Abstract
During DNA replication in Escherichia coli, single-stranded DNA-binding protein (SSB) protects single-stranded DNA from nuclease action and hairpin formation. It is known that the highly conserved C-terminus of SSB contacts the χ subunit of DNA polymerase III. However, there only exists a theoretical model in which the 11 C-terminal amino acids of SSB have been docked onto the surface of χ. In order to refine this model of SSB/χ interaction, we exchanged amino acids in χ and SSB by site-directed mutagenesis that are predicted to be of key importance. Detailed characterization of the interaction of these mutants by analytical ultracentrifugation shows that the interaction area is correctly predicted by the model; however, the SSB C-terminus binds in a different orientation to the χ surface. We show that evolutionary conserved residues of χ form a hydrophobic pocket to accommodate the ultimate two amino acids of SSB, P176 and F177. This pocket is surrounded by conserved basic residues, important for the SSB/χ interaction. Mass spectrometric analysis of χ protein cross-linked to a C-terminal peptide of SSB reveals that K132 of χ and D172 of SSB are in close contact. The proposed SSB-binding site resembles those described for RecQ and exonuclease I.
INTRODUCTION
Bacterial single-stranded DNA-binding (SSB) proteins are essential for cellular viability, as they play an important role in DNA metabolism (1,2). They bind sequence independently and with high affinity to patches of single-stranded DNA (ssDNA), providing protection from nuclease action and hairpin formation, thus stabilizing the nucleic acid in a conformation suitable for replication (3–5), recombination and repair (6–9).
The primary structure of bacterial SSB proteins consists of three different regions. The N-terminal part of the protein harbours the DNA-binding domain (OB-fold) (10–12), which is followed by a mostly unstructured region, rich in proline and glycine residues. The protein sequence is terminated by a highly conserved C-terminal region, containing a consensus sequence of six amino acid residues (DDDIPF).
Apart from their function in stabilizing and protecting ssDNA, SSB proteins interact directly with a number of other proteins. For Escherichia coli, it has been shown that there are at least 14 interaction partners, including exonuclease I, RecQ, primase and the χ subunit of DNA polymerase III (13–17). In all cases investigated so far, protein–protein interaction requires the conserved C-terminal part of SSB (18). Interestingly, SSB proteins present in eukaryotic organisms, for example human mitochondrial SSB, lack this C-terminal region (19). The conserved C-terminal part of the SSB protein from E. coli (EcoSSB) is essential for cellular viability, as SSB truncation variants lacking C-terminal amino acids cannot complement for wild-type protein (20). Moreover, the temperature-sensitive ssb-113 mutant strain, which shows an alteration in the penultimate amino acid residue of SSB (P176S) (21), can no longer replicate DNA at temperatures higher than 30°C (22) and shows hypersensitivity to DNA damage (23,24). SSB-113 and C-terminal truncation variants of SSB are also impaired in interaction with the χ subunit of DNA polymerase III (13,14,25).
The binding of χ to SSB has a modulating function in DNA replication, because it detaches primase from RNA primers (13), increases the affinity of SSB for ssDNA (25) and activates DNA polymerase III holoenzyme at physiological salt concentrations (14,26). Until now it is not entirely clear which amino acid residues of χ and SSB are involved in the interaction and how SSB contacts the surface of χ. So far, the only available model (27) comprises the 3D structure of E. coli χ (Ecoχ) (28) with the 11 C-terminal amino acids of Thermus aquaticus SSB (TaqSSB) docked onto its surface. In 2008, the structure of exonuclease I (ExoI) bound to a peptide including the last nine C-terminal amino acid residues of SSB was determined (29). The structure showed two adjacent SSB-binding sites on the surface of ExoI (A- and B-site), which both contain a hydrophobic pocket to provide space for the ultimate phenylalanine (F177) of SSB. These pockets are flanked by a basic rim to contact the acidic residues of the C-terminus of SSB. More recently, the binding site for the C-terminus of SSB on RecQ was identified by NMR analysis (30) and found to contain very similar structural elements. Upon comparison of these results, it seems likely that the presence of a hydrophobic pocket and a basic patch are common features present on the surface of a wider range of SSB interaction partners. In this study, the model of SSB/χ interaction was refined using a multifaceted approach that includes site-directed mutagenesis, cross-linking, analytical ultracentrifugation and mass spectrometry experiments in order to identify residues directly involved in SSB/χ interaction.
MATERIALS AND METHODS
Buffers and reagents
All materials were of the highest purity available and were obtained from Sigma, Pierce and J.T. Baker. Analytical ultracentrifugation experiments were carried out in a standard buffer containing 20 mM potassium phosphate pH 7.4, 0.3 M NaCl and 0.5 mM DTT (high salt buffer). For cross-linking experiments, the buffer contained 5 mM potassium phosphate pH 7.4, 5 mM NaCl, 8% (w/v) glycerol and 1 mM DTT (low salt buffer).
Protein concentrations were determined using the following extinction coefficients at 280 nm: 29 450 M−1 cm−1 for DNA polymerase III χ subunit wild-type and all its mutants, except χ Y119A, χ Y131A and χ Y131L, for which an extinction coefficient of 27 960 M−1 cm−1 was used; all of these extinction coefficients were calculated from amino acid composition using Sednterp (31,32); 113 000 M−1 cm−1 for EcoSSB wild-type and SSB+Gly (33). SSB concentrations are given in tetramers throughout the text.
Site-directed mutagenesis
Site-directed mutagenesis was performed with the QuikChange site-directed mutagenesis kit of Stratagene (LaJolla, CA, USA) and vectors pET-15b (Novagen) containing χ (pET-15bχ) (25) or pSF1 (34) containing SSB as templates. The point mutations were introduced using the following oligonucleotides (MWG Biotech., Germany); the antisense primers for each mutant have the reverse complementary sequence:
| χ V117F | 5′-CAGAAGTGGTAGACTTCTTTCCGTACGAAGATTCTC-3′ |
| χ V117I | 5′-CAGAAGTGGTAGACTTCATCCCGTACGAAGATTCTC-3′ |
| χ Y119A | 5′-GGTAGACTTCGTTCCTGCAGAAGATTCTCTG-3′ |
| χ K124A | 5′-CTTATGAAGATTCTCTGGCACAGCTGGCGCGCGAAC-3′ |
| χ K124M | 5′-CGTTCCTTATGAAGATTCTCTGATGCAGCTGGCGCGCGAACGC-3′ |
| χ R128A | 5′-CTCTGAAACAACTGGCCGCGGAACGCTATAAAGCCTAC-3′ |
| χ Y131A | 5′-CAACTGGCGCGCGAACGCGCTAAGGCCTACCGCGTGGC-3′ |
| χ Y131L | 5′-CAACTGGCGCGCGAACGCTTAAAGGCCTACCGCGTGGC-3′ |
| χ K132A | 5′-CTGGCGCGCGAGCGCTATGCAGCCTACCGCGTGGC-3′ |
| χ K132M | 5′-CTGGCGCGCGAGCGCTATATGGCCTACCGCGTGGC-3′ |
| SSB+Gly | 5′-GATGACATTCCGTTCGGTTAAGATATCAAAACAATAGGTTATATTG-3′ |
In the case of the χ mutants, the amino acid residue mentioned in the primer name was replaced by a different amino acid (e.g. in the case of χ K124A, the lysine residue at position 124 of χ was replaced by an alanine). However, for the SSB+Gly mutant, the stop codon after F177 of EcoSSB was replaced by a codon for glycine and a new stop codon was inserted downstream. All mutated constructs were checked for errors by sequencing the complete gene (GATC Biotech. AG, Germany).
Protein preparation
Escherichia coli strain BT317 (35) containing the plasmid pRK248, which codes for the thermosensitive λcI857 repressor, was used to produce SSB wild-type and the SSB+Gly mutant. The cells were transformed with pSF1 carrying the ssb gene under the control of the λ pL promoter. Protein expression was induced by a temperature shift from 30°C to 42°C and cells were harvested 3 h after induction. EcoSSB wild-type and SSB+Gly were purified as described by Lohman et al. (36), omitting the ssDNA cellulose affinity column. The purified protein was flash-frozen in N2 (liq) and stored at −80°C.
Vector pET-15bχ carries the gene of the χ subunit of DNA polymerase III under control of the T7 promoter (25) and was used to transform E. coli strain Rosetta (DE3) pLysS (Novagen). Protein expression and purification of χ wild-type and mutants was carried out according to Xiao et al. (37) with the following modifications: the ATP agarose column was omitted; after purification, the protein was precipitated with 500 g l−1 (NH4)2SO4, dissolved in 20 mM HEPES pH 6.9, 0.5 mM EDTA, 1 mM DTT, 10% (v/v) glycerol and then dialysed against 20 mM potassium phosphate pH 7.4, 1 M NaCl, 1 mM EDTA, 1 mM DTT, 60% (v/v) glycerol. The protein was stored at −20°C. After purification, the proteins were checked by analytical ultracentrifugation for homogeneity. All mutants showed the same c(s) distribution as wild-type protein, no hint for changes in tertiary structure due to the introduced mutations could be found.
Analytical ultracentrifugation
Sedimentation velocity experiments were performed in a XL-I analytical ultracentrifuge (Beckman Coulter, USA), using absorption optics (12 mm path length, λ = 280 nm). During the experiment, the absorption of the sample was obtained as a function of the radial position. Programming of the centrifuge and data recording was done using the computer software ProteomeLab™ (Beckman Coulter, USA). Each χ mutant was checked for its affinity to SSB by titrating a constant concentration of SSB (2.5 µM) with increasing concentrations of χ (from 2.5 to 30 µM), representing stoichiometries of 1:1 to 1:12. All experiments were performed under high salt conditions (see ‘Buffers and reagents’ section). Samples with a volume of 400 µl were centrifuged in double sector cells at 20°C and 50 000 rpm in an An50Ti rotor (Beckman Coulter, USA).
The measured data were evaluated with the program package SEDFIT (38). This program provides a model for diffusion-corrected differential sedimentation coefficient distributions [c(s) distributions]. As the areas under the separate peaks in c(s) distributions are a measure of the absorbance of the species represented by the peaks (39), this information can be used to determine binding isotherms (40).
Complementation assay
In E. coli strain RDP268, the chromosomal ssb gene is replaced by a kanamycin cassette (41). These cells carry the essential ssb gene on the additional plasmid pACYCssb, conferring chloramphenicol resistance. Using RDP268, it can be checked whether a mutant of ssb is able to functionally replace the SSB protein of E. coli in vivo.
The cells were transformed with a derivative of the single-copy plasmid pRE432 (42), carrying SSB+Gly under the control of the natural SSB promoter (pRE432PROMSSB+Gly, conferring ampicillin resistance). After transformation, the cells were transferred into a 4 ml culture of minimal medium (43) (1× M9 salts, 0.4% glucose, 60 mg l−1 histidine, 230 mg l−1 threonine, 100 mg l−1 proline, 580 mg l−1 arginine, 230 mg l−1 leucine, 170 mg l−1 thymine, 33.7 mg l−1 thiamine, 2 mM MgSO4, 0.1 mM CaCl2, pH 7.5) supplemented with 40 µg ml−1 ampicillin and 5 µg ml−1 kanamycin and grown overnight at 30°C with heavy shaking. Then 1 µl of this culture was transferred into a new 4 ml culture of minimal medium, containing the respective antibiotics. After six subsequent inoculations, each time allowing the cells to grow for 24 h, the cells were diluted in liquid M9 medium and plated on M9 agar plates containing 40 µg ml−1 ampicillin and 5 µg ml−1 kanamycin. Colonies that had lost pACYCssb were identified by replica plating on M9 agar plates containing 30 µg ml−1 chloramphenicol and 5 µg ml−1 kanamycin. Plasmid DNA was prepared from colonies sensitive to chloramphenicol and the complete ssb gene was sequenced (GATC Biotech. AG, Germany).
Cross-linking experiments
SSB-Carb is a peptide containing the last nine amino acids of EcoSSB and comprises the sequence WMDFDDDIPF (synthesized by Biomatik Corporation, USA). The N-terminal tryptophan residue was added to determine peptide concentration [extinction coefficient 5690 M−1 cm−1 at 280 nm, derived from amino acid composition (32)]. SSB-Carb was used in cross-linking reactions with χ wild-type, χ K124A and χ K132A, using the zero-length cross-linker 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC, Pierce), which is able to covalently link carboxyl groups to closely neighbouring amino groups under the formation of an amide bond. The reactions were performed in low salt buffer (see ‘Buffers and reagents’ section) with 50 µM χ wild-type or mutant, 200 µM SSB-Carb and 20 mM EDC. Samples and controls (lacking either EDC or peptide) were incubated for 2 h at room temperature. Then, the samples were diluted 1:5 in SDS loading buffer [50% (v/v) glycerol, 0.16 M Tris/HCl pH 6.8, 5% (v/v) β-mercaptoethanol, 2% (w/v) SDS, 0.01% (w/v) bromophenol blue] and heated to 95°C for 10 min. For mass spectrometric analysis acrylamide was added to the mixture up to a final concentration of 2%, followed by an incubation for 30 min at room temperature to inactivate free cysteine residues by alkylation. Samples and controls were analysed on a 17% SDS–PAGE (44).
Mass spectrometry
Samples for mass spectrometric analysis were prepared as described by Muetzelburg et al. (45): protein bands were excised from the gel and destained with 25 mM NH4HCO3 in 50% acetonitrile (ACN). After drying the gel pieces with 100% ACN and vacuum evaporation, they were rehydrated with 100 µl of 10 ng µl−1 modified trypsin (Serva) in 25 mM NH4HCO3 and 10% ACN. After incubating for 1 h on ice, residual trypsin solution was removed and substituted by 25 mM NH4HCO3 in 10% ACN. Incubation was carried out over night at 37°C. The supernatant was collected and the gel pieces were washed with 50% ACN in 0.2% trifluoroacetic acid (TFA) for 30 min and then shrunk with 100% ACN. All solutions containing peptides were collected, combined and dried. Before MS analysis, peptides were dissolved in 10 µl 10% ACN in 0.2% TFA and applied to a MALDI target plate (anchor target, Bruker Daltonic) using the dried droplet method: 1 µl sample and 0.8 µl α-hydroxy-cinnamonic acid solution (4 mg ml−1 in 50% ACN in 0.2% TFA) were directly mixed on the target plate and dried at room temperature. MS and MS/MS spectra were generated in an Ultraflex TOF/TOF I (Bruker Daltonic) mass spectrometer. External calibration was done using a peptide calibration standard from Bruker Daltonic. For protein identification a Mascot search was performed. The MS/MS spectra were matched with the Swissprot database. Search parameters for mass tolerance were set to 100 ppm for precursor ions and 0.7 Da for fragment ions, allowing one missed trypsin cleavage.
Modelling
Two binding sites for EcoSSB (A- and B-site) were identified in the X-ray structure of ExoI in the presence of a C-terminal peptide of SSB (29). Comparison of ExoI and the χ subunit of DNA polymerase III of E. coli [pdb code: 1EM8 (28)] revealed significant similarity between structural motifs of the ExoI B-site and the area of the proposed SSB-binding site of χ formed by the helix α4 and strands β1 and β7. The structural comparison of ExoI and χ was performed using the program package Coot (46). The secondary structure matching algorithm (SSM) (47) was used to align the structurally conserved parts of the two proteins. The positions of the last three amino acids of SSB bound to the B-site in the superimposed structure of ExoI complexed with a SSB C-terminal peptide were used as a starting geometry for building the new model of the SSB/χ complex. The amino acid residues D170–D174 of the peptide were built on the proposed SSB-binding area of χ according to stereochemical criteria using the program Coot. The contact between K132 of χ and D172 of SSB detected by mass spectrometry (Figure 4 and Supplementary Figure S2) was accounted for in the initial geometry. This starting model was subsequently subjected to energy minimization, followed by molecular dynamics (MD) simulations during which the positions of the last two peptide residues and the contact between K132 of χ and D172 of SSB were restrained. Thus, the MD simulations involved residues D170–I175 of SSB and the side chains of χ forming the SSB-binding area, whereas the rest of the χ molecule was considered as producing a fixed potential field. The resulting model after 10 ns of MD equilibration at room temperature and final energy minimization displays good stereochemistry. MD simulations and energy minimization procedures were performed using the programs HyperChem (Hypercube, Inc.) and CNS (48).
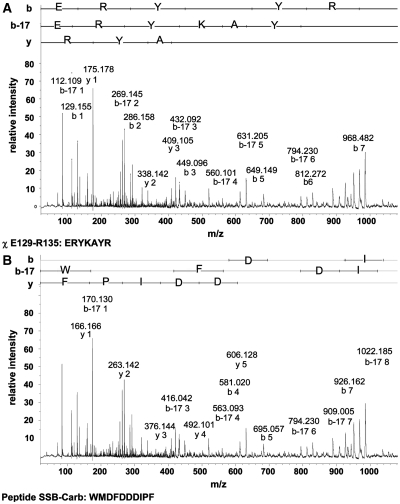
Figure 4.
MS/MS analysis of a peptide consisting of amino acid residues E129-R135 of χ cross-linked to SSB-Carb. Parent mass of 2248.98 was analysed by MALDI TOF/TOF. MS/MS spectra revealed sophisticated fragments of both peptides. MS/MS spectra are shown in the m/z range from 10 to 1100. Larger fragments could not be assigned to the modified peptide. All fragments assigned to y-, b- and b-17 ions are indicated by their m/z values. (A) Fragments originating from the E129–R135 peptide of the χ protein are indicated. (B) Fragments generated from the SSB-Carb peptide are labelled.
RESULTS AND DISCUSSION
In order to test and further refine the interaction model of SSB and the χ subunit of DNA polymerase III of E. coli (27), different mutations were inserted into Ecoχ and EcoSSB by site-directed mutagenesis. In the published model, where the C-terminus of the EcoSSB homologue, TaqSSB, was docked onto the surface of χ, the highly conserved positively charged residues K124, R128, K132 and R135 of χ form hydrogen bonds with the negatively charged part of the C-terminus of TaqSSB. Additionally, the ultimate phenylalanine of TaqSSB makes a stacking interaction with Y119 of χ.
Interaction of EcoSSB and χ
When two proteins interact they form a complex of a larger mass, which usually sediments faster than both proteins alone. Supplementary Figure S1 shows diffusion-corrected sedimentation coefficient distributions [c(s) distributions] obtained from analytical ultracentrifugation experiments for the interaction of wild-type EcoSSB and Ecoχ under high salt conditions. Whereas free Ecoχ showed a sedimentation coefficient of 1.8 S under these conditions, free EcoSSB sedimented with an s-value of 3.8 S, which increased to 5.3 S in the presence of a 12-fold excess of χ, indicating complex formation. As only a single reaction boundary containing EcoSSB and all EcoSSB/χ complexes formed in the presence of an excess of χ, the reaction must be fast compared to the timescale of sedimentation (49). When different ratios of EcoSSB and Ecoχ were applied, the sedimentation coefficient of this reaction boundary increased with increasing amounts of χ. As previously described for the interaction of TaqSSB and Ecoχ (40), integration of the two peaks in the c(s) distribution can be used to determine the concentrations of free and bound χ.
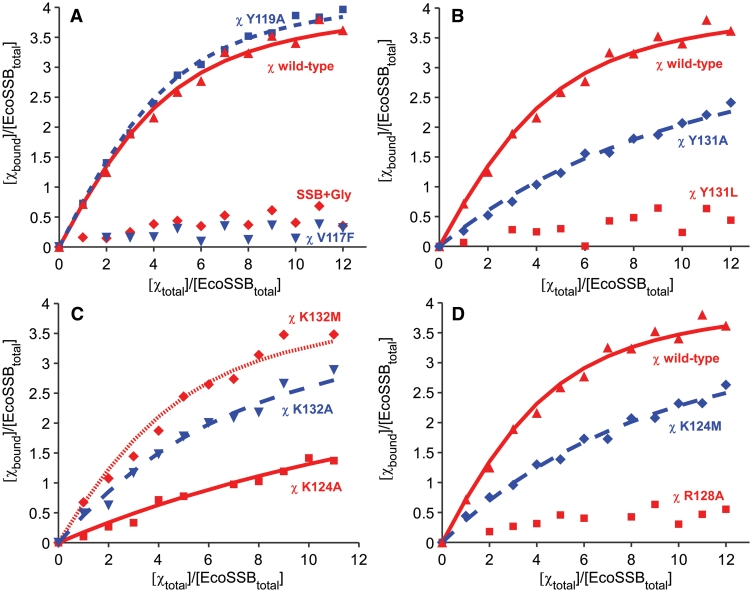
The amount of bound χ per EcoSSB was plotted against the amount of total χ per EcoSSB and a binding isotherm for independent binding of n χ molecules to one EcoSSB molecule was fitted to the data (Figure 1A) (25). The resulting binding isotherm shows that one EcoSSB tetramer can bind up to four molecules of χ with an affinity (KA) of about 3 × 105 M−1 (Figure 1A). These results are in good agreement with previously published data (25,50). An overview of all binding constants determined in this study can be found in Table 1.
Figure 1.
Interaction of different mutants of EcoSSB and χ under high salt conditions. An amount of 2.5 µM of EcoSSB was mixed with different amounts of χ in a buffer containing 20 mM potassium phosphate pH 7.4, 0.3 M NaCl and 0.5 mM DTT and analysed in sedimentation velocity experiments in an analytical ultracentrifuge. In A, B and D, a binding isotherm fitted to the data of the interaction of EcoSSB and χ wild-type (red triangle) with KA = (2.9 ± 1) × 105 M−1 and n = 4.2 is shown for comparison (solid red line). (A) Exchanging V117 to phenylalanine (inverted blue triangle) dramatically lowered the binding affinity between χ and EcoSSB. Also an extension of SSB by a C-terminal glycine (SSB+Gly) disabled interaction with χ (red rhombus). Replacing Y119 by alanine (blue square) did not significantly change the affinity of χ to SSB [dashed blue line: KA = (3.4 ± 1) × 105 M−1 and n = 4.2]. (B) Exchanging tyrosine 131 of χ to leucine dramatically lowered the binding affinity to SSB (red square), whereas χ Y131A retained some of its activity (blue rhombus). The dashed blue line represents a binding isotherm fitted to the data of χ Y131A binding to EcoSSB with KA = (4.8 ± 1) × 104 M−1, n = 4.2. (C) The K124A mutation had a strong effect on the SSB/χ interaction (red square), lowering KA to (2.1 ± 0.5) × 104 M−1 with n = 4.2 (solid red line). The χ K132A mutant (blue triangle) also displayed a lowered binding affinity to SSB [dashed blue line: KA = (9 ± 2.5) × 104 M−1 and n = 4.2]. χ K132M (red rhombus) behaved similar to wild-type protein [fine dashed red line: KA = (2.1 ± 1) × 105 M−1 and n = 4.2]. (D) In order to test the new model of SSB/χ interaction, the K124M and R128A mutants of χ were generated. Both showed effects in accordance with the new model, with K124M (blue rhombus) lowering the binding affinity to SSB to a fifth compared to wild-type [dashed blue line, KA = (6 ± 1) × 104 M−1 and n = 4.2] and R128A (red squares) disabling the interaction with SSB.
Table 1.
Overview of binding constants of examined protein variants as determined by analytical ultracentrifugation
| Protein | Binding constant (KA) |
|---|---|
| χ wild-type | (2.9 ± 1) × 105 M−1 |
| χ V117F | ND |
| χ V117I | ND |
| χ Y119A | (3.4 ± 1) × 105 M−1 |
| χ K124A | (2.1 ± 0.5) × 104 M−1 |
| χ K124M | (6 ± 1) × 104 M−1 |
| χ R128A | ND |
| χ Y131A | (4.8 ± 1) × 104 M−1 |
| χ Y131L | ND |
| χ K132A | (9 ± 2.5) × 104 M−1 |
| χ K132M | (2.1 ± 1) × 105 M−1 |
| SSB+Gly | ND |
In the case of the V117F/I, R128A and Y131L mutants of χ and the C-terminal extension mutant of SSB, SSB+Gly, the binding affinity was so weak that no binding constant could be determined (ND).
V117 mutants confirm the SSB-binding area on the surface of χ
The first two mutations in Ecoχ, V117F and V117I, were selected on the basis of structure analysis and homology modelling of the published SSB/χ interaction (27). According to the model, these mutations should increase the affinity between the hydrophobic part of the C-terminus of SSB and χ. Unexpectedly, the substitution of V117 of Ecoχ by phenylalanine or isoleucine had the contrary effect. Both mutants, χ V117F (Figure 1A) and χ V117I (data not shown) displayed a dramatic loss of affinity to EcoSSB and no binding isotherm could be fitted to the data. Thus, V117 proved to be crucial for the interaction of SSB and χ, showing that the correct binding surface on χ was depicted in the theoretical model (27), but indicating that the orientation of the C-terminus of SSB had to be different.
Tyrosine 119 of χ does not form a stacking interaction with the ultimate phenylalanine of SSB
Due to the unexpected properties of mutants at position V117 of χ in regard to the published model, additional mutations were done to further investigate the orientation of the C-terminus of SSB on χ. Since the model shows that the terminal carboxyl group of SSB is not engaged in an interaction, a C-terminal extension mutant of EcoSSB, SSB+Gly, was checked for its binding properties to wild-type χ. SSB+Gly carries one additional glycine residue at the C-terminus and showed an effect comparable to mutations at position V117 of χ in analytical ultracentrifugation experiments. Again the affinity was so weak that a binding isotherm could not be fitted to the data (Figure 1A). In complementation experiments in RDP268 (see ‘Materials and Methods’ section), SSB+Gly was able to replace EcoSSB wild-type. However, an exchange was only possible under conditions of slow replication (minimal medium and 30°C) and the resulting cells formed smaller colonies than cells complemented with wild-type SSB, indicating that SSB+Gly is also functionally impaired in vivo.
These results disagree with the published model, since the proposed stacking interaction between Y119 of χ and F177 of SSB should position the last amino acids of SSB in such a way that an additional C-terminal glycine residue should not interfere with the binding to χ. In order to check whether Y119 is actually involved in a stacking interaction, it was replaced by alanine. The resulting binding constant of the Y119A mutant was similar to χ wild-type, showing that Y119 is not crucial for the SSB/χ interaction (Figure 1A). This indicates that F177 of SSB and Y119 of χ are not involved in a stacking interaction stabilizing the SSB/χ complex. Thus, the ultimate phenylalanine of SSB must be accommodated elsewhere on the surface of χ, most probably in a well- defined pocket, which would explain the dramatic decrease in affinity of the SSB+Gly mutant.
A conserved hydrophobic pocket on the surface of χ is involved in the interaction with SSB
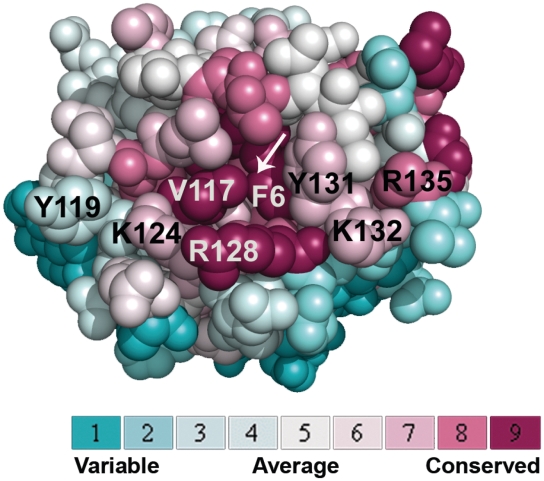
The mutants examined show that the SSB-binding area on the surface of χ presented in the published model is correct (27), but the orientation of the C-terminus of SSB is different. In order to find out which residues of χ apart from V117 are involved in the interaction with SSB, an alignment of homologous χ proteins was projected on the known 3D structure of Ecoχ using the ConSurf server (51) (Figure 2). The results of this analysis revealed that an array of highly conserved residues on the surface of Ecoχ resembles the SSB-binding sites on ExoI and RecQ (29,30). All three contain a hydrophobic pocket surrounded by basic residues.
Figure 2.
Highly conserved hydrophobic and basic residues on the surface of Eco χ. An alignment of homologous χ proteins was projected on the known 3D structure of Ecoχ [pdb code: 1EM8 (28)] using the ConSurf server (51). A hydrophobic pocket, which could accommodate F177 of SSB, is labelled by a white arrow. Like the SSB-binding sites on ExoI and RecQ (29,30), it is surrounded by a stretch of basic residues. The colour legend shows the conservation scores of the respective residues.
The highly conserved hydrophobic pocket present on the surface of χ is created by F6, V117 and Y131 (Figure 2, white arrow). If this pocket accommodated F177 of SSB, it could explain the dramatic decrease in affinity of both the χ V117 and the SSB+Gly mutants (Figure 1A). Since this pocket is formed by the side chains of F6, V117 and Y131, not only an exchange of V117, but also of Y131 should affect the binding affinity. Therefore, tyrosine 131 of χ was replaced by alanine and leucine to address the importance of the identified hydrophobic pocket for the SSB/χ interaction. At this position, a mutation into an alanine (χ Y131A) led to a 6-fold decrease in binding affinity to SSB (Figure 1B). The χ Y131L mutant showed such a small affinity to SSB that no binding isotherm could be fitted to the data (Figure 1B). Since leucine is large and flexible, it could block the access of the C-terminal phenylalanine of EcoSSB to the hydrophobic pocket. On the other hand, the small alanine side chain should not lead to sterical hindrance, but allow less hydrophobic interactions.
K124 and K132 of χ participate in SSB binding
Four highly conserved basic residues are close to the hydrophobic pocket on the surface of χ: K124, R128, K132 and R135 (Figure 2). All of them can possibly interact with the negatively charged aspartates in the C-terminus of SSB. In order to assess the importance of the lysine residue at position 124 of χ, it was substituted by alanine. The binding affinity of the resulting χ K124A mutant to SSB dramatically dropped to less than a tenth compared to wild-type χ (Figure 1C). Since K124 is in close proximity to the supposed hydrophobic binding pocket for F177 of SSB, it may be involved in an electrostatic interaction with the ultimate carboxyl group of the C-terminus of SSB. On the other hand, K124 may interact with one of the aspartates in the C-terminal part of SSB.
Apart from K124, lysine 132 appears to be important for the interaction between SSB and χ. An exchange to alanine at this position lowered the binding constant to a third compared to wild-type (Figure 1C). An exchange to methionine, however, had no significant effect on the binding affinity. Therefore, under high salt conditions as used in the analytical ultracentrifugation experiments, the positive charge of the lysine does not seem to be important at this position, but rather the size of the side chain. Since methionine strongly resembles lysine in size, K132 is most probably involved in hydrophobic interactions with amino acids of the C-terminus of SSB.
K132 of χ is in close contact to D172 of SSB
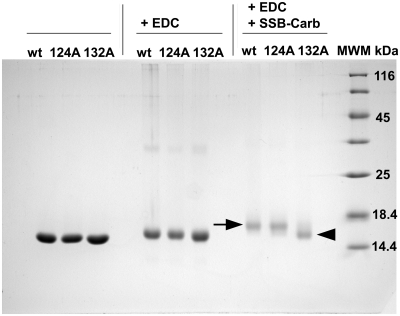
The results from analytical ultracentrifugation showed that V117, K124, Y131 and K132 of χ are involved in SSB binding. However, the interaction partners of these residues in the C-terminus of SSB could not be identified. Therefore, chemical cross-linking reactions with χ and a peptide comprising the last nine C-terminal amino acids of EcoSSB (SSB-Carb) were performed. As the C-terminal sequence of SSB contains many aspartates that may interact with basic residues, EDC was chosen as cross-linking reagent. This chemical compound is a zero-length cross-linker that forms amide bonds between closely neighbouring carboxyl and amino groups (see ‘Materials and Methods’ section). Incubation of wild-type χ with SSB-Carb and EDC resulted in a shift of the protein band of χ in SDS–PAGE (Figure 3, black arrow). Therefore, either a surface lysine of χ is located very close to one of the aspartates in the C-terminus of SSB or it is the C-terminal carboxyl group of the SSB-Carb peptide that interacts with a surface lysine of χ.
Figure 3.
Cross-link of different variants of χ with the SSB-Carb peptide. Reactions were performed in 5 mM potassium phosphate buffer pH 7.4, 5 mM NaCl, 8% (w/v) glycerol and 1 mM DTT, using 50 µM χ wild-type or mutant, 200 µM SSB-Carb and 20 mM EDC. χ wild-type and χ K124A mutant could be cross-linked with the SSB-Carb peptide, using EDC (see black arrow). The band of the χ K132A mutant, however, was not shifted upon reaction with SSB-Carb and EDC (see black arrowhead). wt = wild-type χ; 124A = χ K124A; 132A = χ K132A; 17% SDS–PAGE .
In order to find out which residues had reacted with EDC, the cross-linked χ protein sample and an unmodified χ protein were digested with trypsin and a peptide mass fingerprint was generated (Supplementary Figure S2). Masses corresponding to peptide fragments of χ were detected. Cross-linking with EDC and the SSB-Carb peptide (H2N-WMDFDDDIPF-COOH, Mr = 1299.51) should result in a mass increase of 1281.51 because water is lost during the reaction. In the cross-linked sample, additional masses were identified that corresponded to the cross-linked peptides Y131–R135 and E129–R135 of χ and exhibited masses of 1963.89 (MH+-H2O) and 2248.98 (MH+-H2O), respectively. A neutral loss of water and two intensive oxygen adducts, which could be located on the tryptophan and methionine residues at the N-terminus of the SSB-Carb peptide, were detected for each of the peptides. As K132 is the only lysine residue present in both of the identified sequences of χ, this residue must have been involved in the cross-link. Adduct masses for peptides containing K124 could not be detected. Tandem MS analysis revealed similar MS/MS spectra for the peptides and their corresponding oxygen adducts and displayed a sophisticated analysis of fragments of both the χ and the SSB-Carb peptides. MS/MS analysis of the parent ion of m/z = 2248.98 is shown in Figure 4. This method identified a C-terminal amino acid sequence of the SSB-Carb peptide (DDIPF) and an amino acid sequence of χ (E129–R135) in y and b ion series. Both sequences being detected in the same adduct mass prove that K132 of χ is involved in the interaction. The determined y ion series of SSB (F177–D173) was disrupted after D173, strongly indicating that D172 was cross-linked to K132 of χ (Figure 4B).
The results from mass spectrometric analysis, implying a covalent bond between K132 of χ and D172 of SSB, were checked by performing cross-linking reactions with the K124A and K132A mutants of χ and the SSB-Carb peptide, using EDC. Whereas the χ K124A mutant could be cross-linked to the peptide to the same extent as χ wild-type, shown by a clear decrease in migration velocity for the complete χ band (Figure 3, black arrow), no change in migration velocity was observed in the case of χ K132A (Figure 3, black arrowhead). These findings confirmed the results of mass spectrometric analysis, which could only detect masses for a cross-link involving K132 of χ. Therefore, it is lysine 132 of χ that is close enough to aspartate 172 of EcoSSB to be covalently linked by the zero-length cross-linking reagent EDC.
New model of SSB/χ interaction
The experimental results provided by analytical ultracentrifugation, chemical cross-linking, mass spectrometry and superposition of the structurally conserved parts of ExoI and χ (see ‘Materials and Methods’ section) formed the basis for the construction of a new model of SSB/χ interaction. Superposition of the two structures shows that the hydrophobic pocket on χ, identified by analytical ultracentrifugation, structurally resembles one of the two binding pockets of ExoI (B-site) capable of interaction with the C-terminus of SSB (29). Distances of selected residues determined from the new model of SSB/χ interaction can be found in Supplementary Table S1 and Supplementary Figure S3. In the new SSB/χ interaction model, the two hydrophobic amino acid residues V117 and Y131, which were found to be important for the SSB/χ interaction (Figure 1A and B), are involved in the formation of a pocket on the surface of χ that accommodates the ultimate two amino acid residues of SSB: P176 and F177 (Figure 5). The bottom of the binding pocket is formed by residue F6. Other residues of χ contributing to this pocket are L8 and T143. The hydroxyl group of T143 forms an electrostatic interaction with the carbonyl group of P176 of SSB. The terminal methyl group of T143 makes a hydrophobic contact with F177 of SSB, which in turn is involved in hydrophobic interactions with L8 and V117 of χ (Supplementary Table S1). Replacement of residue V117 of χ with an amino acid carrying a larger side chain, such as isoleucine or phenylalanine, would displace F177 of SSB from its hydrophobic binding site, leading to a significant decrease in the specificity of the SSB/χ interaction. Thus, the model explains the almost complete loss of affinity to SSB that became apparent when both the V117F and V117I mutants of χ were analysed by analytical ultracentrifugation (Figure 1A). Another part of the hydrophobic binding pocket on the surface of χ is formed by Y131 (Figure 5) making contact to P176 and F177 of SSB. These interactions would be lost upon replacement of Y131 with alanine. Replacing Y131 by an amino acid with a more flexible side chain, such as leucine, however, would lead to total displacement of F177 of SSB from the hydrophobic pocket. These findings explain why the Y131L mutant of χ shows a nearly complete loss of binding affinity to SSB, whereas χ Y131A still retains some of its activity (Figure 1B).
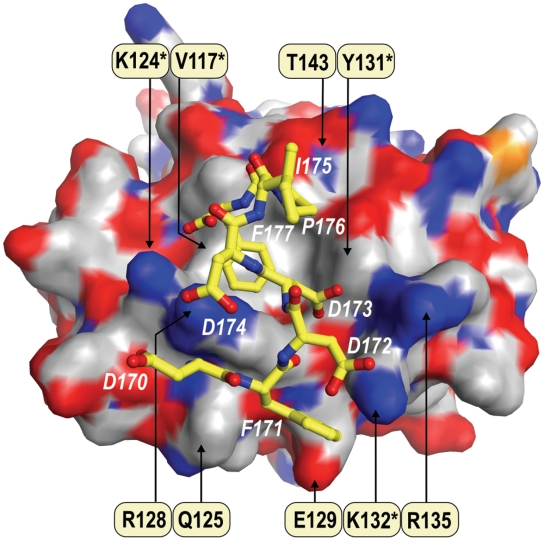
Figure 5.
The proposed model of SSB/χ interaction. The surface of χ [pdb code: 1EM8 (28)] is coloured according to the atomic charge: positive (blue), negative (red) and neutral (light grey). The residues of χ labelled in black are involved in the interaction with SSB. The asterisk indicates residues which were found to be important for the interaction in analytical ultracentrifugation and protein cross-linking experiments. The ball-and-stick model represents the last eight amino acids of the C-terminus of EcoSSB (white labels) bound to χ.
At the rim of the hydrophobic pocket, which accommodates P176 and F177 of SSB, R128 forms a hydrogen bond with D174 of SSB. Upon interaction with the C-terminal peptide of SSB, R128 changes its position by ∼1.4 Å and closes the pocket around the ultimate phenylalanine of SSB. In the case of the SSB+Gly mutant (Figure 1A), the C-terminal glycine would occupy the space between F177 of SSB and R128 of χ, thus sterically hindering the movement of R128 and preventing the closing of the binding pocket. Similar to R128, K124 of χ is involved in both electrostatic and hydrophobic interactions with the C-terminus of SSB (Figure 5). While the amino group of K124 participates in salt bridge formation with the carboxyl group of F177, the aliphatic part of its side chain interacts with the aromatic ring of the phenylalanine, closing one side of the hydrophobic pocket. As the interaction of K124 with the carboxyl group of F177 would also be abolished in the case of the SSB+Gly mutant, the observations concerning K124 and R128 of χ both explain why SSB+Gly has no detectable affinity to χ wild-type in analytical ultracentrifugation experiments (Figure 1A). Additionally, the interaction between F177 and K124 would be lost upon replacement of the latter residue by alanine, thus explaining the dramatic loss of binding affinity in the case of the χ K124A mutant (Figure 1C).
As presented in the model, the carboxyl group of D173 of SSB undergoes electrostatic interactions with R135 of χ. In addition, Cβ of D173 is engaged in hydrophobic interactions with the aliphatic part of R128 of χ and the hydrophobic residues P176 and F177 of SSB. Mass spectrometric analysis of chemically cross-linked SSB/χ complexes under low salt conditions using the zero-length cross-linker EDC had revealed that D172 of SSB and K132 of χ were in close contact (Figures 3 and 4). In the new model of the SSB/χ complex these residues are involved in salt bridge formation (Supplementary Table S1). The aliphatic part of the K132 side chain together with those of Q125, R128 and E129 forms a shallow hydrophobic pocket on the surface of χ. This pocket accommodates the second phenylalanine of the C-terminus of SSB, F171. A large hydrophobic residue such as methionine could easily replace K132 of χ in the shallow pocket without disturbing the interaction with F171. The latter is consistent with the results of analytical ultracentrifugation experiments with mutant K132M of χ that displays no apparent change in affinity compared to wild-type. The lack of effect of the electrostatic interaction on binding affinity is due to the high salt conditions used in this experiment, leading to a strong reduction of the Coulomb contribution to the binding energy. Substituting K132 by an alanine residue, however, diminishes the hydrophobic interactions with F171, explaining the negative effect of the K132A mutation on the binding affinity of χ to SSB (Figure 1C).
In order to test the new model of SSB/χ interaction, two additional mutations, K124M and R128A, were introduced in χ. When the resulting mutant proteins were analysed by analytical ultracentrifugation (Figure 1D), their binding affinities for SSB were found to be in good accordance with our new model (Figure 5). As the model shows, K124 of χ does not only interact with the C-terminal carboxyl group of SSB, but is also involved in hydrophobic interactions with the aromatic ring of F177. Therefore, a mutation of K124 to methionine, a residue which shows a substantially higher hydrophobicity than alanine, should have a lower effect on the binding affinity to SSB than the K124A mutation. Accordingly, the affinity of K124M was 3-fold higher than in the case of K124A (Figure 1C and D). An exchange of R128 to alanine, however, resulted in a nearly complete loss of binding affinity to SSB (Figure 1D). In our model, this residue is not only involved in a hydrogen bond with D174 of SSB but also closes the hydrophobic binding pocket of χ around the ultimate phenylalanine of SSB. Therefore, our model also explains the strong negative effect of the R128A mutation on the binding affinity of χ to SSB.
Our model also explains why SSB-113 (21), a mutant of EcoSSB carrying a serine residue at position 176 instead of a proline, is impaired in the interaction with χ, leading to aberrant DNA replication (22). According to the new SSB/χ interaction model, the stabilizing effect of proline 176 involves a hydrophobic contact with Y131 and the reduction of the conformational flexibility of the C-terminal region, thus, providing a favourable entropic effect for the peptide-binding affinity. Mutation of the proline to a polar serine residue is predicted to increase peptide flexibility and diminish its hydrophobic interactions, making the contact with the surface of χ less favourable.
SUMMARY
SSB proteins are essential for DNA metabolism (18). Apart from their function in DNA recombination and repair, they are involved in replication. In E. coli, SSB contacts the χ subunit of DNA polymerase III, thus activating the replicase at physiological ionic strength (14,26). Although it is known that the highly conserved C-terminus of SSB is needed for this interaction and a preliminary model of SSB/χ interaction has been published (27), the exact residues involved in each of the binding partners were not identified until now.
Results from site-directed mutagenesis, analytical ultracentrifugation, chemical cross-linking and mass spectrometry experiments were combined to refine the model of SSB/χ interaction. The newly proposed SSB-binding site of χ consists of a hydrophobic binding pocket, formed by L8, V117, Y131 and T143, which accommodates the ultimate amino acid residues of SSB, P176 and F177. Similar hydrophobic pockets are present in the other two known structures of SSB-binding sites of ExoI and RecQ (29,30) and could therefore be also implicated in interactions of SSB with other proteins (for a comparison of residues involved in the interaction of ExoI, RecQ and χ with SSB, see Supplementary Table S2). In addition to that, the new model presents a stretch of conserved basic residues, including K124, R128, K132 and R135, on the surface of χ that interacts with negatively charged residues in the C-terminal part of SSB and strongly resembles the basic region also present on the surfaces of ExoI and RecQ.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding to pay the open access charge: Hannover Medical School.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Lidia Litz for excellent technical assistance, Olga Shpigelman for help with mutagenesis and protein preparation and Drs Claus Urbanke and Peter Friedhoff for valuable discussions.
REFERENCES
- 1.Lohman TM, Ferrari ME. Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu. Rev. Biochem. 1994;63:527–570. doi: 10.1146/annurev.bi.63.070194.002523. [DOI] [PubMed] [Google Scholar]
- 2.Mushegian AR, Koonin EV. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl Acad. Sci. USA. 1996;93:10268–10273. doi: 10.1073/pnas.93.19.10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sigal N, Delius H, Kornberg T, Gefter ML, Alberts B. A DNA-unwinding protein isolated from Escherichia coli: its interaction with DNA and with DNA polymerases. Proc. Natl Acad. Sci. USA. 1972;69:3537–3541. doi: 10.1073/pnas.69.12.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Molineux IJ, Pauli A, Gefter ML. Physical studies of the interaction between the Escherichia coli DNA binding protein and nucleic acids. Nucleic Acids Res. 1975;2:1821–1837. doi: 10.1093/nar/2.10.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuzminov A. A mechanism for induction of the SOS response in E. coli: insights into the regulation of reversible protein polymerization in vivo. J. Theor. Biol. 1995;177:29–43. doi: 10.1006/jtbi.1995.0222. [DOI] [PubMed] [Google Scholar]
- 6.Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol. Rev. 1990;54:342–380. doi: 10.1128/mr.54.4.342-380.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuzminov A. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol. Mol. Biol. Rev. 1999;63:751–813. doi: 10.1128/mmbr.63.4.751-813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuzminov A. DNA replication meets genetic exchange: chromosomal damage and its repair by homologous recombination. Proc. Natl Acad. Sci. USA. 2001;98:8461–8468. doi: 10.1073/pnas.151260698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams KR, Spicer EK, LoPresti MB, Guggenheimer RA, Chase JW. Limited proteolysis studies on the Escherichia coli single-stranded DNA binding protein. Evidence for a functionally homologous domain in both the Escherichia coli and T4 DNA binding proteins. J. Biol. Chem. 1983;258:3346–3355. [PubMed] [Google Scholar]
- 11.Casas-Finet JR, Khamis MI, Maki AH, Chase JW. Tryptophan 54 and phenylalanine 60 are involved synergistically in the binding of E. coli SSB protein to single-stranded polynucleotides. FEBS Lett. 1987;220:347–352. doi: 10.1016/0014-5793(87)80844-x. [DOI] [PubMed] [Google Scholar]
- 12.Raghunathan S, Kozlov AG, Lohman TM, Waksman G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat. Struct. Biol. 2000;7:648–652. doi: 10.1038/77943. [DOI] [PubMed] [Google Scholar]
- 13.Yuzhakov A, Kelman Z, O'Donnell M. Trading places on DNA–a three-point switch underlies primer handoff from primase to the replicative DNA polymerase. Cell. 1999;96:153–163. doi: 10.1016/s0092-8674(00)80968-x. [DOI] [PubMed] [Google Scholar]
- 14.Kelman Z, Yuzhakov A, Andjelkovic J, O'Donnell M. Devoted to the lagging strand-the χ subunit of DNA polymerase III holoenzyme contacts SSB to promote processive elongation and sliding clamp assembly. EMBO J. 1998;17:2436–2449. doi: 10.1093/emboj/17.8.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sandigursky M, Mendez F, Bases RE, Matsumoto T, Franklin WA. Protein-protein interactions between the Escherichia coli single-stranded DNA-binding protein and exonuclease I. Radiat. Res. 1996;145:619–623. [PubMed] [Google Scholar]
- 16.Genschel J, Curth U, Urbanke C. Interaction of E. coli single-stranded DNA binding protein (SSB) with exonuclease I. The carboxy-terminus of SSB is the recognition site for the nuclease. Biol. Chem. 2000;381:183–192. doi: 10.1515/BC.2000.025. [DOI] [PubMed] [Google Scholar]
- 17.Shereda RD, Bernstein DA, Keck JL. A central role for SSB in Escherichia coli RecQ DNA helicase function. J. Biol. Chem. 2007;282:19247–19258. doi: 10.1074/jbc.M608011200. [DOI] [PubMed] [Google Scholar]
- 18.Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 2008;43:289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Curth U, Urbanke C, Greipel J, Gerberding H, Tiranti V, Zeviani M. Single-stranded-DNA-binding proteins from human mitochondria and Escherichia coli have analogous physicochemical properties. Eur. J. Biochem. 1994;221:435–443. doi: 10.1111/j.1432-1033.1994.tb18756.x. [DOI] [PubMed] [Google Scholar]
- 20.Curth U, Genschel J, Urbanke C, Greipel J. In vitro and in vivo function of the C-terminus of Escherichia coli single-stranded DNA binding protein. Nucleic Acids Res. 1996;24:2706–2711. doi: 10.1093/nar/24.14.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang TC, Smith KC. Effects of the ssb-1 and ssb-113 mutations on survival and DNA repair in UV-irradiated delta uvrB strains of Escherichia coli K-12. J. Bacteriol. 1982;151:186–192. doi: 10.1128/jb.151.1.186-192.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chase JW, L'Italien JJ, Murphy JB, Spicer EK, Williams KR. Characterization of the Escherichia coli SSB-113 mutant single-stranded DNA-binding protein. Cloning of the gene, DNA and protein sequence analysis, high pressure liquid chromatography peptide mapping, and DNA-binding studies. J. Biol. Chem. 1984;259:805–814. [PubMed] [Google Scholar]
- 23.Greenberg J, Berends LJ, Donch J, Green MH. exrB: a malB-linked gene in Escherichia coli B involved in sensitivity to radiation and filament formation. Genet. Res. 1974;23:175–184. doi: 10.1017/s0016672300014798. [DOI] [PubMed] [Google Scholar]
- 24.Johnson BF. Two-dimensional electrophoretic analysis of the regulation of SOS proteins in three ssb mutants. Arch. Microbiol. 1984;138:106–112. doi: 10.1007/BF00413009. [DOI] [PubMed] [Google Scholar]
- 25.Witte G, Urbanke C, Curth U. DNA polymerase III chi subunit ties single-stranded DNA binding protein to the bacterial replication machinery. Nucleic Acids Res. 2003;31:4434–4440. doi: 10.1093/nar/gkg498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glover BP, McHenry CS. The chi psi subunits of DNA polymerase III holoenzyme bind to single-stranded DNA-binding protein (SSB) and facilitate replication of an SSB-coated template. J. Biol. Chem. 1998;273:23476–23484. doi: 10.1074/jbc.273.36.23476. [DOI] [PubMed] [Google Scholar]
- 27.Fedorov R, Witte G, Urbanke C, Manstein DJ, Curth U. 3D structure of Thermus aquaticus single-stranded DNA-binding protein gives insight into the functioning of SSB proteins. Nucleic Acids Res. 2006;34:6708–6717. doi: 10.1093/nar/gkl1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gulbis JM, Kazmirski SL, Finkelstein J, Kelman Z, O'Donnell M, Kuriyan J. Crystal structure of the chi:psi sub-assembly of the Escherichia coli DNA polymerase clamp-loader complex. Eur. J. Biochem. 2004;271:439–449. doi: 10.1046/j.1432-1033.2003.03944.x. [DOI] [PubMed] [Google Scholar]
- 29.Lu D, Keck JL. Structural basis of Escherichia coli single-stranded DNA-binding protein stimulation of exonuclease I. Proc. Natl Acad. Sci. USA. 2008;105:9169–9174. doi: 10.1073/pnas.0800741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shereda RD, Reiter NJ, Butcher SE, Keck JL. Identification of the SSB binding site on E. coli RecQ reveals a conserved surface for binding SSB's C terminus. J. Mol. Biol. 2009;386:612–625. doi: 10.1016/j.jmb.2008.12.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laue T, Shah B, Ridgeway T, Pelletier S. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding S, Rowe A, Horton J, editors. Cambridge, UK: The Royal Society of Chemistry; 1992. pp. 378–387. [Google Scholar]
- 32.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lohman TM, Overman LB. Two binding modes in Escherichia coli single strand binding protein-single stranded DNA complexes. Modulation by NaCl concentration. J. Biol. Chem. 1985;260:3594–3603. [PubMed] [Google Scholar]
- 34.Bayer I, Fliess A, Greipel J, Urbanke C, Maass G. Modulation of the affinity of the single-stranded DNA-binding protein of Escherichia coli (E. coli SSB) to poly(dT) by site-directed mutagenesis. Eur. J. Biochem. 1989;179:399–404. doi: 10.1111/j.1432-1033.1989.tb14567.x. [DOI] [PubMed] [Google Scholar]
- 35.Landwehr M, Curth U, Urbanke C. A dimeric mutant of the homotetrameric single-stranded DNA binding protein from Escherichia coli. Biol. Chem. 2002;383:1325–1333. doi: 10.1515/BC.2002.151. [DOI] [PubMed] [Google Scholar]
- 36.Lohman TM, Green JM, Beyer RS. Large-scale overproduction and rapid purification of the Escherichia coli ssb gene product. Expression of the ssb gene under lambda PL control. Biochemistry. 1986;25:21–25. doi: 10.1021/bi00349a004. [DOI] [PubMed] [Google Scholar]
- 37.Xiao H, Crombie R, Dong Z, Onrust R, O'Donnell M. DNA polymerase III accessory proteins. III. holC and holD encoding chi and psi. J. Biol. Chem. 1993;268:11773–11778. [PubMed] [Google Scholar]
- 38.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dam J, Schuck P. Sedimentation velocity analysis of heterogeneous protein-protein interactions: sedimentation coefficient distributions c(s) and asymptotic boundary profiles from Gilbert-Jenkins theory. Biophys. J. 2005;89:651–666. doi: 10.1529/biophysj.105.059584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Witte G, Fedorov R, Curth U. Biophysical analysis of Thermus aquaticus single-stranded DNA binding protein. Biophys. J. 2008;94:2269–2279. doi: 10.1529/biophysj.107.121533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Porter RD, Black S, Pannuri S, Carlson A. Use of the Escherichia coli SSB gene to prevent bioreactor takeover by plasmidless cells. Biotechnology. 1990;8:47–51. doi: 10.1038/nbt0190-47. [DOI] [PubMed] [Google Scholar]
- 42.de Vries J, Wackernagel W. Recombination and UV resistance of Escherichia coli with the cloned recA and recBCD genes of Serratia marcescens and Proteus mirabilis: evidence for an advantage of intraspecies combination of P. mirabilis RecA protein and RecBCD enzyme. J. Gen. Microbiol. 1992;138:31–38. doi: 10.1099/00221287-138-1-31. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 44.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 45.Muetzelburg MV, Hofmann F, Just I, Pich A. Identification of biomarkers indicating cellular changes after treatment of neuronal cells with the C3 exoenzyme from Clostridium botulinum using the iTRAQ protocol and LC-MS/MS analysis. J. Chromatogr. B. Analyt Technol. Biomed. Life Sci. 2009;877:1344–1351. doi: 10.1016/j.jchromb.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 46.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 47.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 48.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 49.Dam J, Velikovsky CA, Mariuzza RA, Urbanke C, Schuck P. Sedimentation velocity analysis of heterogeneous protein-protein interactions: Lamm equation modeling and sedimentation coefficient distributions c(s) Biophys. J. 2005;89:619–634. doi: 10.1529/biophysj.105.059568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kozlov AG, Jezewska MJ, Bujalowski W, Lohman TM. Binding specificity of Escherichia coli single-stranded DNA binding protein for the chi subunit of DNA pol III holoenzyme and PriA helicase. Biochemistry. 2010;49:3555–3566. doi: 10.1021/bi100069s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.