Abstract
Like bacterial genes, most plastid (chloroplast) genes are arranged in operons and transcribed as polycistronic mRNAs. Plastid protein biosynthesis occurs on bacterial-type 70S ribosomes and translation initiation of many (but not all) mRNAs is mediated by Shine-Dalgarno (SD) sequences. To study the mechanisms of SD sequence recognition, we have analyzed translation initiation from mRNAs containing multiple SD sequences. Comparing translational efficiencies of identical transgenic mRNAs in Escherichia coli and plastids, we find surprising differences between the two systems. Most importantly, while internal SD sequences are efficiently recognized in E. coli, plastids exhibit a bias toward utilizing predominantly the 5′-most SD sequence. We propose that inefficient recognition of internal SD sequences provides the raison d'être for most plastid polycistronic transcripts undergoing post-transcriptional cleavage into monocistronic mRNAs.
INTRODUCTION
Plastids are descendants of formerly free-living cyanobacteria and, by and large, have retained the prokaryotic gene expression machinery of eubacteria. Most plastid genes are arranged in operons, which are transcribed as polycistronic mRNAs. Translation occurs on bacterial-type 70S ribosomes with the help of the typical set of prokaryotic initiation, elongation and termination factors (1–5). Likewise, the mechanisms of RNA degradation and protein degradation in plastids very much resemble those in eubacteria (6–8).
Plastid gene expression is predominantly controlled at the post-transcriptional level (9,10) and especially at the level of translation (11), although some transcriptional regulation is also known to occur (12–14). Translational regulation is mainly exerted by controlling the translation initiation process via an interaction of sequence elements in the 5′-untranslated region (5′-UTR) of the mRNA and nuclear-encoded translation factors, many of which seem to be mRNA specific (15–18). Ribosome recruitment to the mRNA is often mediated by bacterial-type ribosome-binding sites, which exhibit sequence complementarity to the 3′-end of the 16S ribosomal RNA and are also referred to as Shine-Dalgarno sequences (SD sequences; refs. 3,19). However, not all 5′-UTRs of plastid mRNAs harbor canonical SD sequences in the conserved spacing of 4–9 nt upstream of the start codon (20) and it has been suggested that, in these cases, mRNA-specific translational activator proteins bind to the 5′-UTR and directly guide the ribosomal 30S subunit to the initiation codon (21) or mediate sliding of the ribosome to the next AUG downstream (22). For several plastid mRNAs that possess canonical SD sequences, the function of these sequence elements as ribosome-binding sites was directly demonstrated by mutagenesis experiments (23–25). Recently, strong synthetic or bacteriophage-derived SD sequences have also been employed in chloroplast biotechnology to maximize transgene expression from the plastid genome (26–28).
While in bacteria most polycistronic mRNAs are directly translated, many primary polycistronic transcripts in plastids undergo post-transcriptional cleavage into monocistronic or oligocistronic units, a mechanism referred to as intercistronic processing or RNA cutting (29). Intercistronic processing is believed to be mediated by specific endoribonucleases that may recognize a combination of primary sequence and RNA secondary structure to initiate RNA cleavage at highly specific sites (30–33). At least some polycistronic transcripts are not translatable and therefore, endonucleolytic processing can be a prerequisite for protein biosynthesis to occur. For example, translation of the dicistronic psaC-ndhD precursor transcript is impeded by RNA secondary structure formation between a 8 nt sequence motif within the psaC coding region and a complementary sequence in the 5′-UTR of the downstream ndhD cistron (34). Additional support for translation being dependent on RNA cutting has come from the analysis of nuclear mutants defective in specific intercistronic cleavage reactions. The maize crp1 mutant is defective in intercistronic processing between the petB and petD cistrons and displays a concomitant loss of petD translation (35,36), indicating that the petD mRNA needs to be monocistronic to be translatable. In the Arabidopsis hcf107 mutant, defective processing of the psbH mRNA from the primary pentacistronic transcript of the psbB operon results in loss of psbH translation (37). Similarly, impaired intercistronic RNA processing between the rps7 and ndhB cistrons in the Arabidopsis crr2 mutant leads to loss of the plastid NADH dehydrogenase complex, most probably because the unprocessed ndhB mRNA is not translated (38). In contrast, some polycistronic mRNAs in plastids are translatable and do not undergo post-transcriptional cleavage into monocistronic units. For example, the psbE operon encoding four small genes for subunits of photosystem II (psbE, psbF, psbL and psbJ; ref. 39) is transcribed as a single 1.1 kb mRNA species which remains tetracistronic and is not further processed. Likewise, the psaA/B transcript (encoding the two reaction center proteins of photosystem I, PsaA and PsaB) is not cleaved between the psaA and psaB reading frames (40), indicating that translation must occur from unprocessed transcripts. Unprocessed polycistronic mRNAs often have canonical SD sequences upstream of each individual cistron, suggesting that internal translation initiation can occur.
Why many plastid polycistronic transcripts are processed and at least some must be processed to become translatable is currently not known. Also, how SD sequences are recognized and what determines the efficiency of their utilization is not well understood. Here, we have (i) analyzed the effects of multiple SD sequences on translation initiation efficiency and (ii) tested whether recognition of downstream SD sequences in polycistronic transcripts limits the translation initiation process in plastids. Comparing identical mRNAs in Escherichia coli and plastids, we show that internal SD sequences are less efficiently recognized in plastids than in bacteria. Our data also suggest ways how to manipulate translation initiation efficiency for reverse genetics and transgene expression in plastids.
MATERIALS AND METHODS
Plant material and growth conditions
Tobacco plants (Nicotiana tabacum cv. Petit Havana) were grown under standard greenhouse conditions. Aseptically grown tobacco plants were raised on agar-solidified MS medium containing 30 g l−1 sucrose (41) under a 16:8 day–night cycle. Regenerated shoots from transplastomic lines were rooted and propagated on the same medium. Rooted homoplasmic plants were transferred to soil and grown to maturity in the greenhouse under standard conditions (25°C). Plant material for isolation of nucleic acids and protein was raised from seeds in the greenhouse. After 6 weeks of growth, the first fully expanded leaf from the top was harvested.
Construction of transformation vectors
All constructs are based on the previously described plastid transformation vector pRB95 (42). A GFP expression cassette was constructed in pBluescript SK(+) (Stratagene, La Jolla, CA, USA) by fusing the coding region with the strong rRNA operon promoter and a prfA 5′-UTR-derived leader sequence (43) at the 5′-end and the rps16 3′-UTR from tobacco plastids (44) at the 3′-end. To facilitate manipulation of the 5′-UTR sequence, a unique recognition site for the restriction enzyme BamHI was incorporated (Supplementary Table S1; Table 1). The final cassette was integrated into the multiple cloning site of pRB95 as SacI/HindIII fragment generating plasmid pRB95-GFP.
Table 1.
Sequences of the translation initiation signals in pOD vectors
| Vector | Description | Sequence |
|---|---|---|
| pOD1 | Reference construct | GGATCCAAATACTGCAGTTTAACTTTAAGAAGGAGATATACCCATGG |
| pOD2 | Two spaced SD-sequences | GGATCCAAATACTGCAGCTTAGAATATTGAAGGAGATATACCCATCTATTATAAATAGTGCAGTTTAACTTTAA GAAGGAGATATACCCATGG |
| pOD3 | Three spaced SD-sequences | GGATCCAAATACTGCAGAGATACAATAAGAAGGAGATATACCCATCTGTATTAAATAGTGCAGCTTAGAATATT GAAGGAGATATACCCATCTATTATAAATAGTGCAGTTTAACTTTAAGAAGGAGATATACCCATGG |
| pOD4 | One start-stop without spacer | GGATCCAAATACTGCAGTTTAACTTTAAGAAGGAGATATACATATGTAATAAGAAGGAGATATACCCATGG |
| pOD5 | Two start-stop without spacers | GGATCCAAATACTGCAGTTTAACTTTAAGAAGGAGATATACATATGTAGTAAGAAGGAGATATACATATGTAATAA GAAGGAGATATACCCATGG |
| pOD6 | Two SD-sequences without spacer | GGATCCAAATACTGCAGTTTAACTTTAAGAAGGAGATAGAAGGAGATATACCCATGG |
| pOD7 | Three SD-sequences without spacers | GGATCCAAATACTGCAGTTTAACTTTAAGAAGGAGATAGAAGGAGATAGAAGGAGATATACCCATGG |
| pOD8 | One mini-ORF without spacer | GGATCCAAATACTGCAGTTTAACTTTAAGAAGGAGATATACATATGGGTTAATAAGAAGGAGATATACCCATGG |
| pOD9 | Two mini-ORFs without spacers | GGATCCAAATACTGCAGTTTAACTTTAAGAAGGAGATATACATATGGGATAGTAAGAAGGAGATATACATATGGGTTAATAAGAAGGAGATATACCCATGG |
| pOD11 | Two non-spaced translation initiation sites; peptidase cleavage site | GGATCCAAATACTGCAGTTTAACTTTAAGAAGGAGATATACATATGGGAGAAGGAGATATACCCATGggtgaaaatttatattttcaaagcatgg |
| pOD12 | Three non-spaced translation initiation sites; peptidase cleavage site | GGATCCAAATACTGCAGTTTAACTTTAAGAAGGAGATATACATATGGGAGAAGGAGATATACATATGGGA GAAGGAGATATACCCATGggtgaaaatttatattttcaaagcatgg |
| pOD13 | Reference construct; peptidase cleavage site | GGATCCAAATACTGCAGTTTATCTTTATGAAGGAGATATACCCATGggtgaaaatttatattttcaaagcatgg |
| pOD14 | Two spaced translation initiation sites; peptidase cleavage site | GGATCCAAATACTGCAGCTTACAATATTGAAGGAGATATACATATGGGTATTATAAATAGTGCAGTTTATCTTTAT GAAGGAGATATACCCATGggtgaaaatttatattttcaaagcatgg |
| pOD15 | Three spaced translation initiation sites; peptidase cleavage site | GGATCCAAATACTGCAGCTTACAATATTGAAGGAGATATACATATGGGTATTATAAATAGTGCAGCTTACAATATT GAAGGAGATATACATATGGGTATTATAAATAGTGCAGTTTATCTTTATGAAGGAGATATACCCATGggtgaaaatttatattttcaaagcatgg |
| pOD17 | Two non-spaced translation initiation sites; peptidase cleavage site | GGATCCAAATACTGCAGTTTAACTTTAAGAAGGAGATATACATATGGGAGAAGGAGATATACCCATGG |
| pOD18 | Three non-spaced translation initiation sites; peptidase cleavage site | GGATCCAAATACTGCAGTTTAACTTTAAGAAGGAGATATACATATGGGAGAAGGAGATATACATATGGGA GAAGGAGATATACCCATGG |
| pOD19 | Reference construct without peptidase cleavage site | GGATCCAAATACTGCAGTTTATCTTTATGAAGGAGATATACCCATGG |
| pOD20 | Two spaced translation initiation sites | GGATCCAAATACTGCAGCTTACAATATTGAAGGAGATATACATATGGGTATTATAAATAGTGCAGTTTATCTTTAT GAAGGAGATATACCCATGG |
| pOD21 | Three spaced translation initiation sites | GGATCCAAATACTGCAGCTTACAATATTGAAGGAGATATACATATGGGTATTATAAATAGTGCAGCTTACAATATT GAAGGAGATATACATATGGGTATTATAAATAGTGCAGTTTATCTTTATGAAGGAGATATACCCATGG |
| pOD22 | Four spaced translation initiation sites; peptidase cleavage site | GGATCCAAATACTGCAGCTTACAATATTGAAGGAGATATACATATGGGTATTATAAATAGTGCAGCTTACAATATT GAAGGAGATATACATATGGGTATTATAAATAGTGCAGCTTACAATATTGAAGGAGATATACATGGTATTATAAATAGTGCAGTTTATCTTTATGAAGGAGATATACCCATGggtgaaaatttatattttcaaagcatgg |
Italic letters indicate restriction sites (BamHI, PstI and NcoI). Underlined letters indicate SD-sequences. Bold letters indicate start codons, stop codons and mini-ORFs. Lower case letters indicate TEV peptidase cleavage site.
Vectors of the pOD series were produced by ligating double-stranded oligonucleotides into restriction-enzyme digested plasmid DNA with compatible ends (Supplementary Table S1; Table 1). All oligonucleotide sequences are based on the SD sequence of the phage gene 10 leader and replace the original prfA 5′-UTR-derived leader sequence. Briefly, complementary oligonucleotides were mixed in equimolar amounts (50 µM each), heated to 94°C and annealed by cooling to 20°C (at 3.6 K/min). To generate vector pOD1, plasmid pRB95-GFP was digested with BamHI and NcoI, followed by ligation to the appropriate double-stranded oligonucleotide (Supplementary Table S1; Table 1) with compatible ends (5′-BamHI overhang, 3′-NcoI overhang). Vector pOD2 was produced from pOD1 cleaved with BamHI and PstI. Analogously, pOD3 was produced from pOD2. Similar to pOD2, vectors pOD4-9 were produced from pOD1 (cut with PstI and NcoI).
Likewise, the tobacco etch virus (TEV) peptidase cleavage site was inserted as double-stranded oligonucleotide (into the unique NcoI site of pOD1) generating plasmid pOD10. pOD10 was then used to produce vectors pOD11 and pOD12 (Supplementary Table S1; Table 1). pOD13 was also derived from pOD10 (by exchanging the 5′-UTR after cleavage with BamHI and NcoI). Vector pOD14 was produced from pOD13 (cut with BamHI and PstI) and, similarly, pOD15 was derived from pOD14 and pOD22 from pOD15 (Supplementary Table S1; Table 1). Plasmids pOD11-15 and pOD22 contain a TEV protease cleavage site. Analogously to the construction of these plasmids, pOD1 was used to produce similar constructs without the TEV cleavage site (TEV-containing counterparts in parentheses): pOD17 (pOD11), pOD18 (pOD12), pOD19 (pOD13), pOD20 (pOD14) and pOD21 (pOD15).
Plastid transformation and selection of homoplasmic transformed tobacco lines
Young leaves from aseptically grown tobacco plants were bombarded with plasmid coated 0.6 µm gold particles using a PDS1000He biolistic gun (Bio-Rad, Munich, Germany). Primary spectinomycin-resistant lines were selected on regeneration medium containing 500 mg l−1 spectinomycin (45). Spontaneous antibiotic-resistant lines were eliminated by double selection on medium containing spectinomycin and streptomycin (500 mg l−1 each). For each transformation construct, several independent transplastomic lines were subjected to two to three additional rounds of regeneration on spectinomycin-containing medium to enrich the transformed plastid genome and select for homoplasmic tissue.
Isolation of nucleic acids and hybridization procedures
Total plant DNA was isolated from frozen leaf tissue samples by a rapid cetyltrimethylammoniumbromide-based mini-prep procedure (46). Total plant RNA was extracted using the peqGOLD TriFast™ reagent (Peqlab, Erlangen, Germany) according to the manufacturer’s protocol. Alternatively, RNA was isolated from leaf tissue and E. coli cells using a hot phenol-based protocol from (47). For Southern blot analyses, DNA samples (5 µg total DNA) were digested with the restriction enzyme BglII, separated by gel electrophoresis in 1% agarose gels and transferred onto Hybond XL membranes (GE Healthcare, Freiburg, Germany) by capillary blotting using standard protocols. A psaB-specific probe generated with primer pair P7247/P7244 (Supplementary Table S1) was used in the RFLP analysis of plastid transformants.
Total cellular RNA samples (10 µg RNA) were electrophoresed in formaldehyde containing 1% agarose gels and blotted onto Hybond XL membranes. For detection of gfp and 16S rRNA transcripts, the coding regions of the corresponding genes were amplified from plasmid clones by PCR. A gfp-specific probe was prepared by amplification with primer pair PZF9/PZF10, a plastid 16S rRNA probe with primer pair P16Srrn-F/P16Srrn-R and an E. coli 16S rRNA probe with the same primer pair (Supplementary Table S1). All hybridization probes were purified using the Nucleospin Extract II kit (Macherey-Nagel, Düren, Germany). Probes were radioactively labeled with α32P-dCTP using the MegaPrime kit (GE Healthcare). Hybridizations were performed at 65°C according to published protocols (48).
Protein extraction and immunoblot analyses
Plant total soluble protein (TSP) was extracted from leaf samples homogenized in a buffer containing 50 mM HEPES, 10 mM KAc, 5 mM MgAc, 1 mM EDTA, 1 mM Pefablock (Roth, Karlsruhe, Germany) and 1 mM DTT (pH 7.5). Bacterial TSP was extracted from pelleted E. coli cells homogenized in a buffer containing 50 mM HEPES, 300 mM NaCl and 0.5% SDS (pH 8.0). Extracted protein samples were separated by electrophoresis in 15–20% SDS–polyacrylamide gradient gels and subsequently transferred to polyvinylidene fluoride (PVDF) membranes (GE Healthcare). GFP was detected with a monoclonal mouse anti-GFP antibody (JL-8, Clontech, Mountain View, CA, USA). Immunobiochemical detection was performed using the ECL Plus detection system (GE Healthcare) according to the manufacturer’s instructions. A GFP standard was produced by purifying the protein from pOD1-expressing E. coli cells.
Quantification of RNA and protein accumulation
Quantification of signal intensities in northern blots (phosphorimages) and western blots (X-ray films) was carried out using the ImageQuant software (GE Healthcare). Selected band areas were quantitated using the ‘Volume Report’ which performs an automatic quantification. Background correction was performed with the ‘Object Average’ method by selecting an appropriate background area of the blot. Signal intensities of gfp mRNA bands in northern blots were normalized to the corresponding 16S rRNA signals and finally set in relation to the reference construct pOD1. Likewise, signal intensities of bands in western blots were set in relation to pOD1 signal intensity. Translational efficiencies were calculated by dividing the relative protein accumulation level by the relative RNA accumulation level of a given transgenic bacterial strain or plant line.
Isolation of polysomes
Polysome isolation was performed according to published protocols (49) with the following minor modifications. The gradients contained 4 × 0.9 ml of sucrose at the described concentrations, overlaid with 0.5 ml of sample in extraction buffer. Following centrifugation, each gradient was separated into 15 fractions for northern blot analysis. Comparative analysis of control gradients with puromycin identified fractions that contained only polysomes and were devoid of free mRNA and free ribosomes. Collected fractions were extracted with phenol:chloroform followed by RNA precipitation with ethanol.
RESULTS
Design of reporter gene constructs containing multiple SD sequences
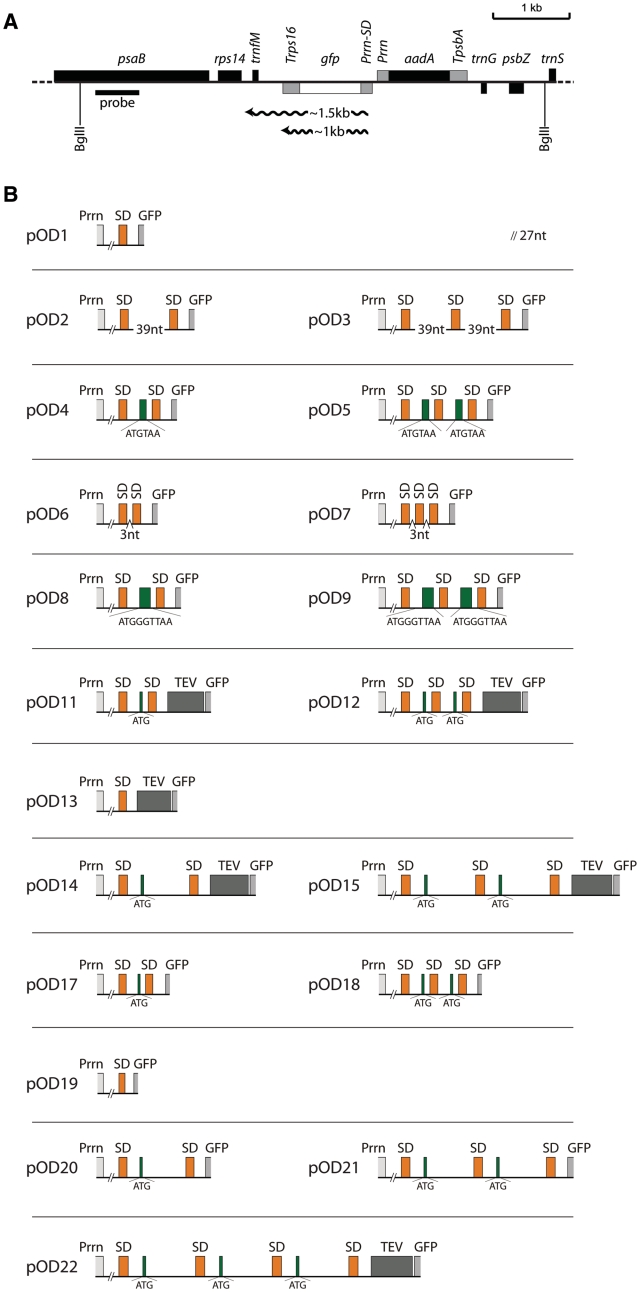
Tobacco plastids and E. coli have identical anti-SD sequences in the 3′-end of their 16S ribosomal RNAs (5′-TGGATCACCTCCTT-3′; anti-SD motif underlined) and, therefore, plastid SD sequences can be recognized in E. coli and vice versa. The SD consensus sequence is GGAGG for both systems. In order to comparatively analyze the principles governing the recognition of SD sequences in plastids and in bacteria, we designed a strategy that allowed us to study translation of identical synthetic transcripts containing multiple SD sequences in E. coli and tobacco plastids. To this end, we constructed a GFP expression cassette driven by the plastid ribosomal operon promoter Prrn, a σ70-type promoter known to be active also in E. coli (27,50,43). A minilinker inserted between the transcriptional start site and the SD sequence upstream of the gfp reading frame allowed the convenient insertion of additional SD sequences, start codons, stop codons and mini-ORFs (open reading frames; Figure 1).
Figure 1.
Construction of vectors to analyze various combinations of plastid translation initiation signals. (A) Physical map of the targeting region in the plastid genome after integration of transformation constructs of the pOD series. The BglII restriction sites used for RFLP analysis are marked. The transgenes are targeted to the intergenic region between the trnfM and trnG genes (42). The GFP expression cassette consists of the ribosomal RNA operon promoter (Prrn) fused to a Shine-Dalgarno (SD) sequence element (see panel B) and the 3′-UTR from the plastid rps16 gene (Trps16). The expected sizes of gfp transcripts are indicated (cf. Figure 3B). The location of the RFLP probe is shown as black bar. The selectable marker gene aadA is driven by a chimeric ribosomal RNA operon promoter (Prrn) and fused to the 3′-UTR from the psbA gene (TpsbA; ref. 45) (B) Schematic maps of the different translation initiation signals tested in this study (pOD vector series). SD sequences are shown in orange, start codons (ATG) and mini-ORFs are indicated in green with the sequence given below the map. TEV: tobacco etch virus peptidase cleavage site; GFP: gene for the green fluorescent protein. The difference between the two basic vectors pOD1 and pOD19 lies in the mutational elimination of an in-frame stop codon upstream of the SD to facilitate translation of GFP from the first SD in constructs pOD20 and pOD21.
Using this basal vector (pOD1; Figure 1), different types of constructs with multiple SD sequences were constructed (Supplementary Table S1; Table 1). Several series of constructs were designed to test the possibility that the presence of multiple SD sequences increases the local concentration of ribosomes and, in this way, recruits more ribosomes to the initiation codon. Constructs pOD6 and pOD7 harbor two and three consecutive SD sequences, respectively. If compared to pOD1, these constructs are suitable to test the possibility that multiple closely spaced SD sequences exert additive effects on ribosome binding to the 5′-UTR (by providing multiple ribosome-binding sites) and, in this way, result in increased rates of translation initiation. The adjacent SD sequences in pOD6 and pOD7 cannot be occupied by ribosomes at the same time, because each 70S ribosome covers an RNA stretch of ∼33–35 nt upstream of the initiation codon (51). Therefore, two constructs were designed (pOD2 and pOD3), in which the distance between two adjacent SD sequences was sufficiently long (39 nt) to allow for simultaneous binding of ribosomes. Together with the SD sequence itself, this provides a sequence stretch of 46 nt, exceeding the minimum footprint of the initiating 30S ribosomal subunit (of ∼40 nt ranging from positions −35 to +5; ref. 52). It should be noted that, in these series of constructs, only a single translation initiation site is present due to the lack of a properly spaced initiation codon downstream of the additional SD sequences. Thus, although these additional SD sequences can potentially increase the local concentration of 30S subunits and, in this way, indirectly increase translation initiation rates, they cannot directly mediate 70S ribosome assembly and translation initiation.
To mimic polycistronic mRNAs not undergoing intercistronic cleavage, two additional series of constructs were produced in which the additional SD sequences were followed by either a start and a stop codon (pOD4 and pOD5) or a mini-ORF containing an additional glycine codon between the start and stop codons (pOD8 and pOD9). In contrast to pOD4 and pOD5, the ribosome performs one elongation cycle after translation initiation at the AUG start codon of the mini-ORFs in pOD8 and pOD9.
We also produced several series of constructs, in which the additional SD sequences were combined with additional in-frame initiation codons. These constructs differ from pOD1-9 in which translation initiation from all SD sequences results in synthesis of GFP. Thus, these constructs allow to directly visualize initiation efficiencies from the different SD sequences, because initiation from the additional SD sequences upstream produces GFPs with short N-terminal extensions which can be separated from the wild-type GFP in polyacrylamide gels. One series (comprising pOD17 and pOD18; Figure 1B) was designed to assess initiation in competition between closely spaced SD sequences, whereas another series (pOD19, pOD20 and pOD21) was analogous to pOD1, pOD2 and pOD3 in that the distance between two adjacent SD sequences was sufficiently long (41 nt) to allow for simultaneous binding of ribosomes to two and three SD sequences, respectively. In order to be able to post-translationally remove the N-terminal extensions resulting from initiation at the upstream SD sequences and start codons, two additional series of constructs contained a TEV protease cleavage site immediately upstream of the GFP sequence. Again, one series harbored closely spaced SD sequences (pOD11 and pOD12), whereas in the second series (pOD14, pOD15 and pOD22), the distances between the SD sequences were large enough to accommodate multiple initiating ribosomes at the same time. To exclude possible effects of the TEV cleavage site on translational efficiencies, construct pOD13 (having one SD sequences and a TEV cleavage site) was generated as an additional control.
Reporter gene expression from mRNAs with multiple SD sequences in bacteria
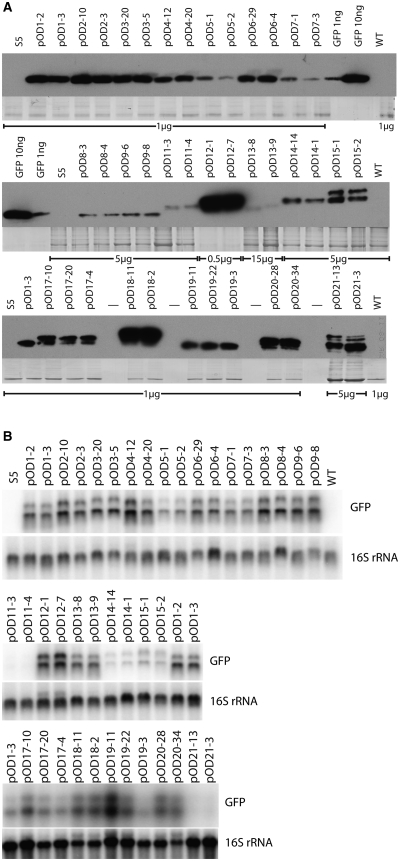
To be able to compare SD sequence recognition in plastids and eubacteria, we first analyzed all 20 constructs in E. coli cells. Determination of GFP accumulation levels revealed strong differences between the different transgenic bacterial strains (Figure 2A). As protein accumulation is the result of both mRNA accumulation levels and translational efficiency, we also determined gfp mRNA accumulation (Figure 2B) and calculated a value for the relative translational efficiency from the RNA accumulation and protein accumulation values (Table 2). RNA accumulation levels varied substantially between the different constructs (Figure 2B) suggesting that the modifications in the 5′-UTR affect mRNA stability. This is unsurprising, because secondary structure, length and sequence of the 5′-UTR are known to represent key determinants of transcript stability in E. coli (53,54). No clear correlation between 5′-UTR sequence, number of SD sequences and/or number of initiation codons and RNA stability was discernable (Table 2; Figures 1B and 2B) confirming that the complex principles that govern mRNA stability in E. coli do not allow to reliably predict transcript stabilities. As the analysis of RNA stability in E. coli was beyond the scope of this study, protein synthesis data were corrected for differences in mRNA abundance by calculating relative translational efficiency values (Table 2) and only these values are considered here. When total translational efficiencies were compared between the 20 constructs, striking differences were seen. First, increasing the spacing between two SD sequences to allow for simultaneous accommodation of two ribosomes increased translational efficiency significantly in the constructs where all GFP translation initiated at the same start codon (cf. pOD2 and pOD3 with pOD6 and pOD7; Table 2). This suggests that additional SD sequences attract more ribosomes, if sufficient spacing permits their simultaneous occupation. Presence of a start and a stop codon or a mini-ORF between two SD sequences resulted in decreased translational efficiency (pOD4, pOD5, pOD8 and pOD9; Table 2), possibly indicating that completion of translation initiation at upstream SD sequences competes with initiation at the downstream SD sequence that controls GFP synthesis.
Figure 2.
Analysis of gfp expression in E. coli. All experiments were repeated at least three times and identical results were obtained. Representative blots are shown here. (A) Western blot analyses to determine GFP accumulation levels in E. coli strains harboring the different pOD plasmids. Loaded amounts of total protein are indicated below the blots. Dashes denote empty lanes. As a control for loading, the high-molecular weight region of the gel (which was not blotted) was stained with Coomassie and is shown below each blot. pRB95: empty vector control. (B) Analysis of gfp mRNA accumulation in pOD strains. As a loading control, the blot was stripped and re-hybridized to a 16S rRNA-specific probe.
Table 2.
Comparison of RNA accumulation, protein accumulation and calculated translational efficiencies (i.e. relative protein accumulation level divided by relative RNA accumulation level) for all constructs tested in this study
| Construct |
Escherichia coli |
Nicotiana tabacum plastids |
||||
|---|---|---|---|---|---|---|
| RNA | Protein | Efficiency | RNA | Protein | Efficiency | |
| pOD1 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| pOD2 | 0.66 | 0.81 | 1.23 | 1.57 | 1.12 | 0.71 |
| pOD3 | 0.56 | 0.78 | 1.39 | 1.36 | 0.92 | 0.68 |
| pOD4 | 0.20 | 0.01 | 0.05 | 1.72 | 0.62 | 0.36 |
| pOD5 | 0.19 | 0.06 | 0.31 | 0.53 | 0.19 | 0.36 |
| pOD6 | 0.24 | 0.12 | 0.49 | 1.06 | 0.71 | 0.67 |
| pOD7 | 0.10 | 0.05 | 0.48 | 0.95 | 0.19 | 0.20 |
| pOD8 | 0.14 | 0.01 | 0.05 | 1.49 | 0.02 | 0.01 |
| pOD9 | 0.11 | 0.05 | 0.47 | 2.00 | 0.03 | 0.02 |
| pOD11 | 0.37 | 0.62 | 1.68 | 0.08 | 0.01 | 0.19 |
| pOD12 | 0.54 | 0.73 | 1.36 | 1.72 | 5.08 | 2.96 |
| pOD13 | 0.55 | 0.06 | 0.10 | 1.21 | 0.00 | 0.00 |
| pOD14 | 1.35 | 1.35 | 1.00 | 0.14 | 0.07 | 0.47 |
| pOD15 | 0.11 | 1.70 | 14.94 | 0.37 | 0.15 | 0.40 |
| pOD17 | 0.39 | 0.72 | 1.84 | 1.21 | 1.29 | 1.07 |
| pOD18 | 0.51 | 1.25 | 2.47 | 2.53 | 4.09 | 1.62 |
| pOD19 | 0.90 | 1.11 | 1.23 | 2.20 | 1.01 | 0.46 |
| pOD20 | 1.12 | 1.26 | 1.13 | 2.23 | 2.01 | 0.90 |
| pOD21 | 1.36 | 1.40 | 1.03 | 0.28 | 0.37 | 1.32 |
| pOD22 | 0.02 | 1.68 | 71.10 | – | – | – |
Values are normalized to the reference construct pOD1 (set to 1.00 in both E. coli and tobacco plastids). Data for plastids represent the means of at least two independently quantitated biological replicas.
Many of the constructs in which recognition of upstream SD sequences produces GFP by reading in frame through the downstream SD sequences showed increased translational efficiencies compared to their respective reference construct with one SD sequence (pOD11–22; Table 2). The increase was extreme in pOD15 (15-fold) and pOD22 (71-fold), constructs with three and four SD sequences, respectively, that were sufficiently spaced to simultaneously accommodate multiple initiating 30S ribosomal subunits. Interestingly, comparison of the construct series with the TEV protease cleavage site (pOD13, pOD11, pOD12, pOD14, pOD15 and pOD22) and the analogous series without it (pOD19, pOD17, pOD18, pOD20 and pOD21) revealed that the presence of the TEV protease cleavage site had a strong effect on GFP accumulation. This could be due to the N-terminal TEV sequence influencing protein stability and/or mRNA folding (55). As these possibilities are difficult to exclude, the appropriate reference construct with one SD sequence was constructed for each series (e.g. pOD13 for all TEV cleavage site-containing constructs) and consequently, comparisons of translational efficiencies should be confined to GFPs with identical N-termini.
Utilization of SD sequences in bacteria
The constructs in which the additional SD sequences were combined with additional in-frame initiation codons allowed us to directly compare the efficiencies of translation initiation at the different SD sequences by quantitating the different GFP length variants (Figure 2A and Table 3). In all of these constructs, all SD sequences present were utilized. However, the relative utilization efficiencies were somewhat variable ranging from about equally efficient utilization (e.g. in pOD12) to an up to 5-fold difference in utilization efficiency (pOD14; Figure 2A and Table 3). These quantitative differences may be due to either the local sequence context surrounding the SD sequence and/or differences in the local RNA secondary structure. Both the neighboring sequences and the folding of the 5′-UTR are known to influence the efficiency of SD sequence recognition and thus of the translation initiation process in E. coli (43,55,56).
Table 3.
Utilization of SD sequences in constructs harboring multiple ribosome-binding sites
| Construct | Initiation from SD No. (3′→5′) | Fraction of GFP accumulation |
|
|---|---|---|---|
| Escherichia coli (%) | Nicotiana tabacum plastids (%) | ||
| pOD11 | 1 | 49.26 | ND |
| 2 | 50.74 | 100 | |
| pOD12 | 1 | 40.41 | 5.78 |
| 2 | 31.08 | 18.34 | |
| 3 | 28.52 | 75.88 | |
| pOD14 | 1 | 14.73 | ND |
| 2 | 85.27 | 100 | |
| pOD15 | 1 | 15.57 | ND |
| 2 | 51.98 | 55.93 | |
| 3 | 32.46 | 44.07 | |
| pOD17 | 1 | 56.14 | 40.01 |
| 2 | 43.86 | 59.99 | |
| pOD18 | 1 | 49.11 | 4.43 |
| 2 | 20.10 | 17.75 | |
| 3 | 30.80 | 77.82 | |
| pOD20 | 1 | 32.20 | 19.90 |
| 2 | 67.80 | 80.10 | |
| pOD21 | 1 | 20.75 | 2.38 |
| 2 | 46.49 | 71.26 | |
| 3 | 32.76 | 26.35 | |
| pOD22 | 1 | 11.71 | – |
| 2 | 54.02 | – | |
| 3 | 11.28 | – | |
| 4 | 22.99 | – | |
Data represent the means of at least two independently quantitated biological replicas. ND, not detectable; –, not analyzed.
Overall, the utilization of all SD sequences in all constructs with multiple SD sequences is consistent with the efficient translation of downstream cistrons in polycistronic mRNAs of eubacteria and confirms that SD sequence recognition and translation initiation are largely independent of the position of the SD sequence in the transcript.
Reporter gene expression from mRNAs with multiple SD sequences in plastids
We next wanted to determine if the relative translational efficiencies from the various constructs in plastids are comparable to those in E. coli and if similar rules for SD sequence recognition apply. To this end, all constructs from pOD1 to pOD21 (Figure 1) were stably integrated into the tobacco plastid genome by chloroplast transformation using the biolistic protocol (45). Chloroplast transformants were selected on plant regeneration medium containing spectinomycin, a chloroplast-specific translational inhibitor to which the aadA selection marker in our transformation vectors (42) confers resistance. Several plastid-transformed (=transplastomic) lines were isolated for each construct and their transgenic status was preliminarily verified by double resistance tests on regeneration medium containing both spectinomycin and streptomycin (45,57). Putative transplastomic lines were subjected to two to three additional rounds of regeneration under selective conditions to eliminate residual wild-type copies of the plastid genome and obtain homoplasmic tissue. Homoplasmy was confirmed by molecular analyses using Southern blotting (Supplementary Figure S1) as well as genetically by conducting inheritance assays and seed tests (45,57). Two to three independently generated homoplasmic transplastomic lines per construct were included in all further analyses.
Analogously to our analyses in E. coli (Figure 2), we first determined GFP accumulation by western blotting using a specific antibody. As in bacteria, comparison of the different constructs revealed strong differences in GFP accumulation levels also in chloroplasts (Figure 3A). To determine to what extent these differences are attributable to differences in mRNA stability, northern blot experiments were carried out (Figure 3B). As in E. coli, these analyses revealed significant (though overall less drastic) differences between the different constructs, confirming that sequence and/or secondary structure of the 5′-UTR influence mRNA stability also in plastids. Interestingly, little if any correlation between transcript stability in plastids and bacteria was detected (cf. Figures 2B and 3B). For example, pOD21, the construct triggering the highest gfp mRNA accumulation in E. coli (Figure 2B and Table 2), yielded relatively low transcript levels in plastids (Figure 3B and Table 2). Conversely, pOD9, a construct conferring high levels of gfp mRNA in plastids, yields only low mRNA levels in E. coli (Table 2). These data suggest that the signals in the 5′-UTR that determine transcript half-life times differ between bacteria and plastids. The molecular mechanisms underlying these striking differences are currently not known.
Figure 3.
Analysis of gfp expression in tobacco plastids. (A) Western blot analyses to determine GFP accumulation levels in transplastomic tobacco plants generated with pOD vectors. Loaded amounts of total protein are indicated below the blots. Dashes denote empty lanes. As a control for loading, the high-molecular weight region of the gel (which was not blotted) was stained with Coomassie and is shown below each blot. WT, wild-type (1 µg protein loaded); S5, transplastomic control (carrying the aadA but no gfp; ref. 71; 1 µg protein loaded); GFP, purified recombinant GFP used as standard for quantitation. (B) Analysis of gfp mRNA accumulation in pOD transplastomic plants. The lower band represents monocistronic gfp mRNA, the upper band is the result of read through transcription and has been observed before in studies using the same plastid transformation vector backbone (pRB95; 28). To control for equal loading, the blot was stripped and re-hybridized to a 16S rRNA-specific probe.
To analyze translation of gfp mRNAs in plastids, mRNA levels and protein levels were quantitated and relative translational efficiencies were calculated (Table 2). As in E. coli, increasing the spacing between adjacent SD sequences to allow for simultaneous binding of two ribosomes increased translational efficiency in the constructs where all GFP translation initiated at the same start codon (cf. pOD2 and pOD3 with pOD6 and pOD7; Table 2). Also, like in E. coli, many of the constructs in which recognition of upstream SD sequences produces GFP by reading in frame through the downstream SD sequences showed increased translational efficiencies compared to their respective reference construct with one SD sequence (cf. e.g. pOD19 with pOD20 and pOD21, or pOD19 with pOD17 and pOD18; Table 2).
A remarkable difference between E. coli and plastids was revealed when the data for pOD4 and pOD5 were compared to those for pOD8 and pOD9. The only difference between pOD4 and pOD8 and pOD5 and pOD9, respectively, lies in the presence of a single glycine codon in the mini-ORF embedded between two SD sequences (Figure 1B). Thus, whereas the ribosome performs a single elongation cycle on the mini-ORF in pOD8 and pOD9, no protein synthesis upstream of the gfp reading frame occurs in pOD4 and pOD5. Interestingly, while in E. coli the occurrence of this single elongation cycle has no effect on translation (in that translational activity for pOD8 is very similar to pOD4 and translational activity for pOD9 very similar to pOD5), it drastically decreases translation in plastids. Whereas gfp is efficiently translated in pOD4 and pOD5, translation is reduced to barely detectable levels in pOD8 and pOD9 (Table 2). This suggests that occurrence of upstream translation, even if limited to a single elongation cycle, has a negative effect on translation initiation in plastids, but not in E. coli. This could introduce a 5′ to 3′ bias in the translational efficiency of the cistrons in (at least some) polycistronic mRNAs.
Utilization of SD sequences in plastids
To explore the potential 5′ to 3′ bias in SD sequence utilization in plastids in greater detail, we analyzed the translation initiation efficiencies at the individual SD sequences in those construct series, in which the additional SD sequences were combined with additional in-frame initiation codons, thus leading to GFP length variants that can be exploited to directly quantitate SD sequence utilization (Figure 3A and Table 3).
Interestingly, SD sequence utilization in plastids and E. coli differed strongly (Table 3). Whereas in E. coli, always all GFP length variants were readily detectable (indicating that all SD sequences are used), identical transcripts in plastids often displayed a preference for the use of a single or two out of three SD sequences. Strikingly, in most constructs the last (i.e. the 3′-most) SD sequence was the least frequently used SD ( = SD sequence number 1 in Table 3) in plastids. This suggests that presence of upstream SD sequences can impair recognition of downstream ones in plastids, but not so much in E. coli. Remarkably, in several constructs (pOD11, pOD14 and pOD15), the 3′-most SD sequence was not used at all in plastids.
In general, whereas no strong tendency to preferentially use a certain SD sequence was recognizable in E. coli and no SD sequence was utilized to <10%, a pronounced bias toward a 5′ to 3′-polarity in SD sequence utilization was seen in plastids. In many constructs (pOD11, pOD12, pOD14, pOD17, pOD18 and pOD20), the translation initiation rates declined progressively in 5′ to 3′ direction (Table 3). The only exceptions were pOD21, one of the construct with three SD sequences, in which the middle SD sequence (Number 2) was more efficiently recognized than the upstream SD sequence (Number 3) and construct pOD15, in which SD sequences Number 2 and 3 are utilized with about the same efficiencies (Table 3). Why these two pairs of SD sequences behave differently is not clear, but aberrant RNA secondary structure formation making the upstream SD sequence less accessible than the downstream one may be one possible explanation. In silico analyses of RNA folding of all constructs provided no obvious indication of masking of individual SD sequences by RNA secondary structure formation. However, there remains a possibility that aberrant structures form in vivo, even though they are not revealed as probable structures in silico. Likewise, we cannot entirely exclude the possibility that RNA folding of some of the gfp mRNAs is slightly different in plastids from E. coli, although this seems not very likely.
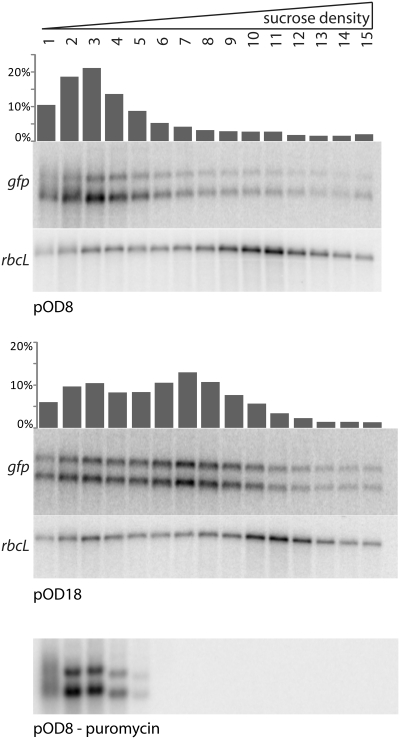
Finally, we wanted to obtain independent confirmation that the differences in translational efficiencies determined from the protein and RNA accumulation data are indeed caused by differences in translation initiation rates. We, therefore, directly compared translational activity for a construct identified as conferring high translation rates (pOD18) and a construct exhibiting poor translatability (pOD8). To this end, polysomes (mRNAs associated with translating ribosomes) were purified from both plant lines and separated in sucrose density gradients. This separation is suitable to resolve differences in the average number of ribosomes translating a given mRNA (49). When the polysome profiles of the pOD8 and pOD18 transplastomic plants were compared, much more gfp mRNA had migrated into the high-density fractions of the gradient in the pOD18 sample than in the pOD8 sample (Figure 4). As the gfp mRNAs in pOD8 and pOD18 plants differ only by their SD sequences in the 5′-UTR, this finding confirms that the measured differences in translational efficiency are indeed due to difference in the rate of translation initiation.
Figure 4.
Confirmation of different translation initiation efficiencies by ribosome loading experiments. Polysome profiles of a weakly GFP accumulating transplastomic tobacco line (pOD8) and a strongly GFP accumulating line (pOD18) are shown. The lines were chosen, because they show similar gfp mRNA accumulation levels, but strong differences in GFP accumulation. Compared to the pOD8 plant, gfp mRNA distribution is shifted toward the bottom of the sucrose gradient in the pOD18 plant, indicating denser coverage with ribosomes and thus, higher translation efficiency. The bar diagrams above each blot show the amount of gfp mRNA present in each fraction (given in percent of the total gfp mRNA in the sample). As an internal control RNA, the distribution of the rbcL mRNA in the gradient was analyzed and, as expected, is identical in the two transplastomic lines. The wedge indicates the increasing sucrose concentration from the top to the bottom of the polysome gradient. For identification of polysome-containing gradient fractions, a puromycin-treated sample is also shown.
DISCUSSION
In this work, we have introduced identical transgenic constructs into E. coli and tobacco plastids to comparatively analyze translation of mRNAs with multiple SD sequences in vivo. As expected from the prokaryotic nature of both systems, a number of similarities were found. For example, the presence of closely spaced SD sequences upstream of a single initiation codon (pOD6 and pOD7) strongly reduced translational efficiency. As the upstream SD sequences are unlikely to contribute significantly to translation initiation due to their inappropriate distance from the initiation codon, this suggests that, rather than being beneficial by increasing the local concentration of ribosomes, multiple closely spaced SD sequences compete for 30S ribosomal subunits and thus exert a negative influence on translation initiation rates. Interestingly, increasing the distance between the SD sequences to allow for simultaneous occupation by 30S subunits alleviated this presumptive competition effect (pOD2 and pOD3) and, in E. coli, even led to higher translational efficiencies than in the construct with a single SD sequence.
A striking difference between E. coli and plastids identified in the course of this work was the decline of translation initiation rates at downstream SD sequences in plastids. While in E. coli, all SD sequences that were followed by a properly spaced initiation codon were utilized, plastid ribosomes displayed a bias toward preferentially (or even exclusively) utilizing the 5′-most SD sequence (Table 3). In most constructs, the 3′-most SD sequence was the least efficiently used one and, in several cases, this SD sequence was not recognized at all (Table 3). This 5′ to 3′ bias in SD sequence utilization could be explained by an mRNA scanning mechanism operating in plastid translation initiation. If confirmed, such a mechanism would be reminiscent of eukaryotic translation initiation in which the capped mRNA 5′-end is recognized by the small subunit of the ribosome (bound to the initiator tRNA and several eukaryotic initiation factors) and then migrates away from the cap along the transcript to identify the AUG start codon (19). Although plastid mRNAs do not carry a 5′-cap modification, it is conceivable that the 30S ribosomal subunit binds to the mRNA 5′-end by recognizing the free 5′-phosphate group (or sequence elements at the 5′-end) and then slides along the mRNA to scan for the presence of a SD sequence and an initiation codon. Interestingly, the presence of a readable codon downstream of the start codon seems to be an additional requirement for cessation of scanning. If the start codon is followed by a stop codon and no elongation cycle occurs (pOD4 and pOD5), the ribosome seems to continue scanning quite efficiently, whereas completion of a single elongation cycle (pOD8 and pOD9) abandons the scanning process, thereby drastically reducing translation initiation at downstream cistrons (Table 2). Interestingly, recent evidence from in vitro translation of atpB, an mRNA lacking a SD sequence, suggests that, prior to initiating translation, the 30S ribosomal subunit interacts with upstream mRNA sequences (22), a finding that would be compatible with ribosome scanning. However, more work is needed to establish whether or not mRNA scanning for SD sequences and initiation codons indeed occurs in plastids.
The striking decline of translation initiation rates at downstream SD sequences in plastids, but not in E. coli, raises important questions about (i) the underlying molecular basis and (ii) the functional significance of this mechanistic difference between the two prokaryotic systems. We propose here that the functional significance of the 5′ to 3′ bias in plastid translation initiation may be related to intercistronic RNA cleavage, a common transcript processing step in plastids, but not in bacteria. If downstream SD sequences in plastid polycistronic mRNAs are difficult to recognize by the translation initiation complex, intercistronic cleavage offers an obvious solution to the problem. Recent work has shown that, after intercistronic processing, specific proteins of the PPR class bind to the 5′-ends of the monocistronic mRNAs and provide protection from exoribonucleolytic degradation (58). Whether or not these proteins also function as translational enhancers by guiding the ribosome to the correct initiation site, remains to be investigated.
It is important to note that the dependence of translation of downstream cistrons on intercistronic processing in plastids is not absolute. Some polycistronic and oligocistronic mRNAs do not undergo intercistronic cleavage into monocistronic units (39,40) and it must be assumed, that in these cases, the downstream cistrons are nonetheless efficiently translated. How in these mRNAs the efficient recognition of the downstream SD sequences is mediated, remains to be established, but the presence of mRNA-specific translational regulator proteins offers an attractive possibility for how this could occur. For example, translation of the downstream psaB cistron in the dicistronic psaA–psaB transcript (40) has been demonstrated to be mediated by a translational regulator protein (Table 2; ref. 59), which is apparently exclusively dedicated to controlling psaB translation. Thus, it seems conceivable that mRNA-specific translational regulator proteins could facilitate the recognition of downstream SD sequences and, in this way, circumvent the requirement for intercistronic cleavage in some plastid transcripts.
What component(s) of the translational machinery could be causally responsible for the observed differences in SD sequence utilization between E. coli and plastids? A possible explanation could lie in the structural differences between the ribosomes in the two systems. In addition to the conserved set of ribosomal proteins typical of all 70S ribosomes, plastid ribosomes also contain a small set of so-called plastid-specific ribosomal proteins (PSRPs; 60–62). The functions of these PSRPs have yet to be determined, but an involvement in SD recognition could potentially explain why these additional ribosomal proteins were acquired in evolution.
A useful by-product of this work is the identification of new plastid expression elements that confer a 4- to 5-fold higher protein accumulation than conventional constructs with a single SD sequence (cf. pOD12 and pOD18 with pOD1; Table 2). While the reference construct pOD1 accumulates GFP to ∼1% of the TSP of the plant, protein accumulation with the best-expressing construct pOD12 is at least 5% of the TSP (Figure 3 and Table 2). Plants with transgenic plastids provide an attractive expression platform to biotechnologists (63–65), due to their high capacity to accumulate foreign proteins (28,66) and their maternal mode of inheritance which greatly reduces unwanted transgene transmission via pollen (67,68). The efficient expression elements identified here provide valuable tools to maximize transgene expression from the plastid genome, which is of particular importance for the high-yield production of pharmaceutical proteins (e.g. antigens and antibodies), an area commonly referred to as molecular farming (69,70).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Max Planck Society (MPG); European Union (FP7 METAPRO 244348). Funding for open access charge: Max Planck Society.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Stefanie Seeger, Claudia Hasse, Annett Kaßner and Dr Stephanie Ruf for help with chloroplast transformations, the MPI-MP Green Team for plant care and cultivation and Claudia Flügel and Britta Ehlert for helpful discussion.
REFERENCES
- 1.Haley J, Bogorad L. Alternative promoters are used for genes within maize chloroplast polycistronic transcription units. Plant Cell. 1990;2:323–333. doi: 10.1105/tpc.2.4.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liere K, Börner T. Transcription and transcriptional regulation in plastids. Top. Curr. Genet. 2007;19:121–174. [Google Scholar]
- 3.Sugiura M, Hirose T, Sugita M. Evolution and mechanism of translation in chloroplasts. Annu. Rev. Genet. 1998;32:437–459. doi: 10.1146/annurev.genet.32.1.437. [DOI] [PubMed] [Google Scholar]
- 4.Beligni MV, Yamaguchi K, Mayfield SP. The translational apparatus of Chlamydomonas reinhardtii chloroplast. Photosynth. Res. 2004;82:315–325. doi: 10.1007/s11120-004-2440-5. [DOI] [PubMed] [Google Scholar]
- 5.Peled-Zehavi H, Danon A. Translation and translational regulation in chloroplasts. Top. Curr. Genet. 2007;19:249–281. [Google Scholar]
- 6.Hayes R, Kudla J, Schuster G, Gabay L, Maliga P, Gruissem W. Chloroplast mRNA 3′-end processing by a high molecular weight protein complex is regulated by nuclear encoded RNA binding proteins. EMBO J. 1996;15:1132–1141. [PMC free article] [PubMed] [Google Scholar]
- 7.Bollenbach TJ, Schuster G, Portnoy V, Stern DB. Processing, degradation, and polyadenylation of chloroplast transcripts. Top. Curr. Genet. 2007;19:175–211. [Google Scholar]
- 8.Adam Z. Protein stability and degradation in plastids. Top. Curr. Genet. 2007;19:315–338. [Google Scholar]
- 9.Schmitz-Linneweber C, Small I. Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 2008;13:663–670. doi: 10.1016/j.tplants.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 10.Stern DB, Goldschmidt-Clermont M, Hanson MR. Chloroplast RNA metabolism. Annu. Rev. Plant Biol. 2010;61:125–155. doi: 10.1146/annurev-arplant-042809-112242. [DOI] [PubMed] [Google Scholar]
- 11.Eberhard S, Drapier D, Wollman F-A. Searching limiting steps in the expression of chloroplast-encoded proteins: relations between gene copy number, transcription, transcript abundance and translation rate in the chloroplast of Chlamydomonas reinhardtii. Plant J. 2002;31:149–160. doi: 10.1046/j.1365-313x.2002.01340.x. [DOI] [PubMed] [Google Scholar]
- 12.Mullet JE, Klein RR. Transcription and RNA stability are important determinants of higher plant chloroplast RNA levels. EMBO J. 1987;6:1571–1579. doi: 10.1002/j.1460-2075.1987.tb02402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein RR, Mullet JE. Light-induced transcription of chloroplast genes. J. Biol. Chem. 1990;265:1895–1902. [PubMed] [Google Scholar]
- 14.Kahlau S, Bock R. Plastid transcriptomics and translatomics of tomato fruit development and chloroplast-to-chromoplast differentiation: chromoplast gene expression largely serves the production of a single protein. Plant Cell. 2008;20:856–874. doi: 10.1105/tpc.107.055202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J, Mayfield SP. The active site of the thioredoxin-like domain of chloroplast protein disulfide isomerase, RB60, catalyzes the redox-regulated binding of chloroplast poly(A)-binding protein, RB47, to the 5′untranslated region of psbA mRNA. Plant Cell Physiol. 2002;43:1238–1243. doi: 10.1093/pcp/pcf129. [DOI] [PubMed] [Google Scholar]
- 16.Choquet Y, Zito F, Wostrikoff K, Wollman F-A. Cytochrome f translation in Chlamydomonas chloroplast is autoregulated by its carboxyl-terminal domain. Plant Cell. 2003;15:1443–1454. doi: 10.1105/tpc.011692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barneche F, Winter V, Crèvecoeur M, Rochaix J-D. ATAB2 is a novel factor in the signalling pathway of light-controlled synthesis of photosystem proteins. EMBO J. 2006;25:5907–5918. doi: 10.1038/sj.emboj.7601472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marín-Navarro J, Manuell AL, Wu J, Mayfield SP. Chloroplast translation regulation. Photosynth. Res. 2007;94:359–374. doi: 10.1007/s11120-007-9183-z. [DOI] [PubMed] [Google Scholar]
- 19.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 20.Ruf M, Kössel H. Occurrence and spacing of ribosome recognition sites in mRNAs of chloroplasts from higher plants. FEBS Lett. 1988;240:41–44. [Google Scholar]
- 21.Stern DB, Higgs DC, Yang J. Transcription and translation in chloroplasts. Trends Plant Sci. 1997;2:308–315. [Google Scholar]
- 22.Hirose T, Sugiura M. Multiple elements required for translation of plastid atpB mRNA lacking the Shine-Dalgarno sequence. Nucleic Acids Res. 2004;32:3503–3510. doi: 10.1093/nar/gkh682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirose T, Kusumegi T, Sugiura M. Translation of tobacco chloroplast rps14 mRNA depends on a Shine-Dalgarno-like sequence in the 5′-untranslated region but not on internal RNA editing in the coding region. FEBS Lett. 1998;430:257–260. doi: 10.1016/s0014-5793(98)00673-5. [DOI] [PubMed] [Google Scholar]
- 24.Hirose T, Suguira M. Functional Shine-Dalgarno-like sequences for translational initiation of chloroplast mRNAs. Plant Cell Physiol. 2004;45:114–117. doi: 10.1093/pcp/pch002. [DOI] [PubMed] [Google Scholar]
- 25.Kuroda H, Suzuki H, Kusumegi T, Hirose T, Yukawa Y, Sugiura M. Translation of psbC mRNAs starts from the downstream GUG, not the upstream AUG, and requires the extended Shine-Dalgarno sequence in tobacco chloroplasts. Plant Cell Physiol. 2007;48:1374–1378. doi: 10.1093/pcp/pcm097. [DOI] [PubMed] [Google Scholar]
- 26.Ye G-N, Hajdukiewicz PTJ, Broyles D, Rodriguez D, Xu CW, Nehra N, Staub JM. Plastid-expressed 5-enolpyruvylshikimate-3-phosphate synthase genes provide high level glyphosate tolerance in tobacco. Plant J. 2001;25:261–270. doi: 10.1046/j.1365-313x.2001.00958.x. [DOI] [PubMed] [Google Scholar]
- 27.Kuroda H, Maliga P. Complementarity of the 16S rRNA penultimate stem with sequences downstream of the AUG destabilizes the plastid mRNAs. Nucleic Acids Res. 2001;29:970–975. doi: 10.1093/nar/29.4.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou F, Badillo-Corona JA, Karcher D, Gonzalez-Rabade N, Piepenburg K, Borchers A-MI, Maloney AP, Kavanagh TA, Gray JC, Bock R. High-level expression of HIV antigens from the tobacco and tomato plastid genomes. Plant Biotechnol. J. 2008;6:897–913. doi: 10.1111/j.1467-7652.2008.00356.x. [DOI] [PubMed] [Google Scholar]
- 29.Sugiura M. The chloroplast genome. Plant Mol. Biol. 1992;19:149–168. doi: 10.1007/BF00015612. [DOI] [PubMed] [Google Scholar]
- 30.Westhoff P, Herrmann RG. Complex RNA maturation in chloroplasts. Eur. J. Biochem. 1988;171:551–564. doi: 10.1111/j.1432-1033.1988.tb13824.x. [DOI] [PubMed] [Google Scholar]
- 31.Sugita M, Sugiura M. Regulation of gene expression in chloroplasts of higher plants. Plant Mol. Biol. 1996;32:315–326. doi: 10.1007/BF00039388. [DOI] [PubMed] [Google Scholar]
- 32.Herrin DL, Nickelsen J. Chloroplast RNA processing and stability. Photosynth. Res. 2004;82:301–314. doi: 10.1007/s11120-004-2741-8. [DOI] [PubMed] [Google Scholar]
- 33.Zhou F, Karcher D, Bock R. Identification of a plastid Intercistronic Expression Element (IEE) facilitating the expression of stable translatable monocistronic mRNAs from operons. Plant J. 2007;52:961–972. doi: 10.1111/j.1365-313X.2007.03261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirose T, Sugiura M. Both RNA editing and RNA cleavage are required for translation of tobacco chloroplast ndhD mRNA: a possible regulatory mechanism for the expression of a chloroplast operon consisting of functionally unrelated genes. EMBO J. 1997;16:6804–6811. doi: 10.1093/emboj/16.22.6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barkan A, Walker M, Nolasco M, Johnson D. A nuclear mutation in maize blocks the processing and translation of several chloroplast mRNAs and provides evidence for the differential translation of alternative mRNA forms. EMBO J. 1994;13:3170–3181. doi: 10.1002/j.1460-2075.1994.tb06616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fisk DG, Walker MB, Barkan A. Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J. 1999;18:2621–2630. doi: 10.1093/emboj/18.9.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Felder S, Meierhoff K, Sane AP, Meurer J, Driemel C, Plücken H, Klaff P, Stein B, Bechtold N, Westhoff P. The nucleus-encoded HCF107 gene of Arabidopsis provides a link between intercistronic RNA processing and the accumulation of translation-competent psbH transcripts in chloroplasts. Plant Cell. 2001;13:2127–2141. doi: 10.1105/TPC.010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hashimoto M, Endo T, Peltier G, Tasaka M, Shikanai T. A nucleus-endoded factor, CRR2, is essential for the expression of chloroplast ndhB in Arabidopsis. Plant J. 2003;36:541–549. doi: 10.1046/j.1365-313x.2003.01900.x. [DOI] [PubMed] [Google Scholar]
- 39.Willey DL, Gray JC. Two small open reading frames are co-transcribed with the pea chloroplast genes for the polypeptides of cytochrome b-559. Curr. Genet. 1989;15:213–220. doi: 10.1007/BF00435508. [DOI] [PubMed] [Google Scholar]
- 40.Meng BY, Tanaka M, Wakasugi T, Ohme M, Shinozaki K, Sugiura M. Cotranscription of the genes encoding two P700 chlorophyll a apoproteins with the gene for ribosomal protein CS14: determination of the transcriptional initiation site by in vitro capping. Curr. Genet. 1988;14:395–400. doi: 10.1007/BF00419998. [DOI] [PubMed] [Google Scholar]
- 41.Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue culture. Physiol. Plant. 1962;15:473–497. [Google Scholar]
- 42.Ruf S, Hermann M, Berger IJ, Carrer H, Bock R. Stable genetic transformation of tomato plastids and expression of a foreign protein in fruit. Nature Biotechnol. 2001;19:870–875. doi: 10.1038/nbt0901-870. [DOI] [PubMed] [Google Scholar]
- 43.Neupert J, Karcher D, Bock R. Design of simple synthetic RNA thermometers for temperature-controlled gene expression in Escherichia coli. Nucleic Acids Res. 2008;36:e124. doi: 10.1093/nar/gkn545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wurbs D, Ruf S, Bock R. Contained metabolic engineering in tomatoes by expression of carotenoid biosynthesis genes from the plastid genome. Plant J. 2007;49:276–288. doi: 10.1111/j.1365-313X.2006.02960.x. [DOI] [PubMed] [Google Scholar]
- 45.Svab Z, Maliga P. High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc. Natl Acad. Sci. USA. 1993;90:913–917. doi: 10.1073/pnas.90.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- 47.Verwoerd TC, Dekker BMM, Hoekema A. A small-scale procedure for the rapid isolation of plant RNA. Nucleic Acids Res. 1989;17:2362. doi: 10.1093/nar/17.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Church GM, Gilbert W. Genomic sequencing. Proc. Natl Acad. Sci. USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barkan A. Approaches to investigating nuclear genes that function in chloroplast biogenesis in land plants. Methods Enzymol. 1998;297:38–57. [Google Scholar]
- 50.Suzuki JY, Sriraman P, Svab Z, Maliga P. Unique architecture of the plastid ribosomal RNA operon promoter recognized by the multisubunit RNA polymerase in tobacco and other higher plants. Plant Cell. 2003;15:195–205. doi: 10.1105/tpc.007914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Borisova GP, Volkova TM, Berzin V, Rosenthal G, Gren EJ. The regulatory region of MS2 phage RNA replicase cistron. IV. Functional activity of specific MS2 RNA fragments in formation of the 70 S initiation complex of protein biosynthesis. Nucleic Acids Res. 1979;6:1761–1774. doi: 10.1093/nar/6.5.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hüttenhofer A, Noller HF. Footprinting mRNA-ribosome complexes with chemical probes. EMBO J. 1994;13:3892–3901. doi: 10.1002/j.1460-2075.1994.tb06700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bouvet P, Belasco JG. Control of RNase E-mediated RNA degradation by 5′-terminal base pairing in E. coli. Nature. 1992;360:488–491. doi: 10.1038/360488a0. [DOI] [PubMed] [Google Scholar]
- 54.Feng Y, Cohen SN. Unpaired terminal nucleotides and 5′ monophosphorylation govern 3′ polyadenylation by Escherichia coli poly(A) polymerase I. Proc. Natl Acad. Sci. USA. 2000;97:6415–6420. doi: 10.1073/pnas.120173797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Studer SM, Joseph S. Unfolding of mRNA secondary structure by the bacterial translation initiation complex. Mol. Cell. 2006;22:105–115. doi: 10.1016/j.molcel.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 56.Vimberg V, Tats A, Remm M, Tenson T. Translation initiation region sequence preferences in Escherichia coli. BMC Mol. Biol. 2007;8:100. doi: 10.1186/1471-2199-8-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bock R. Transgenic chloroplasts in basic research and plant biotechnology. J. Mol. Biol. 2001;312:425–438. doi: 10.1006/jmbi.2001.4960. [DOI] [PubMed] [Google Scholar]
- 58.Pfalz J, Bayraktar OA, Prikryl J, Barkan A. Site-specific binding of PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. 2009;28:2042–2052. doi: 10.1038/emboj.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dauvillée D, Stampacchia O, Girard-Bascou J, Rochaix J-D. Tab2 is a novel conserved RNA binding protein required for translation of the chloroplast psaB mRNA. EMBO J. 2003;22:6378–6388. doi: 10.1093/emboj/cdg591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamaguchi K, von Knoblauch K, Subramanian AR. The plastid ribosomal proteins. Identification of all the proteins in the 30 S subunit of an organelle ribosome (chloroplast) J. Biol. Chem. 2000;275:28455–28465. doi: 10.1074/jbc.M004350200. [DOI] [PubMed] [Google Scholar]
- 61.Yamaguchi K, Subramanian AR. The plastid ribosomal proteins. Identification of all the proteins in the 50 S subunit of an organelle ribosome (chloroplast) J. Biol. Chem. 2000;275:28466–28482. doi: 10.1074/jbc.M005012200. [DOI] [PubMed] [Google Scholar]
- 62.Manuell AL, Quispe J, Mayfield SP. Structure of the chloroplast ribosome: Novel domains for translation regulation. PLoS Biol. 2007;5:1785–1797. doi: 10.1371/journal.pbio.0050209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maliga P. Plastid transformation in higher plants. Annu. Rev. Plant Biol. 2004;55:289–313. doi: 10.1146/annurev.arplant.55.031903.141633. [DOI] [PubMed] [Google Scholar]
- 64.Bock R. Structure, function, and inheritance of plastid genomes. Top. Curr. Genet. 2007;19:29–63. [Google Scholar]
- 65.Koop H-U, Herz S, Golds TJ, Nickelsen J. The genetic transformation of plastids. Top. Curr. Genet. 2007;19:457–510. [Google Scholar]
- 66.Tregoning JS, Nixon P, Kuroda H, Svab Z, Clare S, Bowe F, Fairweather N, Ytterberg J, van Wijk KJ, Dougan G. Expression of tetanus toxin fragment C in tobacco chloroplasts. Nucleic Acids Res. 2003;31:1174–1179. doi: 10.1093/nar/gkg221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ruf S, Karcher D, Bock R. Determining the transgene containment level provided by chloroplast transformation. Proc. Natl Acad. Sci. USA. 2007;104:6998–7002. doi: 10.1073/pnas.0700008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Svab Z, Maliga P. Exceptional transmission of plastids and mitochondria from the transplastomic pollen parent and its impact on transgene containment. Proc. Natl Acad. Sci. USA. 2007;104:7003–7008. doi: 10.1073/pnas.0700063104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ma JK-C, Barros E, Bock R, Christou P, Dale PJ, Dix PJ, Fischer R, Irwin J, Mahoney R, Pezzotti M. Molecular farming for new drugs and vaccines. EMBO Rep. 2005;6:593–599. doi: 10.1038/sj.embor.7400470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bock R, Warzecha H. Solar-powered factories for new vaccines and antibiotics. Trends Biotechnol. 2010;28:246–252. doi: 10.1016/j.tibtech.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 71.Bock R, Kössel H, Maliga P. Introduction of a heterologous editing site into the tobacco plastid genome: the lack of RNA editing leads to a mutant phenotype. EMBO J. 1994;13:4623–4628. doi: 10.1002/j.1460-2075.1994.tb06784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.