Abstract
Wnts comprise a family of 20 lipid-modified glycoproteins in mammals and play critical roles during embryological development and organogenesis of several organ systems, including the heart. They are required for mesoderm formation and have been implicated in promoting cardiomyogenic differentiation of mammalian embryonic stem cells, but the underlying mechanisms regulating Wnt signaling during cardiomyogenesis remain poorly understood. In this report, we show that in a pluripotent mouse embryonal carcinoma stem cell line, SFRP2 inhibits cardiomyogenic differentiation by regulating Wnt3a transcription. SFRP2 inhibited early stages of cardiomyogenesis, preventing mesoderm specification and maintaining the cells in the undifferentiated state. Using a gain- and loss-of-function approach, we demonstrate that although addition of recombinant SFRP2 decreased Wnt3a transcription and cardiomyogenic differentiation, silencing of Sfrp2 led to enhanced Wnt3a transcription, mesoderm formation, and increased cardiomyogenesis. We show that the inhibitory effects of SFRP2 on Wnt transcription are secondary to interruption of a positive feedback effect of Wnt3a on its own transcription. Wnt3a increased its own transcription via the canonical pathway and TCF4 family of transcription factors, and the inhibitory effects of SFRP2 on Wnt3a transcription were associated with disruption of downstream canonical Wnt signaling. The inhibitory effects of Sfrp2 on Wnt3a expression identify Sfrp2 as a “checkpoint gene,” which exerts its control on cardiomyogenesis through regulation of Wnt3a transcription.
Keywords: Myogenesis, P19, Wnt3a, Teratocarcinoma
Introduction
The Wnt signaling system in mammals, comprising 20 lipid-modified glycoproteins, exerts pleiotropic effects on a wide variety of cellular processes, including proliferation, apoptosis, self-renewal, and differentiation [1, 2]. These proteins play a crucial role during embryological development [3] and organogenesis of several organ systems, including cardiac morphogenesis [4]. In birds and amphibians, Wnts inhibit heart formation [5–7], whereas canonical Wnt signaling is required for mesoderm specification [8, 9] and promotes cardiomyogenic differentiation of pluripotent mouse embryonic stem cells [10, 11]. Classically, Wnts have been divided into two groups, canonical and noncanonical. Canonical Wnt signaling pathways involve binding of Wnt to its Frizzled receptor in association with LRP5/6, with subsequent translocation of Axin to the cell membrane and inhibition of GSK3β. GSK3β, in the absence of Wnt ligand, phosphorylates β-catenin, directing it for subsequent degradation via the ubiquitin proteasomal pathway. In the presence of Wnt ligand, GSK3β is inhibited. This leads to accumulation of cytoplasmic β-catenin, which subsequently translocates to the nucleus, binds to corepressors such as Groucho, and initiates target gene transcription via the TCF/LEF family of transcription factors. Wnt signaling is finely tuned through the presence of Wnt antagonists, which are divided into two classes, the Dickkopf (Dkk) class and the SFRP class [12]. Whereas Dkk specifically antagonizes the canonical pathway by interrupting the binding of Wnt/Fz to LRP5/6 coreceptor, SFRPs bind to Wnts and potentially antagonize both the canonical and the noncanonical pathways. In mammals, the SFRP class mainly comprises the family of SFRP proteins, of which five have been identified so far. On the basis of sequence homology, the family can be divided into two groups; SFRP1, SFRP2, and SFRP5 form one group, whereas SFRP3 and SFRP4 form another [12]. SFRPs, in the N-terminal half of the protein, possess a cysteine-rich region very similar to one present on the Frizzled receptor and are thought to function as extracellular Wnt antagonists by binding Wnts [13].
Despite the existence of extracellular modulators of Wnt signaling, factors regulating Wnt transcription and downstream signaling in mammalian cardiomyogenesis are largely unknown. SFRPs are present in the heart-forming fields and adjoining regions in the developing embryo [4]. For instance, SFRP2 is expressed in a broad ectodermal territory overlapping with the heart-forming fields, whereas SFRP1 and SFRP3 are expressed in all three germ layers [5, 14, 15]. Despite their presence in these regions, the interaction of SFRPs with Wnts, the mechanisms of modulation of Wnt effect, and their role in regulating cardiac morphogenesis in mammals are unclear.
In this study, we investigated transcriptional regulation and modulation of Wnt signaling during cardiomyogenic differentiation. We show for the first time that SFRP2, although an extracellular Wnt antagonist, inhibits Wnt3a transcription and regulates cardiomyogenic differentiation. We studied this interaction in a pluripotent mouse embryonal carcinoma stem cell line (P19CL6), which has served as a well-validated model for the dissection of pathways determining cardiomyogenic differentiation [16]. We demonstrate that (a) Wnt3a exerts positive feedback on its own transcription and (b) SFRP2, by interrupting this positive feedback loop, inhibits Wnt3a transcription and regulates cardiomyogenic differentiation. Cells treated with recombinant SFRP2 protein had dramatically decreased cardiomyogenic differentiation and failed to express Wnt3a. Conversely, silencing of SFRP2 led to enhanced Wnt3a transcription and cardiomyogenic differentiation. Although it belongs to the same family as SFRP2, SFRP1 did not exert these inhibitory effects on differentiation. These data describe a novel function of SFRP2 in regulating Wnt transcription during cardiomyocyte specification and differentiation.
Materials and Methods
Cells and Reagents
P19CL6 and stable transfectant MLC2v-GFP P19CL6 cell lines were kindly provided by Dr. Richard Kitsis (Albert Einstein College of Medicine, Bronx, NY) and Dr. Christine Mummery (The Netherlands Institute of Developmental Biology, Utrecht, The Netherlands). Recombinant WNT3A, recombinant SFRP1, SFRP2, and Dkk-1 were purchased from R&D Systems Inc. (Minneapolis, http://www.rndsystems.com). Quantitative polymerase chain reaction (PCR) primers for Wnt3a, Nanog, Sfrp2, GATA4, MHC, MLC2a, TBX5, and cardiac actin were bought from Applied Bio-systems (Foster City, CA, http://www.appliedbiosystems.com). TOP (a luciferase reporter plasmid driven by 4 × copies of TCF-binding sequence), FOP flash (the same vector containing mutated TCF4-binding domain) vectors, Wnt3a cDNA in pUSE-amp, and dominant negative TCF4 plasmid were purchased from Upstate Biotechnology (Billerica, MA, http://www.upstate.com). Short interfering RNA (siRNA) oligonucleotides targeting Wnt3a and SFRP2 were bought from Ambion (Foster City, CA, http://www.ambion.com). Vector backbone (pcDNA 6.2-GW/EmGFP-micro-RNA [miR]) for micro-RNA expression for gene silencing was bought from Invitrogen (Carlsbad, CA, http://www.invitrogen.com). β-Catenin antibody was purchased from Cell Signaling Technology (Boston, http://www.cellsignal.com), and antibodies against glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and TBP and cardiac troponin T were bought from Abcam (Cambridge, MA, http://www.abcam.com). MF20 antibody was bought from Developmental Studies Hybridoma Bank (Iowa City, IA, http://www.uiowa.edu/~dshbwww).
Cell Culture and Differentiation
P19CL6 cells were grown in 10-cm dishes in the presence of α Minimum Essential Medium (Gibco, Grand Island, NY, http://www.invitrogen.com) supplemented with 10% fetal bovine serum, penicillin, and streptomycin (growth medium). For induction of myogenic differentiation, cells were initially seeded on 6-cm dishes (4 × 103–4 × 104 cells per dish) and grown in the presence of growth medium and 1% dimethyl sulfoxide (DMSO) for 14 days (differentiation medium). Stably transfected P19CL6 cells expressing MLC-2v-GFP were grown in the presence of Dulbecco's modified Eagle's medium:Ham's F-12 medium (Invitrogen) supplemented with 7.5% fetal calf serum, essential amino acids, and 600 μg/ml G418.
Conventional and Quantitative Reverse Transcription-PCR
RNA extraction was performed with the help of the RNeasy Plus kit (Qiagen, Hilden, Germany, http://www1.qiagen.com), and cDNA synthesis was carried out with Superscript III (Invitrogen) according to the manufacturer's instructions. Conventional PCR was performed with 100 ng of cDNA, each reaction being run for 30 cycles. GAPDH was run for 20 cycles. For real-time PCR, 100 ng of cDNA was used as a template, and gene expression was analyzed using the ΔCT method with light cycler (ABI Prism 7700; Applied Biosystems). GAPDH was used for internal control to normalize for equal RNA loading. Controls without reverse transcription (RT) (cDNA synthesis without adding reverse transcriptase) were used for each gene analyzed to exclude genomic DNA contamination. Conventional PCR primer sequences are available upon request.
Western Blotting and Immunofluorescent Staining
Western blotting and immunofluorescent staining was performed using standard protocols as previously described [10]. For detection of nuclear and cytoplasmic β-catenin, the nuclear and cytoplasmic fractions were separated using the Fraction Prep cell fractionation kit (Biovision Inc., Mountain View, CA, http://www.biovision.com). In each case, signal was detected using a chemiluminescent detection system (ECL; Amersham Biosciences, Piscataway, NJ, http://www.amersham.com). Area densitometry analysis was performed using Fluostar software (UVP Bioimaging Systems, Upland, CA, USA). For immunofluorescent staining, cells following a 14-day treatment in differentiation medium (DM) with or without SFRP2 were trypsinized, reseeded onto two-well chamber slides (Lab-Tek II; Nalge Nunc International, Rochester, NY, http://www.nalgenunc.com), fixed with 4% paraformaldehyde, stained with the primary and secondary antibodies for 1 hour each, and visualized using a Zeiss Axiovert 200 microscope (Carl Zeiss, Jena, Germany, http://www.zeiss.com). For MF20 staining, cells were fixed with methanol (precooled at –20°C) for 15 minutes.
Luciferase Assay
Cells were seeded onto six-well plates overnight at a seeding density of 1 × 105 cells per well and were transfected the next day using Lipofectamine 2000 (Invitrogen) in low-serum Opti-MEM medium (Gibco) for 6 hours. Each well contained 4 μg of TOP or FOP Flash vector and 100 ng of pRL-CMV as the cotransfected control. Medium containing Wnt, lithium, or differentiating medium was added to the wells 6 hours after transfection. Cells were harvested and lysed after overnight treatment with Wnt/lithium and after a 48-hour treatment with differentiating medium. Firefly luciferase activity (TOP and FOP) was measured and normalized to Renilla luciferase activity for each sample using the Dual luciferase system (Promega, Madison, WI, http://www.promega.com).
Vector-Based Micro-RNA Production, siRNA, and Dominant Negative TCF4 Transfection Protocols
For creating a stably transfected P19CL6 cell line expressing a vector encoding a micro-RNA against SFRP2, we used the pcDNA 6.2-GW/EmGFP-miR vector backbone (Invitrogen) that contains enhanced green fluorescent protein (EGFP) and a blasticidin resistance cassette. Micro-RNA sequences targeting SFRP2 were generated using computer-based algorithms (Invitrogen) and cloned into the vector backbone. Cells were transiently transfected with several constructs and were screened by RT-PCR to identify one with the highest efficiency in silencing SFRP2. Owing to cocistronic expression of EGFP, cells transiently transfected with the chosen SFRP2 micro-RNA construct were sorted by flow cytometry with green fluorescent protein (GFP) expression and subsequently used for differentiation experiments. As the vector backbone contains a blasticidin resistance cassette, the cells were grown in differentiation medium or growth medium containing blasticidin for maintenance of selected cells. As a control, cells transfected with the same vector encoding a micro-RNA targeting the β-galactosidase gene were used and grown in medium containing blasticidin. For transient knockdown of SFRP2 expression, we also used siRNA oligos. Cells were seeded onto six-well plates at a density of 1 × 105 cells per well and transfected using Lipofectamine 2000 and 50 nM siRNA. A scrambled siRNA was used in each case at the same concentration. Cells were incubated overnight with the transfection mixture followed by addition of fresh medium the next day. Cells were transfected with dominant negative TCF4 or empty vector using a similar protocol and were used for differentiation experiments 48 hours following transfection.
Results
SFRP2 Inhibits Myogenic Differentiation of P19CL6 Mouse Embryonic Stem Cells
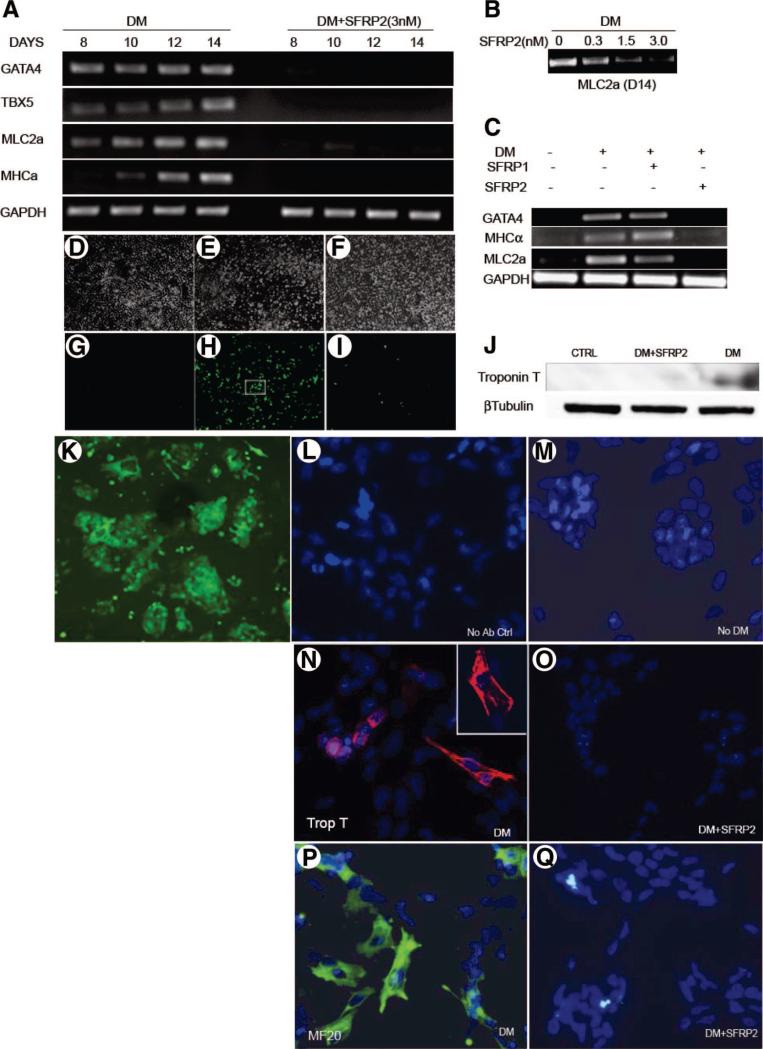
We first determined the effects of SFRP1 and SFRP2 on myogenic differentiation. We treated P19CL6 cells with cardiomyogenic differentiation medium containing 1% DMSO for 14 days in the absence or presence of SFRP2 (3 nM) (with cells being replenished with fresh medium every other day). Previous studies have demonstrated that these cells at baseline do not express myocyte-specific transcription factors or structural proteins but can be induced to differentiate into cardiomyocytes when treated with medium containing DMSO [10, 11]. Consistent with these reports, using RT-PCR we observed induction of expression of cardiomyocyte-specific transcription factors, such as GATA4 and TBX5, and structural proteins, such as myosin heavy chain (MHC)α and myosin light chain (MLC)2a, following 8 days of treatment with differentiation medium (Fig. 1A). In the presence of differentiation medium, GATA4, TBX5, MHC, and MLC2a mRNA expression increased over the course of 14 days, consistent with cardiomyogenic differentiation (Fig. 1A). However, concomitant treatment with recombinant SFRP2 protein dramatically inhibited the expression of these transcription factors and structural proteins (Fig. 1A). The expression of MLC2a following treatment with differentiation medium for 14 days was inhibited by SFRP2 in a concentration-dependent fashion (Fig. 1B). Moreover, when we treated cells with SFRP1, a closely related family member, no significant inhibitory effects on myogenesis were observed, suggesting that the effects are specific to SFRP2 rather than a property exhibited by this family of molecules. SFRP1, in contrast to SFRP2, did not decrease the expression of transcription factors and structural proteins characteristic of myogenic differentiation (Fig. 1C). To further quantitate the degree of inhibition exerted by SFRP2, we treated P19CL6 cells expressing GFP driven by a MLC2v promoter (a kind gift from Dr. Mummery, Utrecht, The Netherlands) with differentiation medium. Following treatment with DM for 14 days, approximately 20% of the cells expressed GFP, whereas a dramatic reduction in GFP expression was observed in cells treated concomitantly with SFRP2 (Fig. 1G–1I, 1K). Using Western blotting and immunofluorescent staining techniques, we also observed dramatic reduction in expression of cardiac-specific structural proteins. Cells treated with DM for 14 days expressed cardiac troponin T protein, but troponin T expression was dramatically inhibited with concomitant treatment with SFRP2 (3 nM) (Fig. 1J). Similarly, differentiated P19Cl6 cells stained for cardiac troponin (Fig. 1N) with characteristic striations (Fig. 1N, inset) and MF20 (sarcomeric myosin) (Fig. 1P) compared with minimal differentiation observed when cells were grown in the absence of DM (Fig. 1M). Concomitant treatment with SFRP2 resulted in a dramatic reduction in expression of these proteins (Fig. 1O, 1Q). Finally, we did not observe any beating cardiomyocytes following treatment with DM and SFRP2, but typical beating clusters (supplemental online Fig. 1A, 1B) were seen 14 days following treatment with DM alone.
Figure 1.
SFRP2 inhibits myogenic differentiation of P19Cl6 cells. Representative reverse transcription-polymerase chain reaction (n = 3) demonstrating (A): expression of myogenic transcription factors and structural proteins in P19Cl6 cells treated with differentiation medium (DM) in the absence or presence of SFRP2 (3 nM), (B): expression of MLC2a in P19Cl6 cells treated with DM for 14 days in the absence or presence of increasing concentrations of SFRP2, and (C) expression of myocyte-specific structural proteins and transcription factors in P19Cl6 cells treated with DM for 14 days in the presence of SFRP1 (3 nM) or SFRP2 (3 nM). (D–I): Phase-contrast views (×100) and green fluorescent protein (GFP) expression in MLC2v-GFP P19Cl6 cells at (D, G) baseline; (E, H) treated with DM; (F, I) DM and SFRP2 (3 nM) for 14 days (n = 3). (J): Western blotting for cardiac Troponin T expression in P19Cl6 cells treated with DM ± SFRP2; CTRL represents untreated cells. (K): High-power view (×200) of (H) inset showing GFP-positive MLV 2v cells. (L–Q): Immunofluorescent staining for Troponin T (red) and MF20 (green) in P19Cl6 cells treated with DM for 14 days. (L): Cells stained without adding primary antibody. Cardiac Troponin T staining of cells: (M): grown in growth medium without induction of differentiation, (N): treated with DM (N inset) zoomed image of Troponin T-positive cardiomyocyte, (O) treated with DM + SFRP2 (3 nM). MF20 staining of cells; (P) treated with DM; (Q) treated with DM + SFRP2 (3 nM). (Nuclei are stained with DAPI for L–Q; total magnification of ×400 for fluorescent images). Abbreviations: Ab, antibody; CTRL, control (untreated cells); DM, differentiation medium; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; MHCα, myosin heavy chain; MLC2a, myosin light chain; Trop T, troponin T.
SFRP2 Prevents Mesodermal Commitment and Aggregate Formation of P19CL6 Cells Treated with Cardiomyogenic Differentiation Medium
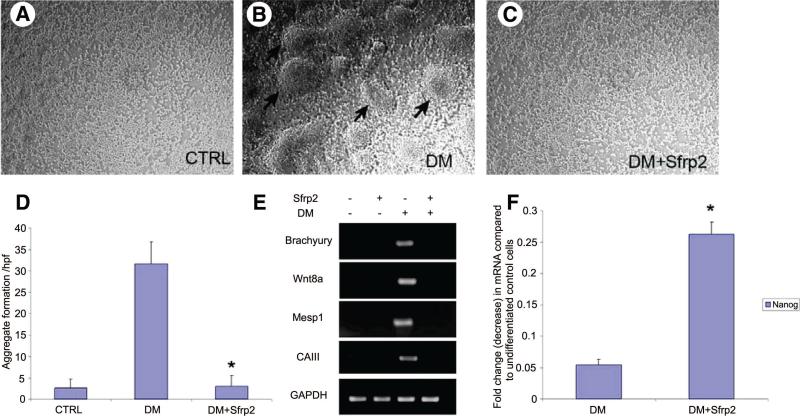
We next investigated the stage at which SFRP2 inhibits cardiomyogenic commitment and differentiation. One of the early events that take place during cardiomyogenic differentiation of P19 embryonic stem cells is aggregate formation and differentiation into mesodermal lineage [17]. No distinct embryoid body-like structures were observed in P19CL6 cells in the absence of differentiating medium (Fig. 2A); however, when these cells were treated with differentiating medium, cellular aggregates resembling embryoid body-like structures formed by 4 – 6 days of treatment (Fig. 2B). However, concomitant treatment of cells with differentiation medium and SFRP2 significantly decreased embryoid body formation (p < .05), and the cells were morphologically indistinguishable from those grown in the absence of differentiating medium (Fig. 2C, 2D). In fact, SFRP2 decreased the number of cellular aggregates in cells treated with differentiation medium to that seen in control cells (Fig. 2D). As the appearance of embryoid bodies has been suggested to be associated with mesodermal commitment, we determined the expression of various mesodermal markers by RT-PCR. Following 6 days of treatment with differentiation medium, we observed dramatic upregulation of the mesodermal marker gene Brachyury. Expression of mesp1, a marker of cardiac-specific mesoderm, as well as expression of Wnt8a, also significantly increased. We also noted induction of carbonic anhydrase III expression, an early marker of mesoderm specification and one expressed in developing mouse embryonic muscle [18]. However, addition of recombinant SFRP2 (3 nM) to cells treated with differentiation medium completely abolished upregulation of genes associated with mesodermal or myogenic specification, such as brachyury, mesp1, carbonic anhydrase III, and Wnt8a (Fig. 2E). These data suggest that SFRP2 inhibits myogenic commitment of P19CL6 cells very early during the process of differentiation by preventing aggregation and mesodermal commitment of these cells. Differentiation of these cells into other related myogenic lineages, such as skeletal muscle, in the presence of SFRP2 was also excluded by measuring expression of key skeletal muscle regulatory genes, such as myogenin, MRF4, and MYF5 (data not shown). We next investigated whether SFRP2, by inhibiting cardiomyogenic differentiation, maintains the cells in the undifferentiated state. We determined Nanog expression in cells treated with differentiation medium for 6 days. Nanog is a transcription factor that is associated with the pluripotent state in embryonic stem cells and is downregulated with the onset of differentiation [19]. Using quantitative PCR, we showed that treatment with differentiating medium led to near complete disappearance of Nanog expression in P19CL6 cells, whereas cells concomitantly treated with SFRP2 exhibited fivefold higher Nanog expression compared with cells treated with differentiation medium alone (Fig. 2F) (p < .05).
Figure 2.
SFRP2 inhibits aggregate formation and mesodermal specification in P19Cl6 cells treated with DM for 6 days (n = 6). (A–C): Bright-field images (×200) showing morphology of P19Cl6 cells grown as a monolayer at baseline (A), following treatment with DM (B), or following treatment with DM and SFRP2 (3 nM) (C). (D): Number of aggregates per high power field in P19Cl6 cells following treatment with DM in the presence or absence of SFRP2. (*, p < .01 between DM-treated and DM + Sfrp2-treated groups.) (E): Reverse transcription-polymerase chain reaction (PCR) demonstrating expression of genes associated with mesodermal specification in P19Cl6 cells treated with DM for 6 days in the absence or presence of SFRP2 (3 nM). (F): Nanog expression by quantitative PCR in P19Cl6 cells treated with DM for 6 days with or without SFRP2 (3 nM) (*, p < .05; n = 3; bars represent SD). Abbreviations: CTRL, control; DM, differentiation medium; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Silencing of SFRP2 in P19CL6 Cells Enhances Cardiomyogenesis
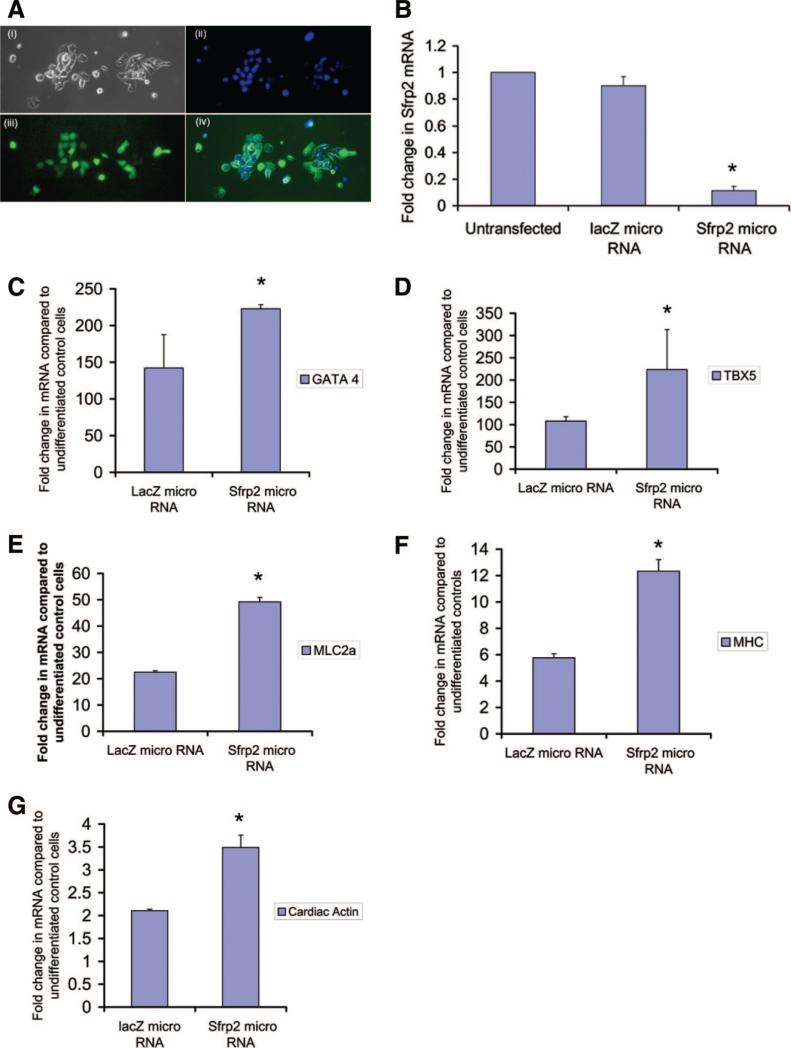
As P19CL6 cells express SFRP2, we next investigated whether silencing of SFRP2 expression leads to enhanced cardiomyogenic differentiation. For silencing of Sfrp2 expression, we transfected cells with a plasmid vector encoding a micro-RNA targeting Sfrp2. We obtained transfection efficiencies of 60%–70%, and cells expressing the micro-RNA targeting Sfrp2 also possessed cocistronic expression of EmGFP (Fig. 3Ai–3Aiv). Cells transfected with a vector construct encoding a micro-RNA directed against the β-galactosidase gene served as controls. We obtained nearly 90% knockdown of Sfrp2 gene expression with this technique (Fig. 3B). As the vector backbone also contained a blasticidin resistance cassette, transfected cells (expressing lacZ miR or SFRP2 miR) following transient transfection were selected on the basis of GFP expression by flow cytometry and treated with differentiation medium containing blasticidin (200 μg/ml) for a period of 14 days. Cells were harvested at the end of 14 days and analyzed by quantitative PCR for the expression of transcription factors and structural proteins characteristic of myogenic differentiation. We also confirmed, by RT-PCR, persistent knockdown of SFRP2 expression at the end of 14 days (data not shown). As shown in Figure 3C–3G, differentiation medium in control cells (lacZ micro-RNA-transfected) induced an increase of nearly 100-fold in expression of transcription factors such as GATA4 and TBX5 compared with undifferentiated cells. However, cells harboring a knockdown of SFRP2 expression exhibited a further significant increase of 2–2.5-fold (p < .05) in gene expression of transcription factors such as GATA4 and TBX5, as well as structural proteins such as MHC and MLC2a. These results, taken together with our gain-of-function experiments, suggest that SFRP2 regulates cardiomyogenesis by exerting an inhibitory effect.
Figure 3.
Knockdown of SFRP2 enhances myogenic differentiation of P19Cl6 cells. (A): P19Cl6 cells transfected with a plasmid vector possessing cocistronic expression of GFP and micro-RNA targeting SFRP2 showing phase-contrast (Ai), Hoechst-stained nuclei (Aii), GFP (Aiii), and merged figure (Aiv). (B): Quantitative polymerase chain reaction (QPCR) demonstrating knockdown of SFRP2 gene expression in P19Cl6 cells following transfection with vector encoding SFRP2 micro-RNA or lacZ (control) micro-RNA (n = 3; *, p < .05). (C–G): QPCR demonstrating gene expression of cardiomyogenic transcription factors and structural proteins in P19Cl6 cells treated with differentiation medium for 14 days, following knockdown of either SFRP2 or LacZ expression using vector-based micro-RNA (n = 3; *, p < .05 for all groups; bars represent SD). Abbreviations: MHC, myosin heavy chain; MLC, myosin light chain.
SFRP2 Inhibits Canonical Wnt Signaling During Cardiomyogenic Differentiation
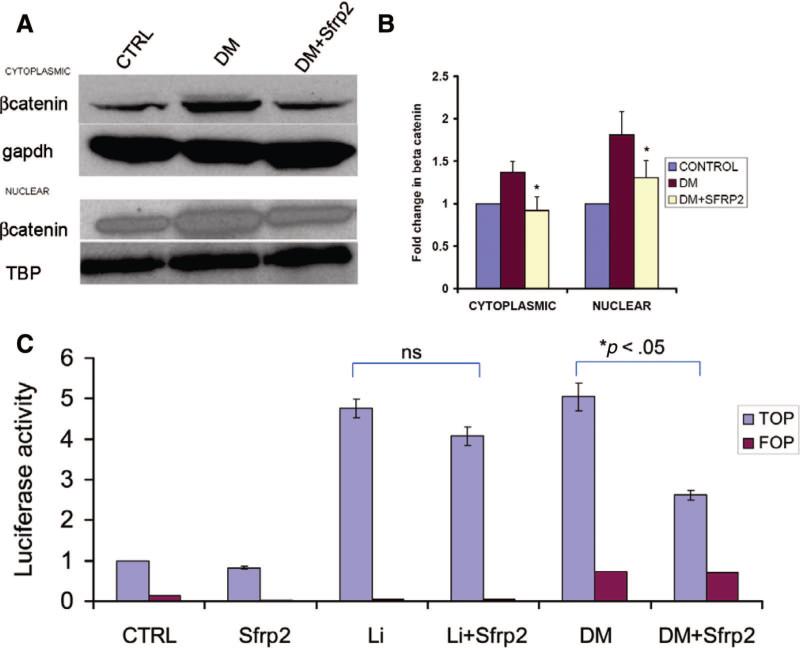
Canonical Wnt signaling has been demonstrated to be necessary for primitive streak formation in vivo and embryonic stem cell-derived mesoderm formation in vitro [8]. Moreover, cardiomyogenic differentiation of P19CL6 cells is associated with activation of canonical Wnt signaling. As SFRP2 is a known extracellular Wnt antagonist [12], we hypothesized that it could be exerting inhibitory effects on cardiomyogenic differentiation by disruption of downstream canonical Wnt signaling. We initially determined cytoplasmic and nuclear β-catenin levels by Western blotting in P19CL6 cells that had been treated with differentiation medium for 4 days in the absence or presence of SFRP2. As previously reported [10], we noted increased β-catenin in the cytoplasmic fraction of the lysate of cells treated with differentiation medium. Western blotting also demonstrated increased accumulation of nuclear β-catenin, consistent with translocation of cytoplasmic β-catenin to the nucleus. In contrast, SFRP2 significantly decreased the accumulation of both nuclear and cytoplasmic β-catenin (Fig. 4A, 4B; p < .05).
Figure 4.
SFRP2 interrupts canonical Wnt signaling in P19Cl6 cells treated with DM. (A): Representative Western blotting (n = 3) for β-catenin in nuclear and cytoplasmic fractions of cell lysates following treatment with DM for 48 hours in the absence or presence of SFRP2 (3 nM). GAPDH and Tata-binding proteins served as loading CTRLs. (B): Densitometry analysis of Western blots in (A) (*, p < .05 for DM + SFRP2 groups compared with groups treated with DM alone in both nuclear and cytoplasmic fractions; bars represent SE). (C): Luciferase activity (fold change compared with untreated CTRLs) in P19Cl6 cells treated with DM or LiCl (20 mM) for 48 hours in the absence or presence of SFRP2 (3 nM) (n = 6; each experiment performed in triplicate; *, p < .05). FOP activity (red) served as negative CTRL. Abbreviations: CTRL, control; DM, differentiation medium; gapdh, glyceraldehyde-3-phosphate dehydrogenase; ns, not significant.
To further confirm the inhibitory effects of SFRP2 on canonical Wnt signaling in differentiating P19CL6 cells, we used a reporter assay measuring canonical Wnt-dependent transcriptional activity. P19CL6 cells were transfected with the TOP and FOP Flash vectors and then treated with differentiation medium for 48 hours. The TOPFlash vector has luciferase expression driven by 4 × TCF4 DNA-binding sequence. Cells transfected with the FOPFlash vector containing mutated TCF4-binding sequences served as negative control to demonstrate specificity of the reporter assay. We observed a fivefold increase in lucif-erase activity in P19CL6 cells treated with cardiomyogenic differentiating medium, whereas addition of recombinant SFRP2 protein significantly attenuated the increase in luciferase activity by almost 50% (p < .05) (Fig. 4C). Canonical Wnt signaling results in inhibition of GSK3β with subsequent inhibition of ubiquitin-mediated proteolytic degradation of cytoplasmic β-catenin. To demonstrate that SFRP2 did not exert any inhibitory effects downstream of GSK3β, we also treated cells with lithium chloride (20 mM). Lithium chloride has been shown to inhibit GSK3β and to simulate canonical Wnt signaling. As shown in Figure 4C, lithium chloride-treated cells exhibited an increase of nearly fivefold in reporter activity, similar to cells treated with differentiation medium. Interestingly, cells treated with SFRP2 alone in the absence of lithium chloride had a 18% decrease in reporter activity, suggesting the possible presence of autocrine canonical Wnt signaling. However, in contrast to its effects on reporter activity in cells treated with differentiation medium, SFRP2 did not significantly decrease the lithium chloride-induced increase in reporter activity (Fig. 4C). The 15% attenuation of LiCl-induced reporter activity was similar to the decrease in reporter activity seen in cells treated with SFRP2 alone (p > .05). Taken together, these observations suggest that SFRP2 inhibits canonical Wnt signaling in differentiating P19CL6 cells upstream of GSK3β.
SFRP2 Regulates Wnt3a Transcription by Inhibiting Wnt3a Autopositive Feedback
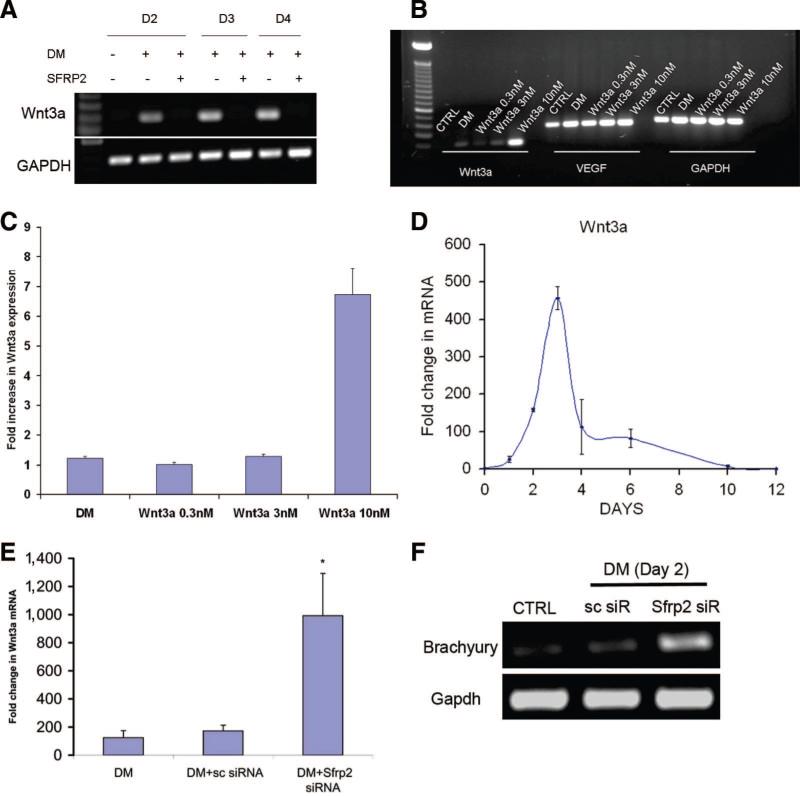
We next focused on the interaction of SFRP2 with Wnts that are potentially critical for successful cardiomyogenic differentiation. Canonical Wnts such as Wnt3a are expressed in the first few days following treatment with differentiation medium. Naito et al. have previously demonstrated that Wnt3a is sufficient to induce myogenesis in P19CL6 cells even in the absence of differentiation medium [11]. Moreover, mice lacking Wnt3 fail to develop primitive streak and mesoderm. We thus explored the interaction of Wnt3a and SFRP2 and unexpectedly observed that SFRP2 inhibited transcription of Wnt3a mRNA within 2 days of treatment with differentiation medium. P19CL6 cells were treated with differentiation medium for 4 days in either the presence or the absence of SFRP2. Cells were harvested at 2, 3, and 4 days and analyzed for expression of Wnt3a mRNA by RT-PCR. In contrast to untreated control cells, cells treated with SFRP2 (3 nM) failed to exhibit any expression of Wnt3a mRNA (Fig. 5A). It is interesting to note that even though SFRP2 completely suppressed DM-induced Wnt3a expression between day (D)2 and D4 following induction of differentiation, maximal suppression of expression of mesodermal marker genes, such as Brachyury and Mesp1, occurred at a slightly later time point (D6). The expression of mesodermal genes at D2–D4 in the absence of Wnt3a expression suggests that SFRP2's inhibition of Wnt3a expression is independent of its effects on mesoderm formation (supplemental online Fig. 5).
Figure 5.
SFRP2 regulates Wnt3a transcription. (A): Representative reverse transcription-polymerase chain reaction (RT-PCR) (n = 3) for Wnt3a in P19Cl6 cells treated with DM in the absence or presence of SFRP2 (3 nM) for 2, 3, and 4 D. (B): Representative RT-PCR (n = 3) for Wnt3a in P19Cl6 cells treated with DM or recombinant WNT3A protein in increasing concentrations. VEGF expression serves as a positive CTRL. GAPDH expression demonstrates equal loading. (C): Semiquantitative analysis of Wnt3a mRNA expression in cells treated with DM or increasing concentrations of Wnt3a, as seen in (B). Untreated cells served as CTRLs. (D): Temporal profile of Wnt3a expression by quantitative polymerase chain reaction (QPCR) in P19Cl6 cells treated with DM compared with untreated CTRLs (n = 3; each experiment performed in triplicate). (E): QPCR demonstrating Wnt3a expression in P19Cl6 cells following transient transfection with SFRP2 siRNA or sc siRNA (cells treated with DM for 48 hours) (n = 3; *, p < .05 between Sfrp2 siRNA-treated and sc siRNA-treated groups; bars represent SD). (F): Representative RT-PCR (n = 3) for Brachyury expression in cells treated with SFRP2 siRNA compared with cells treated with sc siRNA, 48 hours following addition of DM. CTRL cells were not treated with DM. Abbreviations: CTRL, control; D, day; DM, differentiation medium; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; sc, scrambled; siR, short interfering RNA; siRNA, short interfering RNA; VEGF, vascular endothelial growth factor.
As SFRP2 is considered to be an extracellular Wnt antagonist and possesses strong binding affinity for Wnt3a [20], we hypothesized that its inhibitory effects on Wnt3a transcription are potentially secondary to interruption of a positive feedback effect of Wnt3a on its own transcription. To demonstrate that Wnt3a increases its own mRNA, we treated P19CL6 cells with different concentrations of recombinant Wnt3a protein for 48 hours. Cells were subsequently harvested and examined for Wnt3a mRNA expression. As shown in Figure 5B, P19CL6 cells do not express Wnt3a mRNA at baseline. However, treatment with 10 nM Wnt3a protein dramatically increased Wnt3a mRNA. Using semiquantitative analysis, we showed that the increase in Wnt3a mRNA following treatment with 10 nM Wnt3a protein was significantly higher than the induction of Wnt3a mRNA following treatment with differentiation medium for the same duration of time (Fig. 5C). As a positive control, we also noted an increase of vascular endothelial growth factor (VEGF) mRNA following treatment with Wnt3a protein, VEGF being a known downstream target of Wnt3a.
To further demonstrate that SFRP2 not only inhibits but regulates Wnt3a transcription, we used a loss-of-function approach to determine the effects of silencing SFRP2 expression on Wnt3a expression. P19CL6 cells in the undifferentiated state do not express Wnt3a, but induction of Wnt3a expression occurs early on during cardiomyogenic differentiation. As illustrated in Figure 5D, Wnt3a expression peaks by 3 days and disappears by 10 days. Again, P19CL6 cells harboring knockdown of Sfrp2 were subsequently treated with differentiation medium for 2 days, at which time the cells were harvested and analyzed for Wnt3a expression by quantitative PCR. We chose this time point to be on the ascending limb of the curve for Wnt3a transcription (Fig. 5D). Untransfected controls, as well as cells transfected with scrambled RNA, exhibited a 200-fold increase in Wnt3a expression. SFRP2 siRNA-transfected cells had a further fivefold significant (p < .05) increase in Wnt3a mRNA expression compared with untransfected or scrambled RNA-transfected controls (Fig. 5E). Taken together, these data strongly support the notion that Wnt3a exerts a positive feedback effect on its own transcription that is regulated by SFRP2. As SFRP2 silencing leads to increased DM-induced Wnt3a expression, we next investigated whether SFRP2 silencing leads to increased expression of mesodermal marker Brachyury, a known Wnt3a target gene [21]. Indeed, we did observe dramatic upregulation of Brachyury expression in cells harboring SFRP2 knockdown just 2 days following treatment with DM (Fig. 5F). This observation is consistent with our earlier finding of increased cardiomyogenesis in cells harboring SFRP2 knockdown; enhanced cardiomyogenesis may be mediated by increased Brachyury expression and mesoderm formation.
Wnt3a Activates Canonical Signaling to Exert a Positive Feedback on Its Own Transcription
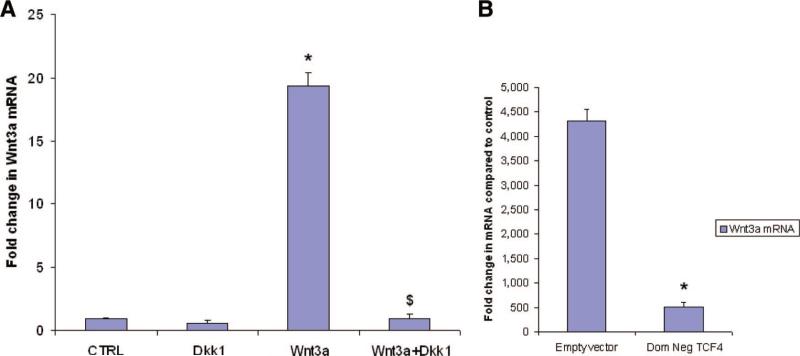
Having demonstrated that Wnt3a increases its own transcription in a positive feedback effect, we next investigated the mechanism of Wnt3a-induced increase in Wnt3a mRNA expression. We hypothesized that Wnt3a activates the canonical signaling pathway to increase its own transcription. We treated P19CL6 cells with Wnt3a (10 nM) in the presence or absence of Dkk1 (500 ng/ml) for 48 hours. Dkk1 binds to LRP5/6 and is thought to be a specific inhibitor of canonical Wnt signaling, leaving noncanonical Wnt signaling intact. Cells were subsequently harvested and examined for Wnt3a mRNA expression by quantitative PCR. As shown in Figure 6A, recombinant WNT3A protein increased Wnt3a mRNA in P19CL6 cells by almost 20-fold after 48 hours (p < .05). However, concomitant treatment with Dkk-1 completely abolished the increase in Wnt3a mRNA (p < .05).
Figure 6.
WNT3A induces Wnt3a mRNA expression via the canonical pathway. (A): Quantitative polymerase chain reaction (QPCR) for Wnt3a in P19Cl6 cells treated with Wnt3a (10 nM) for 48 hours in either the absence or presence of DKK-1 (cells were preincubated with Dkk-1 for 1 hour before adding Wnt3a)(n = 3; each experiment in triplicate; *, p < .05 compared with CTRL cells; $, p < .05 compared with Wnt3a-treated group; bars represent SD). (B): QPCR for Wnt3a expression in differentiation medium-treated P19Cl6 cells transfected with either empty vector or Dom Neg TCF4 (n = 3; each experiment in triplicate; *, p < .05). Abbreviations: CTRL, control; Dkk, Dickkopf; Dom Neg, dominant negative.
Activation of canonical Wnt signaling results in translocation of β-catenin to the nucleus, where it initiates target gene transcription, mediated by the TCF/LEF group of transcription factors. We next determined whether transcription of Wnt3a is mediated by the TCF group of transcription factors. In particular, previous reports have demonstrated the importance of TCF4 in β-catenin signaling. We therefore transfected cells with a dominant negative form of TCF4 and treated them with differentiating medium for 48 hours. Cells transfected with the empty vector served as control. As shown in Figure 6B, differentiating medium induced a dramatic upregulation of Wnt3a in cells transfected with empty vector. However, cells transfected with the dominant negative form of TCF4 experienced a 90% reduction of Wnt3a mRNA expression (p < .05), demonstrating the critical importance of the canonical system and TCF family of transcription factors in exerting this feedback.
Discussion
In this study, we report a novel form of interaction between Wnts and the Sfrp family of Wnt signaling modulators. We demonstrate that SFRP2 inhibits Wnt3a transcription by interrupting a positive feedback effect of Wnt3a on its own transcription. Wnt3a-induced positive feedback is intriguing, as Wnt3a is not known to be its own target gene. Using gain- and loss-of-function approaches, we show that SFRP2, by regulating this positive feedback, modulates cardiomyogenic differentiation. Teleologically, a positive feedback effect of Wnt3a on its own transcription is advantageous, as it provides the cells with a mechanism to rapidly amplify critical differentiation signals. During the preparation of this article, Ueno et al. reported Wnt3a-induced transcriptional inhibition of Wnt1 and Wnt3a in differentiating embryoid bodies of mouse embryonic stem cells [22]. Although our results are apparently discrepant with the conclusions drawn by Ueno et al. [22] in regard to Wnt3a-induced transcriptional feedback, on closer inspection, the existence of temporally separated, both early positive and late negative feedback loops, appears to be a strong possibility. Unchecked Wnt signaling during later stages of myogenesis inhibits myogenesis and directs differentiation along a hematopoietic lineage [17, 22]. Thus, an early positive and late negative regulation of Wnt3a transcription would be optimal for cardiomyocyte specification and differentiation. In our study, silencing Sfrp2 expression in P19Cl6 cells alone did not induce Wnt3a transcription but required concomitant treatment with DMSO. Moreover, Wnt3a expression, although significantly higher in cells lacking SFRP2, was not sustained and followed a temporal profile similar to that of control cells, suggesting the presence of other regulatory molecules. Nevertheless, the transient but significant increase in Wnt3a transcription was sufficient to enhance cardiomyogenic differentiation.
Although our study suggests the role of SFRP2 as a cardiomyogenic “checkpoint” molecule regulating cell differentiation via modulation of key Wnt feedback loops, it raises other questions. What are the factors that determine whether Wnts exert negative or positive feedback on a cell? What are the other checkpoints for control of a Wnt-positive feedback within the cell? Differences in the ability of Wnts to mount positive feedback loops and of SFRPs to control such transcriptional networks might explain divergent or opposing effects of Wnts in development and disease. For instance, a recently published report demonstrates much higher binding affinity of Wnt3a for SFRP2 than SFRP1 [20]; this might explain the differences between SFRP1 and SFRP2 with regard to inhibition of cardiomyogenesis and Wnt3a transcription as observed in our study. The opposite roles of Wnt in heart formation in mammalian systems and chick/Xenopus have been attributed to species differences and biphasic effects of Wnt signaling [17]. Our data suggest that transcriptional regulation of Wnts by Wnt modulators is another novel area of Wnt/Wnt antagonist interaction that should be considered to explain such differences. In this regard, we have recently described an antiapoptotic effect of SFRP2 on cardiomyocytes [23]. The contrasting effects of SFRP2 on cardiomyogenesis and cell survival may be related to its antagonistic effects on expression of different Wnts.
It is also tempting to speculate whether transcriptional regulation of Wnts by SFRPs, as noted in this study, is important in regulation of differentiation in other cell systems or in aberrant cell growth and dedifferentiation seen in tumors. SFRPs are considered to have tumor-suppressive properties, and epigenetic silencing of Sfrps including Sfrp2 has been observed in colorectal and esophageal carcinomas [24]. It is thought that Sfrp silencing leads to constitutively active autocrine Wnt signaling, a phenomenon shown to exist in human cancer cells [25] and thought to have a causal relation with development of cancer. Our study raises the questions of whether Sfrps mediate tumor-suppressive properties by potentially regulating Wnt transcription and whether silencing of Sfrps leads to enhanced Wnt transcription and a cycle of positive feedback signaling. Clearly, future studies need to address the role of regulators of Wnt transcription in development and diseased states.
Conclusion
In summary, our study provides evidence for the existence of Wnt3a-positive feedback signaling in cardiomyogenic differentiation of murine P19 embryonal carcinoma stem cells and shows a novel form of transcriptional cross-talk between SFRP2 and WNT3A. With recent observations that Wnts regulate stem cell renewal [26], identification of Wnt feedback loops, their principal molecular regulators and downstream effects, may be key to understanding mechanisms determining cell choices of self-renewal versus differentiation.
Supplementary Material
Acknowledgments
We thank Dr. Richard Kitsis (Albert Einstein College of Medicine, New York, NY) for P19CL6 cells, Dr. Christine Mummery (The Netherlands Institute of Developmental Biology, Utrecht, The Netherlands) for the generous gift of MLC2v P19CL6 cells, and the Developmental Studies Hybridoma Bank for the MF20 antibody. We also thank Dr. Blanche Capel (Duke University, Durham, NC) and Dr. Tannishtha Reya (Duke University, Durham, NC) for helpful suggestions and critical reading of the manuscript. This study was supported by NIH Grants R01-HL081744, R01-HL073219, R01-HL072010, and R01-HL035610 and by grants from the Mandel foundation (to V.J.D.). A.D. was supported by the NIH K99 HL088317 Pathway to Independence Award and by a Young Investigator Award from the GlaxoSmithKline Beecham Cardiovascular Research and Education Foundation. A.D. is currently affiliated with the Carolina Cardiovascular Biology Center, University of North Carolina, Chapel Hill, NC.
Footnotes
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
References
- 1.Gordon MD, Nusse R. Wnt signaling: Multiple pathways, multiple receptors, and multiple transcription factors. J Biol Chem. 2006;281:22429–22433. doi: 10.1074/jbc.R600015200. [DOI] [PubMed] [Google Scholar]
- 2.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 3.Logan CY, Nusse R. The Wnt Signaling Pathway in Development and Disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 4.Eisenberg LM, Eisenberg CA. Wnt signal transduction and the formation of the myocardium. Dev Biol. 2006;293:305. doi: 10.1016/j.ydbio.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 5.Marvin MJ, Di Rocco G, Gardiner A, et al. Inhibition of Wnt activity induces heart formation from posterior mesoderm. Genes Dev. 2001;15:316–327. doi: 10.1101/gad.855501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schneider VA, Mercola M. Wnt antagonism initiates cardiogenesis in Xenopus laevis. Genes Dev. 2001;15:304–315. doi: 10.1101/gad.855601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tzahor E, Lassar AB. Wnt signals from the neural tube block ectopic cardiogenesis. Genes Dev. 2001;15:255–260. doi: 10.1101/gad.871501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindsley RC, Gill JG, Kyba M, et al. Canonical Wnt signaling is required for development of embryonic stem cell-derived mesoderm. Development. 2006;133:3787–3796. doi: 10.1242/dev.02551. [DOI] [PubMed] [Google Scholar]
- 9.Gadue P, Huber TL, Paddison PJ, et al. Wnt and TGF-beta signaling are required for the induction of an in vitro model of primitive streak formation using embryonic stem cells. Proc Natl Acad Sci U S A. 2006;103:16806–16811. doi: 10.1073/pnas.0603916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakamura T, Sano M, Songyang Z, et al. A Wnt- and beta-catenin-dependent pathway for mammalian cardiac myogenesis. Proc Natl Acad Sci U S A. 2003;100:5834–5839. doi: 10.1073/pnas.0935626100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naito AT, Akazawa H, Takano H, et al. Phosphatidylinositol 3-kinase-Akt pathway plays a critical role in early cardiomyogenesis by regulating canonical Wnt signaling. Circ Res. 2005;97:144–151. doi: 10.1161/01.RES.0000175241.92285.f8. [DOI] [PubMed] [Google Scholar]
- 12.Kawano Y, Kypta R. Secreted antagonists of the Wnt signalling pathway. J Cell Sci. 2003;116:2627–2634. doi: 10.1242/jcs.00623. [DOI] [PubMed] [Google Scholar]
- 13.Jones SE, Jomary C. Secreted Frizzled-related proteins: Searching for relationships and patterns. Bioessays. 2002;24:811–820. doi: 10.1002/bies.10136. [DOI] [PubMed] [Google Scholar]
- 14.Chapman SC, Brown R, Lees L, et al. Expression analysis of chick Wnt and frizzled genes and selected inhibitors in early chick patterning. Dev Dyn. 2004;229:668–676. doi: 10.1002/dvdy.10491. [DOI] [PubMed] [Google Scholar]
- 15.Baranski M, Berdougo E, Sandler JS, et al. The dynamic expression pattern of frzb-1 suggests multiple roles in chick development. Dev Biol. 2000;217:25–41. doi: 10.1006/dbio.1999.9516. [DOI] [PubMed] [Google Scholar]
- 16.van der Heyden MAG, Defize LHK. Twenty-one years of P19 cells: What an embryonal carcinoma cell line taught us about cardiomyocyte differentiation. Cardiovasc Res. 2003;58:292–302. doi: 10.1016/s0008-6363(02)00771-x. [DOI] [PubMed] [Google Scholar]
- 17.Naito AT, Shiojima I, Akazawa H, et al. Developmental stage-specific biphasic roles of Wnt/beta-catenin signaling in cardiomyogenesis and hematopoiesis. Proc Natl Acad Sci U S A. 2006;103:19812–19817. doi: 10.1073/pnas.0605768103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lyons GE, Buckingham ME, Tweedie S, et al. Carbonic anhydrase III, an early mesodermal marker, is expressed in embryonic mouse skeletal muscle and notochord. Development. 1991;111:233–244. doi: 10.1242/dev.111.1.233. [DOI] [PubMed] [Google Scholar]
- 19.Mitsui K, Tokuzawa Y, Itoh H, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 20.Wawrzak D, Metioui M, Willems E, et al. Wnt3a binds to several sFRPs in the nanomolar range. Biochem Biophys Res Commun. 2007;357:1119–1123. doi: 10.1016/j.bbrc.2007.04.069. [DOI] [PubMed] [Google Scholar]
- 21.Yamaguchi TP, Takada S, Yoshikawa Y, et al. T (Brachyury) is a direct target of Wnt3a during paraxial mesoderm specification. Genes Dev. 1999;13:3185–3190. doi: 10.1101/gad.13.24.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ueno S, Weidinger G, Osugi T, et al. From the cover: Biphasic role for Wnt/beta-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104:9685–9690. doi: 10.1073/pnas.0702859104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mirotsou M, Zhang Z, Deb A, et al. Secreted frizzled related protein 2 (Sfrp2) is the key Akt-mesenchymal stem cell-released paracrine factor mediating myocardial survival and repair. Proc Natl Acad Sci U S A. 2007;104:1643–1648. doi: 10.1073/pnas.0610024104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki H, Watkins DN, Jair K-W, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36:417. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 25.Bafico A, Liu G, Goldin L, et al. An autocrine mechanism for constitutive Wnt pathway activation in human cancer cells. Cancer Cell. 2004;6:497–506. doi: 10.1016/j.ccr.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 26.Reya T, Duncan AW, Ailles L, et al. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003;423:409–414. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.