Abstract
The NAD(P)H cytochrome b5 oxidoreductase, Ncb5or (previously named b5+b5R), is widely expressed in human tissues and broadly distributed among the animal kingdom. NCB5OR is the first example of an animal flavohemoprotein containing cytochrome b5 and cytochrome b5 reductase domains. We initially reported human NCB5OR to be a 487-residue soluble protein that reduces cytochrome c, methemoglobin, ferricyanide, and molecular oxygen in vitro. Bioinformatic analysis of genomic sequences suggested the presence of an upstream start codon. We confirm that endogenous NCB5OR indeed has additional NH2-terminal residues. By performing fractionation of subcellular organelles and confocal microscopy, we show that NCB5OR colocalizes with calreticulin, a marker for endoplasmic reticulum. Recombinant NCB5OR is soluble and has stoichiometric amounts of heme and flavin adenine dinucleotide. Resonance Raman spectroscopy of NCB5OR presents typical signatures of a six-coordinate low-spin heme similar to those found in other cytochrome b5 proteins. Kinetic measurements showed that full-length and truncated NCB5OR reduce cytochrome c actively in vitro. However, both full-length and truncated NCB5OR produce superoxide from oxygen with slow turnover rates: kcat = ~0.05 and ~1 s−1, respectively. The redox potential at the heme center of NCB5OR is −108 mV, as determined by potentiometric titrations. Taken together, these data suggest that endogenous NCB5OR is a soluble NAD(P)H reductase preferentially reducing substrate(s) rather than transferring electrons to molecular oxygen and therefore not an NAD(P)H oxidase for superoxide production. The subcellular localization and redox properties of NCB5OR provide important insights into the biology of NCB5OR and the phenotype of the Ncb5or-null mouse.
The NAD(P)H cytochrome b5 oxidoreductase, Ncb5or (previously named b5+b5R), was first identified in human cell lines and tissues by mining the expressed sequence tag data base (1). This is the first example in animals of a flavohemoprotein containing a cytochrome b5-like domain at the NH2 terminus and a cytochrome b5 reductase (b5R)1-like domain at the COOH terminus (1). NCB5OR is encountered in a broad range of vertebrates and invertebrates and is expressed in a variety of cells and tissues. Similar to the classical single-domain cytochrome b5 and b5R counterparts, the cytochrome b5 domain in rat NCB5OR has a six-coordinate b-type heme (2), and the b5R domain binds flavin adenine dinucleotide (FAD) and NAD(P)H prosthetic groups as found in all members of the ferredoxin-NADP+ reductase superfamily (3). The classical cytochrome b5 and b5R proteins form a complex and reside on endoplasmic reticulum (ER) membranes with Δ9 stearoyl-CoA desaturase to catalyze fatty acid desaturation. To anchor on ER membrane, both cytochrome b5 and b5R single-domain proteins use a stretch of hydrophobic residues at their COOH- and NH2-end, respectively, and b5R has additional N-myristoylation (4). Two pairs of charged residues are responsible for the proper docking between these two proteins to allow electron flow from the b5R to cytochrome b5 protein (5). In contrast, NCB5OR is a soluble flavohemoprotein, and its hinge apparently mediates the docking between the cytochrome b5 and b5R domains (1). Moreover, the hinge sequences show a well-conserved CHORD and SGT1 (CS) motif as found in p23/HSP20, SGT1, CHP, and melusin, where it functions as a mediator for protein-protein interaction (6). NCB5OR expressed in Escherichia coli reduces a variety of artificial substrates in vitro, including cytochrome c, ferricyanide, methemoglobin, and molecular oxygen (1). Because of its ubiquitous expression and capability for superoxide production, we speculated that NCB5OR might function as an oxygen sensor (7).
Since our original publication on human Ncb5or, new sequence information has been generated from genome data bases suggesting the presence of an upstream start codon (8). This new start codon would result in a full-length transcript encoding 34 additional amino acid residues at the NH2 terminus of the truncated NCB5OR. There has been significant interest regarding whether NCB5OR could produce superoxide in vivo. To clarify these issues, we report in this article the identification of the Ncb5or gene family in animals by bioinformatic analysis. We also confirm by Western analysis that endogenous NCB5OR in mouse tissues is indeed the full-length protein. We show here by both confocal microscopy and subcellular fractionation that endogenous NCB5OR colocalizes with calreticulin, an ER marker. Both full-length and truncated recombinant NCB5OR proteins have been generated in a bacterial system. Kinetic measurements have been performed to analyze their NAD(P)H oxidase activity in superoxide production, as well as their NAD(P)H reductase activity in cytochrome c reduction. These studies of cellular localization, enzymology, and redox properties have considerably enhanced our understanding of the biological function of NCB5OR and the phenotype of the Ncb5or-null mouse.2
EXPERIMENTAL PROCEDURES
Fractionation of Subcellular Organelles
Equal-sized livers from wild-type or Ncb5or-null mice (5-week-old females)2 were rapidly excised, rinsed briefly with phosphate-buffered saline, and minced using a sterile razor blade. Tissue slices were transferred to a 50-ml tube, washed with ~30 ml of phosphate-buffered saline, and allowed to settle to the bottom. This step was repeated once, and the tissue was then resuspended in 7.5 ml of homogenization medium (0.25 m sucrose, 1 mm EDTA, and 10 mm HEPES, pH 7.4) containing Complete proteinase inhibitor mixture (Roche Applied Science) at the recommended concentration. Cells were disrupted by Dounce homogenization (15 turns) and then centrifuged at 3000 × g for 15 min to obtain a post-nuclear supernatant. The post-nuclear supernatant for each sample was transferred to a polyallomer centrifuge tube (Seton Scientific) and centrifuged at 100,000 × g for 40 min to pellet light organelles. This pellet was gently resuspended in homogenization buffer and made up to a volume of 0.6 ml with a density of 5% Nycodenz (5-(N-2,3-dihydroxypropylacetamido)-2,4,6-triiodo-N,N,-bis(2,3-dihydroxypropyl)-isophthalamide; Accurate Chemical). Each sample was loaded on top of a 9-ml continuous 10–30% Nycodenz gradient and then centrifuged for 18 h at 100,000 × g in an SW41Ti rotor on a Beckman L8–55M ultracentrifuge. Twelve 0.75-ml fractions were collected by tube puncture. Aliquots of each fraction (40 µl) were loaded onto a 10% denaturing gel for SDS-PAGE and Western blot analysis.
Western Blot Analysis
Primary antibodies were rabbit polyclonal antibodies against NCB5OR NH2-terminal sequence (NH2-LN-VPSQAFPAPGSQQ-COOH; Genemed Synthesis Inc.), cytochrome c (H104; Santa Cruz Biotechnology), or calreticulin (PA3–900; Affinity BioReagents), all at a 1:1000 dilution. Horseradish peroxidase-conjugated goat anti-rabbit IgG (1:1000; Pierce) was used as the secondary antibody, and enhanced chemiluminescence detection was performed with SuperSignal West Dura Extended Duration Substrate (Pierce).
Two-photon Confocal Microscopy of HepG2 Cells
HepG2 cells (American Type Culture Collection) were grown to subconfluence on poly-d-lysine-coated glass coverslips. They were fixed by ice-cold methanol/acetone (1:1) for 10 min (−20 °C) and then blocked with 3% bovine serum albumin in phosphate-buffered saline. Primary antibodies were rabbit polyclonal antibody against human NCB5OR (1:200 dilution; Genemed Synthesis Inc.) and chicken polyclonal antibody against human calreticulin (1:100 dilution; Affinity BioReagents). Secondary antibodies were the Alexa-568-conjugated goat anti-rabbit IgG (1:400 dilution; Molecular Probes) and the Cy2-conjugated rabbit anti-chicken IgG (1:200 dilution; Jackson ImmunoResearch). Visualization was performed with a Nikon E1000 microscope (Nikon, Düsseldorf, Germany), equipped with a charge-coupled digital camera (Optronics; Visitron Systems, Puchheim, Germany) and the image acquisition software EZ2000 (Coord, Utrecht, Netherlands). Two-photon images of single immunostained cells were captured by a converted Nikon TE300 microscope employing a Ti:sapphire laser that was tuned to 850 nm and delivered 110-fs pulses at a repetition rate of 76 MHz and a 590–650 nm (Alexa) and a 500–530 nm (Cy2) emission bandpass filter. Two different fluorescence channels were recorded simultaneously. Image stacks (512 × 512 × 64 pixels) were obtained by optical sectioning with a 60× water objective (Nikon, Plan Apochromat DIC H, 1.2 NA). The spatial data were three dimensionally reconstructed from image stacks with the Application Visualization System (Waltham, MA). To enable comparative analysis of the different fluorescent structures, the fluorescence emission was normalized. Areas of equal intensity of fluorescence emission were rendered as isosurfaces and encoded in false colors (red and blue).
Confocal Microscopy of Transfected COS7 Cells
Subconfluent COS7 cells were transfected with expression constructs of human full-length or truncated NCB5OR (8 µg DNA/8 million cells on a 10-cm dish) with LipofectAMINE Plus reagents (Invitrogen) in Dulbecco’s modified Eagle’s medium containing 2% fetal bovine serum for 5 h. Cells were then grown in 10% fetal bovine serum medium overnight and split into 6-well plates to grow on glass cover slides. Subconfluent cells were treated as described previously before labeling with antibodies (1). Primary antibodies were rabbit polyclonal antibody against human calreticulin (1: 250 dilution; Affinity BioReagents) and mouse monoclonal antibody 9E10 against c-myc epitope (1:100 dilution; Sigma). Secondary antibodies were the fluorescein isothiocyanate conjugated goat anti-rabbit IgG (1:200 dilution; Jackson ImmunoResearch) and the rhodamine-conjugated goat anti-mouse IgG (1:200 dilution; Roche Applied Science). Confocal microscopy was performed using a Zeiss LSM 510 system (Carl Zeiss, Inc.) at Harvard Center for Neurodegeneration and Repair. The confocal images of transfected COS7 cells were obtained on a Zeiss Axiope 2 microscope with a 560–615 nm (rhodamine) and a 505–530 nm (fluorescein isothiocyanate) emission bandpass filter. Two different fluorescence channels were recorded sequentially with a 63× oil objective (1.4 NA).
Preparation of Recombinant NCB5OR
The full-length human and mouse NCB5OR, as well as truncated human NCB5OR, were prepared with a bacterial expression system (1). The coding sequence was cloned into NdeI and BamHI cloning sites of the pET19b vector (Novagen), and NCB5OR proteins were produced in BL21(DE3) host cells under isopropyl-β-d-thiogalactopyranoside induction at 8 °C to ensure proper folding. The poly-His NCB5OR was purified to homogeneity with nick-el-nitrilotriacetic acid chelation (Qiagen) plus ion-exchange Sepharose Q column chromatography (Amersham Biosciences), all performed at 4 °C. NCB5OR holo-proteins, with an A410/A274 of ~1.0 (explained below), were eluted from Q column with a salt gradient starting with a low-salt buffer (20 mm potassium phosphate, pH 7.6, 50 mm KCl) and concentrated with YM10 centricon (Amicon). Small aliquots were made, flash-frozen in liquid nitrogen, and stored at −80 °C. All NCB5OR samples used in biochemical assays are 90% pure on SDS-PAGE gel and contain ~95% holo-protein with a > 85% NADH-reducible heme content (explained below). Pure cytochrome b5-core proteins with an A410/A274 of ~2.5 were generated by using the same system.
UV-visible Spectroscopy for Recombinant NCB5OR
Approximately 0.5–1.0 µm (heme) NCB5OR protein in 0.3 ml of low-salt buffer (see above) was analyzed as follows on a Hitachi dual-beam U-2000 spectrophotometer at room temperature. Spectrum 1, a wavelength scan from 650 to 250 nm, was recorded for the oxidized NCB5OR as purified under nonreducing conditions (see above). NCB5OR was reduced instantaneously upon mixing with 3 µl of 10 mm NADH (0.1 mm final concentration) within the sample cuvette, and spectrum 2 was recorded immediately. The difference spectrum [2−1] represents “NADH-reducible” heme. A few granules of solid dithionite powder were added to the above sample to generate spectrum 3. The difference spectrum [3−1] represents “total” heme including both NCB5OR and its degradation product, cytochrome b5. The concentration of heme can be calculated from its difference spectrum, either [2−1] or [3−1], with ε(424–410) = 185 mm−1 cm−1 (1), or from spectrum 1 of 100% oxidized NCB5OR with ε410 = 138 mm−1 cm−1. All NCB5OR proteins were analyzed for NADH-reducible heme contents and for their heme/protein ratios. The percentage of NADH-reducible heme was calculated from [heme]reduced (2−1/[heme]reduced (3−1). This value reflects the content of NCB5OR among the total hemoproteins (NCB5OR+b5), as well as the electron flow within NCB5OR, which is intrinsically determined by protein folding. Pure and fully native NCB5OR flavohemoprotein is expected to have 100% NADH-reducible heme. The heme/protein ratio, as calculated from their corresponding absorbance A410 and A274, was used to estimate the total heme content of proteins. Pure NCB5OR holo-protein has an A410/A274 ratio of ~1.0, indicating one heme per protein. This value has been confirmed by our reverse phase HPLC analysis, which simultaneously quantifies prothestic groups and polypeptides of the same protein (see below).
Reverse Phase HPLC
Approximately 500 pmol of (heme) recombinant human NCB5OR (truncated) were loaded onto a Discovery C8 column (15 cm × 4.6 mm; 5 µm; Supelco) connected to a Ranin Rabbit HP system (Varian). The FAD, heme, and polypeptides were eluted with a water and acetonitrile gradient containing 0.1% trifluoroacetic acid and monitored at 220 nm with Dynamax UV-C detector (Varian). The absolute amounts of FAD, heme, and polypeptides were determined from individual peak area against standard curves of 320–8400 pmol of FAD and hemin (Sigma) and 4–50 µg of bovine serum albumin (Pierce). Peak area from the chromatogram was integrated with Dynamax Method Manager 1.4.3 (Varian) and used to quantify FAD, heme, cytochrome b5, and NCB5OR fractions. All major peaks were collected, lyophilized, and identified by color and SDS-PAGE. The concentration of FAD standard solution was determined by its A375 or A450 with ε375 = 9.4 mm−1 cm−1 or ε450 = 11.3 mm−1 cm−1, respectively. Alkaline hematin D575 was used to determine the concentration of hemin standard solution (10).
Resonance Raman Spectroscopy
Resonance Raman spectra were acquired at ambient temperature using 413.1 nm excitation from Kr+ lasers (11). The 10-mW laser beam was focused to a line on the sample, and UV-visible spectra were recorded before and after resonance Raman experiments to be certain that laser-induced degradation of neither the heme nor the flavin had occurred during Raman acquisition. Samples ranging in concentration from 0.05 to 0.30 µm were contained in 5-mm NMR tubes and spun at ~20 Hz using a homemade NMR tube spinner. Spectra were recorded using f1 collection, holographic notch filters, a 0.64-m spectrograph fitted with a 110 × 110-mm, 2400-groove/mm holographic grating, and a 1100-pixel liquid N2-cooled charge-coupled device detection system. The spectrometer was calibrated with toluene, dimethylformamide, and CH2Br2 as frequency standards.
Kinetics of Superoxide Production
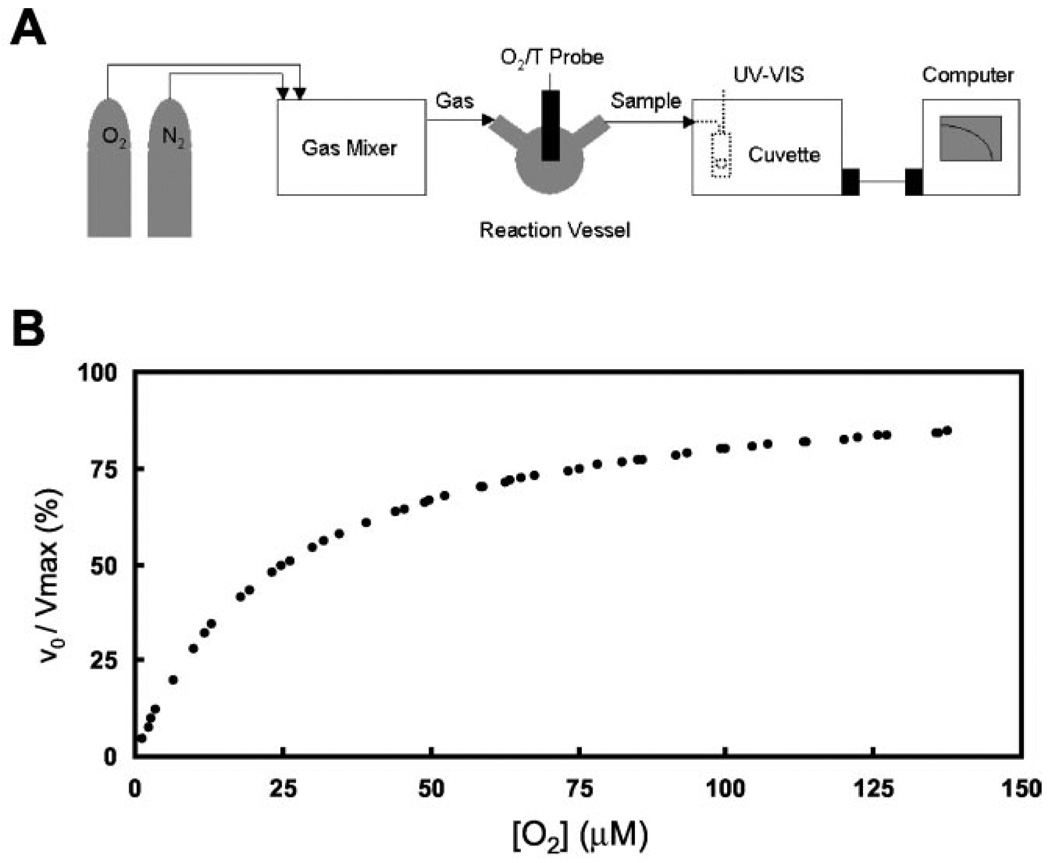
The spectrophotometric-based steady-state kinetics apparatus is specifically designed for this experiment (see Fig. 6A). Oxygen (O2) and nitrogen (N2) gasses (BOC gases) were mixed to the desired concentration using a computerized gas mixer (Environics Series 200) before being delivered at 120 ml/min to the gas-tight reaction vessel (The Custom Glass Shop) with Viton tubing (Masterflex L/S 16; Cole-Parmer). Sample dissolved oxygen concentration and temperature were monitored using a temperature compensated oxygen and temperature electrode and meter (OM2000 from Cameron Instrument Company or inoLab Oxi DO WTW from Fisher Scientific). Before use, the electrode was zeroed by exposure to 100% N2 gas and spanned by exposure to 20% O2 gas or room air. The UV-visible spectrophotometer was interfaced via RS-232C to a Macintosh IIsi computer running FutureBasic II (STAZ Software, Inc.) to provide remote control, data acquisition, and data storage. All chemicals were from Sigma. Stock solutions of 10 mm NADH/1 mm NAD or 10 mm NADPH/1 mm NADP and 2000 units/15 µl superoxide dismutase in 50 mm potassium phosphate buffer, pH 7.8, were freshly prepared and kept on ice under N2 and protected from ambient light until use. Each reaction mixture was prepared on ice immediately before individual kinetics runs. Samples included 10–40 nm (heme) for truncated NCB5OR or 0.5–1 µm (heme) for full-length NCB5OR plus 1000 units of superoxide dismutase in a final volume of 1.5 ml of 50 mm potassium phosphate buffer, pH 7.8. The reaction mixture was equilibrated to the desired dissolved O2 concentration at 20 °C with gentle stirring before initiating the reaction with 15 µl of NADH/NAD or NADPH/NADP solution. The final reaction mixture was stirred for an additional 5 s and subsequently delivered under gas pressure to the flow cell (10-mm path length; Starna). A340 was measured as a function of time from 0 to 600 s in 6-s intervals. Data analysis was performed on a Mac G4 computer (Apple) running Excel (Microsoft) with Solver. dA340/dt was fit to a linear function, the slope of which was used to calculate the initial velocity of NAD(P)H consumption or superoxide production, υo, using ε340 = 6.22 mm−1 cm−1 for NAD(P)H and assuming the rate of [NAD(P)H] consumption is equal to twice the rate of superoxide production. υo versus [O2] was fit to the Michaelis-Menten equation to determine Km and Vmax and to calculate kcat and the enzyme efficiency kcat/Km. For all calculations, 100% O2 = 1.3 mm (20°C, 1 atm).
FIG. 6. Superoxide production of re-combinant NCB5OR.
A, the experimental apparatus is designed according to “Experimental Procedures” to monitor NADH consumption as a function of oxygen tension, υo versus [O2]. B, the υo versus [O2] curve of superoxide production by human truncated NCB5OR is shown as a representative of similar curves for full-length human and mouse NCB5OR. To compile different series of data points, all the individual data points are shown in υo/Vmax values, where Vmax is obtained from each series and refitted to the Michaelis-Menten equation.
Kinetic Measurement of Cytochrome c Reduction
The spectrophotometric-based steady-state kinetics apparatus, similar to that outlined in Fig. 6A, was used to monitor cytochrome c reduction. The reaction vessel and gas control devices were omitted in the NAD(P)H reductase assay for the following reason. When compared with the rate of cytochrome c reduction directly by NCB5OR, the rate as contributed from superoxide, a product from the NAD(P)H oxidase activity of NCB5OR, was negligible under ambient air. The measurement was initiated immediately after NADH or NADPH was added to a quartz cuvette and mixed thoroughly with 25 nm (heme) NCB5OR plus oxidized cytochrome c (Sigma) in a final volume of 0.3 ml of low-salt buffer (20 mm potassium phosphate, pH 7.6, 50 mm KCl). To measure the Km for cytochrome c, the substrate was mixed at a final concentration of 0–40 µm with 0.1 mm NADH or NADPH, and the absorbance change at 340 nm was recorded. To measure the Km for NADH or NADPH, the reductant was mixed at a final concentration of 0–10 µm with 20 µm cytochrome c, and the absorbance change at 550 nm was recorded. Data analysis was performed as described above. The υo versus [substrate] curve was fitted to the Michaelis-Menten equation to generate kinetics constants. υo is defined as the rate of electron transfer in all calculations.
| (Eq.1) |
| (Eq.2) |
Measurement of Oxidation-Reduction Potentials
Potentiometric titrations were performed as described previously (12). 20–50 pmol (10–30 nm) of human NCB5OR (truncated) or cytochrome b5 core proteins in 1.8 ml of 100 mm KCl, 50 mm MOPS, pH 7.0, were used for each measurement. This buffer was supplemented with Complete proteinase inhibitor mixture (Roche Applied Science) to prevent degradation of the sample during the analysis (3–4 h). Spectra were recorded between 600 and 520 nm at a series of electrode potentials, using a Perkin-Elmer Lambda 18 spectrophotometer. An Orion 720A meter (Orion Research Inc.) was used to measure the half-cell potential relative to a saturated calomel reference electrode. The potential was adjusted by the addition of <5 µl aliquots of solutions of sodium dithionite (reductive titrations) and potassium ferricyanide (oxidative titrations). The accuracy of the apparatus was checked by titration of a 5 µm solution of phenosafranin (E = −252 mV). The following mediators were used at 10 µm: phenazine methosulfate; phenazine ethosulfate; anthraquinone; anthraquinone 2-sulfonate; anthraquinone 2,6-disulfonate; 2-hydroxy-1,4-naphthoquinone; and 2,3,5,6-tetramethylphenylenediamine. Pyocyanine was added at 6 µm. The degree of reduction of NCB5OR or cytochrome b5 core was estimated from the height of the α absorbance band at 558 nm. The values of absorbance were plotted against electrode potential, and the best fit to the data points was calculated using the GraphPad Prism software package (San Diego, CA) using the Nernst equation.
RESULTS
Ncb5or Gene Family
The Ncb5or gene, first cloned from human liver cells (1), has now been identified in all animals tested, including mammals, chicken, Xenopus, fish, tunicate, Drosophila, nematodes, sea urchin, and algae. The amino acid sequences of all currently identified NCB5OR proteins are aligned in Supplementary Fig. 1. As reported previously by our group (1), NCB5OR proteins contain a cytochrome b5-like domain at the NH2 terminus (~130 residues), a b5R-like domain at the COOH terminus (~300 residues), and a hinge (~90 residues) linking the two domains. The primary sequence of the cytochrome b5 domain is very conserved in all NCB5OR proteins. Likewise, the b5R domains share conserved FAD- and NAD(P)H-binding sites. NCB5OR family members can be distinguished by their overall structural motifs, as well as their signature sequences, which are different from those of the closely related cytochrome b5 and b5R. Namely, NCB5OR is one residue shorter between the two axial histidines in the cytochrome b5 domain and fifteen residues shorter between FAD2 and NAD(P)H1 sites in the b5R domain (Fig. 1 in Ref. 1).
FIG. 1. The Kozak sequences in vertebrate Ncb5or genes.
Nucleotide sequences at the two potential start sites (uppercase letters) are shown, with arrows pointing to the −3 sites. GenBank™ accession numbers are as follows: human, AL832113 and BC025380; mouse, AK090159 and XM135010; rat, CB711872 and CK366963; cow, BI537309, CB441027, and CB456333; chicken, AJ452144; Xenopus, AL680691 and AL956329; zebrafish, BU808429; zebrafish-Singapore, CK674731; rice fish, BJ006309; and tunicate, BW240834.
NH2 Terminus of Endogenous NCB5OR
All Ncb5or genes currently identified in animals have an additional start codon upstream of the one previously assigned. This start codon was not detected during our initial cloning for 5′-untranslated region sequence by rapid amplification of cDNA ends-PCR. Moreover, our previous sequence alignment of 5′-untranslated region presented no consensus among all expressed sequence tag entries available at that time.
Our bioinfomatic analysis supports the assignment of an upstream start codon. The Ncb5or genes of zebrafish-Singapore and algae have only the 5′ start codon. Their second putative start site is absent due to a single-base change resulting in an amino acid substitution from methionine to leucine in zebrafish-Singapore or from methionine to valine in algae (shown in Supplementary Fig. 1). In addition, the Kozak sequences in all vertebrate Ncb5or genes have a better match to the optimal sequences at the 5’ ATG site (Fig. 1) because, according to a survey of 699 vertebrate genes (13), A/G is preferred over C/T at the −3 site.
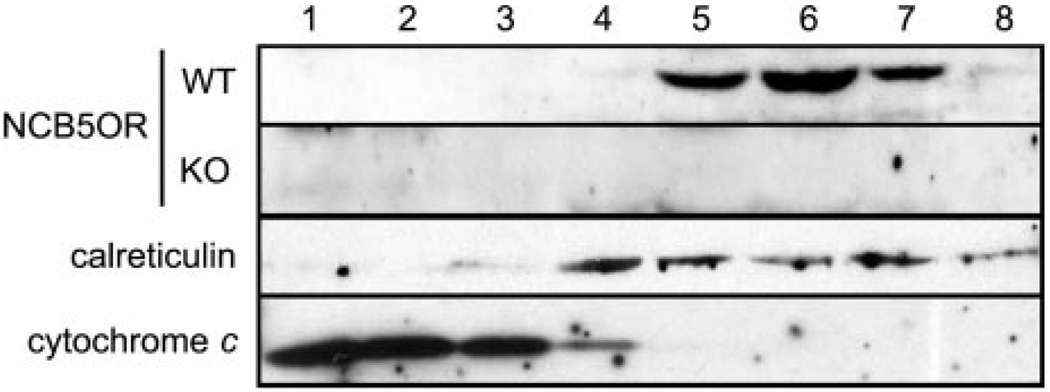
To confirm this NH2-terminal assignment, we have performed Western blot analysis to detect endogenous NCB5OR with a polyclonal antibody against the NH2 terminus of NCB5OR. We took advantage of the Ncb5or-null mice that have been recently generated by gene-specific targeting.2 By comparing equal amounts of protein samples from wild-type and Ncb5or-null mice, a protein band of the expected size was present in wild-type liver, but not in Ncb5or-null tissue (Fig. 2). The same pattern was observed in mouse pancreata with the same antibody (data not shown) or a polyclonal antibody against the whole mouse NCB5OR.2 Therefore, endogenous NCB5OR is translated from the upstream initiation site in these tissues.
FIG. 2. Western analysis of endogenous NCB5OR in fractionated subcellular organelles from livers of wild-type (WT) and Ncb5or-null (KO) mice.
The endogenous NCB5OR is enriched in intact subcellular organelles containing calreticulin, not cytochrome c. Lanes 1 (high density) through 8 (low density) represent intact subcellular organelles from wild-type and knockout livers after separation on a continuous density gradient. The epitope of primary antibody against NCB5OR is its NH2-terminal fifteen-residue sequence (NH2-LNVPSQ-AFPAPGSQQ-COOH).
Subcellular Localization
Our initial studies of subcellular localization of NCB5OR were based on confocal microscopy on COS7 (monkey kidney) cells expressing exogenous human truncated NCB5OR with a c-myc epitope tag. To define the subcellular localization of endogenous NCB5OR in more detail, we have employed two different methods. As shown in Fig. 2, after the removal of nuclei and cytosol, fractionation of intact subcellular organelles of mouse liver on a continuous density gradient established that endogenous mouse NCB5OR is present in isolated fractions that are enriched with the ER marker calreticulin (lanes 4–8) but is not found in the fractions enriched with the mitochondrial marker cytochrome c (lanes 1–3). Because recombinant NCB5OR is soluble, endogenous NCB5OR is likely to reside in the lumen of isolated ER.
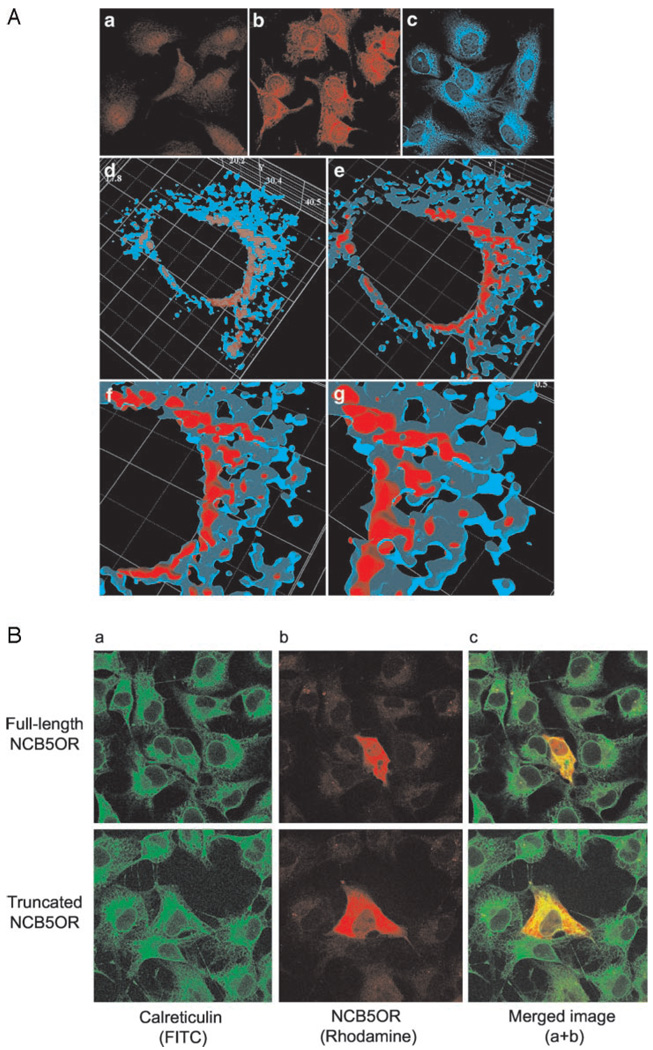
In parallel, we have performed two-photon confocal microscopy on endogenous human NCB5OR in HepG2 cells (Fig. 3A), as well as confocal microscopy on exogenous human NCB5OR in COS7 cells (Fig. 3B), to study the colocalization of NCB5OR with ER. Using antibodies against calreticulin and NCB5OR or its epitope tag, we have colocalized NCB5OR with calreticulin in both HepG2 and COS7 cells at the same perinuclear space as we previously reported (1). Because the highest red fluorescence is surrounded by the light blue limits of the ER signals (Fig. 3A), NCB5OR most likely resides within the ER. The same distribution and ER colocalization pattern is observed for both truncated and full-length NCB5OR (Fig. 3B). Both calreticulin and NCB5OR signals are antigen specific because there is no cross-reaction between each second antibody and the primary antibody against the other antigen (data not shown). Similar confocal experiments using antibody against cytochrome c have shown no colocalization between mitochondria and NCB5OR in either HepG2 or COS7 cells (data not shown). This finding agrees with Western blot results on intact subcellular organelles as shown in Fig. 2.
FIG. 3. Confocal microscopy for (A) endogenous NCB5OR in HepG2 cells and (B) exogenous NCB5OR in transfected COS7 cells.
A, fluorescence (a–c) and two-photon confocal images (d–g) were obtained from fixed HepG2 cells stained with (a) pre-immune serum (negative control; 1:200 dilution), (b) anti-NCB5OR post-immune serum (1:200 dilution), and (c) anti-calreticulin antibody (1: 100 dilution). d, three-dimensional colocalization of signals is shown for ER (blue) and NCB5OR (red). e, signals in one plane are obtained from optical slicing through the three-dimensional structure; f and g, signals in the same plane at higher magnification, with NCB5OR (red) most likely residing within the ER (blue), of which the limiting isosurfaces are depicted in light blue. B, confocal images are shown for COS7 cells transfected with either full-length (top panels) or truncated (bottom panels) human NCB5OR. Images represent (a) fluorescein isothiocyanate (FITC) (calreticulin), (b) rhodamine (NCB5OR), and (c) merged (colocalization) signals.
Stoichiometry of NCB5OR
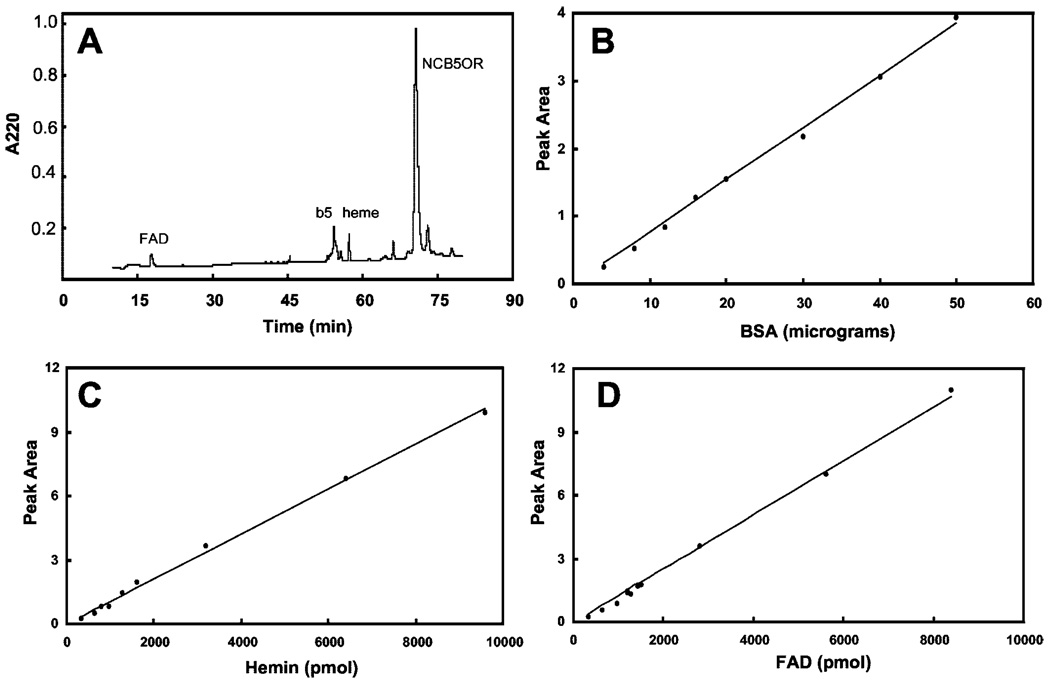
To study the biochemical properties of NCB5OR, we have generated high-quality recombinant NCB5OR from a bacterial overexpression system as described previously (1). All our recombinant NCB5OR are native flavohemoproteins containing both FAD and heme. These prosthetic groups have been identified by comparing their retention time with that of FAD and heme standards on reverse phase HPLC (Fig. 4). The color and lack of Coomassie Blue staining of FAD and heme peaks further confirm their identities. Flavin mononucleotide has a longer retention time and thus can be easily distinguished from FAD. NCB5OR proteins have been routinely purified to near homogeneity containing ~85% NADH-reducible heme as estimated from Coomassie Blue staining and UV-visible spectroscopy. Our reverse phase HPLC analysis supports this conclusion.
FIG. 4. Reverse phase HPLC of recombinant NCB5OR.
The FAD, heme, cytochrome b5, and NCB5OR are eluted from C8 column as shown in the chromatogram (A). The identified FAD, heme, cytochrome b5, and NCB5OR peaks are quantified by their peak area against those of bovine serum albumin (B), FAD (C), and hemin (D). All peak areas, in arbitrary units, were obtained under the same conditions, e.g. elution gradient, flow rate, detector range, and integration method. This NCB5OR sample has ~450 pmol of FAD, ~500 pmol of heme, <3.0 µg (<190 pmol) of cytochrome b5 polypeptides, and ~28 µg (490 pmol) of NCB5OR polypeptides.
Heme Environment
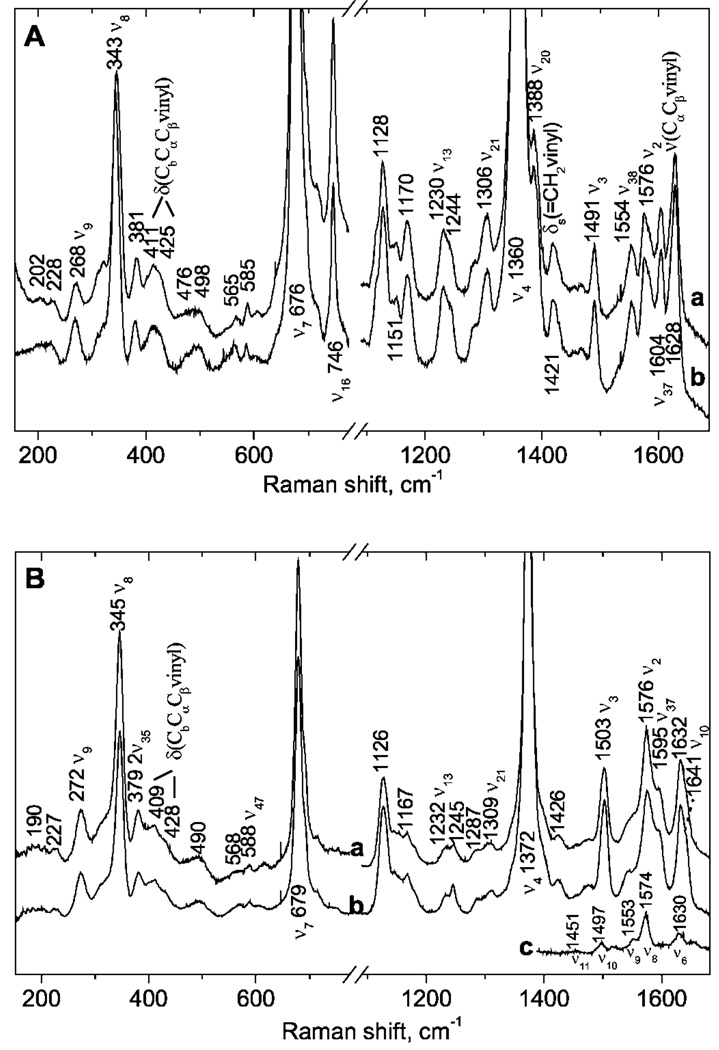
The heme environment in the native NCB5OR has been probed by resonance Raman spectroscopy. The 413.1-nm resonance Raman spectra of ferrous dithionite-reduced NCB5OR (human truncated) and its cytochrome b5 core domain are shown in Fig. 5A, a and b, respectively. These spectra are indistinguishable from those obtained from the NADH-reduced NCB5OR proteins. The frequencies of core size marker bands, ν2 and ν3, are consistent with six-coordinate, low-spin ferrous heme. The vibrational bands in Fig. 5A have been assigned by analogy to cytochrome b5 (14), flavocytochrome b2 (15), and FeII (PPIX)(ImH)2 (16). The heme resonance Raman frequencies for NCB5OR and cytochrome b5 are virtually identical, consistent with similar heme environments in the presence and absence of the reductase domain. The 202 cm−1 bands in the NCB5OR and cytochrome b5 spectra are tentatively assigned to the symmetric stretching mode, ν(His-FeII-His), by analogy to 203- and 205-cm−1 bands in the spectra of FeII(PPIX)(ImH)2 (16, 17) and ferrous flavocytochrome b2 (15), respectively. Hence, in the ferrous form, NCB5OR and cytochrome b5 appear to have FeII-His bond strengths typical of b-type cytochromes with neutral imidazole ligands. The band at 228 cm−1 is in a frequency range consistent with axial imidazole ligands serving as H-bond donors (16). Thus it is possible that there are multiple forms of the low-spin six-coordinate heme.
FIG. 5. Resonance Raman spectra of recombinant human NCB5OR obtained with 413.1-nm excitation.
A, resonance Raman spectra of (a) ferrous human truncated NCB5OR and (b) ferrous cytochrome b5 core protein. Reduced proteins were generated under anaerobic conditions with addition of a 10-fold excess (based on heme + flavin) of buffered sodium dithionite or NADH. B, resonance Raman spectra of (a) ferric human truncated NCB5OR and (b) ferric cytochrome b5 core protein. The difference spectrum, NCB5OR-cytochrome b5 core, is shown in c. The difference features correspond to the indicated resonance Raman bands from FAD. All samples were in 20 mm phosphate buffer, pH 7.6, 50 mm KCl.
The resonance Raman spectra for both ferric NCB5OR and cytochrome b5 proteins (Fig. 5B) exhibit v2, v3, and v10 bands at frequencies consistent with six-coordinate, low-spin ferric hemes (17). Because FAD in aqueous solution does not exhibit strong Raman bands at 1372 cm−1, the heme band at 1372 cm−1 was used as an internal subtraction standard for generation of a “NCB5OR–b5” difference spectrum. The bands shown in Fig. 5B, c correspond to the indicated Raman modes of oxidized flavin (15, 18, 19). These bands, which are attributable to the presence of oxidized flavin in the NCB5OR spectrum, account for the slight differences in the high-frequency regions of Fig. 5B, a and b. Thus, there are no apparent differences in the heme contributions to the spectra of NCB5OR and the cytochrome b5 core domain. Given the breadth and low intensity of the band(s) near 200 cm−1, no assignment of a symmetric His-FeIII-His stretching band is suggested here. However, there is Raman intensity in the 200–230 cm−1 region of the spectrum that may be attributable to modes with Fe-His stretching character.
Superoxide Production
A spectrophotometric assay was established to elucidate the kinetics of superoxide production by recombinant NCB5OR. Individual data points from seven independent series using human truncated NCB5OR and NADH, normalized by their respective Vmax, are displayed in Fig. 6B. Using the Michaelis-Menten model, kcat = ~1 s−1, and Km = ~25 mm (O2). Similar results were obtained when NADPH was used as the reductant (data not shown). As shown in Table I, human and mouse full-length NCB5OR proteins have almost identical kinetic constants: kcat = ~0.05 s−1 and Km = ~12 mm (O2). Their kcat is 20-fold lower than that of truncated human NCB5OR. Because physiological enzymes have kcat values ranging from 1 to 109 s−1 (20), the kcat value of NCB5OR is either at the low limit of an enzyme-based reaction (truncated NCB5OR) or far beyond the low limit (full-length NCB5OR). The same conclusion can be reached by using their catalytic efficiency, kcat/Km (20). Therefore, full-length NCB5OR by itself would not be expected to function as an NAD(P)H oxidase in vitro or in vivo.
TABLE I.
Kinetic constants of NCB5OR
| Substrate | Kinetic constants | Human full-length | Human truncated | Mouse full-length |
|---|---|---|---|---|
| Molecular oxygen | kcat (s−1) | 0.055 | 1.0 | 0.054 |
| Fixed [NADH] | Km (µm) | 13 | 25 | 11 |
| kcat/Km (µm−1 s−1) | 0.004 | 0.04 | 0.005 | |
| Cytochrome c | kcat(s−1) | 8.0 | 14 | 8.0 |
| Fixed [NADH] | Km (µm) | 3.0 | 4.0 | 3.2 |
| kcat/Km (µm−1 s−1) | 2.7 | 3.5 | 2.5 | |
| Cytochrome c | kcat (s−1) | 10 | 7.2 | 15 |
| Fixed [NADPH] | Km (µm) | 12 | 2.5 | 13 |
| kcat/Km (µm−1 s−1) | 0.84 | 2.9 | 1.2 | |
| NADH | kcat (s−1) | 5.0 | 10 | 3.5 |
| Fixed [cytochrome c] | Km (µm) | 1.8 | 1.4 | 1.3 |
| kcat/Km (µm−1 s−1) | 2.8 | 7.2 | 2.7 | |
| NADPH | kcat (s−1) | 2.4 | 2.4 | 6.4 |
| Fixed [cytochrome c] | Km (µm) | 1.4 | 0.91 | 2.0 |
| kcat/Km (µm−1 s−1) | 1.7 | 2.7 | 3.2 |
Cytochrome c Reduction
We have used cytochrome c to measure the NADH reductase activity of NCB5OR. As shown in Table I, both full-length and truncated NCB5OR are able to reduce cytochrome c with NADH or NADPH. Overall, the two full-length NCB5OR proteins from human and mouse have very similar kinetic behavior. Each NCB5OR protein can use both NADH and NADPH as a reductant with similar catalytic efficiency.
The catalytic efficiency with artificial substrates, cytochrome c (Table I), and ferricyanide (data not shown) for all the full-length and truncated NCB5OR proteins tested in this study is in the physiological range (20) and at least 68-fold higher than that with molecular oxygen. Therefore, both full-length and truncated NCB5OR function as a NAD(P)H reductase in vitro.
Redox Potential
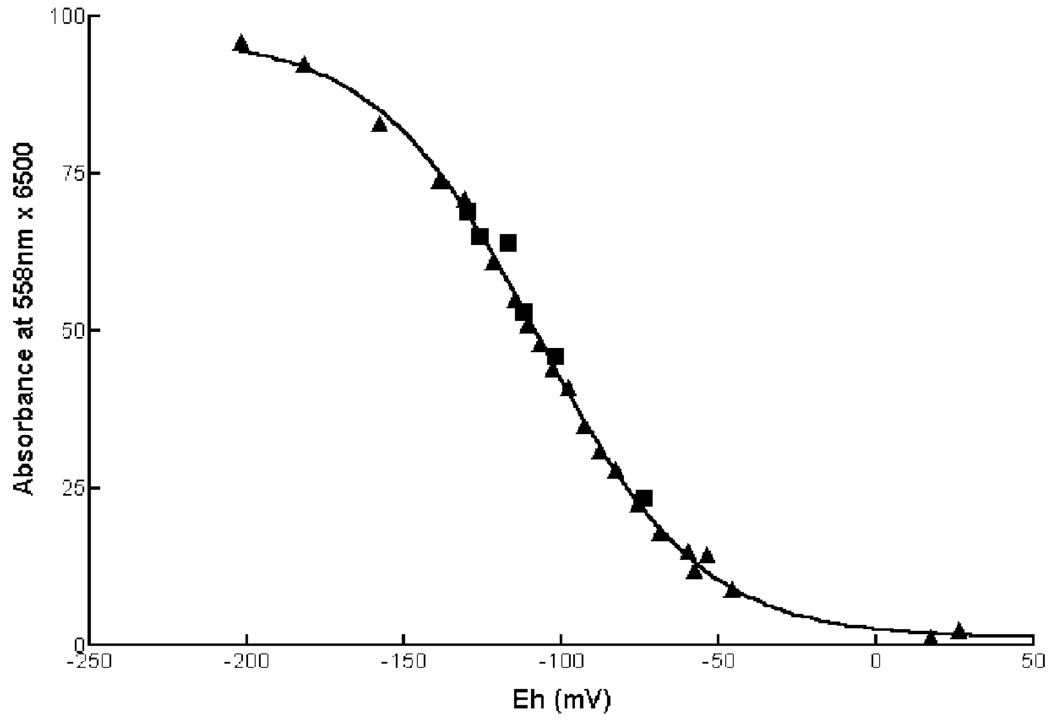
We have measured the redox potential of the heme center in truncated human NCB5OR with potentiometric titrations. The midpoint potential was calculated to be −108 mV for truncated human NCB5OR (Fig. 7), as well as for its cytochrome b5 core (data not shown). As shown in Table II, this value is almost identical to that of solubilized cytochrome b5 on outer mitochondrial membrane (−102 mV). Despite sharing the same cytochrome b5 global structure, both the cytochrome b5 core of nitrate reductase and solubilized cytochrome b5 on ER membrane have a similar value (~0 mV), whereas cytochrome b5 core of yeast cytochrome b2 has a value between −10 and −30 mV. Although the redox potential of the heme in NCB5OR is sufficiently high to permit its reduction by a number of biological reductants, in view of the marked similarity of this system to the classic cytochrome b5 reductase/cytochrome b5 system, it seems highly probable that the physiological reductant(s) of NCB5OR is NADH or NADPH, both of which have redox potentials around −320 mV (20).
FIG. 7. Oxidation-reduction potential measurements of recombinant NCB5OR.
The absorbance at 558 nm (y axis) is used to monitor the reduced heme and plotted against electrode potential (x axis) to calculate the midpoint redox potential at the heme center of human truncated NCB5OR. The solid line represents the best-fit theoretical line for a single-species, one-electron transfer process with a midpoint potential of −108 mV. Triangles represent oxidative titrations with potassium ferricyanide; squares represent reductive titrations with sodium dithionite.
TABLE II.
Redox potential of heme center in NCB5OR and related heme proteins
| Protein | E (mV) | Reference |
|---|---|---|
| NCB5OR | ||
| Human, whole | −108 | This study |
| Human, b5 core | −108 | This study |
| Solubilized outer mitochondrial membrane b5 | ||
| Human and rat, whole | −102 | 21 |
| Nitrate reductase | ||
| Spinach, b5 core | 15 | 22 |
| Green algae, b5 core | 16 | 23 |
| Solubilized ER membrane b5 | ||
| Human | −3 | 24 |
| Human | −10 | 25 |
| Human | 4 | 26 |
| Cytochrome b2 | ||
| Saccharomyces cerevisiae | −31 | 27 |
| Hansenula anomala | −10 | 28 |
| Respiratory burst oxidase b558 | ||
| Human, 2 heme centers | −225, −265 | 12 |
DISCUSSION
Impact of NH2 Terminus on NCB5OR Structure and Function
The NH2 termini of NCB5OR from mammals to Xenopus have the same length and very similar amino acid sequences. As the comparison is extended into invertebrates, there is less similarity both in length and in primary sequence. Compared with the other regions of NCB5OR, the NH2 terminus is most divergent, which implies a less important functional role. Nevertheless the NH2-terminal exon clearly has functional significance when NAD(P)H-dependent superoxide production and cytochrome c reduction are monitored. Truncated NCB5OR has a slightly higher catalytic efficiency than human and mouse full-length NCB5OR in both reactions. The only exception is the NADPH-dependent cytochrome c reduction by mouse full-length NCB5OR.
The NH2-terminal sequences appear to have no impact on the solubility or subcellular localization of NCB5OR. Both full-length and truncated recombinant NCB5OR proteins are soluble. The same distribution and ER colocalization pattern is observed for both truncated and full-length NCB5OR by confocal analysis. When overexpressed in COS7 cells, both forms of NCB5OR are found in the lysate, not in membrane fractions (data not shown).
Lack of NAD(P)H Oxidase Activity
Superoxide production by the full-length NCB5OR is not favored due to its extremely low kcat value. Moreover, the redox potential at the heme center of NCB5OR is at least 100 mV higher than the values −225 and −265 mV (12) of the two heme centers in the respiratory burst oxidase cytochrome b558. This NAD(P)H oxidase is best known for its robust H2O2 production with a kcat of ~300 s−1 (29). Because Em7 = −160 mV for O2 → (30), the superoxide production at the heme centers of cytochrome b558 is greatly favored thermodynamically, whereas reduction of oxygen to superoxide would not be favored at the NCB5OR heme center.
Until recently, neutrophil cytochrome b558 was the only mammalian b-type cytochrome known to be capable of converting oxygen into superoxide as a natural product through six-coordinate low-spin hemes (31, 32). In cytochrome b558, the electron transfer from heme to oxygen is thought to be an outer-sphere process from the heme edge and not directly from the heme iron. A number of homologues of flavocytochrome b558 have been discovered recently (the NAD(P)H oxidase family), and it is likely that some or all of them produce superoxide at the expense of NAD(P)H. These proteins are likely to play important roles in cell signaling (reviewed in Ref. 33). Other mammalian hemeproteins have also been shown to generate superoxide at low rates by autoxidation of heme; these include nitric-oxide synthase (34) and other members of the cytochrome P450 family, hemoglobin/myoglobin, and cytochrome b5 (reviewed in Ref. 35). In all these cases, the production of superoxide is thought to be an unwanted side reaction. Because both rat and human NCB5OR have a six-coordinate low-spin heme as in other cytochrome b5 proteins and, more importantly, the redox potential at the heme center is higher than that needed to favor the oxygen conversion to superoxide, it would not be expected for NCB5OR to function as an NAD(P)H oxidase. The NAD(P)H-cytochrome c reductase activity of NCB5OR is likely mediated by electron channel through side chains of charged residues, similar to those found in classical single-domain cytochrome b5 and cytochrome c complex (36–38).
In the low frequency resonance Raman spectrum of NAD(P)H oxidase cytochrome b558, the ferric Fe-His stretching frequency has been tentatively assigned to a band at 269 cm−1. If this assignment is correct, it would be consistent with anionic (imidazolate) character of the axial His side chains (17). This is consistent with the relatively negative redox potential of cytochrome b558 because anionic axial ligands generally stabilize the ferric state of the heme preferentially. By contrast, the resonance Raman spectra of NCB5OR and its cytochrome b5 core domain only show the ν9 bands of the ferrous and ferric hemes in this region of the spectrum. Although it is possible that a FeIII-His band could be overlapping the 272 cm−1 band, this would be a very high frequency and seems unlikely. It is more likely that the resonance Raman intensity between 200 and 230 cm−1 is attributable in full or in part to vibrations with Fe-His stretching contributions. Interestingly, there seems to be little or no difference between the frequencies of these bands in the ferrous (Fig. 5A) and ferric (Fig. 5B) spectra. This apparent similarity in the Fe-His bond strengths of the ferrous and ferric hemes, as reflected by Fe-His stretching frequencies, is consistent with the more positive redox potential of −108 mV relative to that observed in the NAD(P)H oxidase cytochrome b558. Thus, the lack of NAD(P)H oxidase activity may be correlated with relative invariance in the Fe(II) and Fe(III) axial ligand bond strengths in NCB5OR.
The coexistence of the cytochrome b5 and cytochrome b5 reductase activities in the same peptide begs the question of whether interaction of the heme and reductase domains or variations therein modulate the heme environment and therefore the redox potential of the heme. The similarities in both resonance Raman vibrational signatures of the heme and heme redox potential for truncated NCB5OR and the cytochrome b5 core domain suggest that the redox potential is an intrinsic property of the cytochrome b5 domain. Hence, the thermodynamic driving force for heme electron transfer reactions and the immediate heme environment would appear to be insensitive to the reductase.
ER Localization
An important clue to understanding the biological function of NCB5OR is its subcellular localization. As a soluble protein, the full-length NCB5OR appears to reside in the lumen of ER, as documented by two independent methods. Because NCB5OR protein has a slow turnover in cells,3 its presence in the ER is unlikely to be due to rapid synthesis and degradation. NCB5OR is distributed closer to nuclei and more focused than calreticulin, probably because calreticulin, as a member of the ER molecular chaperones, can associate with incompletely folded protein intermediates in pre-Golgi compartments (39, 40).
The ER localization of NCB5OR is somewhat unexpected because the NH2-terminal sequence in full-length NCB5OR lacks a canonical signal peptide sequence. NCB5OR also lacks a COOH-terminal KDEL motif (41) as well as NH2-terminal or COOH-terminal KKXX and KXKX motifs (42, 43). Some ER proteins lack these classic addressins (44). All vertebrate NCB5OR share a SQQR sequence just downstream from an NH2-terminal hydrophobic stretch, consistent with the (S/T)-X-(Q/E)-(R/K) motif that directs glycotransferases to the Golgi (45). Alternatively, a thiol-mediated protein retention mechanism through ERp44 may be involved (46).
Function of NCB5OR
Based on circumstantial evidence that a cytochrome b-type NAD(P)H oxidase might regulate target gene expression through oxidative modification of hypoxia-inducible factor 1 (47–49), we initially suggested that NCB5OR might function as a candidate oxygen sensor. The truncated human NCB5OR has kinetic properties consistent with a weak oxidase with a Km of ~2%, responding to oxygen in a physiological range. However, the markedly lower kcat of full-length protein indicates that the endogenous NCB5OR cannot function as an NAD(P)H oxidase.
The biological function of NCB5OR remained elusive until we prepared mice with targeted inactivation of the gene. The predominant phenotypic defect is insulin-deficient diabetes.2 The pancreatic β cell is particularly susceptible to ER stress (50). Thus, the phenotype of the Ncb5or-null mouse suggests that this enzyme plays a critical role in the ER stress response pathway.2 It is noteworthy that the phenotype of the Ncb5or-null mouse bears a striking resemblance to that of two mouse models of ER stress in the β cell, the Akita mouse (Ins2 C96Y) (51) and the Perk-null mouse (52, 53). All three have normal pancreatic and β-cell development, but at age 4–6 weeks, they develop progressive loss of β-cell mass and insulin-deficient diabetes. Perinuclear space in various cells has been shown to stain heavily for hydroxyl free radicals (49), which correlates with the oxidative environment within ER lumen, a necessary condition for protein folding (54). As disulfides bonds are formed in the ER lumen, electrons are passed first to protein disulfide isomerase and then to Ero1, a soluble FAD-binding protein (9). Oxygen appears to be the terminal electron acceptor. Disulfide bond formation may account for about 25% of the superoxide generated in most cells (54). Oxidative stress in the β-cell ER is likely to be even greater because of the high level of insulin synthesis and the high content of disulfide bonds in a relatively small protein. It will be interesting to see whether NCB5OR, as a novel soluble NAD(P)H reductase localized in the ER lumen, interacts with the redox proteins protein disulfide isomerase and Ero1 and whether NCB5OR attenuates superoxide formation inside the ER lumen.
Supplementary Material
Acknowledgments
K. Malecka provided technical assistance in protein preparation and enzymatic assays and M. Ocana (Harvard Center for Neurodegeneration and Repair) provided technical support for confocal microscopy.
Footnotes
This work was supported by National Institutes of Health Grants RO1 DK56050 (to H. F. B), K01 DK59901 (to H. Z.), and RO1 AI24838 (to A. R. C); Juvenile Diabetes Research Foundation International Innovative Grant 5-2003-589 (to H. F. B.); United States Department of Agriculture Grant 97-5305-5158 (to G. S. L.-R. and K. R. R.); Hermann Frasch Foundation Grant 466-HF97 (to G. S. L.-R. and K. R. R.); Germany Bundesministerium für Bildung und Forschung Grant 13N7447/5 (to H. A.); Deutsche Forschungsgemeinschaft Grant FA 225/19-1 (to J. F.), and NSERC Canada (K. L.) and France INSERM (A. L.).
The on-line version of this article (available at http://www.jbc.org) contains Supplementary Fig. 1.
The abbreviations used are: b5R, cytochrome b5 reductase; ER, endoplasmic reticulum; FAD, flavin adenine dinucleotide; MOPS, 4-morpholinepropanesulfonic acid; HPLC, high performance liquid chromatography.
Xie, J., Zhu, H., Larade, K., Ladoux, A., Seguritan, A., Chu, M., Ito, S., Bronson, R. T., Leiter, E. H., Zhang, C.-Y., Rosen, E. D., and Bunn, H. F. (2004) Proc. Nat’l. Acad. Sci. U. S. A., in press.
H. Zhu, unpublished data.
REFERENCES
- 1.Zhu H, Qiu H, Yoon H, Huang S, Bunn H. Proc. Natl. Acad. Sci. U. S. A. 1999;96:14742–14747. doi: 10.1073/pnas.96.26.14742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis CA, Dhawan IK, Johnson MK, Barber MJ. Arch. Biochem. Biophys. 2002;400:63–75. doi: 10.1006/abbi.2002.2783. [DOI] [PubMed] [Google Scholar]
- 3.Karplus PA, Daniels MJ, Herriott JR. Science. 1991;251:60–66. [PubMed] [Google Scholar]
- 4.Borgese N, Aggujaro D, Carrera P, Pietrini G, Bassetti M. J. Cell Biol. 1996;135:1501–1513. doi: 10.1083/jcb.135.6.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawano M, Shirabe K, Nagai T, Takeshita M. Biochem. Biophys. Res. Commun. 1998;245:666–669. doi: 10.1006/bbrc.1998.8502. [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Ranea JA, Mirey G, Camonis J, Valencia A. FEBS Lett. 2002;529:162–167. doi: 10.1016/s0014-5793(02)03321-5. [DOI] [PubMed] [Google Scholar]
- 7.Zhu H, Bunn HF. Respir. Physiol. 1999;115:239–247. doi: 10.1016/s0034-5687(99)00024-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curry BJ, Roman SD, Wallace CA, Scott R, Miriami E, Aitken RJ. Genomics. 2004;83:425–438. doi: 10.1016/j.ygeno.2003.08.020. [DOI] [PubMed] [Google Scholar]
- 9.Tu BP, Weissman JS. Mol. Cell. 2002;10:983–994. doi: 10.1016/s1097-2765(02)00696-2. [DOI] [PubMed] [Google Scholar]
- 10.Zander R, Lang W, Wolf HU. J. Clin. Chem. Clin. Biochem. 1989;27:185–189. doi: 10.1515/cclm.1989.27.4.185. [DOI] [PubMed] [Google Scholar]
- 11.Lukat-Rodgers GS, Wengenack NL, Rusnak F, Rodgers KR. Biochemistry. 2000;39:9984–9993. doi: 10.1021/bi0006870. [DOI] [PubMed] [Google Scholar]
- 12.Cross AR, Rae J, Curnutte JT. J. Biol. Chem. 1995;270:17075–17077. doi: 10.1074/jbc.270.29.17075. [DOI] [PubMed] [Google Scholar]
- 13.Kozak M. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitagawa T, Sugiyama T, Yamano T. Biochemistry. 1982;21:1680–1686. doi: 10.1021/bi00536a032. [DOI] [PubMed] [Google Scholar]
- 15.Desbois A, Tegoni M, Gervais M, Lutz M. Biochemistry. 1989;28:8011–8022. doi: 10.1021/bi00446a007. [DOI] [PubMed] [Google Scholar]
- 16.Desbois A, Lutz M. Eur. J. Biochem. 1992;20:321–335. [Google Scholar]
- 17.Hurst JK, Loehr TM, Curnutte JT, Rosen H. J. Biol. Chem. 1991;266:1627–1634. [PubMed] [Google Scholar]
- 18.Cohen JD, Bao W, Renganathan V, Subramaniam SS, Loehr TM. Arch. Biochem. Biophys. 1997;341:321–328. doi: 10.1006/abbi.1997.9987. [DOI] [PubMed] [Google Scholar]
- 19.Sugiyama T, Nisimoto Y, Mason HS, Loehr TM. Biochemistry. 1985;24:3012–3019. doi: 10.1021/bi00333a031. [DOI] [PubMed] [Google Scholar]
- 20.Voet D, Voet JG. Biochemistry. 2nd Ed. New York: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 21.Seetharaman R, White SP, Rivera M. Biochemistry. 1996;35:12455–12463. doi: 10.1021/bi9529723. [DOI] [PubMed] [Google Scholar]
- 22.Ratnam K, Shiraishi N, Campbell WH, Hille R. J. Biol. Chem. 1997;272:2122–2128. doi: 10.1074/jbc.272.4.2122. [DOI] [PubMed] [Google Scholar]
- 23.Cannons AC, Barber MJ, Solomonson LP. J. Biol. Chem. 1993;268:3268–3271. [PubMed] [Google Scholar]
- 24.Wang ZQ, Wang YH, Wang WH, Xue LL, Wu XZ, Xie Y, Huang ZX. Biophys. Chem. 2000;83:3–17. doi: 10.1016/s0301-4622(99)00119-2. [DOI] [PubMed] [Google Scholar]
- 25.Xue LL, Wang YH, Xie Y, Yao P, Wang WH, Qian W, Huang ZX, Wu J, Xia ZX. Biochemistry. 1999;38:11961–11972. doi: 10.1021/bi990893b. [DOI] [PubMed] [Google Scholar]
- 26.Barker PD, Ferrer JC, Mylrajan M, Loehr TM, Feng R, Konishi Y, Funk WD, MacGillivray RT, Mauk AG. Proc. Natl. Acad. Sci. U. S. A. 1993;90:6542–6546. doi: 10.1073/pnas.90.14.6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brunt CE, Cox MC, Thurgood AG, Moore GR, Reid GA, Chapman SK. Biochem. J. 1992;283:87–90. doi: 10.1042/bj2830087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silvestrini MC, Brunori M, Tegoni M, Gervais M, Labeyrie F. Eur. J. Biochem. 1986;161:465–472. doi: 10.1111/j.1432-1033.1986.tb10467.x. [DOI] [PubMed] [Google Scholar]
- 29.Cross AR, Erickson RW, Curnutte JT. Biochem. J. 1999;341(Pt 2):251–255. [PMC free article] [PubMed] [Google Scholar]
- 30.Wood PM. Biochem. J. 1988;253:287–289. doi: 10.1042/bj2530287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Isogai Y, Iizuka T, Shiro Y. J. Biol. Chem. 1995;270:7853–7857. doi: 10.1074/jbc.270.14.7853. [DOI] [PubMed] [Google Scholar]
- 32.Fujii H, Johnson MK, Finnegan MG, Miki T, Yoshida LS, Kakinuma K. J. Biol. Chem. 1995;270:12685–12689. [PubMed] [Google Scholar]
- 33.Lambeth JD, Cheng G, Arnold RS, Edens WA. Trends Biochem. Sci. 2000;25:459–461. doi: 10.1016/s0968-0004(00)01658-3. [DOI] [PubMed] [Google Scholar]
- 34.Stuehr D, Pou S, Rosen GM. J. Biol. Chem. 2001;276:14533–14536. doi: 10.1074/jbc.R100011200. [DOI] [PubMed] [Google Scholar]
- 35.Cross AR, Jones OTG. Biochim. Biophys. Acta. 1991;1057:281–298. doi: 10.1016/s0005-2728(05)80140-9. [DOI] [PubMed] [Google Scholar]
- 36.Northrup SH, Thomasson KA, Miller CM, Barker PD, Eltis LD, Guillemette JG, Inglis SC, Mauk AG. Biochemistry. 1993;32:6613–6623. doi: 10.1021/bi00077a014. [DOI] [PubMed] [Google Scholar]
- 37.Guillemette JG, Barker PD, Eltis LD, Lo TP, Smith M, Brayer GD, Mauk AG. Biochimie (Paris) 1994;76:592–604. doi: 10.1016/0300-9084(94)90136-8. [DOI] [PubMed] [Google Scholar]
- 38.Sun YL, Wang YH, Yan MM, Sun BY, Xie Y, Huang ZX, Jiang SK, Wu HM. J. Mol. Biol. 1999;285:347–359. doi: 10.1006/jmbi.1998.2295. [DOI] [PubMed] [Google Scholar]
- 39.Zuber C, Fan JY, Guhl B, Parodi A, Fessler JH, Parker C, Roth J. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10710–10715. doi: 10.1073/pnas.191359198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang J, Herscovitz H. J. Biol. Chem. 2003;278:7459–7468. doi: 10.1074/jbc.M207976200. [DOI] [PubMed] [Google Scholar]
- 41.Lewis MJ, Pelham HR. Cell. 1992;68:353–364. doi: 10.1016/0092-8674(92)90476-s. [DOI] [PubMed] [Google Scholar]
- 42.Letourneur F, Gaynor EC, Hennecke S, Demolliere C, Duden R, Emr SD, Riezman H, Cosson P. Cell. 1994;79:1199–1207. doi: 10.1016/0092-8674(94)90011-6. [DOI] [PubMed] [Google Scholar]
- 43.Teasdale RD, Jackson MR. Annu. Rev. Cell Dev. Biol. 1996;12:27–54. doi: 10.1146/annurev.cellbio.12.1.27. [DOI] [PubMed] [Google Scholar]
- 44.Suokas M, Lampela O, Juffer AH, Myllyla R, Kellokumpu S. Biochem. J. 2003;370:913–920. doi: 10.1042/BJ20021533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bendiak B. Biochem. Biophys. Res. Commun. 1990;170:879–882. doi: 10.1016/0006-291x(90)92173-w. [DOI] [PubMed] [Google Scholar]
- 46.Anelli T, Alessio M, Bachi A, Bergamelli L, Bertoli G, Camerini S, Mezghrani A, Ruffato E, Simmen T, Sitia R. EMBO J. 2003;22:5015–5022. doi: 10.1093/emboj/cdg491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goldberg MA, Dunning SP, Bunn HF. Science. 1988;242:1412–1415. doi: 10.1126/science.2849206. [DOI] [PubMed] [Google Scholar]
- 48.Cross AR, Henderson L, Jones OT, Delpiano MA, Hentschel J, Acker H. Biochem. J. 1990;272:743–747. doi: 10.1042/bj2720743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Porwol T, Ehleben W, Zierold K, Fandrey J, Acker H. Eur. J. Biochem. 1998;256:16–23. doi: 10.1046/j.1432-1327.1998.2560016.x. [DOI] [PubMed] [Google Scholar]
- 50.Oyadomari S, Araki E, Mori M. Apoptosis. 2002;7:335–345. doi: 10.1023/a:1016175429877. [DOI] [PubMed] [Google Scholar]
- 51.Oyadomari S, Koizumi A, Takeda K, Gotoh T, Akira S, Araki E, Mori M. J. Clin. Investig. 2002;109:525–532. doi: 10.1172/JCI14550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harding HP, Zeng H, Zhang Y, Jungries R, Chung P, Plesken H, Sabatini DD, Ron D. Mol. Cell. 2001;7:1153–1163. doi: 10.1016/s1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- 53.Zhang P, McGrath B, Li S, Frank A, Zambito F, Reinert J, Gannon M, Ma K, McNaughton K, Cavener DR. Mol. Cell. Biol. 2002;22:3864–3874. doi: 10.1128/MCB.22.11.3864-3874.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tu BP, Weissman JS. J. Cell Biol. 2004;164:341–346. doi: 10.1083/jcb.200311055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.