Rev. 2009;227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCartney SA, Colonna M. Viral sensors: diversity in pathogen recognition. Immunol. Rev. 2009;227:87–94. doi: 10.1111/j.1600-065X.2008.00726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, Colonna M. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl Acad. Sci. USA. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 17.Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 18.Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, Coch C, Janke M, Mihailovic A, Wardle G, et al. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt A, Schwerd T, Hamm W, Hellmuth JC, Cui S, Wenzel M, Hoffmann FS, Michallet MC, Besch R, Hopfner KP, et al. 5′-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc. Natl Acad. Sci. USA. 2009;106:12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlee M, Hartmann G. The chase for the RIG-I ligand-recent advances. Mol. Ther. 2010;18:1254–1262. doi: 10.1038/mt.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rehwinkel J, Tan CP, Goubau D, Schulz O, Pichlmair A, Bier K, Robb N, Vreede F, Barclay W, Fodor E, et al. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell. 2010;140:397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 22.Fujita T. A nonself RNA pattern: tri-p to panhandle. Immunity. 2009;31:4–5. doi: 10.1016/j.immuni.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 23.Marques JT, Devosse T, Wang D, Zamanian-Daryoush M, Serbinowski P, Hartmann R, Fujita T, Behlke MA, Williams BR. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat. Biotechnol. 2006;24:559–565. doi: 10.1038/nbt1205. [DOI] [PubMed] [Google Scholar]
- 24.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malathi K, Dong B, Gale M, Jr, Silverman RH. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahasi K, Yoneyama M, Nishihori T, Hirai R, Kumeta H, Narita R, Gale M, Jr, Inagaki F, Fujita T. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol. Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 27.Li X, Lu C, Stewart M, Xu H, Strong RK, Igumenova T, Li P. Structural basis of double-stranded RNA recognition by the RIG-I like receptor MDA5. Arch. Biochem. Biophys. 2009;488:23–33. doi: 10.1016/j.abb.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 28.Cui S, Eisenacher K, Kirchhofer A, Brzozka K, Lammens A, Lammens K, Fujita T, Conzelmann KK, Krug A, Hopfner KP. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 29.Pippig DA, Hellmuth JC, Cui S, Kirchhofer A, Lammens K, Lammens A, Schmidt A, Rothenfusser S, Hopfner KP. The regulatory domain of the RIG-I family ATPase LGP2 senses double-stranded RNA. Nucleic Acids Res. 2009;37:2014–2025. doi: 10.1093/nar/gkp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu C, Xu H, Ranjith-Kumar CT, Brooks MT, Hou YT, Hu F, Herr AB, Strong RK, Kao C, Li P. The structural basis of 5′ triphosphate double-stranded RNA recognitiion by RIG-I C-terminal domain. Structure. 2010;18:1032–1043. doi: 10.1016/j.str.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Ludwig J, Schuberth C, Goldeck M, Schlee M, Li H, Juranek S, Sheng G, Micura R, Tuschl T, et al. Structural and functional insights into 5′-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat. Struct. Mol. Biol. 2010;17:781–787. doi: 10.1038/nsmb.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otwinowski Z, Minor W. Processing of X-ray diffraction data by collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 33.CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 34.Jones TA, kjeldgaard M. Electron-density map interpretation. Methods Enzymol. 1997;277:173–208. doi: 10.1016/s0076-6879(97)77012-5. [DOI] [PubMed] [Google Scholar]
- 35.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 36.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 37.Schlee M, Hartmann E, Coch C, Wimmenauer V, Janke M, Barchet W, Hartmann G. Approaching the RNA ligand for RIG-I? Immunol. Rev. 2009;227:66–74. doi: 10.1111/j.1600-065X.2008.00724.x. [DOI] [PubMed] [Google Scholar]
- 38.Takahasi K, Kumeta H, Tsuduki N, Narita R, Shigemoto T, Hirai R, Yoneyama M, Horiuchi M, Ogura K, Fujita T, et al. Solution structures of cytosolic RNA sensors MDA5 and LGP2 C-terminal domains: Identification of the RNA recognition loop in RIG-I like receptors. J. Biol. Chem. 2009;284:17465–17474. doi: 10.1074/jbc.M109.007179. [DOI] [PMC free article] [PubMed] [Google Scholar]