Abstract
Many bacteria and archaea protect themselves from viruses and other invasive genomes through a genetic interference pathway. The small RNAs that guide this defence specify the direct cleavage of foreign DNA.
All cells invest heavily in maintaining their genetic identities against invasion by foreign nucleic acids such as viral genomes. This problem is particularly acute for bacteria and archaea because of the pervasive presence of bacteriophages, plasmids and other mobile genetic elements in the biosphere. Genetic entities called clustered regularly interspaced short palindromic repeats (CRISPRs) have been revealed as adaptive, genomically encoded immune systems1 that protect bacteria and archaea from phages2, plasmids3 and probably other forms of foreign DNA.
This system is sequence-directed, like the well-known RNA interference (RNAi) pathway4 that operates in organisms such as plants and animals. If a portion of the CRISPR locus matches a sequence from the invasive genome, the invader is thwarted. This mechanism requires the action of CRISPR RNAs (crRNAs) that render the pathway addressable5, presumably by delivering an interference machinery to target nucleic acids that are recognized by the complementarity of Watson–Crick base pairing. On page 67 of this issue, Garneau et al.6 now show that the core CRISPR machinery protects cells in the most direct way conceivable: by crRNA-specified cleavage of the foreign DNA (Fig. 1).
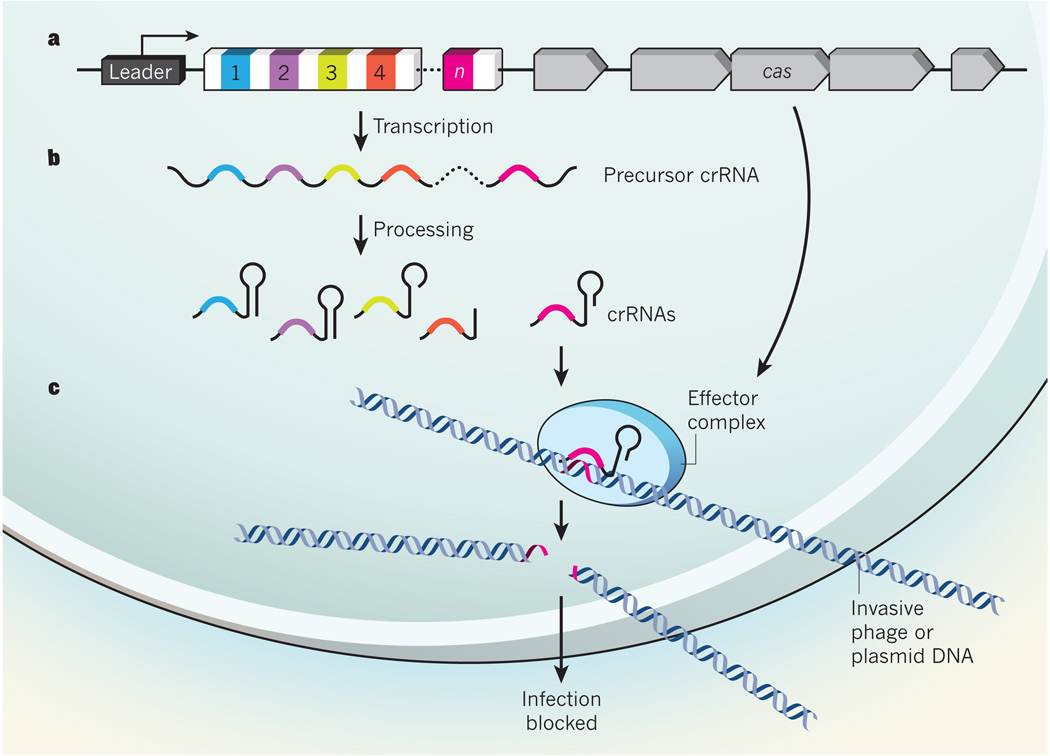
Figure 1. RNA-guided DNA cleavage by the core CRISPR machinery.
a, CRISPR loci include variable numbers of repeat and spacer sequences, with the latter derived from invasive DNAs previously encountered by the cell or its ancestors. The repeat/spacer region is flanked by a ‘leader’ sequence and by cas genes1. b, CRISPR transcription and processing of precursor crRNA yields mature crRNAs. c, These presumably associate with an effector complex that includes protein products of the cas genes. Garneau et al.6 report that the CRISPR pathway in Streptococcus thermophilus induces double-strand breaks at cognate protospacers embedded within invasive DNAs, implying that the core effector complex achieves genetic interference, and so infection blockage, by crRNA-guided DNA cleavage.
The basic features of any CRISPR system include the CRISPR locus itself, which consists of short repeats that are separated by non-repetitive sequences called spacers. These spacers match ‘proto-spacer’ sequences from phage genomes, plasmids and other previously encountered foreign DNAs, providing a genomically recorded memory of past invasions. This memory is expressed in the form of crRNAs. CRISPR loci are also flanked by a set of CRISPR-associated (cas) genes that encode protein components of the interference machinery.
When cells are challenged by a phage with no match to any CRISPR spacer, they are unprotected and (if no other anti-phage defence pathways are operative) die. However, a small fraction of infected cells can hijack pieces of the phage genome for the purpose of ‘immunization’: one or a few new phage-derived spacers are inserted at one end of the CRISPR locus, conferring immunity to that phage and allowing the adapted cell and its progeny to flourish even in the face of repeated attacks.
After the CRISPR pathway was uncovered as an adaptive, RNA-directed immune system2, an immediate issue was the identity of the molecular target. Does CRISPR interference (like RNAi) target RNAs transcribed from invasive genomes, or does it target invasive DNA itself? DNA was quickly recognized as a prime suspect in the bacterium Escherichia coli5 because crRNAs were found to be single-stranded entities without an antisense bias, making it difficult (but not impossible)7 to envisage RNA as the target. More definitive evidence for DNA targeting was reported shortly there-after in Staphylococcus epidermidis3. In spite of the genetic evidence for DNA recognition, however, the precise molecular mechanism of interference was still not proven. Formal possibilities included DNA cleavage (the leading contender), DNA sequestration, and inhibition of any number of essential DNA-based processes, such as transcription or replication.
Enter Garneau and colleagues6. In their efforts to establish that plasmids can induce CRISPR adaptation and interference in Streptococcus thermophilus, they made a surprising observation: one adapted strain with only partial plasmid resistance accumulated a linear plasmid form that was easily lost under non-selective conditions. Only the circular form was found in non-adapted strains or in a derivative of the adapted strain with a key cas gene disrupted. Sequencing of the linear plasmid revealed a break at a specific site near one end of the matching protospacer, suggesting a crRNA-based measuring mechanism.
This phenomenon applies to phage interference as well, because Southern blotting revealed the presence of phage genome fragments in multiple infected strains carrying matching CRISPR spacers. Crucially, when strains carrying distinct CRISPR spacers were tested, the cleavage site moved in concert with the site of the cognate protospacer in the phage genome. During generation of sequencing templates for cleavage-site mapping, the protospacer ends that were cleaved in vivo could ligate to an opposite blunt end of the DNA fragment generated by a restriction enzyme, indicating that CRISPR cleavage generates blunt ends as well, in agreement with the sequencing data.
Multiple categories of CRISPR/Cas loci exist with overlapping but distinct sets of Cas proteins1, raising the likelihood of mechanistic differences between the subtypes. It is likely that DNA cleavage is a general mechanism that does not apply only to S. thermophilus, but such generality remains to be established. Intriguingly, proteins encoded by ‘RAMP module’ genes, which accompany core CRISPR/Cas loci in a subset of mostly archaeal species, have been shown to catalyse crRNA-guided RNA cleavage in vitro7. Thus, as with RNAi, multiple branches of the CRISPR interference pathway probably coexist. It will be a fascinating task to disentangle the in vivo relationships between the RNA-targeting RAMP module and the DNA-targeting core Cas machinery.
With crRNA-directed double-stranded DNA cleavage now established, the race is on to identify and characterize the DNA cleavage effector enzymes in S. thermophilus as well as representatives of other CRISPR/Cas subtypes. A strong suspect (Cas3) with a likely double-strand-specific HD nuclease domain8 has already been implicated in E. coli5. Interestingly, the CRISPR/Cas loci in S. thermophilus6 and S. epidermidis3 — where the evidence for DNA targeting is strongest — do not include obvious cas3 genes, indicating that multiple, subtype-specific effectors remain to be identified.
Finally, Garneau and colleagues’ demonstration6 of crRNA-directed DNA cleavage could have practical as well as biological import. As we have noted previously3, the potential to specify RNA-guided, sequence-specific DNA cleavage via simple Watson–Crick pairing rules, rather than through complex protein–DNA interactions, opens up many enticing practical possibilities.
Contributor Information
Erik J. Sontheimer, Email: erik@northwestern.edu, Department of Molecular Biosciences, Northwestern University, Evanston, Illinois 60208, USA..
Luciano A. Marraffini, Email: marraffini@mail.rockefeller.edu, Laboratory of Bacteriology, Rockefeller University, New York, New York 10065, USA..
References
- 1.Karginov FV, Hannon GJ. Mol. Cell. 2010;37:7–19. doi: 10.1016/j.molcel.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrangou R, et al. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 3.Marraffini LA, Sontheimer EJ. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Q, Paroo Z. Annu. Rev. Biochem. 2010;79:295–319. doi: 10.1146/annurev.biochem.052208.151733. [DOI] [PubMed] [Google Scholar]
- 5.Brouns SJJ, et al. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garneau JE, et al. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 7.Hale CR, et al. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han D, Krauss G. FEBS Lett. 2009;583:771–776. doi: 10.1016/j.febslet.2009.01.024. [DOI] [PubMed] [Google Scholar]