Abstract
3MC syndrome has been proposed as a unifying term to integrate the overlapping Carnevale, Mingarelli, Malpuech and Michels syndromes. These rare autosomal recessive disorders of unknown cause comprise a spectrum of developmental features including characteristic facial dysmorphism, cleft lip and/or palate, craniosynostosis, learning disability, and genital, limb and vesicorenal anomalies. In a cohort of eleven 3MC families, we identified two mutated genes COLEC11 and MASP1 both of which encode proteins within the lectin complement pathway (CL-K1 and MASP-1 & −3 respectively). CL-K1 is highly expressed in embryonic murine craniofacial cartilage, heart, bronchi, kidney, and vertebral bodies. Zebrafish morphants develop pigment defects and severe craniofacial abnormalities.
Here, we show that CL-K1 serves as a key guidance cue for neural crest cell migration thus demonstrating for the first time, a role for complement pathway factors in fundamental developmental processes and the origin of 3MC syndrome.
Introduction
The Carnevale, Mingarelli, Malpuech and Michels syndromes are four rare autosomal recessive disorders1-4 that were recently postulated to be part of the same clinical entity termed the “3MC syndrome”5,6. The main features are facial dysmorphic traits including hypertelorism, blepharophimosis, blepharoptosis, and highly arched eyebrows present in 70 to 95% of patients (Suppl Fig 1). Cleft lip and palate, post-natal growth deficiency, cognitive impairment, and hearing loss are also consistent findings, occurring in 40 to 68% of patients. Craniosynostosis, radioulnar synostosis, genital and vesicorenal anomalies occur in 20 to 30% of patients. Rarely occurring features include anterior chamber defects, cardiac anomalies, caudal appendage, umbilical hernia/omphalocele, and diastasis recti. Thirty two patients from 20 families have been described so far6,7. The causes of 3MC have thus far remained elusive.
Here we report the identification of two genes, mutations in which, cause disorders eponymously described as Carnevale, Malpuech, Michels and Mingarelli syndromes. COLEC11 and MASP1, encode components of the lectin complement pathway thus implicating this diverse inflammation/chemotaxis cascade in the aetiology of human developmental disorders. Furthermore, we show that one of these secreted proteins, CL-K1 serves as a guidance cue for migrating neural crest and other cell types during embryogenesis.
Results
Genetic studies reveal mutations in COLEC11 cause 3MC
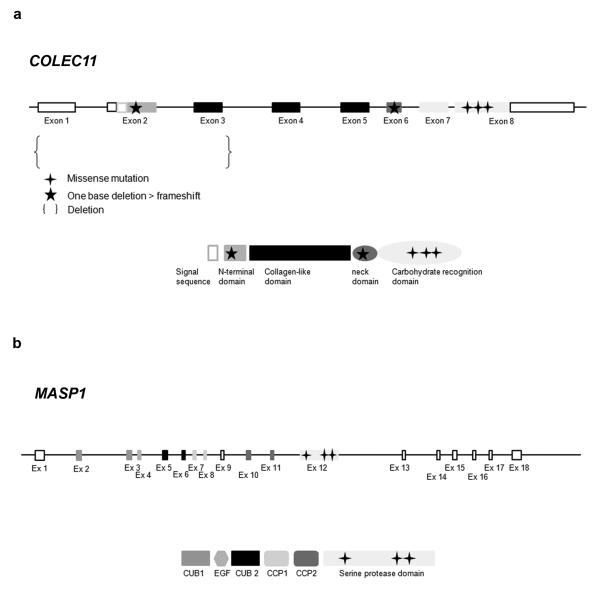
We collected a cohort of patient DNA samples comprising diagnoses of Carnevale, Mingarelli, Michels and Malpuech syndromes. The families were of Asian and Middle Eastern origin and four pedigrees were consanguineous in which we genotyped (Illumina SNP6.0/Affymetrix 250k) all available members revealing two regions of homozygosity shared by affected individuals. Pedigrees MC1 (Tunisian), MC2 (Bangladeshi), MC4 (Afghani) and MC8 (Saudi)(not shown) shared a homozygous region of more than 2.2 Mb on 2p25.3 (Suppl Fig 2, Table 1). Two further pedigrees, MC3 (Greek) and MC5 (Italian) shared a common region of homozygosity on 3q27.3 (2.2 Mb). In the 2p25 interval we sequenced the open reading frames of 9/15 candidate transcripts (Suppl Fig 2) and discovered three homozygous non-synonymous mutations and one in-frame deletion in COLEC11; MC1 - c.496T>C/p.Ser169Pro (exon 8)(Suppl Fig 3); MC2 - c.45delC/p.Phe16SerfsX85 (exon 2)(Suppl Fig 3); MC4 - c.610G>A/p.Gly204Ser (exon 8)(Table1; Suppl Fig 3); and MC8 - c.648-650delCTC (p.Ser217del)(exon 8)(not shown). Sequencing COLEC11 in further 3MC patients revealed mutations in two probands described in the original Carnevale and Mingarelli papers1,2. The first of these, MC11 harbored a 27 kb homozygous deletion encompassing exons 1-3 of COLEC11, predicted to result in a complete loss of the N-terminal and partial loss of the collagen-like domains of the protein (Fig 1, Table 1). Sequence analysis of the proximal and distal breakpoints of the junction fragment detected low copy repeats (LCRs) sequences corresponding to long interspersed nuclear elements (LINE: respectively L1MA1 and L1MD1). This suggests that non-allelic homologous recombination (NAHR) between these LCRs is the most likely mechanism mediating this microdeletion in MC11. The other case, MC10 harbored a homozygous single base deletion, c.300delT/G101VfsX113 (exon 6) which predicts premature termination of CL-K1. The localization of mutations in the COLEC11 gene and protein is summarized in Fig 1. Each mutation segregated with the disease in every family. Furthermore, none of these base changes were found in ethnically matched control chromosomes (Table 1) and none are listed in either dbSNP or 1000 Genomes database. Moreover, the wild-type amino acid residues are highly conserved in evolution (Suppl Fig 4) and the 27 kb deletion was not present in 286 control chromosomes, nor reported in the latest release of the database of genomic variants (DGV)(http://projects.tcag.ca/variation/). Taken together, these data suggest they are causative mutations for 3MC.
Figure 1.
Mutation summary. Location of the 5 different mutations identified in COLEC11 gene and protein. Location of the 3 different mutations identified in MASP1 gene and protein.
Table 1.
Molecular data in 11 families presenting with 3MC syndrome. Number of control chromosomes tested is included. dark grey = mutations in COLEC11 / light grey = mutations in MASP1.
| Family | Patient | Diagnosis | Origin | Nucleotide change |
Protein change | Control chromosomes |
|---|---|---|---|---|---|---|
| MC1 | 1.1 | Carnevale | Tunisia | c.496T>C | p.Ser169Pro | 60 Tunisian 284 north- European |
| 1.2 | Carnevale | Tunisia | c.496T>C | p.Ser169Pro | 60 Tunisian 284 north- European |
|
| MC2 | 2.1 | Malpuech | Banglades h |
c.45delC | p.Phe16SerfsX 85 |
94 Bangladesh 284 north- European |
| 2.2 | Malpuech | Banglades h |
c.45delC | p.Phe16SerfsX 85 |
94 Bangladesh 284 north- European |
|
| MC4 | 4.1 | Malpuech/ Michels |
Afghanista n |
c.610G>A | p.Gly204Ser | 72 Bangladesh 284 north- European |
| 4.2 | Malpuech/ Michels |
Afghanista n |
c.610G>A | p.Gly204Ser | 72 Bangladesh 284 north- European |
|
| MC8 | 8.1 | Carnevale | Saudi Arabia |
c.648_650delCT C |
p.Ser217del | 192 Saudi |
| MC9 | 9.1 | Malpuech | Pakistan | c.610G>A | p.Gly204Ser | 72 Bangladesh 284 north- European |
| MC10 | 10 | Mingarelli | Italy | c.300delT | p.Gly101ValfsX 113 |
334 north- European |
| MC11 | 11 | Carnevale | Italy | ex 1,2,3 deletion |
Non-functional protein? |
286 north- European |
| MC3 | 3.1 | Carnevale | Greece | c.1489 C>T | p.His497Tyr | 572 north- European |
| MC5 | 5.1 | Malpuech | Italy | c.1888T>C | p.Cys630Arg | 506 north- European |
| 5.2 | Malpuech | Italy | c.1888T>C | p.Cys630Arg | 506 north- European |
|
| MC6 | 6.1 | 3MC | Brazil | c.1997G>A | p.Gly666Glu | 506 north- European |
| 6.2 | 3MC | Brazil | c.1997G>A | p.Gly666Glu | 506 north- European |
|
| MC7 | 7.1 | Michels | Brazil | c.1997G>A | p.Gly666Glu | 506 north- European |
Expression of COLEC11 in craniofacial cartilage and other tissues
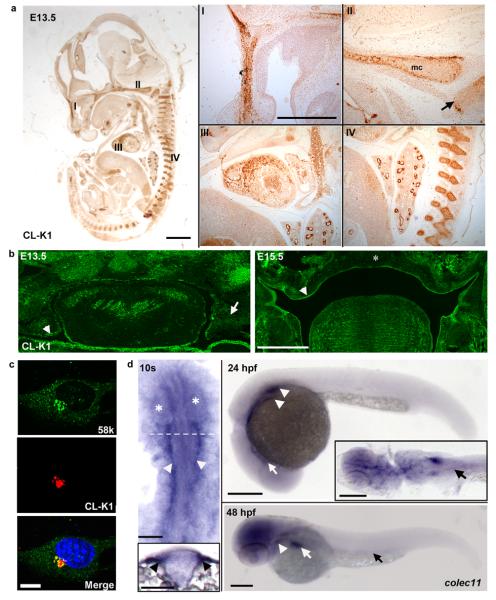
CL-K1 is a member of the protein family of C-type lectins, which contain a collagen-like domain and a carbohydrate recognition domain, thought to play a role in host-defense8 (PubMed 17179669). CL-K1 is highly conserved with homologs in chimpanzee, dog, cow, mouse, chicken, and zebrafish (Suppl Fig 5). Combined data from Serial Analysis of Gene Expression (SAGE) and eNorthern analyses suggest that human tissue expression is highest in brain, liver, kidney, spleen, lung, skin, breast, ovary, testis and placenta. We demonstrated broadly distributed expression in mouse tissues including craniofacial cartilage (nasal septum, meckel's cartilage and posterior palate), heart, bronchi, kidney, vertebral bodies at E13.5 (Fig 2a). Expression was also observed in palatal structures at E13.5 and E15.5 (Fig 2b).
Figure 2.
Immunolocalization of CL-K1 protein with polyclonal antibody a. in mouse embryo whole sections (scale bar: main panel 1 mm, inset panels 500 μm); CL-K1 (DAB) is expressed in the developing murine nasal septum (I), cartilage primordium of the basisphenoid bone (II, arrow), Meckel's cartilage (II, mc), myocardium (III), bronchioles and vertebrae (IV) at E13.5. b. CL-K1 is highly expressed in the palatal mesenchyme (arrow) and epithelium (arrow head) at E13.5. Later, at E15.5 CL-K1 is downregulated in the fused palatal shelf (asterisk) while expression is maintained in the palatal shelf epithelium (arrow head) (scale bar: 500 μm). c. ATDC5 cells showing immuno-colocalisation of endogenous CL-K1 and Golgi marker 58K (scale bar: 10 μm). d. colec11 in situ hybridisation at 10s, 24 hpf, and 48 hpf stages. At 10s, expression is localised to the cranial paraxial mesendoderm (arrow heads and transverse section, inset). The eyes are demarcated by asterisks and the relative position of the section marked by a dotted line. Scale bar: 100 μm. At 24hpf, transcripts are detected in the glomeruli (arrow heads) and cranial ventral midline (arrow). Inset shows a dorsal flatmount at 24 hpf highlighting expression in the glomeruli and pronephric ducts (PNDs, black arrow). At 48 hpf, colec11 remains in the glomeruli (arrow head), weakly in the PNDs (black arrow), and strongly in the liver (white arrow). Scale bar: 200 μm.
Next we evaluated the cellular localization of CL-K1 in a murine chondrocyte cell line, ATDC5. Here we observed specific localization in the golgi apparatus consistent with a secreted peptide (Fig 2c).
Previous studies reported detection of CL-K1 in the serum8. Using western blotting, we failed to detect any secreted CL-K1 in the serum of two patients (Gly204Ser), in contrast to control samples (Suppl Fig 6). This suggests that the missense substitution in these patients leads to cellular retention of the protein.
Loss of colec11 in zebrafish embryos causes truncal shortening and craniofacial abnormalities
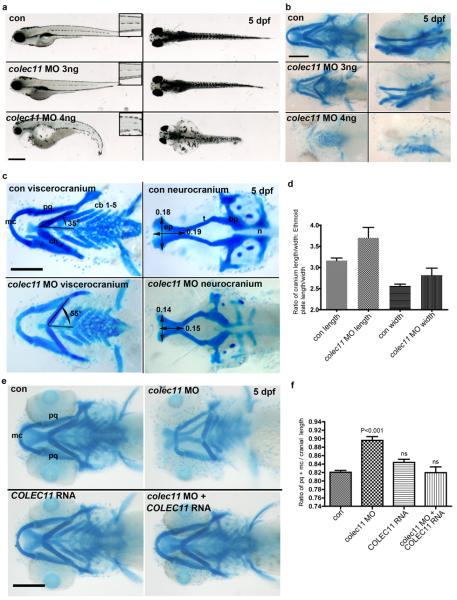
As nothing is known of the specific role of COLEC11 during embryonic development and in the absence of an available mouse model, we demonstrated expression in zebrafish embryos in the pronephric duct, lateral hindbrain and liver (Fig 2d). Next we sought to determine the effects of loss of function of this protein during zebrafish embryogenesis. Two antisense morpholinos were designed; one directed against the initiation site (colec11-ATG-MO) and another against the exon 2-intron 2 splice site (colec11-SPL-MO)(Suppl Fig 7). Injection of both MOs from 1 to 8 ng doses at the one-cell stage gave rise to morphological abnormalities which could be reversed upon co-injection with full-length COLEC11 mRNA (Fig 3e,f). This effect was dose-dependent including, for higher doses (4 ng), heart edema, pronephric cyst formation, curved body axis, disorganised pigment distribution and high mortality (Fig 3a). To ensure that these phenotypes were not the result of morpholino off-target effects (e.g up regulation of p53), we co-injected p53 morpholino with colec11 MO (4 ng) but could not rescue the phenotypes (Suppl Fig 8 and not shown).
Figure 3.
a. General morphology of colec11 zebrafish morphants at higher (4 ng) and lower (3 ng) doses of colec11 MO. Higher doses give rise to pronephric cysts, curved body axis, and cardiac oedema not present at 3 ng doses. A dose dependent loss of medial trunk pigmentation (insets – 1.5x magnification ) was observed (scale bar is 500 μm) b. Alcian blue cartilage staining in colec11 morphants (3 and 4 ng) at 5 dpf showing generalised cartilage defects (scale bar: 200 μm) c. Palate measurements in the colec11 morphants (3 ng) demonstrate shortening of length and width of the ethmoid plate, abnormal Meckel's cartilage (mc) and abnormal angulation of the ceratohyal cartilage (ch). Scale bar: 200 μm. d. Graph of ethmoid plate length and width measurements expressed as a ratio against cranial length and width, respectively, in the presence or absence of colec11 morpholino. There is clear evidence of significant morphological effects on both length and width, indicating shortening in both axes. (con length 3.164 ± 0.062 N=8, colec11 MO length 3.702 ± 0.245 N=8, con width 2.563 ± 0.0441 N=8, colec11 MO width 2.819 ± 0.168 N=8, each value indicates the mean ± SEM). e. Alcian blue cartilage staining of colec11 morphants rescued with human RNA COLEC11 (75 pg) compared to colec11 morphants (3ng), control uninjected and COLEC11 RNA injected alone embryos (scale bar: 200 μm). pq: palatoquadrate, mc: Meckel's cartilage. f. Graph showing the length ratio of both pq cartilages plus mc standardised over the cranial length. Con (0.821 ± 0.004, n=51), colec11 MO (0.896 ± 0.009, n=50), COLEC11 RNA (0.844 ± 0.007, n=21), colec11 MO plus COLEC11 RNA (0.820 ± 0.014, n=16). Values shown as mean ± SEM. Data was analysed by one way ANOVA with Tukey's Multiple Comparison Test comparing column data against control measurements. Morphants have a significantly shorter pq+mc length compared to uninjected controls (p<0.001), whilst no significant (ns) difference was observed in controls versus colec11MO plus COLEC11 RNA or versus COLEC11 RNA injected alone.
Key angles and measures are indicated to illustrate cartilage defects. ep = ethmoid plate, t=trabeculae, n=notochord, bp=basal plate, pq=palatoquadrate, cb=ceratobranchials
Next we assessed the craniofacial morphology of colec11 zebrafish morphants by staining the cartilage with alcian blue at 5 dpf with lower (3 ng) doses of MOs to circumvent the high mortality. Striking differences in the craniofacial skeleton were observed compared with uninjected/standard morpholino injected embryos, such as reduced mandibular length (e.g. shortened Meckel and palatoquadrate cartilages), malformation of the anterior neurocranium with shortening of the trabeculae and the ethmoid plate, shortening and abnormal angulation of the ceratohyal cartilage (Fig 3b-d).
A second complement-related gene on 3q27 also causes 3MC
Homozygosity mapping in two further 3MC families not mutated for COLEC11, share a region of identity-by-descent on 3q27 spanning 2.2 Mb, which contained 16 candidate genes (Suppl Fig 9). One of these, MASP1 (Mannose-associated serine protease 1) codes for a protein also in the lectin complement pathway. MASP-1 binds to MBL (Mannose binding lectin) and serves to form C3 convertase, C4bC2b by cleaving C29. Another isoform (referred to as MASP-3) contains the same heavy chain as MASP-1 but a completely different serine protease domain10 encoded by a single exon (exon 12).
On screening MASP1 (NM_139125, NCBI) in the two 3q27-linked families we discovered two homozygous missense mutations in the affected patients from MC3 (c.1489 C>T/p.His497Tyr)(Suppl Fig 10) and MC5 (c.1888T>C/p.Cys630Arg)(Suppl Fig 10), alleles of which segregated with the disease in each pedigree. Additionally, we screened two more families from Brazil6,11 (previously reported as having Michels/3MC syndrome) and found the same missense mutation in both MC6 and MC7: c.1997G>A/p.Gly666Glu - homozygous in each affected person (Suppl Fig 10). These mutations reside in exon 12, in well conserved amino acid residues (Suppl Fig 4). In addition all these variants are predicted to be damaging (Polyphen – data not shown)) and were not found in at least 506 ethnically matched control chromosomes (Table 1). The localization of mutations in MASP1 gene and protein are summarized in Fig 1.
Loss of masp1 causes craniofacial abnormalities and pigment defects
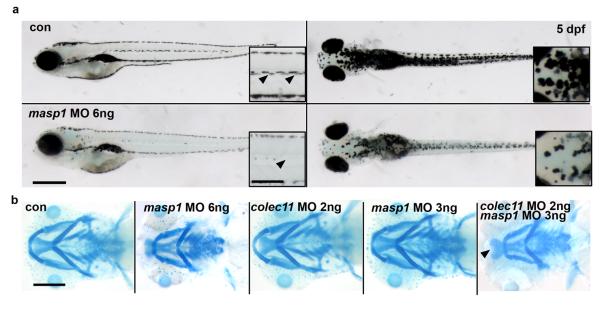
Next, we examined in zebrafish the effects of loss of masp1 in vivo. Two antisense morpholinos were designed; one directed against the initiation site (masp1-ATG-MO) and another against the exon 3-intron 3 splice site (masp1-SPL-MO) (Suppl Fig 7). Injection of both ATG and splice MOs at the one-cell stage gave rise to pigment (Fig 4a) and craniofacial cartilage defects (Fig 4b) similar to those seen in colec11 morphants.
Figure 4.
a. General morphology of masp1 zebrafish morphants showing pigmentation defect (arrows; scale bar; main panel = 500 μm; inset = 200 μm) b. Alcian blue cartilage staining at 5 dpf showing cartilage defects in masp1 morphants (6 ng) (panel second from left). Alcian blue staining in morphants double-injected with suboptimal doses of colec11 (2 ng) and masp1 (3ng) MO (panel far right). We observed a cartilage defect only in double-injected morphants compared with single suboptimal injections of either colec11 or masp1 MO (panels third and fourth from left).
Combinatorial loss of colec11 and masp1 reveals epistasis
Given the similar phenotypes arising from mutation of either COLEC11 or MASP1, we tested for epistasis between the two genes. Suboptimal doses (a dose below which no phenotype is observed when injected alone) of both colec11 and masp1 MOs were injected into one cell-stage zebrafish embryos and we found that ~73% (16/22) of embryos had severe craniofacial abnormalities and some developed clefts in the anterior neurocranium/ethmoid plate (Fig 4b). These results suggest that both gene products likely function in the same pathway consistent with a recent study in which these proteins directly interact12.
COLEC11 and MASP1 are likely guidance cues for neural crest cell migration
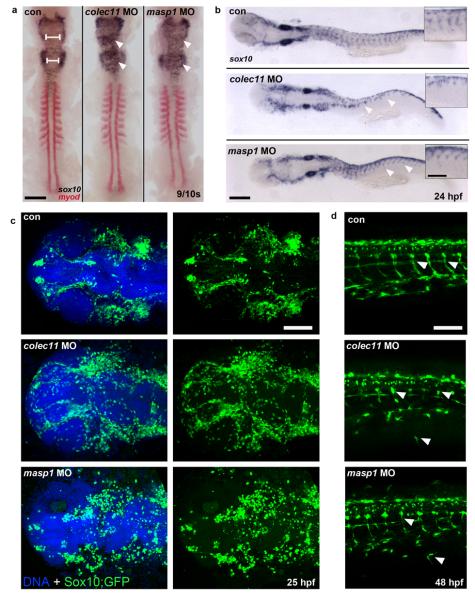
Owing to the pigmentation defect observed in the zebrafish morphants, and because the majority of the head skeleton derives from cranial neural crest cells (CNCC) that migrate from the dorsal aspect of the neural tube into the frontonasal process and the pharyngeal arches, we next investigated whether CL-K1 and MASP-1 proteins were involved in CNCC migration. We performed sox10 in situ hybridization in colec11 and masp1 zebrafish morphant embryos at 10 somites and 24 hpf respectively. An abnormal distribution of the CNCC in the hindbrain was observed in 10 somite-stage embryos with massive expansion of cells across the midline compared to controls (Fig 5a). At 24 hpf, the organisation of streaming NCC through the somites is severely disrupted in the colec11 morphants and obviously truncated in the masp1 morphants (Fig 5b). In addition, we injected SOX10-eGFP zebrafish with colec11 and masp1 MO and observed at 48 hpf, ectopic NCC in the head, trunk and periphery (Fig 5c,d). Together these data suggest that CL-K1 and MASP-1 likely behave as early guidance cues to direct the migration of neural crest cells during embryonic development.
Figure 5.
a. SOX10/MyoD in situ experiments on zebrafish colec11 and masp1 morphants showing an abnormal distribution of the CNCC in the hindbrain of 10 somite-stage embryos, with a massive expansion of cells across the midline compared to controls (left panel, white arrowheads). b. Streaming of NCCs into the head and through the somites (arrow heads) is also disrupted in colec11 and masp1 morphants at 24 hpf, as indicated by aberrant sox10 expression. (scale bar: main panels 200 μm, inset panels 100 μm) c. Expression of Sox10:eGFP (green) in the head at 24 hpf highlights disorganised NCCs in colec11 and masp1 morphants (scale bar: 100 μm). d. At 48 hpf, clearly defined tracks of Sox10:eGFP (white arrow heads) are detected migrating through tail somites of uninjected control embryos, however both colec11 and masp1 morphants show abnormal and ectopic migration of NCCs throughout the tail. Scale bar: 100 μm.
COLEC11 likely functions as a chemoattractant
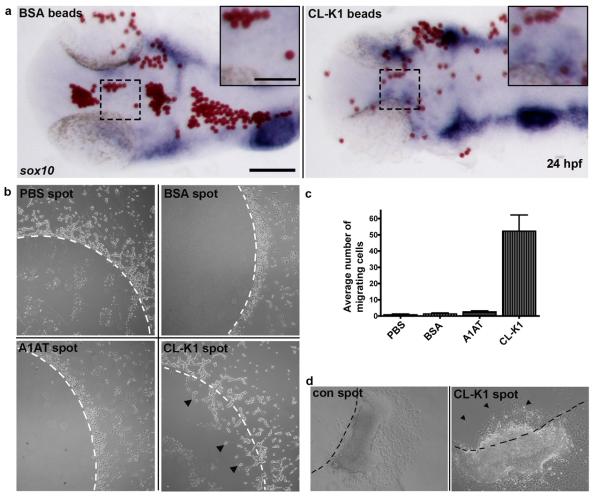
To further substantiate our CNCC migration observations in fish and to understand the role of CL-K1 on cell migration we injected 10μm beads soaked in recombinant CL-K1 into the head region of zebrafish at 18 somites (n=30). At 24 hpf, we observed (by in situ hybridization) NCCs associated with CL-K1 beads (Fig 6a) but not with control beads.
Figure 6.
Cell migration assays. a. Zebrafish in vivo NCC chemo-attraction assay. Embryos were implanted with either BSA or CL-K1 coated micro beads (red) into the head of 18 somite stage embryos that were subsequently grown to 24 hpf. In situ hybridisation for sox10 (blue) reveals preferential migration of NCC towards beads coated with CL-K1 but not BSA (inset). Scale bars: main panels 100 μm, inset panels 50 μm. b. Recombinant protein assays: representative images of control agarose spots containing either PBS, BSA, alpha-1 antitrypsin (A1AT), or CL-K1. White dashed lines represent the border of the spot. No cells migrated under the control agarose spots. In contrast HeLa cells were attracted to the CL-K1 containing spot, clearly migrating through the agarose (black arrowheads). Scale bar: 200 μm. c. Chart showing the average number of cells moving across the agarose border in five independent experiments. d. Quail neural tube assay: CL-K1 and PBS control agarose spots were placed on coverslips to which explanted neural tubes were carefully laid adjacent to (but not touching) the spot. NCCs migrating away from the neural tubes actively invaded CL-K1- but not PBS-containing spots, of which there was little to no migration. Scale bar: 200 μm.
Next we investigated the behaviour of human cells in culture by seeding flasks with HeLa cells containing ~3 mm disks of LMP agarose mixed with recombinant CL-K1. For controls, BSA and recombinant α1-anti-trypsin (A1AT) was substituted for CL-K1. After 24 hours of growth all CL-K1 agarose disks were invaded by streams of migrating HeLa cells (Fig 6b,c). In contrast, none of the control disks had significant cell invasion (Fig 6b,c,).
To confirm the effect of CL-K1 on migrating neural crest cells we utilised a quail neural tube explant assay13. Fertilised quail embryos (n=4; 16-20 somites) were dissected, the neural tubes isolated and carefully placed onto glass coverslips adjacent to which recombinant CL-K1-agarose was spotted as described above. The explants were incubated for 24 hours whereupon streams of NCCs (labelled with HNK1; Suppl Figure 11) were observed to preferentially migrate into protein-containing agarose spots but not into controls. These results demonstrate that CL-K1 likely behaves as a chemoattractant molecule with effects on migrating NCCs (Fig 6d).
DISCUSSION
We have identified two genes COLEC11 and MASP1 which when mutated cause four syndromes: Carnevale, Mingarelli, Malpuech and Michels. Mutations in MASP1 have also recently been reported in two families with Carnevale syndrome14. Both these genes were mutated in all 16 patients (from 11 families) tested. We identified six different mutations in COLEC11 and three different mutations in MASP1, all of which were found in the homozygous state. For each gene, there were patients presenting with each of the four diagnoses (Suppl Table 1), thus confirming that these disorders are allelic variants of the same disease category and should henceforth be referred to as the 3MC syndrome or the more descriptive Craniofacial-Ulnar-Renal Syndrome. Furthermore, we could not discern any consistent differences in the presenting phenotype between patients carrying the respective gene mutations (Table 1 and Suppl Table 1).
COLEC11 was first identified in 2006 as a new member of the collectin family, named CL-K1 (collectin kidney 1)8,9, and reported to be ubiquitously expressed. It is a secreted protein that contains two characteristic domains; a collagen-like domain and a carbohydrate recognition domain similar to another member of the collectin family, MBL (mannan-binding lectin), a serum protein15 that binds to various ligands leading to opsonization and activation of the complement cascade eventually forming the membrane attack complex for cell lysis. CL-K1 could act through the lectin complement pathway, similar to MBL. However, measurement of serum complement (C2, C3 and C4) levels in two unrelated patients were found to be normal indicating that downstream complement factor processing remains intact possibly through activation by the classical and alternative pathways. Moreover, we have observed that a mutation in COLEC11 caused depletion of CL-K1 in the sera of two patients, indicating that it must have direct roles, independent of downstream complement cascade activation, in 3MC.
MASP-1 is a serine protease that binds to MBL, as well as MASP2, and triggers complement activation by cleaving C2 to form C3 convertase, C4bC2b9. MASP1 codes for three different isoforms: The long MASP-1 is isoform 1 with a light chain containing the serine protease domain encoded by exons 13 to 18. MASP1 isoform 2, also known as MASP-3, has the same heavy chain as isoform 1 but a different serine protease domain encoded by the single exon 1210. It is also purported to process IGFBP516,17. A third isoform has been recently identified and is composed of a single shortened heavy chain18,19. All our MASP1 3MC patients had mutations in exon 12. It has been proposed that this isoform lacks processing abilities such that upon binding to MBL it does not cleave C2 or C420, or even inhibits formation of C3 convertase9. Compared to the other isoforms, MASP-3 expression appears ubiquitous18,21. CL-K1 is highly conserved between vertebrate species which clearly share a common origin (Suppl Fig 5).
The identification of mutations in COLEC11 and MASP1 was surprising as no complement-related components have previously been implicated in the pathogenesis of developmental disorders. This raises the possibility of a role for further constituent proteins in embryonic development. In contrast to MASP-1, little is known of the precise function of CL-K1 and we therefore sought to understand its role in early developmental processes. CL-K1 expression is widespread and seemingly abundant in ectodermal tissues. Strikingly, the craniofacial disruption observed in zebrafish morphants (and humans) was reminiscent of neural crest migration disorders. As we observed expansion of migrating streams of CNCC in colec11 and masp1 morphants this indicates that these proteins probably act as guidance cues. To this end, we show that CL-K1 has chemoattractant properties affecting migrating NCCs. It remains to be determined how CL-K1 influences migrating cell populations and whether or not it directionally stabilizes cell protrusions promoted by cell contact as recently published for the chemokine Sdf122.
CL-K1 and MASP-3, members of the lectin activation pathway are clearly important secreted proteins with novel functions which we now demonstrate here for the first time, include the orchestration of cell migration during vertebrate embryogenesis. This is further supported by the expression of colec11 along the path of migration of NCCs. The consequence of absence of these proteins during development culminates in multisystem abnormalities including craniofacial defects, skeletal, renal and neuronal aberrations in humans. It is important that we now turn our attentions to the more fundamental roles for this complex cascade in regulating embryogenesis in general and craniofacial development in particular.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported in part by grants from NEWLIFE (PLB, ADF, CR), the Wellcome Trust (PLB), Dubai Harvard Foundation for Medical Research (FSA), the University Hospital of Bordeaux (CR) and the Medical Research Council (AW). EU-FP7 (201804-EUCILIA)(VHH, DJ, DO). PLB is a Wellcome Trust Senior Research Fellow.
REFERENCES
- 1.Carnevale F, Krajewska G, Fischetto R, Greco MG, Bonvino A. Ptosis of eyelids, strabismus, diastasis recti, hip defect, cryptorchidism, and developmental delay in two sibs. Am J Med Genet. 1989;33:186–9. doi: 10.1002/ajmg.1320330210. [DOI] [PubMed] [Google Scholar]
- 2.Mingarelli R, Castriota Scanderbeg A, Dallapiccola B. Two sisters with a syndrome of ocular, skeletal, and abdominal abnormalities (OSA syndrome) J Med Genet. 1996;33:884–6. doi: 10.1136/jmg.33.10.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malpuech G, Demeocq F, Palcoux JB, Vanlieferinghen P. A previously undescribed autosomal recessive multiple congenital anomalies/mental retardation (MCA/MR) syndrome with growth failure, lip/palate cleft(s), and urogenital anomalies. Am J Med Genet. 1983;16:475–80. doi: 10.1002/ajmg.1320160405. [DOI] [PubMed] [Google Scholar]
- 4.Michels VV, Hittner HM, Beaudet AL. A clefting syndrome with ocular anterior chamber defect and lid anomalies. J Pediatr. 1978;93:444–6. doi: 10.1016/s0022-3476(78)81154-8. [DOI] [PubMed] [Google Scholar]
- 5.Al Kaissi A, et al. Asymmetrical skull, ptosis, hypertelorism, high nasal bridge, clefting, umbilical anomalies, and skeletal anomalies in sibs: is Carnevale syndrome a separate entity? Am J Med Genet A. 2007;143:349–54. doi: 10.1002/ajmg.a.31610. [DOI] [PubMed] [Google Scholar]
- 6.Leal GF, Silva EO, Duarte AR, Campos JF. Blepharophimosis, blepharoptosis, defects of the anterior chamber of the eye, caudal appendage, radioulnar synostosis, hearing loss and umbilical anomalies in sibs: 3MC syndrome? Am J Med Genet A. 2008;146A:1059–62. doi: 10.1002/ajmg.a.32252. [DOI] [PubMed] [Google Scholar]
- 7.Priolo M, et al. Malpuech syndrome: broadening the clinical spectrum and molecular analysis by array-CGH. Eur J Med Genet. 2007;50:139–43. doi: 10.1016/j.ejmg.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 8.Keshi H, et al. Identification and characterization of a novel human collectin CL-K1. Microbiol Immunol. 2006;50:1001–13. doi: 10.1111/j.1348-0421.2006.tb03868.x. [DOI] [PubMed] [Google Scholar]
- 9.Moller-Kristensen M, Thiel S, Sjoholm A, Matsushita M, Jensenius JC. Cooperation between MASP-1 and MASP-2 in the generation of C3 convertase through the MBL pathway. Int Immunol. 2007;19:141–9. doi: 10.1093/intimm/dxl131. [DOI] [PubMed] [Google Scholar]
- 10.Dahl MR, et al. MASP-3 and its association with distinct complexes of the mannan-binding lectin complement activation pathway. Immunity. 2001;15:127–35. doi: 10.1016/s1074-7613(01)00161-3. [DOI] [PubMed] [Google Scholar]
- 11.Leal GF, Baptista EV. Three additional cases of the Michels syndrome. Am J Med Genet A. 2007;143A:2747–50. doi: 10.1002/ajmg.a.32029. [DOI] [PubMed] [Google Scholar]
- 12.Hansen S, et al. Collectin 11 (CL-11, CL-K1) Is a MASP-1/3-Associated Plasma Collectin with Microbial-Binding Activity. J Immunol. 2010;185:6096–104. doi: 10.4049/jimmunol.1002185. [DOI] [PubMed] [Google Scholar]
- 13.Delalande JM, et al. The receptor tyrosine kinase RET regulates hindgut colonization by sacral neural crest cells. Dev Biol. 2008;313:279–92. doi: 10.1016/j.ydbio.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 14.Sirmaci A, et al. MASP1 mutations in patients with facial, umbilical, coccygeal, and auditory findings of Carnevale, Malpuech, OSA, and Michels syndromes. Am J Hum Genet. 2010;87:679–86. doi: 10.1016/j.ajhg.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ezekowitz RA, Day LE, Herman GA. A human mannose-binding protein is an acute-phase reactant that shares sequence homology with other vertebrate lectins. J Exp Med. 1988;167:1034–46. doi: 10.1084/jem.167.3.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cortesio CL, Jiang W. Mannan-binding lectin-associated serine protease 3 cleaves synthetic peptides and insulin-like growth factor-binding protein 5. Arch Biochem Biophys. 2006;449:164–70. doi: 10.1016/j.abb.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Degn SE, Thiel S, Jensenius JC. New perspectives on mannan-binding lectin-mediated complement activation. Immunobiology. 2007;212:301–11. doi: 10.1016/j.imbio.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 18.Degn SE, et al. MAp44, a human protein associated with pattern recognition molecules of the complement system and regulating the lectin pathway of complement activation. J Immunol. 2009;183:7371–8. doi: 10.4049/jimmunol.0902388. [DOI] [PubMed] [Google Scholar]
- 19.Skjoedt MO, et al. A novel mannose-binding lectin/ficolin-associated protein is highly expressed in heart and skeletal muscle tissues and inhibits complement activation. J Biol Chem. 2010;285:8234–43. doi: 10.1074/jbc.M109.065805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zundel S, et al. Characterization of recombinant mannan-binding lectin-associated serine protease (MASP)-3 suggests an activation mechanism different from that of MASP-1 and MASP-2. J Immunol. 2004;172:4342–50. doi: 10.4049/jimmunol.172.7.4342. [DOI] [PubMed] [Google Scholar]
- 21.Lynch NJ, et al. Composition of the lectin pathway of complement in Gallus gallus: absence of mannan-binding lectin-associated serine protease-1 in birds. J Immunol. 2005;174:4998–5006. doi: 10.4049/jimmunol.174.8.4998. [DOI] [PubMed] [Google Scholar]
- 22.Theveneau E, et al. Collective chemotaxis requires contact-dependent cell polarity. Dev Cell. 2010;19:39–53. doi: 10.1016/j.devcel.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.