In estrogen receptor (ER) positive breast cancer cells, insulin-like growth factor signaling activates S6K1 to initiate ER-mediated gene transcription and cell growth.
Abstract
The IGF pathway stimulates malignant behavior of breast cancer cells. Herein we identify the mammalian target of rapamycin (mTOR)/S6 kinase 1 (S6K1) axis as a critical component of IGF and estrogen receptor (ER)α cross talk. The insulin receptor substrate (IRS) adaptor molecules function downstream of IGF-I receptor and dictate a specific biological response, in which IRS-1 drives proliferation and IRS-2 is linked to motility. Although rapamycin-induced mTOR inhibition has been shown to block IGF-induced IRS degradation, we reveal differential effects on motility (up-regulation) and proliferation (down-regulation). Because a positive correlation between IRS-1 and ERα expression is thought to play a central role in the IGF growth response, we investigated the potential role of ERα as a downstream mTOR target. Small molecule inhibition and targeted knockdown of S6K1 blocked the IGF-induced ERαS167 phosphorylation and did not influence ligand-dependent ERαS118 phosphorylation. Inhibition of S6K1 kinase activity consequently ablated IGF-stimulated S6K1/ERα association, estrogen response element promoter binding and ERα target gene transcription. Moreover, site-specific ERαS167 mutation reduced ERα target gene transcription and blocked IGF-induced colony formation. These findings support a novel link between the IGF pathway and ERα, in which the translation factor S6K1 affects transcription of ERα-regulated genes.
The clinical benefit of targeting estrogen receptor (ER)α in breast tumor subsets is well established (1–3). The interplay between ERα and numerous growth factor pathways may represent a mechanism of resistance to ERα-directed systemic therapies (e.g. selective ER modulators, aromatase inhibitors) (4, 5). Survival and growth of breast cancer cells through the positive reciprocal action between IGF and ERα cross talk is well documented (6–9). A number of studies demonstrated that ERα can drive IGF-mediated biology via transcriptional up-regulation of key IGF pathway genes [e.g. IGF ligands, IGF-I receptor (IGF1R), insulin receptor substrate (IRS)] (10–12). Moreover, loss of ERα expression and/or function results in decreased IGF-induced growth and survival (13–15). Similarly, IGF has been shown to stimulate multiple facets of ERα activity (e.g. serine phosphorylation, promoter binding, and target gene transcription) (16–19). Although this biochemical augmentation of ERα is associated with cellular proliferation and growth, a direct linkage of IGF to ERα functional up-regulation has not been established.
The IGF pathway stimulates the malignant phenotype during breast cancer initiation and progression (20–23). After IGF ligand stimulation, IRS adaptor molecules bind the IGF-1R to facilitate the recruitment of various downstream signaling factors critical to aberrant cellular growth. As an intrinsic means of endogenous autoregulation, prolonged pathway activation triggers downstream components of the phosphatidylinositol 3-kinase (PI3K) axis, namely ribosomal S6 kinase 1 (S6K1), resulting in serine phosphorylation of the IRS proteins and subsequent proteasomal degradation (24). This negative feedback loop is effectively blocked by rapamycin-mediated inhibition of mammalian target of rapamycin (mTOR) and leads to a potentiation of IGF-induced Akt phosphorylation (25). Transient transfection models suggest that Akt directly phosphorylates ERα in a site-specific manner at serine 167 within the A/B domain (26, 27). Akt-induced ERα phosphorylation has been shown to both stimulate and repress ERα function (28, 29). Therefore, it is plausible that elevated levels of Akt phosphorylation found in breast tumors resistant to the rapamycin analogs may be linked to changes in IGF/ERα cross talk (30). However, the biological repercussions of rapamycin-induced IGF signal potentiation have yet to be determined.
The overall aim of our study was to elucidate the molecular mechanism responsible for IGF-induced changes in ERα function. Because negative feedback inhibition is known to potentiate IGF-induced Akt phosphorylation, we hypothesized that ERα phosphorylation and function would be altered in corresponding fashion. Herein we reveal a novel mechanism of growth factor and steroid receptor cross talk, whereby the IGF pathway regulates ERα function in an mTOR/S6K1-dependent manner. Furthermore, these data provide additional rationale to target the IGF pathway in rapamycin-resistant, ERα-positive breast tumors.
Results
Rapamycin blocks IGF-induced proliferation and ERα phosphorylation
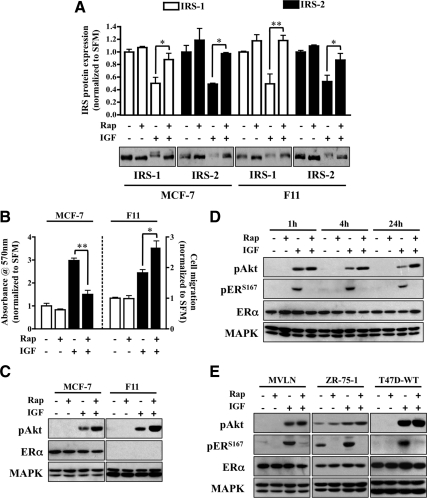
The initial objective was to determine whether IGF-induced negative feedback was linked to specific IRS isoforms. IRS-1 and IRS-2 are coexpressed in both MCF-7 and MDA-231-BO (F11) breast cancer cell lines. In MCF-7 cells, IGF1R phosphorylates IRS-1 and stimulates proliferation through IRS-1, whereas in F11 cells, IRS-2 is the predominant substrate and links IGF signaling to motility (31–33). The inhibition of mTOR by rapamycin reverses the IGF-induced down-regulation of IRS proteins. Cells stimulated with IGF in the absence or presence of rapamycin were assessed for IRS protein levels (Fig. 1A). The down-regulation and rescue of IRS species appeared identical across cell lines. However, when IGF-stimulated cell behavior was examined, phenotypic differences were observed (Fig. 1B). Although rapamycin increased IGF-induced transwell migration in F11 cells, IGF-induced proliferation in MCF-7 cells was largely inhibited. It should be noted that IGF does not alter the proliferation of F11 cells (data not shown) or motility in MCF-7 cells (34). In addition, the rapamycin concentration used for these studies did not affect basal growth or migration, suggesting that results were not due to nonspecific effects of the drug.
Fig. 1.
Rapamycin blocks IGF-induced proliferation and ERα phosphorylation. A, MCF-7 and MDA-231BO (F11) cells were serum starved overnight and pretreated with rapamycin (Rap; 10 nm) for 30 min before IGF stimulation (4 h). Lysates were collected and resolved by SDS-PAGE for immunoblot analysis against the proteins of interest. The graph represents IRS protein levels as fold change of treated vs. nontreated control. B, Monolayer proliferation was measured by MTT assay in MCF-7 cells (left axis), and motility was determined by transwell Boyden chamber in F11 cells (right axis) in response to rapamycin and IGF treatment. The graph is presented as fold change response vs. nontreated control. C, After rapamycin pretreatment, MCF-7 (left panel) and F11 (right panel) cells were exposed to IGF (1 h) and immunoblot analysis performed. D, MCF-7 cells were serum starved overnight, pretreated with rapamycin for 30 min, and treated with IGF for the indicated time points. Lysates were resolved by SDS-PAGE and immunoblotted against the proteins of interest. E, Western blot analysis of MVLN, ZR-75-1, and T47D cells exposed to IGF (1 h) was performed after 30 min of rapamycin pretreatment. Error bars represent sd on all graphs. The MAPK (anti-MAPK) served as a loading control for immunoblot experiments, and all results are representative of at least three independent replicates.
To determine how rapamycin disruption of the negative feedback pathways differentially altered biology, Akt phosphorylation was measured in response to IGF stimulation. Immunoblot analysis confirmed that rapamycin potentiated IGF-induced Akt phosphorylation (Fig. 1C) in a manner consistent with previous studies (25). Moreover, Akt hyperphosphorylation occurred in an ERα-independent manner. Although increasing levels of Akt phosphorylation appear to correlate directly with motility in F11 cells, this was not the case in MCF-7 cells and suggests that signaling factors downstream of both Akt and mTOR play a role in IGF-induced proliferation.
The substantial evidence linking ERα to breast cancer cell proliferation implies that rapamycin disrupted a potential influence of mTOR over ERα function. In MCF-7 cells (Fig. 1D), both short (1 and 4 h) and long-term exposure (24 h) to IGF increased Akt phosphorylation and resulted in increased ERαS167 phosphorylation. Whereas pretreatment with rapamycin had little effect on Akt phosphorylation, the abrogation of ERα phosphorylation was largely unexpected because Akt activation has been shown to directly result in ERα phosphorylation of S167 (26, 27). These results were confirmed in additional ERα-positive breast cancer cells (Fig. 1E). Rapamycin and IGF did not affect total levels of ERα, but IGF-mediated S167 phosphorylation was consistently inhibited by rapamycin. To confirm that mTOR signaling to ERα was not limited to IGF-specific Akt activation, we used the MCF-7 variant myr-Akt3 line (Supplemental Fig. 1, published on The Endocrine Society's Journals Online web site at http://mend.endojournals.org). These cells were previously selected for stable expression of a myristylated Akt3 to result in constitutive activation of Akt (35). In myr-Akt3 cells, ERαS167 phosphorylation was independent of the level of activity of Akt phosphorylation but suppressed by rapamycin. As an additional control, the ERα mRNA message remained static throughout the experiment, regardless of treatment (Supplemental Fig. 2). Taken together, these results implicate molecules downstream of Akt responsible for IGF-induced ERα phosphorylation and proliferation.
IGF evokes an estrogen-like response
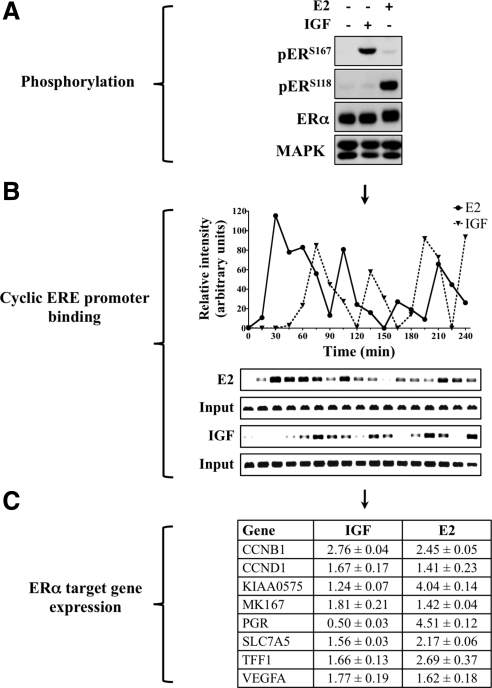
Cross talk between the IGF system and ERα has long been known to positively regulate cell cycle entry, growth, proliferation, and resistance to apoptosis (36, 37). Although Akt is a critical component of IGF biology, evidence suggests that IGF-independent Akt activation serves as a negative regulator of ERα function (38). Therefore, to better understand the role that mTOR plays during IGF regulation of ERα, we determined whether IGF regulation of ERα function is similar to its cognate ligand estradiol (E2). In accordance with aforementioned and published data, IGF treatment stimulated ERαS167 phosphorylation and E2 increased ERαS118 phosphorylation (Fig. 2A). Under these conditions, cross-activation was not observed because both IGF and E2 induced specific phosphorylation of discrete serine sites. Interestingly, although IGF-induced MAPK phosphorylation can reportedly signal through ERαS118 phosphorylation, this effect was not observed here (39). As a means of validation, additional cell lines and time points were tested and the MCF-7 results confirmed (data not shown).
Fig. 2.
IGF evokes an estrogen-like response. A, MCF-7 cells were cultured in estrogen-depleted conditions for 3 d, serum starved overnight, treated with IGF or E2 (1 h), and immunoblot analysis performed. B, ChIP was performed after IGF or E2 exposure. Protein/chromatin complexes were collected every 15 min, ERα-bound DNA fragments isolated, and the pS2 ERE amplified by PCR. The graph is a measure of relative intensity. C, After 24 h of IGF or E2 exposure, mRNA was isolated and RT-qPCR performed. Target gene expression was normalized to RPLP0 gene expression and is presented as fold change of treatment vs. serum-free conditions with sd included. All results are representative of at least three independent replicates.
Both IGF and E2 have been shown to independently regulate ERα-dependent chromatin binding, target gene transcription, and cell proliferation. However, Akt activation has also been shown to negatively regulate ERα through increased promoter and genome-wide chromatin binding (28). These inconsistencies likely stem from differences in both model systems and conclusions based on sampling the interactions at limited time points. In an attempt to explain this contradiction and delineate the effect of IGF-induced ERα phosphorylation, efforts were focused on the dynamic relationship between ERα and chromatin because the biological effects of ERα require cycling on and off DNA. As a result, MCF-7 cells were treated with IGF or E2 and assessed for binding of ERα to the pS2 estrogen response element (ERE) by chromatin immunoprecipitation (ChIP) assay (Fig. 2B). The IGF and E2 response was measured over the course of 4 h, in 15-min increments. Consistent with previous reports, E2 treatment resulted in the rapid association and dissociation of ERα to the pS2 promoter (40). Although IGF-induced binding occurred later than that of E2, it matched the cyclical pattern of promoter binding and dissociation. In the absence of E2 or IGF stimulation, promoter binding was not apparent (data not shown). Thus, IGF stimulation induced cyclical binding of ERα to chromatin at the ERE promoter in a manner similar to E2-induced activation.
The pattern of IGF-induced chromatin binding closely mirrors that of E2 and lends further evidence that ligand-independent activation of ERα DNA binding downstream of IGF1R activation may positively regulate gene transcription. Therefore, the expression of a number of ERα-regulated genes was measured after IGF or E2 treatment by reverse transcription quantitative-PCR (RT-qPCR) analysis (Fig. 2C). All of the genes examined have been linked to ERα-positive breast tumors and are vital to cellular proliferation. IGF treatment increased the expression of many ERα-linked genes, but there were important differences. For example, IGF1R activation has been shown to decrease progesterone receptor gene (PGR) expression (41). It should be noted that although IGF is known to down-regulate PGR expression, absolute steroid receptor levels (e.g. ERα, progesterone receptor) do not always correlate directly with levels of target gene transcription (42, 43). Taken together, these results suggest that IGF promotes E2-like regulation of ERα at the level of chromatin and that this activity may play a role in IGF-induced proliferation.
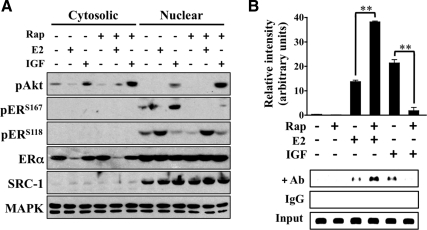
Rapamycin inhibits nuclear ERα activity
Because IGF is a positive regulator of ERα promoter binding and transcription, the effect of rapamycin on ERα phosphorylation in reference to cellular localization was evaluated. MCF-7 cells were pretreated with rapamycin before IGF or E2 exposure, the cytosolic and nuclear extracts were separated by fractionation, and lysates were subjected to immunoblot analysis (Fig. 3A). In accordance with previous reports, cytosolic ERα levels were markedly decreased in response to E2 with a corresponding increase in nuclear ERαS118 phosphorylation (44). IGF did not grossly alter ERα localization and Akt phosphorylation was detected in both fractions, suggesting that IGF treatment resulted in the phosphorylation of a smaller, yet significant portion of nuclear ERα. As predicted, rapamycin blocked IGF-induced nuclear ERαS167 phosphorylation and had no effect on ERαS118 phosphorylation. Furthermore, ChIP assays confirmed that mTOR inhibition blocked IGF-stimulated ERα promoter binding (Fig. 3B). Interestingly, rapamycin increased E2-induced chromatin binding independent of ERαS118 phosphorylation. These data confirm that mTOR facilitates IGF-induced ERα phosphorylation and promoter occupancy.
Fig. 3.
Rapamycin inhibits nuclear ERα activity. A, MCF-7 cells were cultured in the absence of hormone for 3 d, serum starved overnight, and pretreated with rapamycin (Rap) for 30 min before IGF or E2 (1 h) exposure. Nuclear and cytosolic extracts were isolated by fractionation, resolved by SDS-PAGE, and immunoblotted against the indicated proteins. Steroid receptor coactivator (SRC)-1 (anti-SRC-1) and MAPK (anti-MAPK) were used as nuclear and total lysate loading controls. B, ChIP was performed on cells pretreated with rapamycin for 30 min before IGF or E2 treatment (4 h). For ChIP assays, IgG and input lanes were included for each sample to control for nonspecificity and ensure equal DNA loading. The graph is a measure of relative intensity in which black bars indicate IGF treatment and error bars represent sd. All results are representative of at least three independent replicates.
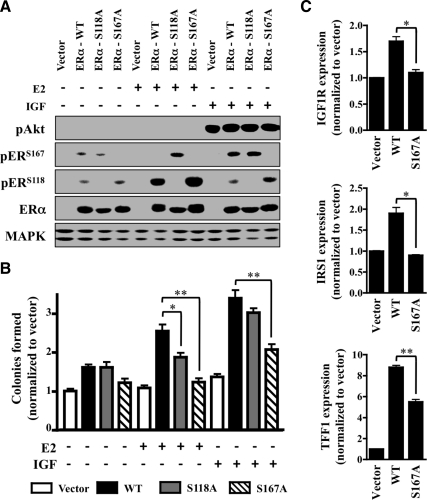
ERαS167 mediates IGF-induced phosphorylation, transcription, and growth
To better establish the link between site-specific IGF-induced ERα phosphorylation and proliferation, the effects of IGF and E2 on wild-type (WT) and mutant phosphoserine ERα constructs were tested. As previously described, the C4–12 cell line was clonally selected from MCF-7 cells for a total loss of ERα expression (45). Early passage C4–12 cells were transiently transfected with ERα-WT, a serine to alanine mutant of ERα (ERα-S167A or ERα-S118A) or empty vector (vector), and the response to IGF or E2 exposure was measured (Fig. 4A). Immunoblot analysis confirmed that C4–12 cells were negative for endogenous ERα expression, and an equal rate of both transfection and expression was attained by the ERα constructs. IGF stimulated ERαS167 phosphorylation in ERα-WT and ERα-S118A mutants but failed to do so in the ERα-S167A mutant. In addition, E2 stimulated ERαS118 phosphorylation in ERα-WT and ERα-S167A mutants but not in the ERα-S118A mutant. The low levels of ERα phosphorylation detected under basal conditions (IGF and E2 free) likely result from high amounts of transient ERα overexpression. Interestingly, IGF induced ERαS118 phosphorylation in the ERα-S167A mutant and E2 induced ERαS167 phosphorylation in the ERα-S118A mutant. Although this rapid cross-phosphorylation suggests a potential compensatory mechanism by IGF and E2, it was not observed in any of the additional cell lines tested (data not shown) and is in all probability a unique artifact of the C412 transfectant model. Nonetheless, further exploration is currently underway. In addition, detection of ERα phosphorylation in the vector transfectants was negligible under basal or any treatment conditions.
Fig. 4.
ERαS167 mediates IGF-induced phosphorylation, transcription, and growth. A, ERα-negative C4-12 cells were transiently transfected (48 h) with empty vector (Vector), ERα-WT, point-mutated ERαS118A (S118A), or ERαS167A (S167A) and serum starved overnight. Cells were then exposed to IGF or E2 (1 h) and immunoblotted against the proteins of interest. ERα (anti-ERα) was used to ensure equal transfection rates, and MAPK (anti-MAPK) served as a loading control. B, C4-12 cells were transfected with empty vector (Vector), ERα-WT or a mutant form of ERα (ERα-S118A or ERα-S167A), and anchorage-independent growth in response to IGF or E2 was assessed by soft agar assay. Colony-forming ability was expressed as fold change over vector-transfected cells. C, IGF1R, IRS1, and TFF1 gene expression of C4-12 cells transfected with empty vector, ERα-WT, or ERα-S167A was measured by RT-qPCR. Target gene expression was normalized to RPLP0 gene expression and presented as fold change over vector-transfected control. All error bars depict sd and all results are representative of at least three independent replicates.
To determine the biological consequences of impaired signaling, anchorage-independent growth in response to IGF or E2 was assessed in the transiently transfected C4–12 cells described above (Fig. 4B). Initial observation confirmed that expression of WT-ERα or either of the mutants improved basal growth levels as compared with vector alone, albeit less strikingly in the ERα-S167A mutant. This decrease in basal growth paralleled ERα target gene transcription because expression of TFF1, IGF1R, and IRS1 was markedly reduced in the ERα-S167A mutant as compared with WT-ERα (Fig. 4C). In conjunction with prior reports, response to E2 was significantly decreased in both of the mutants (46). Moreover, IGF responsiveness was markedly reduced in the ERα-S167A mutant, whereas the ERα-S118A mutant was unaffected. These data confirm that ERαS167 phosphorylation plays an integral role during hormone- and growth factor-induced cell growth.
IGF regulates ERα through S6K1
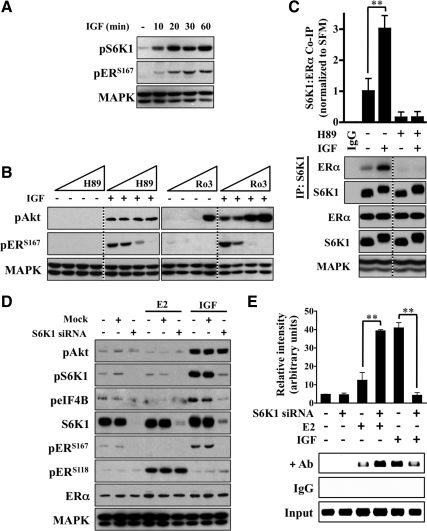
A number of growth factors [IGF, epidermal growth factor (EGF), fibroblast growth factor, platelet-derived growth factor] induce both gene transcription and translation (47). IGF has been shown to regulate cap-dependent translation via S6K1 and 4E-binding protein 1 (4EBP1) as a result of signaling through the IRS/PI3K/Akt/mTOR pathway (48). The emergence of mTOR as a regulator of IGF-induced ERα phosphorylation provided rationale to investigate additional factors that are both downstream of mTOR and classically involved in translation. In MCF-7 cells, S6K1 phosphorylation preceded that of maximal ERαS167 phosphorylation (Fig. 5A). Furthermore, inhibition of S6K1 kinase activity via small molecule inhibition (H-89 and Ro 31-8220) reduced IGF-induced ERαS167 phosphorylation in a dose-dependent manner (Fig. 5B) (49). Although both compounds effectively blocked ERαS167 phosphorylation, higher concentrations of Ro 31-8220 triggered IGF-induced Akt phosphorylation, suggesting nonspecific effects and prompted the use of H-89 for the remainder of the study.
Fig. 5.
IGF regulates ERα through S6K1. A, MCF-7 cells were exposed to IGF for the indicated time points and Western blot analysis performed. B, Beginning at 1 nm, cells were exposed to logarithmic dose increases of H89 or Ro31-8220 for 30 min before IGF exposure (1 h) and immunoblot analysis performed. C, Cells were pretreated with H89 for 30 min before IGF (4 h), and the association between S6K1 and ERα was measured by coimmunoprecipitation analysis. The graph represents the amount of immunoprecipitated ERα in IGF treated relative to nontreated. A nonspecific IgG control and total S6K1 and ERα levels are presented as loading controls. D, Cells were transiently transfected with either mock control or S6K1-targeting siRNA, treated with IGF or E2(1 h), and assessed for knockdown efficiency by immunoblot analysis. E, ChIP of the pS2 ERE was performed after S6K1 knockdown and exposure to IGF and E2 (4 h). The graph is a measure of relative intensity, and error bars represent sd. The MAPK (anti-MAPK) served as a loading control for immunoblot experiments, and all results are representative of at least three independent replicates.
Next, a potential IGF-dependent physical interaction between S6K1 and ERα was investigated (Fig. 5C). Immunoprecipitation analysis confirmed an interaction and revealed that IGF stimulation increased an S6K1/ERα association. Furthermore, inhibition of S6K1 via H-89 ablated both basal and IGF-induced levels of S6K1/ERα association.
To further demonstrate that IGF-induced ERα regulation was specific to S6K1, small interfering RNA (siRNA) directed against S6K1 was used and the effect on ERα activity measured (Fig. 5D). After transient knockdown, MCF-7 cells were treated with IGF or E2 and immunoblot analysis performed. Mock-transfected cells remained indistinguishable from nontransfected cells. An efficient S6K1 knockdown did not alter levels of E2-induced ERαS118 phosphorylation or Akt phosphorylation after IGF stimulation. However, levels of both basal and IGF-induced ERαS167 phosphorylation were markedly reduced in the S6K1 knockdown cells. Total ERα levels remained consistent throughout the experiment to rule out changes in ER levels as a potential confounder and further support the hypothesis that S6K1 specifically mediates IGF-induced ERαS167 phosphorylation. In addition, these findings were confirmed at a later (4 h) time point (data not shown).
The effect of S6K1 knockdown on ligand and ligand-independent ERα promoter binding was further examined. ChIP assays confirmed that after IGF treatment, S6K1 knockdown reduced pS2 ERE promoter binding (Fig. 5E). Coupled to a loss of ERαS167 phosphorylation, this observation suggests that S6K1-mediated ERα phosphorylation regulates promoter binding in response to IGF. Interestingly, although E2-induced ERαS118 phosphorylation remained constant, levels of E2-induced ERα-chromatin binding actually increased after S6K1 knockdown. Regardless, these results support that S6K1 associates with ERα to facilitate IGF-regulated ERα phosphorylation and promoter binding.
S6K1 axis facilitates IGF/ERα cross talk through target gene transcription
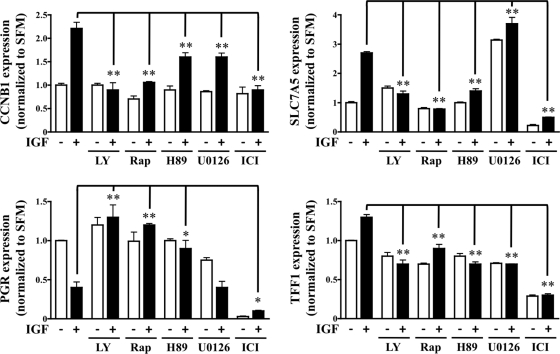
Binding of steroid receptors to gene promoter elements serves as a precursor to induction or repression of gene transcription (50). Therefore, gene expression changes of the following four ERα-regulated genes were measured: CCNB1, PGR, SLC7A5, and TFF1 (Fig. 6). As previously described, MCF-7 cells were pretreated with inhibitor for 30 min before IGF stimulation and mRNA species assessed by RT-qPCR analysis. The IGF-induced up-regulation of CCNB1, SLC7A5, and TFF1 and down-regulation of PGR was markedly reduced or completely blocked as a result of PI3K/mTOR/S6K1 pathway inhibition. Inhibition of PI3K via LY294002 and mTOR via rapamycin blocked IGF-induced up-regulation (CCNB1, SLC7A5, TFF1) and down-regulation (PGR) consistently across all samples. Albeit less strikingly in the case of CCNB1, S6K1 inhibition via H89 significantly repressed IGF-regulated gene expression of all four genes. Although no consistent pattern emerged, inhibition of MAPK affected both basal and IGF-induced gene expression. This was not unexpected because MAPK and ERα are known to cooperate in a number of ligand-independent respects (51). The pure antiestrogen fulvestrant (ICI 182780), which targets ERα for degradation and subsequent transcriptional inhibition, served as a negative control and resulted in reduced basal and IGF-regulated gene expression (52). In addition, IGF-minus and IGF-plus samples were compared directly as an additional measure of confirmation that drug alone did not significantly alter basal gene expression (data not shown).
Fig. 6.
S6K1 axis facilitates IGF/ERα cross talk through target gene transcription. MCF-7 cells were pretreated with LY294002 (LY; 10 μm), rapamycin (Rap; 10 nm), H89 (10 μm), U0126 (10 μm), or ICI 182780 (ICI; 1 μm) for 30 min before IGF (black bars) exposure. After 24 h of IGF exposure, mRNA was isolated and RT-qPCR performed. Target gene expression was normalized to RPLP0 gene expression and is presented as fold change of treatment vs. serum-free conditions. Error bars represent sd, and all results are representative of at least three independent replicates.
IGF1R phosphorylates ERα via the mTOR/S6K1 axis
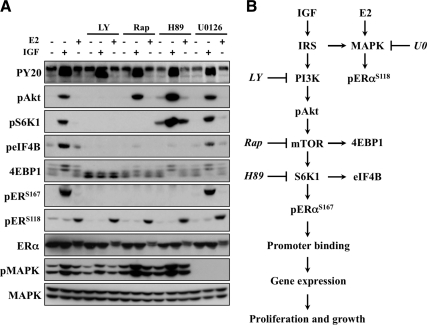
Finally, efforts were aimed at determining whether the signaling machinery responsible for IGF-mediated ERα phosphorylation occurred independently of ligand-dependent ERαS118 phosphorylation. MCF-7 cells were stimulated with IGF or E2 and cell lysates isolated for immunoblot analysis (Fig. 7A). IGF treatment resulted in the rapid phosphorylation of a number of PI3K signaling molecules [PY20, phosphorylated Akt, phosphorylated S6K1, phosphorylated eukaryotic initiation factor 4B (eIF4B), 4EBP1]. Conversely, ERαS118 phosphorylation occurred independently of the PI3K/mTOR pathway. MAPK kinase inhibition (U0126) did not affect IGF-induced ERα phosphorylation. Interestingly, E2-induced ERαS118 phosphorylation persisted in response to all of the inhibitors, including U0126. This was unexpected as phosphorylated MAPK levels were abolished in response to U0126. An alternative MAPK inhibitor (PD-98059) and cell line (T47D) yielded similar results (data not shown). Although these data demonstrate that rapid E2-induced ERαS118 phosphorylation can occur via a MAPK-independent mechanism, it is important to note that alternate kinases have been shown to regulate ERαS118 phosphorylation and may play a role (53–55).
Fig. 7.
IGF phosphorylates ERα via the mTOR/S6K1 axis. A, MCF-7 cells were cultured under hormone-free conditions for 3 d before overnight serum starvation. Cells were pretreated with LY294002 (LY; 10 μm), rapamycin (Rap; 10 nm), H89 (10 μm), or U0126 (10 μm) for 30 min before IGF or E2 (1 h) exposure and subjected to immunoblot analysis. MAPK (anti-MAPK) served as a loading control for all experiments, and results are representative of at least three independent replicates. B, Model of proposed pathway.
More importantly, PI3K inhibition (LY294002) blocked IGF signaling downstream of IRS (PY20) independently of ligand-induced ERαS118 phosphorylation. It is known that mTOR-mediated 4EBP1 phosphorylation results in an accumulation of the lower-molecular-weight α- and β-species with a concordant decrease in the higher-molecular-weight γ-isoform (56). As a result, 4EBP1 phosphorylation was appropriately blocked by LY294002 and rapamycin but remained IGF sensitive in the presence of H89 to suggest that in the context of IGF to PI3K signaling, H89 is specific. It also reveals that functional mTOR was insufficient to mediate IGF-induced ERαS167 phosphorylation. Lastly, IGF-induced ERαS167 and eIF4B phosphorylation, a direct S6K1 substrate, was blocked by LY294002, rapamycin, and H89, therein further confirming that S6K1 is a necessary signaling component of IGF-induced ERα phosphorylation (Fig. 7B). The constitutively active myr-Akt3 line provided additional evidence of S6K1/ERα cross talk because PI3K/mTOR/S6K1 inhibitors selectively blocked constitutive ERαS167 phosphorylation in a ligand-independent and MAPK-independent manner (S1).
Discussion
The use of mTOR inhibitors in clinical trials, coupled to the observation that Akt hyperphosphorylation results from negative feedback inhibition of IRS degradation, prompted us to investigate if the IGF pathway serves as a potential mechanism of resistance/susceptibility in breast cancer cells (57–59). Because blockade of IRS degradation occurs in an isoform-independent manner, we postulated that mTOR inhibition via rapamycin would similarly affect both IRS-1-mediated proliferation and IRS-2-driven motility. However, we were surprised to discover that rapamycin differentially altered IGF-stimulated behavior independent of IRS isoform expression in ERα-positive breast cancer cells. Although the observation that rapamycin potentiates IGF-induced motility in ERα-negative breast cancer cells warrants further investigation, it is beyond the scope of this study. Herein we sought to define the governing mechanism responsible for rapamycin-mediated blockade of IGF-induced proliferation in ERα-positive breast cancer cells.
IRS-1 and ERα cross talk is a well-established attribute of aberrant proliferation and growth (60, 61). We hypothesized that mTOR may function in both transcriptional and translational capacities to biochemically link IGF signaling to ERα function. To better understand the impact of mTOR inhibition, we sought first to identify how ERα activity is altered by IGF in our model. Nuclear ERα function is largely predicated on its ability to freely bind and dissociate chromatin at the promoter level (62). As the gold standard of antiestrogen therapy and endocrine treatment of choice for hormone positive breast cancer, tamoxifen functions as a selective ER modulator by repressing ERα dissociation from DNA (63, 64). To our knowledge, this is the first study to demonstrate that IGF stimulates cyclical ERα promoter binding in an E2-like fashion. Furthermore, because transcriptional regulation of IGF mirrored E2, it suggests that IGF-mediated Akt activation does not negatively repress ERα function.
A number of growth factors have been shown to regulate ERα function via posttranslational modification of different serine residues (65, 66). Although individual kinases have been linked to multiple and overlapping sites of regulation on ERα, the converse is also true because pp90RSK and CK2 have both been shown to directly stimulate ERαS167 phosphorylation (67, 68). Our data reveal that IGF regulates ERα phosphorylation in a site-specific and ligand-independent fashion. The observed lack of E2-induced ERαS167 phosphorylation seemingly contradicts earlier findings that identified S167 as the major E2-regulated site of ERα phosphorylation (69). This difference is likely due to primary vs. secondary ligand effects of E2 stimulation because we observed a nearly 7-fold increase in IGF ligand expression after E2 stimulation (data not shown). Moreover, our data suggest Akt, although functioning as an intermediate signaling component, is not directly required for ERα phosphorylation because inhibition of PI3K signaling downstream of Akt abolished IGF-induced ERαS167 phosphorylation. It is plausible that differences in Akt signaling to ERα likely stem from the use of transient transfection model systems as a method to link ERα to Akt as a direct substrate. Accordingly, we conducted our experiments using cells in which ERα and IGF1R function was well characterized.
Our study demonstrates that S6K1 facilitates cross talk between the IGF pathway and ERα to promote posttranslational modification, chromatin binding, target gene transcription, and breast cancer cell growth. Although a number of reports demonstrate that growth factor pathways regulate ERα expression and posttranslational modification, our study focused on linking IGF-induced ERα phosphorylation to ERα-dependent behavior and growth. It has been previously reported that insulin stimulates ERαS167 phosphorylation in an S6K1-dependent manner (70, 71). However, these experiments were conducted in the presence of estrogen-containing serum. In our experimental system, we examined the effects of IGF signaling in the absence of E2 to more clearly define the effect of growth factor signaling on ERα regulation via S6K1 (e.g. substrate binding, phosphorylation, plasmid transcription, and growth). For example, IGF ligand expression is markedly up-regulated in response to chronic estrogen exposure (72, 73). In an effort to circumvent the influence of estrogen on growth factor-specific ERα regulation in our experiments, all cells were subjected to appropriate periods of hormone deprivation before IGF exposure. We further confirmed that basal levels of ERα phosphorylation and promoter binding were accordingly abrogated as a direct result of prolonged culture in the absence of hormone (data not shown). Both insulin and IGF effects on breast cancer cells are likely important, but there are differences between these pathways in some cancer cell lines (74, 75). To assume that IGF and insulin regulate S6K1 in a synonymous and overlapping fashion is not appropriate because numerous examples of differential IGF and insulin regulation have been shown (76). However, IGF and insulin signal through numerous overlapping mechanisms. Therefore, it is highly likely that IGF and insulin regulate ERα in a similar fashion. Given the clinical role for estrogen deprivation of ERα-expressing breast cancers, defining the role for other pathways that influence ERα function in the absence of E2 might have clinical relevance.
The finding that S6K1 knockdown increased E2-induced chromatin binding and that this effect was closely mirrored by rapamycin in the presence of endogenous S6K1 expression was largely unexpected. This enhancement of E2-induced binding may alter one or more of the following ERα-related functions: target gene transcription, cofactor dynamics, promoter selectivity, protein-protein interaction, and degradation. Alternatively, it is possible that inhibition of IGF-induced ERα phosphorylation triggers compensation by E2. Although not reflected in growth patterns, loss of the ERαS167 phosphorylation site by S167A transfected C4-12 cells resulted in markedly enhanced E2-induced ERαS118 phosphorylation in comparison with ERα-WT. These data support the mounting evidence behind IGF/ERα interdependent cross talk (77).
Taken together, data presented here demonstrate that the IGF pathway alters ERα function through an mTOR/S6K1-dependent mechanism and that phosphorylation is an integral step to facilitate chromatin binding, transcription, growth, and proliferation (Fig. 7B). Results from a recent clinical study found that everolimus significantly improved the efficacy of letrozole in neoadjuvant therapy for patients with ERα-positive breast cancer (30). Coupled to our findings, these data suggest that targeting the IGF pathway in combination with ERα and/or mTOR presents as a promising therapeutic strategy in the treatment of breast cancer.
Materials and Methods
Reagents
Culture media, supplements, and 17β-E2 were purchased from Invitrogen (Carlsbad, CA). IGF-I was purchased from GroPep (Adelaide, Australia). LY294002 and U0126 were purchased from Calbiochem (Luzern, Switzerland). LY294002, rapamycin, and UO126 were purchased from Sigma (St. Louis, MO). H89 and Ro 31-8220 were purchased from Upstate (Lake Placid, NY).
Antibodies
Horseradish peroxidase-conjugated antiphosphotyrosine (PY-20) was purchased from BD Biosciences (Sparks, MD). The ERα antibody used for Western blot analysis was purchased from Neomarkers Lab Vision (Fremont, CA) and the ERα antibody used for immunoprecipitation from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies for phosphorylated and total Akt, p70S6K, eIF4B, 4EBP, ERα, and p44/42 (MAPK/ERK) were purchased from Cell Signaling Technology (Beverly, MA). Antirabbit and antimouse horseradish peroxidase-conjugated secondary antibodies were purchased from Pierce (Rockford, IL). Effectene transfection reagent was purchased from QIAGEN (Valencia, CA). ON-TARGETplus SMARTpool siRNA targeting p70S6K1 and protein G agarose/salmon sperm DNA was purchased from Millipore (Watford, UK).
Cell culture
All cells were grown at 37 C in a humidified atmosphere containing 5% CO2 and supplemented with fetal bovine serum (FBS), 100 U/ml penicillin, 100 μg/ml streptomycin, and 11.25 nm insulin. MCF-7 cells were provided by C. Kent Osborne (Baylor College of Medicine, Houston, TX) and maintained in improved MEM Richter's modification medium (zinc option) supplemented with 5% FBS. MDA-231BO (F11) cells were provided by Toshiyuki Yoneda (University of Texas Health Science Center, San Antonio, TX) and maintained in DMEM supplemented with 10% FBS (78). MVLN cells were maintained as previously described (79). ZR-75–1 cells were maintained in RPMI 1640 medium containing phenol red and supplemented with 5% FBS. T47D-WT cells were provided by Kathryn Horwitz (University of Colorado Health Science Center, Denver, CO) and maintained in Eagle's MEM supplemented with 5% FBS, 6 ng/liter insulin and 20 μl/ml nonessential amino acids (100 times). C4-12 cells were maintained in phenol red-free αMEM and supplemented with 5% charcoal/dextran-treated FBS. MCF-7 myr-Akt3 cells were described previously (35). Briefly, MCF-7 cells were stably transfected with a constitutively active Akt3 (myr-Akt3) containing a myristoylation membrane-targeting sequence and HA tag and were selected by 600 μg/ml G418. Stable clones with hyperactive Akt were later verified by Western blot analysis with antibodies against Akt phosphorylation and HA tag. Before IGF and E2 stimulation, all cells were serum starved in phenol red-free IMEM supplemented with 20 mmol/liter HEPES, 1× trace elements, 2 μg/ml transferring, and 2 μg/ml fibronectin.
Immunoblot
Cells were plated at a density of 3 × 105 in 60-mm-diameter dishes and allowed to equilibrate overnight. Full medium was replaced with dextran-coated-charcoal (DCC)-treated fetal calf serum for the next 3–5 d, after which cells were switched to serum-free medium (SFM) for 24 h. Upon reaching 70% confluency, cells were treated, placed on ice, washed twice with ice-cold PBS, and lysed with lysis buffer of 50 mm Tris-Cl (pH 7.4), 1% Nonidet P-40, 2 mm EDTA (pH 8.0), 100 mm NaCl, 10 mm sodium orthovanadate, 1 mm phenylmethylsulfonyl fluoride, 20 μg/ml leupeptin, and 20 μg/ml aprotinin). Lysates were clarified by centrifugation at 12,00 × g for 15 min at 4 C. Protein concentrations were determined using the bicinchoninic acid protein assay reagent kit (Pierce). Cellular protein (40 μg) was separated by SDS-PAGE, transferred to a nitrocellulose membrane, and immunoblotted according to manufacturer guidelines. Fractionation experiments were performed as previously described (80).
Immunoprecipitation
Cell lysates (1000 μg) were isolated, precleared with 60 μl of protein A agarose for 60 min, incubated with S6K1 antibody overnight and 60 μl of protein A agarose added for 4 h at 4 C. Immunoprotein complexes were washed three times with buffer of 50 mm Tris-Cl (pH 7.4), 1% Nonidet P-40, 2 mm EDTA (pH 8.0), 100 mm NaCl, 10 mm sodium orthovanadate, 1 mm phenylmethylsulfonyl fluoride, 20 μg/ml leupeptin, and 20 μg/ml aprotinin; Laemmli sample buffer (1 time) added; samples ran on an SDS-PAGE gel; and transferred to a nitrocellulose membrane for immunoblot analysis.
siRNA and transfections
ON-TARGETplus SMARTpool siRNA targeting RPS6KB1 (S6K1; Millipore) was transfected into MCF-7 cells using the Effectene transfection kit. Cells were exposed to siRNA (100 nm) for the 3-d duration of DCC medium starvation. Mock transfections were performed with transfection reagent alone. Alternatively, C4-12 cells were transfected with plasmids encoding for ERα (vector, WT, S118A, S167A) for 2 d in DCC growth medium before SFM starvation. The ERα plasmids were a generous gift from Benita Katzenellenbogen (University of Illinois at Urbana-Champaign, Urbana-Champaign, IL). All transfections were performed with Effectene transfection reagent (QIAGEN). After treatment, cells were lysed and Western blotting performed.
Chromatin immunoprecipitation
Cells were plated at a density of 4 × 106 in 150-mm-diameter dishes, allowed to equilibrate overnight, hormone starved for 3 d, and incubated overnight in SFM. ChIP assays were performed as previously described (40). After treatment, cells were cross-linked with an 8-min incubation in 1% formaldehyde and the reaction quenched by glycine treatment for 5 min at room temperature. Cells were washed three times and sonicated using the Bioruptor from Diagenode (Liege, Belgium) for 15 min at 4 C (30 sec burst cycles every minute). Input controls were collected and lysate protein concentration was determined by bicinchoninic acid assay. Five hundred micrograms of protein were incubated with 40 μg of protein A agarose/salmon sperm DNA before overnight incubation at 4 C with an ERα antibody, a nonspecific mouse IgG, or no antibody control. Protein/chromatin complexes were immunoprecipitated with 40 μg of protein A agarose/salmon sperm DNA for 2 h, washed, eluted from beads, and reverse cross-linked in a 0.3 m NaCl solution at 65 C for 6 h (input controls added). After proteinase K treatment for 1 h at 45 C, DNA was ethanol precipitated at −20 C overnight. DNA was purified (QIAGEN DNA cleanup kit), PCR amplified with primers specific to the pS2 ERE promoter site, and amplified products ran on a 1% agarose gel.
Reverse transcription-quantitative real-time polymerase chain reaction
Cells were plated at a density of 1 × 106 in 100-mm-diameter dishes, allowed to equilibrate overnight, DCC starved for 3 d, and incubated overnight in SFM. Cellular RNA was isolated using the 5 Prime PerfectPure RNA tissue kit according to the manufacturer (Fisher Scientific, Fair Lawn, NJ). For quality control and to determine concentration, a 260:280 assay was performed on a spectrophotometer. Forward and reverse primers were designed to target the following transcripts: CCNB1, CCND1, ESR1, IRS1, IGF1R, KIAA0575, PGR, MK167, SLC7A5, TFF1, and VEGFA. A total of 2 μg of RNA was reverse transcribed using the Quantitect reverse transcriptase kit, and quantitative PCR was performed using the Quantifast SYBR Green kit according to the manufacturer's recommended protocol (QIAGEN) on an Eppendorf Mastercycler Realplex4 machine (Hamburg, Germany). The relative concentration of mRNA was calculated using cycle threshold values that were derived from a standard curve and normalized to ribosomal protein, large, PO as an internal control.
Monolayer proliferation
Cells were plated in 24-well plates at a density of 10,000 cells/well, allowed to equilibrate overnight, and starved in SFM for 24 h. After 3 d of treatment, growth was assessed via the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay as described previously (81). Sixty microliters of 5 mg/ml MTT solution in SFM were added to each well. After incubation for 4 h at 37 C, wells were aspirated and formazan crystals were lysed with 500 μl of solubilization solution (95% dimethylsulfoxide + 5% improved minimal essential media). Absorbance was measured with a plate reader at 570 nm using a 650-nm differential filter to assess growth.
Anchorage-independent growth
Anchorage-independent growth assays were performed as previously described (8). A 1-ml layer of 0.8% SeaPlaque-agarose (BioWhittaker, Rockland, ME) in 1% FBS-containing growth media was solidified into each well of a six-well plate. The bottom layer was overlaid with 0.8 ml of a 0.45% top agar mixture for 10,000 cells per well with appropriate treatment. All plates were incubated at 37 C. After 12 d colony number was assessed on a light microscope with an ocular grid. Five random fields were counted per well and only colonies exceeding two thirds of a grid square were scored.
Statistical analysis
Statistical significance between two groups was tested using Student's t test, and ANOVA with Bonferroni's post hoc test was used for multiple-comparison analysis. Error bars represent sd and asterisks denote statistical significance: *, P < 0.05; **, P < 0.01.
Acknowledgments
This work was supported by the Department of Defense Predoctoral Traineeship Award BC073039 (to M.A.B.), National Institutes of Health Grant R01CA74285 (to D.Y.), and National Cancer Institute Cancer Center Support Grant P30077598.
Disclosure Summary: The authors have nothing to disclose.
NURSA Molecule Pages:
Nuclear Receptors: ER-α;
Ligands: 17β-estradiol | Fulvestrant.
Footnotes
- ChIP
- Chromatin immunoprecipitation
- DCC
- dextran-coated charcoal
- SFM
- serum-free medium
- E2
- estradiol
- EGF
- epidermal growth factor
- 4EBP1
- 4E-binding protein 1
- eIF4B
- eukaryotic initiation factor 4B
- ER
- estrogen receptor
- ERE
- estrogen response element
- FBS
- fetal bovine serum
- IGF1R
- IGF-I receptor
- IRS
- insulin receptor substrate
- mTOR
- mammalian target of rapamycin
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PGR
- progesterone receptor gene
- PI3K
- phosphatidylinositol 3-kinase
- RT-qPCR
- reverse transcription quantitative-PCR
- siRNA
- small interfering RNA
- S6K1
- S6 kinase 1
- WT
- wild type.
References
- 1. Kiang DT, Frenning DH, Goldman AI, Ascensao VF, Kennedy BJ. 1978. Estrogen receptors and responses to chemotherapy and hormonal therapy in advanced breast cancer. N Engl J Med 299:1330–1334 [DOI] [PubMed] [Google Scholar]
- 2. Ward HW. 1973. Anti-oestrogen therapy for breast cancer: a trial of tamoxifen at two dose levels. Br Med J 1:13–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Knight WA, Livingston RB, Gregory EJ, McGuire WL. 1977. Estrogen receptor as an independent prognostic factor for early recurrence in breast cancer. Cancer Res 37:4669–4671 [PubMed] [Google Scholar]
- 4. Mohamood AS, Gyles P, Balan KV, Hollis VW, Eckberg WR, Asseffa A, Han Z, Wyche JH, Anderson WA. 1997. Estrogen receptor, growth factor receptor and protooncogene protein activities and possible signal transduction cross talk in estrogen dependent and independent breast cancer cell lines. J Submicrosc Cytol Pathol 29:1–17 [PubMed] [Google Scholar]
- 5. Arpino G, Wiechmann L, Osborne CK, Schiff R. 2008. Cross talk between the estrogen receptor and the HER tyrosine kinase receptor family: molecular mechanism and clinical implications for endocrine therapy resistance. Endocr Rev 29:217–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pratt SE, Pollak MN. 1994. Insulin-like growth factor binding protein 3 (IGF-BP3) inhibits estrogen-stimulated breast cancer cell proliferation. Biochem Biophys Res Commun 198:292–297 [DOI] [PubMed] [Google Scholar]
- 7. Zugmaier G, Knabbe C, Fritsch C, Simpson S, Ennis B, Lippman M, Dickson RB. 1991. Tissue culture conditions determine the effects of estrogen and growth factors on the anchorage independent growth of human breast cancer cell lines. J Steroid Biochem Mol Biol 39:681–685 [DOI] [PubMed] [Google Scholar]
- 8. Figueroa JA, Sharma J, Jackson JG, McDermott MJ, Hilsenbeck SG, Yee D. 1993. Recombinant insulin-like growth factor binding protein-1 inhibits IGF-I, serum, and estrogen-dependent growth of MCF-7 human breast cancer cells. J Cell Physiol 157:229–236 [DOI] [PubMed] [Google Scholar]
- 9. Lee AV, Jackson JG, Gooch JL, Hilsenbeck SG, Coronado-Heinsohn E, Osborne CK, Yee D. 1999. Enhancement of the insulin-like growth factor pathway by estrogen in human breast cancer cells. Mol Endocrinol 13:787–796 [DOI] [PubMed] [Google Scholar]
- 10. Umayahara Y, Kawamori R, Watada H, Imano E, Iwama N, Morishima T, Yamasaki Y, Kajimoto Y, Kamada T. 1994. Estrogen regulation of the insulin-like growth factor i gene transcription involves an AP-1 enhancer. J Biol Chem 269:16433–16442 [PubMed] [Google Scholar]
- 11. Stewart AJ, Johnson MD, May FE, Westley BR. 1990. Role of insulin-like growth factors and the type I insulin-like growth factor receptor in the estrogen-stimulated proliferation of human breast cancer cells. J Biol Chem 265:21172–21178 [PubMed] [Google Scholar]
- 12. Salerno M, Sisci D, Mauro L, Guvakova MA, Ando S, Surmacz E. 1999. Insulin receptor substrate 1 is a target for the pure antiestrogen ICI 182,780 in breast cancer cells. Int J Cancer 81:299–304 [DOI] [PubMed] [Google Scholar]
- 13. Tong GM, Pento JT, Rajah TT. 1999. Influence of antiestrogens on EGF- and IGF-I-mediated proliferation of human breast cancer cells (Letter). In Vitro Cell Dev Biol Animal 35:19–21 [DOI] [PubMed] [Google Scholar]
- 14. Varma H, Conrad SE. 2002. Antiestrogen ICI 182,780 decreases proliferation of insulin-like growth factor I (IGF-I)-treated MCF-7 cells without inhibiting IGF-I signaling. Cancer Res 62:3985–3991 [PubMed] [Google Scholar]
- 15. Zhang S, Li X, Burghardt R, Smith R, 3rd, Safe SH. 2005. Role of estrogen receptor (ER) α in insulin-like growth factor (IGF)-I-induced responses in MCF-7 breast cancer cells. J Mol Endocrinol 35:433–447 [DOI] [PubMed] [Google Scholar]
- 16. Lee AV, Weng CN, Jackson JG, Yee D. 1997. Activation of estrogen receptor-mediated gene transcription by IGF-I in human breast cancer cells. J Endocrinol 152:39–47 [DOI] [PubMed] [Google Scholar]
- 17. Cascio S, Bartella V, Garofalo C, Russo A, Giordano A, Surmacz E. 2007. Insulin-like growth factor 1 differentially regulates estrogen receptor-dependent transcription at estrogen response element and AP-1 sites in breast cancer cells. J Biol Chem 282:3498–3506 [DOI] [PubMed] [Google Scholar]
- 18. Kato S, Endoh H, Masuhiro Y, Kitamoto T, Uchiyama S, Sasaki H, Masushige S, Gotoh Y, Nishida E, Kawashima H, Metzger D, Chambon P. 1995. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science 270:1491–1494 [DOI] [PubMed] [Google Scholar]
- 19. Le Goff P, Montano MM, Schodin DJ, Katzenellenbogen BS. 1994. Phosphorylation of the human estrogen receptor: identification of hormone regulated sites and examination of their influence on transcriptional activity. J Biol Chem 269:4458–4466 [PubMed] [Google Scholar]
- 20. Burgaud JL, Resnicoff M, Baserga R. 1995. Mutant IGF-I receptors as dominant negatives for growth and transformation. Biochem Biophys Res Commun 214:475–481 [DOI] [PubMed] [Google Scholar]
- 21. Arteaga CL, Kitten LJ, Coronado EB, Jacobs S, Kull FC, Jr., Allred DC, Osborne CK. 1989. Blockade of the type I somatomedin receptor inhibits growth of human breast cancer cells in athymic mice. J Clin Invest 84:1418–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dickson RB, McManaway ME, Lippman ME. 1986. Estrogen-induced factors of breast cancer cells partially replace estrogen to promote tumor growth. Science 232:1540–1543 [DOI] [PubMed] [Google Scholar]
- 23. Yee D, Paik S, Lebovic GS, Marcus RR, Favoni RE, Cullen KJ, Lippman ME, Rosen N. 1989. Analysis of IGF-I gene expression in malignancy, evidence for a paracrine role in human breast cancer. Mol Endocrinol 3:509–517 [DOI] [PubMed] [Google Scholar]
- 24. Harrington LS, Findlay GM, Gray A, Tolkacheva T, Wigfield S, Rebholz H, Barnett J, Leslie NR, Cheng S, Shepherd PR, Gout I, Downes CP, Lamb RF. 2004. The TSC1–2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J Cell Biol 166:213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shi Y, Yan H, Frost P, Gera J, Lichtenstein A. 2005. Mammalian target of rapamycin inhibitors activate the AKT kinase in multiple myeloma cells by up-regulating the insulin-like growth factor receptor/insulin receptor substrate-1/phosphatidylinositol 3-kinase cascade. Mol Cancer Ther 4:1533–1540 [DOI] [PubMed] [Google Scholar]
- 26. Campbell RA, Bhat-Nakshatri P, Patel NM, Constantinidou D, Ali S, Nakshatri H. 2001. Phosphatidylinositol 3-kinase/AKT-mediated activation of estrogen receptor α: a new model for anti-estrogen resistance. J Biol Chem 276:9817–9824 [DOI] [PubMed] [Google Scholar]
- 27. Martin MB, Franke TF, Stoica GE, Chambon P, Katzenellenbogen BS, Stoica BA, McLemore MS, Olivo SE, Stoica A. 2000. A role for Akt in mediating the estrogenic functions of epidermal growth factor and insulin-like growth factor I. Endocrinology 141:4503–4511 [DOI] [PubMed] [Google Scholar]
- 28. Bhat-Nakshatri P, Wang G, Appaiah H, Luktuke N, Carroll JS, Geistlinger TR, Brown M, Badve S, Liu Y, Nakshatri H. 2008. AKT alters genome-wide estrogen receptor α binding and impacts estrogen signaling in breast cancer. Mol Cell Biol 28:7487–7503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ma Y, Hu C, Riegel AT, Fan S, Rosen EM. 2007. Growth factor signaling pathways modulate BRCA1 repression of estrogen receptor-α activity. Mol Endocrinol 21:1905–1923 [DOI] [PubMed] [Google Scholar]
- 30. Baselga J, Semiglazov V, van Dam P, Manikhas A, Bellet M, Mayordomo J, Campone M, Kubista E, Greil R, Bianchi G, Steinseifer J, Molloy B, Tokaji E, Gardner H, Phillips P, Stumm M, Lane HA, Dixon JM, Jonat W, Rugo HS. 2009. Phase II randomized study of neoadjuvant everolimus plus letrozole compared with placebo plus letrozole in patients with estrogen receptor-positive breast cancer. J Clin Oncol 27:2630–2637 [DOI] [PubMed] [Google Scholar]
- 31. Byron SA, Horwitz KB, Richer JK, Lange CA, Zhang X, Yee D. 2006. Insulin receptor substrates mediate distinct biological responses to insulin-like growth factor receptor activation in breast cancer cells. Br J Cancer 95:1220–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jackson JG, Zhang X, Yoneda T, Yee D. 2001. Regulation of breast cancer cell motility by insulin receptor substrate-2 (IRS-2) in metastatic variants of human breast cancer cell lines. Oncogene 20:7318–7325 [DOI] [PubMed] [Google Scholar]
- 33. Nolan MK, Jankowska L, Prisco M, Xu S, Guvakova MA, Surmacz E. 1997. Differential roles of IRS-1 and SHC signaling pathways in breast cancer cells. Int J Cancer 72:828–834 [DOI] [PubMed] [Google Scholar]
- 34. Zhang X, Kamaraju S, Hakuno F, Kabuta T, Takahashi S, Sachdev D, Yee D. 2004. Motility response to insulin-like growth factor-I (IGF-I) in MCF-7 cells is associated with IRS-2 activation and integrin expression. Breast Cancer Res Treat 83:161–170 [DOI] [PubMed] [Google Scholar]
- 35. Kim HJ, Cui X, Hilsenbeck SG, Lee AV. 2006. Progesterone receptor loss correlates with human epidermal growth factor receptor 2 overexpression in estrogen receptor-positive breast cancer. Clin Cancer Res 12:1013s–1018s [DOI] [PubMed] [Google Scholar]
- 36. Mawson A, Lai A, Carroll JS, Sergio CM, Mitchell CJ, Sarcevic B. 2005. Estrogen and insulin/IGF-1 cooperatively stimulate cell cycle progression in MCF-7 breast cancer cells through differential regulation of c-Myc and cyclin D1. Mol Cell Endocrinol 229:161–173 [DOI] [PubMed] [Google Scholar]
- 37. Gooch JL, Van Den Berg CL, Yee D. 1999. Insulin-like growth factor (IGF)-I rescues breast cancer cells from chemotherapy-induced cell death—proliferative and anti-apoptotic effects. Breast Cancer Res Treat 56:1–10 [DOI] [PubMed] [Google Scholar]
- 38. Stoica GE, Franke TF, Moroni M, Mueller S, Morgan E, Iann MC, Winder AD, Reiter R, Wellstein A, Martin MB, Stoica A. 2003. Effect of estradiol on estrogen receptor-α gene expression and activity can be modulated by the ErbB2/PI 3-K/Akt pathway. Oncogene 22:7998–8011 [DOI] [PubMed] [Google Scholar]
- 39. Santen RJ, Fan P, Zhang Z, Bao Y, Song RX, Yue W. 2009. Estrogen signals via an extra-nuclear pathway involving IGF-1R and EGFR in tamoxifen-sensitive and -resistant breast cancer cells. Steroids 74:586–594 [DOI] [PubMed] [Google Scholar]
- 40. Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843–852 [DOI] [PubMed] [Google Scholar]
- 41. Cui X, Zhang P, Deng W, Oesterreich S, Lu Y, Mills GB, Lee AV. 2003. Insulin-like growth factor-I inhibits progesterone receptor expression in breast cancer cells via the phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin pathway: progesterone receptor as a potential indicator of growth factor activity in breast cancer. Mol Endocrinol 17:575–588 [DOI] [PubMed] [Google Scholar]
- 42. Shen T, Horwitz KB, Lange CA. 2001. Transcriptional hyperactivity of human progesterone receptors is coupled to their ligand-dependent down-regulation by mitogen-activated protein kinase-dependent phosphorylation of serine 294. Mol Cell Biol 21:6122–6131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lonard DM, Nawaz Z, Smith CL, O'Malley BW. 2000. The 26S proteasome is required for estrogen receptor-α and coactivator turnover and for efficient estrogen receptor-α transactivation. Mol Cell 5:939–948 [DOI] [PubMed] [Google Scholar]
- 44. Garola RE, McGuire WL. 1977. An improved assay for nuclear estrogen receptor in experimental and human breast cancer. Cancer Res 37:3333–3337 [PubMed] [Google Scholar]
- 45. Oesterreich S, Zhang P, Guler RL, Sun X, Curran EM, Welshons WV, Osborne CK, Lee AV. 2001. Re-expression of estrogen receptor α in estrogen receptor α-negative MCF-7 cells restores both estrogen and insulin-like growth factor-mediated signaling and growth. Cancer Res 61:5771–5777 [PubMed] [Google Scholar]
- 46. Joel PB, Smith J, Sturgill TW, Fisher TL, Blenis J, Lannigan DA. 1998. pp90rsk1 regulates estrogen receptor-mediated transcription through phosphorylation of Ser-167. Mol Cell Biol 18:1978–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schiff R, Massarweh SA, Shou J, Bharwani L, Mohsin SK, Osborne CK. 2004. Cross-talk between estrogen receptor and growth factor pathways as a molecular target for overcoming endocrine resistance. Clin Cancer Res 10:331S–336S [DOI] [PubMed] [Google Scholar]
- 48. Gingras AC, Raught B, Sonenberg N. 2001. Regulation of translation initiation by FRAP/mTOR. Genes Dev 15:807–826 [DOI] [PubMed] [Google Scholar]
- 49. Davies SP, Reddy H, Caivano M, Cohen P. 2000. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem J 351:95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Glass CK, Franco R, Weinberger C, Albert VR, Evans RM, Rosenfeld MG. 1987. A c-erb-A binding site in rat growth hormone gene mediates trans-activation by thyroid hormone. Nature 329:738–741 [DOI] [PubMed] [Google Scholar]
- 51. Coutts AS, Murphy LC. 1998. Elevated mitogen-activated protein kinase activity in estrogen-nonresponsive human breast cancer cells. Cancer Res 58:4071–4074 [PubMed] [Google Scholar]
- 52. Wakeling AE, Dukes M, Bowler J. 1991. A potent specific pure antiestrogen with clinical potential. Cancer Res 51:3867–3873 [PubMed] [Google Scholar]
- 53. Chen D, Riedl T, Washbrook E, Pace PE, Coombes RC, Egly JM, Ali S. 2000. Activation of estrogen receptor α by S118 phosphorylation involves a ligand-dependent interaction with TFIIH and participation of CDK7. Mol Cell 6:127–137 [PubMed] [Google Scholar]
- 54. Joel PB, Traish AM, Lannigan DA. 1998. Estradiol-induced phosphorylation of serine 118 in the estrogen receptor is independent of p42/p44 mitogen-activated protein kinase. J Biol Chem 273:13317–13323 [DOI] [PubMed] [Google Scholar]
- 55. Medunjanin S, Hermani A, De Servi B, Grisouard J, Rincke G, Mayer D. 2005. Glycogen synthase kinase-3 interacts with and phosphorylates estrogen receptor alpha and is involved in the regulation of receptor activity. J Biol Chem 280:33006–33014 [DOI] [PubMed] [Google Scholar]
- 56. Beretta L, Gingras AC, Svitkin YV, Hall MN, Sonenberg N. 1996. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J 15:658–664 [PMC free article] [PubMed] [Google Scholar]
- 57. Noh WC, Kim YH, Kim MS, Koh JS, Kim HA, Moon NM, Paik NS. 2008. Activation of the mTOR signaling pathway in breast cancer and its correlation with the clinicopathologic variables. Breast Cancer Res Treat 110:477–483 [DOI] [PubMed] [Google Scholar]
- 58. Hynes NE, Boulay A. 2006. The mTOR pathway in breast cancer. J Mammary Gland Biol Neoplasia 11:53–61 [DOI] [PubMed] [Google Scholar]
- 59. Yu K, Toral-Barza L, Discafani C, Zhang WG, Skotnicki J, Frost P, Gibbons JJ. 2001. mTOR, a novel target in breast cancer: the effect of CCI-779, an mTOR inhibitor, in preclinical models of breast cancer. Endocr Relat Cancer 8:249–258 [DOI] [PubMed] [Google Scholar]
- 60. Molloy CA, May FE, Westley BR. 2000. Insulin receptor substrate-1 expression is regulated by estrogen in the MCF-7 human breast cancer cell line. J Biol Chem 275:12565–12571 [DOI] [PubMed] [Google Scholar]
- 61. Jackson JG, White MF, Yee D. 1998. Insulin receptor substrate-1 is the predominant signaling molecule activated by insulin-like growth factor-I, insulin, and interleukin-4 in estrogen receptor-positive human breast cancer cells. J Biol Chem 273:9994–10003 [DOI] [PubMed] [Google Scholar]
- 62. Metivier R, Penot G, Hübner MR, Reid G, Brand H, Kos M, Gannon F. 2003. Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115:751–763 [DOI] [PubMed] [Google Scholar]
- 63. Jordan VC, Dowse LJ. 1976. Tamoxifen as an anti-tumour agent: effect on oestrogen binding. J Endocrinol 68:297–303 [DOI] [PubMed] [Google Scholar]
- 64. Jordan VC. 1997. Tamoxifen treatment for breast cancer: concept to gold standard. Oncology (Williston Park) 11:7–13 [PubMed] [Google Scholar]
- 65. Bunone G, Briand PA, Miksicek RJ, Picard D. 1996. Activation of the unliganded estrogen receptor by EGF involves the MAP kinase pathway and direct phosphorylation. EMBO J 15:2174–2183 [PMC free article] [PubMed] [Google Scholar]
- 66. Aronica SM, Katzenellenbogen BS. 1993. Stimulation of estrogen receptor-mediated transcription and alteration in the phosphorylation state of the rat uterine estrogen receptor by estrogen, cyclic adenosine monophosphate, and insulin-like growth factor-I. Mol Endocrinol 7:743–752 [DOI] [PubMed] [Google Scholar]
- 67. Lannigan DA. 2003. Estrogen receptor phosphorylation. Steroids 68:1–9 [DOI] [PubMed] [Google Scholar]
- 68. Arnold SF, Obourn JD, Jaffe H, Notides AC. 1995. Phosphorylation of the human estrogen receptor by mitogen-activated protein kinase and casein kinase II: consequence on DNA binding. J Steroid Biochem Mol Biol 55:163–172 [DOI] [PubMed] [Google Scholar]
- 69. Arnold SF, Obourn JD, Jaffe H, Notides AC. 1994. Serine 167 is the major estradiol-induced phosphorylation site on the human estrogen receptor. Mol Endocrinol 8:1208–1214 [DOI] [PubMed] [Google Scholar]
- 70. Yamnik RL, Digilova A, Davis DC, Brodt ZN, Murphy CJ, Holz MK. 2009. S6 kinase 1 regulates estrogen receptor α in control of breast cancer cell proliferation. J Biol Chem 284:6361–6369 [DOI] [PubMed] [Google Scholar]
- 71. Yamnik RL, Holz MK. 2010. mTOR/S6K1 and MAPK/RSK signaling pathways coordinately regulate estrogen receptor α serine 167 phosphorylation. FEBS Lett 584:124–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yee D, Cullen KJ, Paik S, Perdue JF, Hampton B, Schwartz A, Lippman ME, Rosen N. 1988. Insulin-like growth factor II mRNA expression in human breast cancer. Cancer Res 48:6691–6696 [PubMed] [Google Scholar]
- 73. Murphy LJ, Murphy LC, Friesen HG. 1987. Estrogen induces insulin-like growth factor-I expression in the rat uterus. Mol Endocrinol 1:445–450 [DOI] [PubMed] [Google Scholar]
- 74. Zhang H, Pelzer AM, Kiang DT, Yee D. 2007. Down-regulation of type I insulin-like growth factor receptor increases sensitivity of breast cancer cells to insulin. Cancer Res 67:391–397 [DOI] [PubMed] [Google Scholar]
- 75. Zhang H, Fagan DH, Zeng X, Freeman KT, Sachdev D, Yee D. 2010. Inhibition of cancer cell proliferation and metastasis by insulin receptor downregulation. Oncogene 29:2517–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Entingh-Pearsall A, Kahn CR. 2004. Differential roles of the insulin and insulin-like growth factor-I (IGF-I) receptors in response to insulin and IGF-I. J Biol Chem 279:38016–38024 [DOI] [PubMed] [Google Scholar]
- 77. Jackson JG, Yee D. 1999. IRS-1 expression and activation are not sufficient to activate downstream pathways and enable IGF-I growth response in estrogen receptor negative breast cancer cells. Growth Horm IGF Res 9:280–289 [DOI] [PubMed] [Google Scholar]
- 78. Yoneda T, Williams PJ, Hiraga T, Niewolna M, Nishimura R. 2001. A bone-seeking clone exhibits different biological properties from the MDA-MB-231 parental human breast cancer cells and a brain-seeking clone in vivo and in vitro. J Bone Miner Res 16:1486–1495 [DOI] [PubMed] [Google Scholar]
- 79. Pons M, Gagne D, Nicolas JC, Mehtali M. 1990. A new cellular model of response to estrogens: a bioluminescent test to characterize (anti) estrogen molecules. Biotechniques 9:450–459 [PubMed] [Google Scholar]
- 80. Qiu M, Olsen A, Faivre E, Horwitz KB, Lange CA. 2003. Mitogen-activated protein kinase regulates nuclear association of human progesterone receptors. Mol Endocrinol 17:628–642 [DOI] [PubMed] [Google Scholar]
- 81. Twentyman PR, Luscombe M. 1987. A study of some variables in a tetrazolium dye (MTT) based assay for cell growth and chemosensitivity. Br J Cancer 56:279–285 [DOI] [PMC free article] [PubMed] [Google Scholar]