Glucocorticoids repress the oncogenic microRNA cluster miR-17∼92 as a mechanism to induce Bim, a pro-apoptotic protein in a therapeutically important cell death pathway.
Abstract
Synthetic glucocorticoids were one of the first effective treatments for lymphoid malignancies because of their ability to induce apoptosis and are still used in combination with other chemotherapeutic agents. Up-regulation of Bim, a proapoptotic member of the B-cell lymphoma-2 family, is an important mediator of glucocorticoid-induced apoptosis. Although glucocorticoids are known to elevate Bim mRNA and protein, little is known about the mechanism. Here, we report that glucocorticoids repress the expression of the microRNA cluster miR-17∼92, which results in elevated Bim protein expression as a mechanism by which glucocorticoids induce Bim. Using a luciferase-Bim 3′ untranslated region construct, we demonstrate that glucocorticoids mediate Bim induction posttranscriptionally after miR-17∼92 repression, resulting in increased Bim protein expression. Overexpression of miR-17∼92 microRNAs decreases Bim induction and attenuates glucocorticoid-mediated apoptosis. Conversely, knockdown of miR-17∼92 increases Bim protein expression and glucocorticoid-mediated apoptosis. These findings indicate that endogenous levels of miR-17∼92 repress Bim expression in T-cell lymphoid malignancies and that glucocorticoids induce Bim expression via down-regulation of the miR-17∼92 microRNA cluster. Our findings present a novel mechanism that contributes to the up-regulation of Bim and induction of apoptosis in lymphocytes after glucocorticoid treatment. Furthermore, our work demonstrating that inhibition of miR-17∼92 increases glucocorticoid-induced apoptosis highlights the potential importance of miR-17∼92 as a therapeutic target in leukemias and lymphomas.
Glucocorticoid hormones play an important role in the development and maintenance of the immune system (1). Because of their systemic immunosuppressive effects, glucocorticoids are invaluable therapeutic agents used in the treatment of a wide variety of inflammatory conditions. In addition, high-dose synthetic glucocorticoids, including prednisone and dexamethasone, remain a mainstay in the treatment of lymphoid malignancies, such as acute lymphoblastic leukemia (ALL), by virtue of their ability to induce apoptosis (2, 3).
Glucocorticoids induce apoptosis in immature lymphoblasts by binding to the glucocorticoid receptor (GR), a transcription factor and member of the nuclear steroid hormone receptor superfamily. Upon binding by glucocorticoids, the GR disassociates from its cytosolic complex, translocates to the nucleus, and alters gene transcription by binding directly to DNA response elements or through interactions with other transcription factors (4, 5). Early studies in the WEHI7.2 and S49 murine T-cell lines demonstrated that glucocorticoid-induced apoptosis is GR dependent, because GR antagonism by RU-486 blocks glucocorticoid-induced apoptosis. This suggests a requirement for glucocorticoid bound GR-mediated gene changes (6). Additional studies, some using the CEM human T-cell leukemia cell lines, further characterized the mechanism of glucocorticoid-induced apoptosis and mechanisms of resistance to glucocorticoids (7). Due to the complexity of GR-mediated transcriptional changes, the gene changes responsible for induction of apoptosis are not completely understood.
Previous studies identified that glucocorticoids induce a proapoptotic member of the B-cell lymphoma 2 (Bcl-2) protein family, Bim (Bcl-2-interacting mediator of cell death) (8, 9). Proteins in the Bcl-2 family contain at least one of four conserved Bcl-2 homology (BH) domains and regulate the intrinsic apoptotic pathway (10, 11). Antiapoptotic family members, such as Bcl-2 and Bcl-xL (Bcl-2-related gene, long isoform), contain all four BH domains. Proapoptotic family members Bax (Bcl-2-associated x protein) and Bak (Bcl-2 antagonist killer 1) contain three BH domains (BH1–BH3) and, upon activation, are the final effectors of the Bcl-2 family, leading to cytochrome c release from the mitochondria. The proapoptotic BH3-only proteins (12, 13), including Bim, comprise the largest subgroup of the Bcl-2 family and function either by directly activating Bax and Bak (14) or inhibiting antiapoptotic family members (15).
Complementing the initial discovery that glucocorticoids induce Bim, other groups also have implicated Bim in glucocorticoid-induced apoptosis. Bim knockdown by small interfering RNA (siRNA) prevents glucocorticoid-induced apoptosis in B-cell models (16), and Bim−/− mouse thymocytes display reduced apoptosis after glucocorticoid treatment (17, 18). Also, malignant cells isolated from childhood ALL patients resistant to glucocorticoids did not elevate Bim to the same levels as in patients sensitive to glucocorticoids (19). Taken together, these findings suggest that Bim protein levels in immature lymphocyte models are elevated after glucocorticoid treatment and serve as an important mediator of apoptosis.
Although the role of Bim in glucocorticoid-induced apoptosis is established, the mechanism leading to Bim's elevation currently is unknown. Previous reports have established that Bim is regulated at the transcriptional (20–23), posttranscriptional (24), and posttranslational (25, 26) levels. Recent reports demonstrated that an oncogenic cluster of microRNAs (miRNAs), miR-17∼92, regulates Bim in lymphocytes (27, 28). miRNAs are 18–24 nucleotide single-stranded RNAs that induce gene silencing by binding to target mRNAs with partial complementarity (29, 30). RNA polymerase II transcribes the primary miRNA transcript, which is cleaved into an approximately 70-nucleotide stem-loop intermediate (pre-miRNA). The pre-miRNA is exported from the nucleus and further processed into the mature miRNA. The miR-17∼92 cluster contains six highly conserved miRNAs and is frequently overexpressed in lymphoid malignancies (31), and enforced expression can accelerate tumor development in mouse models (32).
Using a luciferase-Bim 3′ untranslated region (UTR) construct, we find that glucocorticoid-mediated up-regulation of Bim is dependent upon the 3′UTR in the T-cell leukemia model WEHI7.2, suggesting posttranscriptional regulation. This finding led us to the hypothesis that glucocorticoids induce Bim by down-regulating miR-17∼92. Using quantitative real-time PCR (qPCR) and ribonuclease (RNase) protections assays, we demonstrate that glucocorticoids repress expression of both pri-miR-17∼92 and the resulting mature miRNAs. Additionally, overexpression of miR-17∼92 partially inhibits both the induction of Bim and apoptosis. Inhibition of miR-17∼92 increases Bim protein levels and enhances glucocorticoid-induced apoptosis. Using our luciferase-Bim3′UTR construct, we also identify the individual miRNAs from the miR-17∼92 cluster that repress Bim.
Overall, there are two major conclusions from our work. First, glucocorticoids repress miR-17∼92, and glucocorticoid-mediated miR-17∼92 contributes to the induction of apoptosis. Second, we provide substantial experimental evidence that glucocorticoid-mediated repression of miR-17∼92 contributes to the elevation of Bim, providing novel insight into the mechanism by which glucocorticoids induce expression of this proapoptotic protein.
Results
Glucocorticoid-mediated induction of Bim leads to apoptosis
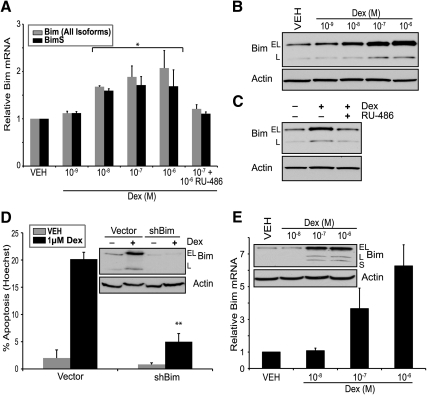
The murine T-cell lymphoma cell line WEHI7.2 corresponds to double positive (CD4+ and CD8+) cortical stage immature thymocytes. This model system has been used extensively for the study of glucocorticoid-induced apoptosis by others (33, 34) and is one of the cell systems we previously used in finding a glucocorticoid-induced expressed sequence tag corresponding to Bim (9). To build on these experiments, we used qPCR to determine the level of Bim mRNA up-regulation. Consistent with our microarray findings, the WEHI7.2 cell line demonstrates modest dexamethasone-mediated up-regulation of Bim mRNA (Fig. 1A). To confirm that Bim protein also was induced after dexamethasone treatment, we measured Bim protein levels via Western blotting (Fig. 1B). The Bim gene is alternatively spliced into three predominant isoforms and all three are induced: BimEL (Fig. 1B, top band), BimL (Fig. 1B, bottom band), and BimS (data not shown due to low expression). Addition of the GR antagonist RU-486 inhibited dexamethasone-induced Bim up-regulation, suggesting a GR-dependent mechanism (Fig. 1C). Although it previously has been shown that siRNA-mediated inhibition of Bim blocks glucocorticoid-induced apoptosis in B-cell lymphoma models (16), we wanted to determine whether a similar reduction in glucocorticoid-mediated apoptosis was observed in T-cell models. Transduction of lentiviral constructs containing short hairpin RNAs (shRNAs) targeted to Bim reduced dexamethasone-mediated Bim induction and apoptosis (Fig. 1D). Bim mRNA and protein also are induced in the human T-ALL cell line CEMC7 (Fig. 1E). Taken together, these findings demonstrate that Bim is an important mediator of glucocorticoid-induced apoptosis.
Fig. 1.
Bim is induced by dexamethasone (Dex) and is necessary for glucocorticoid-induced apoptosis. A, qPCR for Bim mRNA expression in WEHI7.2 cells cultured with increasing concentrations of dexamethasone for 24 h. Gray bars represent expression of all Bim isoforms, and black bars represent expression of BimS. B, Immunoblot of Bim protein in WEHI7.2 cells treated with increasing concentrations of dexamethasone for 24 h. C, Immunoblot of Bim in WEHI7.2 cells treated with vehicle (VEH) (0.1% ethanol), dexamethasone (100 nm), or dexamethasone (100 nm) and RU-486 (100 nm). D, WEHI7.2 cells transduced with a lentiviral shRNA targeting Bim. Cells were transduced with empty vector or Bim shRNA, treated with (1 μm) or without (0.1% ethanol) dexamethasone for 24 h and stained with Hoechst 33342. The percentage of cells displaying typical apoptotic nuclear morphology was determined by microscopy in which at least 200 cells were counted. Inset, Immunoblot for Bim. E, qPCR for Bim mRNA expression in CEMC7 (T-ALL) cells treated with increasing concentrations of dexamethasone for 24 h. Inset, Immunoblot of Bim protein. Error bars represent mean ± se of at least three independent experiments. All immunoblots are representative of at least three independent experiments. *, P < 0.05; **, indicates P < 0.01.
Glucocorticoids elevate Bim via 3′UTR elements
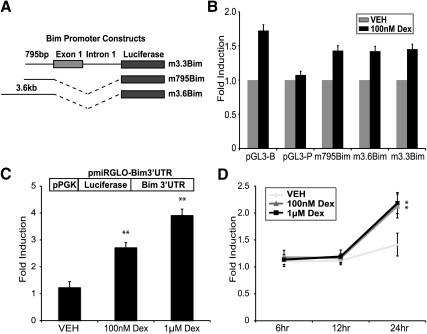
In an effort to determine the mechanism by which glucocorticoids elevate Bim, we initially investigated direct transcriptional regulation of Bim. We performed luciferase assays with several mouse and human 5′ Bim promoter constructs previously used to identify transcriptional regulators of Bim (Fig. 2A) (20, 23). Mouse Bim promoter constructs were transfected into the WEHI7.2 cell line. Compared with a renilla internal control (pGL4.74), we observe mild up-regulation of luciferase from both the Bim promoter containing luciferase vectors and the empty control luciferase vector (pGL3-B) (Fig. 2B). Therefore, we are not able to determine whether dexamethasone is inducing the Bim promoters directly or through an off-target effect on the vector. To determine whether a regulatory element in the 3′UTR of Bim is responsible for dexamethasone-mediated up-regulation, we cloned the 3′UTR downstream of the Renilla luciferase coding region in the psiCheck2 vector. Due to dexamethasone-mediated transcriptional changes of psiCheck2, the Bim 3′UTR was subcloned downstream of firefly luciferase in pmiRGLO. After transfection of pmiRGLO-Bim3′UTR into WEHI7.2 cells and treatment with dexamethasone, luciferase activity was elevated significantly (Fig. 2C). To remove potential off-target effects of dexamethasone on transcription of the pmiRGLO vector, WEHI7.2 cells were transfected with either empty pmiRGLO or pmiRGLO-Bim3′UTR and treated with 10 and 100 nm dexamethasone for 6, 12, and 24 h. After normalizing pmiRGLO-Bim3′UTR to empty pmiRGLO, dexamethasone significantly increases Bim 3′UTR-regulated luciferase levels (Fig. 2D). These results demonstrate that full induction of Bim after glucocorticoid treatment requires the Bim 3′UTR and suggest that glucocorticoids induce Bim, at least in part, through a posttranscriptional mechanism.
Fig. 2.
Dexamethasone (Dex)-mediated induction of Bim occurs through the 3′UTR. A, Representation of Bim promoter luciferase constructs. B, Fold induction of luciferase in WEHI7.2 cells transfected with pGL3-B (empty luciferase vector), pGL3-P (SV40 promoter-driven luciferase vector), or Bim promoter-driven luciferase vectors (m795Bim-pGL3, m3.6Bim-pGL3, and m3.3Bim-pGL3) after 24 h of treatment with vehicle (VEH) (0.1% ethanol) or dexamethasone (100 nm). C, Fold induction of luciferase in WEHI7.2 cells transfected with pmiRGLO-Bim3′UTR treated for 24 h with vehicle (0.1% ethanol) or dexamethasone (100 nm or 1 μm). Inset, pmiRGLO-Bim3′UTR vector schematic. D, Fold induction of luciferase in WEHI7.2 cells transfected with pmiRGLO-Bim3′UTR. Lysates were taken at 6, 12, and 24 h. The Bim3′UTR-mediated fold induction by dexamethasone was normalized to identically treated samples transfected with empty pmiRGLO vector to eliminate the effect of off-target transcriptional changes by dexamethasone. In all experiments, firefly luciferase was normalized to Renilla luciferase. Error bars represent mean ± se of at least three independent experiments. *, P < 0.05; **, P < 0.01.
Glucocorticoids repress expression of the miR-17∼92 cluster of miRNAs
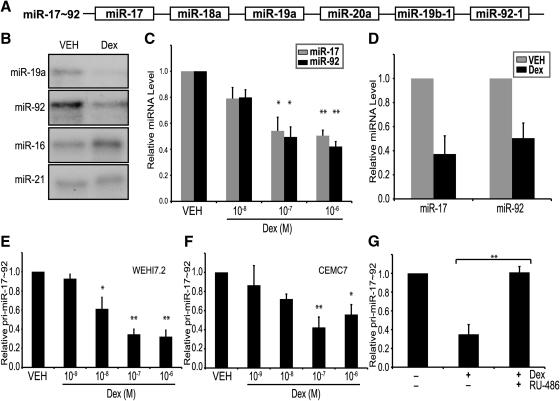
The miR-17∼92 cluster encodes six miRNAs, and at least three of these have been reported to regulate Bim mRNA by binding to its 3′UTR and inducing posttranscriptional repression (Fig. 3A) (27, 28, 35, 36). Measuring several miRNAs as representative of the cluster, we sought to test whether glucocorticoids repress miR-17∼92, because this would explain the 3′UTR-mediated up-regulation of Bim. Using an RNase protection assay, we determined that dexamethasone decreased the expression of miR-19a and miR-92 in WEHI7.2 cells, using miR-16 and miR-21 as controls (Fig. 3B). To ensure that glucocorticoids down-regulate the mature miRNAs, we used reverse transcription primers and qPCR probes specific for miR-17 and miR-92. MiR-17 and miR-92 were down-regulated in the WEHI7.2 cell line in a dose-dependent manner after dexamethasone treatment (Fig. 3C). Intraperitoneal injection of dexamethasone into mice also decreased miR-17 and miR-92 levels in primary thymocytes (Fig. 3D). Using custom designed primers and a TaqMan probe, we determined that pri-miR-17∼92 is repressed by dexamethasone in WEHI7.2 (Fig. 3E) and CEMC7 (Fig. 3F). Addition of RU-486 inhibited dexamethasone-mediated down-regulation of pri-miR-17∼92 in WEHI7.2 cells (Fig. 3G). Taken together, these data demonstrate that glucocorticoids down-regulate both mature miRNAs from the miR-17∼92 cluster and pri-miR-17∼92.
Fig. 3.
Dexamethasone (Dex) down-regulates miR-17∼92. A, Schematic representation of miR-17∼92. B, RNase protection assay of WEHI7.2 cells treated with vehicle (VEH) (0.1% ethanol) or dexamethasone (1 μm). RNA hybridized with 32P-labeled probes for miR-19a, miR-92, miR-16, and miR-21. C, qPCR for miR-17 (gray bars) and miR-92 (black bars) in WEHI7.2 cells cultured with increasing concentrations of dexamethasone for 24 h. D, qPCR for miR-17 and miR-92 in primary thymocytes isolated from mice 18 h after an ip injection of vehicle (500 ml PBS, gray bars) or dexamethasone (250 μg in 500 ml PBS, black bars). E and F, qPCR for pri-miR-17∼92 in WEHI7.2 cells (E) and CEM C7 cells (F) treated with increasing concentrations of dexamethasone for 24 h. G, qPCR for pri-miR-17∼92 in WEHI7.2 cells treated with vehicle (0.1% ethanol), dexamethasone (100 nm), or dexamethasone (100 nm) and RU-486 (100 nm) for 24 h. qPCR values for individual miRNAs were determined using Applied Biosystems primer and probe sets and normalized to Sno202RNA. qPCR values for pri-miR-17∼92 were determined using custom primer and MGB probes and normalized to 18S RNA and actin. Error bars represent mean ± se of three independent experiments. *, P < 0.05; **, P < 0.01.
The miR-17∼92 overexpression attenuates glucocorticoid-mediated Bim induction and apoptosis
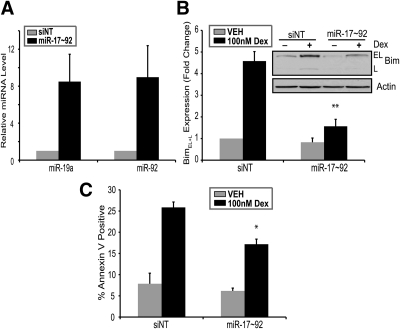
Previous reports demonstrate that miRNAs from the miR-17∼92 cluster function as oncogenes by altering tumor formation and angiogenesis (32, 37) and regulate Bim levels posttranscriptionally (27, 28). Next, we sought to test whether miR-17∼92 overexpression would decrease glucocorticoid-mediated Bim induction and apoptosis. To directly test whether miR-17∼92 levels alter glucocorticoid-mediated Bim induction and apoptosis, we transfected WEHI7.2 cells with miR-17∼92 miRNA mimics or nontargeting siRNAs (Fig. 4A). In WEHI7.2 cells transfected with miR-17∼92 mimics, dexamethasone does not up-regulate Bim levels to the same degree as in cells transfected with nontargeting oligonucleotides (Fig. 4B). Additionally, transfection of miR-17∼92 miRNA mimics reduces glucocorticoid-induced apoptosis in WEHI7.2 cells (Fig. 4C). This indicates that preventing the down-regulation of miR-17∼92 by glucocorticoids partially inhibits the up-regulation of Bim protein and the induction of apoptosis. Overexpression of miR-17∼92 did not inhibit glucocorticoid-induced apoptosis or Bim induction to the level achieved by shRNAs targeting Bim (Fig. 1D). This could be due to several factors: 1) miRNAs do not typically exert as strong an inhibitory influence as siRNAs (29) or 2) it is possible that Bim is also induced by an additional yet unknown mechanism. Collectively, these data indicate that the expression level of miR-17∼92 is an important regulator of glucocorticoid-induced apoptosis, and overexpression can partially prevent glucocorticoid-mediated induction of Bim.
Fig. 4.
The miR-17∼92 overexpression attenuates Bim induction and apoptosis. A, Transfection of WEHI7.2 cells with nontargeting siRNA (siNT) (1200 nm) or miRNA mimics for miR-17∼92 (200 nm each); 18 h after transfection, cells were harvested for qPCR of miR-19a and miR-92. B, WEHI7.2 cells were transfected with nontargeting siRNA (1200 nm) or miRNA mimics for miR-17∼92 (200 nm each); 4 h after transfection, cells were treated with (100 nm) or without (0.1% ethanol) dexamethasone (Dex) for 24 h. Densitometry was used to determine signal intensity of BimEL and BimL. BimEL+l signal intensity was normalized to the signal intensity of actin. Inset, Representative immunoblot for Bim. C, WEHI7.2 cells treated as described in B were stained with Annexin V and propidium iodide and measured by flow cytometric analysis. Error bars represent mean ± se of three independent experiments. *, P < 0.05; **, P < 0.01. VEH, Vehicle.
The miR-17∼92 inhibition elevates Bim levels and increases glucocorticoid-induced apoptosis
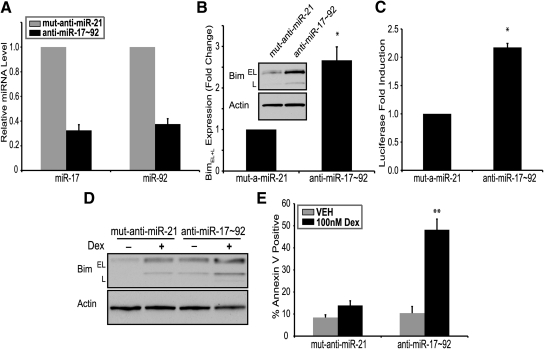
Next, we sought to determine whether inhibition of miR-17∼92 would elevate Bim expression and glucocorticoid-induced apoptosis. To determine whether miR-17∼92 constitutively regulates Bim levels in WEHI7.2 cells, we transfected anti-miRNA oligonucleotides (AMOs) of miR-17∼92 miRNAs. Compared with a mutated miR-21 AMO used as a control, inhibition of miR-17∼92 with AMOs resulted in reduction of miR-17∼92 expression (Fig. 5A) and increased Bim protein levels approximately 2.5-fold (Fig. 5B). To demonstrate that inhibition of miR-17∼92 regulates Bim through its 3′UTR, WEHI7.2 cells expressing psiCheck2-Bim3′UTR were transfected with AMOs targeting miR-17∼92. Compared with controls, inhibition of miR-17∼92 increased Bim 3′UTR-regulated luminescence (Fig. 5C). Taken together, these data suggest that reducing expression of miR-17∼92 miRNAs removes a posttranscriptional inhibition of Bim, mediated through its 3′UTR. Although overexpression of miR-17∼92 partially inhibited Bim induction and glucocorticoid-induced apoptosis, we wanted to determine whether inhibition of miR-17∼92 would have an opposite effect. Indeed, when miR-17∼92 is inhibited by AMOs, dexamethasone treatment of WEHI7.2 cells leads to elevated Bim protein levels (Fig. 4D) and glucocorticoid-induced apoptosis, compared with controls (Fig. 4E). Dexamethasone treatment of miR-17∼92 AMO-transfected WEHI7.2 cells led to almost complete cell death at 24 h (data not shown). Therefore, in Fig. 4, D and E, experiments were performed for 18 h. As compared with the 20–25% apoptosis that dexamethasone induces in control WEHI7.2 cells (Figs. 1D and 4C) at 24 h, transfection of AMOs targeting miR-17∼92 leads to approximately 50% apoptosis at 18 h. This suggests that inhibition of miR-17∼92 leads to significantly stronger induction of glucocorticoid-induced apoptosis.
Fig. 5.
The miR-17∼92 inhibition elevates glucocorticoid-induced apoptosis. A and B, Nucleofection of WEHI7.2 cells with mutated anti-miR-21 (1000 nm) or anti-miRNAs targeting miR-17∼92 (200 nm each); 18 h after transfection, cells were harvested for qPCR of miR-17 and miR-92 (A). B, BimEL and BimL signal intensity was determined using densitometry and all signal intensities were normalized to actin. Inset, Representative immunoblot of Bim. C, WEHI7.2 cells expressing a psiCheck2-Bim3′UTR plasmid were transfected with mutated anti-miR-21 (1000 nm) or anti-miRNAs targeting miR-17∼92 (200 nm each). Cells were analyzed for luciferase levels 18 h after transfection, and fold induction of Renilla luciferase was normalized to the internal control firefly luciferase. D, Immunoblot for Bim in WEHI7.2 cells transfected with mutated anti-miR-21 (1000 nm) or anti-miRNAs targeting miR-17∼92 (200 nm each). Cells were treated with vehicle (VEH) (0.1% ethanol) or dexamethasone (Dex) (10–7 m) for 18 h. E, WEHI7.2 cells treated as described in D were stained with Annexin V and propidium iodide, and apoptosis was measured by flow cytometric analysis. Error bars represent mean ± se of three independent experiments. *, P < 0.05; **, P < 0.01.
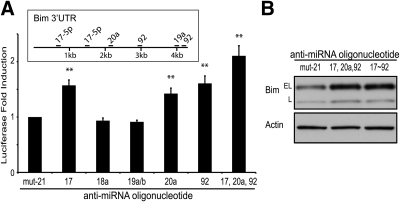
Although the inhibition of the entire miR-17∼92 cluster leads to induction of Bim, we investigated which individual miRNAs from the miR-17∼92 cluster regulate Bim expression. Using partial fragments of the Bim 3′UTR, several groups have previously implicated a role for miR-19 and miR-92, observing that miR-92 has the dominant effect (27, 28). Using several bioinformatic algorithms (38, 39), we determined several potential binding sites for miRNAs from the miR-17∼92 cluster (Fig. 6A, inset). We transfected individual AMOs for the miR-17∼92 cluster into WEHI7.2 cells along with a full-length Bim 3′UTR luciferase vector. Our findings demonstrate that miR-17, miR-20a, and miR-92 individually regulate Bim through its 3′UTR (Fig. 6A). Also, if we transfect AMOs targeting miR-17, miR-20a, and miR-92, we observe Bim up-regulation similar to that found after transfection of AMOs targeting the whole cluster (Fig. 6B). This suggests that inhibition of miR-17, miR-20a, and miR-92 is sufficient to maximally repress miR-17∼92's effect on Bim. Jointly, these data indicate that the expression level of miR-17∼92 is an important regulator of glucocorticoid-induced apoptosis, and miR-17∼92 levels may be an additional therapeutic target for lymphomas.
Fig. 6.
Effect of individual miRNAs of the miR-17∼92 cluster on Bim expression. A, WEHI7.2 cells expressing a psiCheck2-Bim3′UTR plasmid were transfected with mutated anti-miR-21 (200 nm) or anti-miRNAs targeting miR-17∼92 (200 nm each). Cells were analyzed for luciferase levels 18 h after transfection, and fold induction of Renilla luciferase was normalized to the internal control firefly luciferase. Inset, Schematic of predicted miR-17∼92 binding sites in the Bim3′UTR. B, WEHI7.2 cells were transfected with mutated anti-miR-21 (1000 nm); anti-miRNAs targeting miR-17, miR-20a, and miR-92; or anti-miRNAs targeting miR-17∼92 (200 nm each). Error bars represent mean ± se of three independent experiments. **, P < 0.01.
Discussion
Glucocorticoids were one of the earliest effective agents used for the treatment of lymphoid malignancies and continue to be used today as part of combination chemotherapy regimens (2). Microarray technology led to the discovery that glucocorticoids induce the proapoptotic BH3-only protein Bim in multiple lymphocyte models (8, 9). Other groups have further demonstrated the importance of Bim up-regulation to glucocorticoid-induced apoptosis (16, 18, 19). These reports, along with our findings that lentiviral-mediated Bim repression inhibits glucocorticoid-induced apoptosis, demonstrate the importance of Bim to the chemotherapeutic function of glucocorticoids (Fig. 1D). However, to our knowledge, there have been no reports on the mechanism by which glucocorticoids induce Bim in lymphocytes.
Here, we report that the repression of miR-17∼92 after glucocorticoid treatment alleviates an inhibitory effect on Bim through its 3′UTR. Additionally, glucocorticoids repress both the primary transcript and mature miRNAs of the miR-17∼92 cluster. Similar to the findings in other reports (27, 28), inhibition of miR-17∼92 leads to the 3′UTR-dependent up-regulation of Bim in our lymphoma models. We also present evidence that modulation of miR-17∼92 expression alters glucocorticoid-induced apoptosis, because overexpression of miR-17∼92 partially inhibits apoptosis, whereas inhibition of miR-17∼92 increases apoptosis. Using a luciferase-Bim 3′UTR construct, we demonstrate that Bim levels are posttranscriptionally elevated after 24 h of dexamethasone treatment (Fig. 2, C and D). Interestingly, we do not see Bim 3′UTR-mediated up-regulation of luciferase until 24 h, suggesting the posttranslational up-regulation occurs between 12 and 24 h. The regulation of miRNA turnover is an expanding area of study, and recently, Hwang et al. (40) demonstrated that miR-29b has a shorter half-life in cycling cells when compared with arrested cells. As it is known that glucocorticoids decrease cell cycle progression in lymphocytes, with future experiments we hope to determine whether dexamethasone can regulate miRNA half-life either globally or individually. Also, the regulation of glucocorticoid-induced apoptosis by miR-17∼92 occurs, at least in part, through regulation of Bim protein levels. Although our findings do not rule out other potential mechanisms by which glucocorticoids regulate Bim, such as a remote glucocorticoid response element, to our knowledge, this is the first report detailing a mechanism for glucocorticoid-mediated induction of Bim.
The BH3-only protein Bim is an important mediator of apoptosis in hematopoietic cells in response to some stimuli but not others. When compared with wild-type lymphocytes, Bim−/− murine immature lymphocytes display delayed apoptosis in response to γ-irradiation and glucocorticoid treatment, yet display similar levels of apoptosis in response to phorbol 12-myristate 13-acetate and Fas ligand (17). In other models, cAMP, but not UV radiation, induces Bim before apoptosis (41). Also, cytokine withdrawal from hematopoietic progenitor cells can lead to the up-regulation of Bim and induction of apoptosis (20, 42). Consistent with these previous findings, we found that inhibition of Bim by shRNAs partially inhibits glucocorticoid-induced apoptosis in the T-cell lymphoma model WEHI7.2 (Fig. 1D).
Bim is regulated at the transcriptional, posttranscriptional and posttranslational levels. In response to imatinib (21), paclitaxel (43), and cytokine withdrawal (20), the transcription factor forkhead box class O 3a induces Bim transcription. Also, endoplasmic reticulum stress can induce Bim transcription through the C/EBPα (CCAAT/enhancer binding protein) and CHOP (C/EBP homologous protein) transcription factors (23). Several kinases, including ERK (25, 26) and protein kinase B (44), can phosphorylate BimEL, decreasing its apoptotic activity and leading to proteasomal degradation (45). Conversely, protein phosphatase 2A can dephosphorylate Bim increasing its apoptotic activity (23). In addition to regulating the transcription of Bim, cytokines can regulate Bim posttranscriptionally by directing the binding of heat shock protein 70 to AU-rich elements in Bim's 3′UTR (24). Recently, there have been several articles demonstrating that miR-17∼92 regulates Bim at the posttranscriptional level (27, 28). In this study, we demonstrate that overexpression of miR-17∼92 partially blocks the up-regulation of Bim and induction of apoptosis (Fig. 4). Also, inhibition of miR-17∼92 increases basal expression of Bim protein and increases glucocorticoid-induced apoptosis (Figs. 5 and 6).
There have been several other reports investigating the role of individual miRNAs from the miR-17∼92 cluster in the regulation of Bim (27, 28). Using partial fragments of the Bim 3′UTR, these groups propose a role for miR-19a and miR-92 in the posttranscriptional regulation of Bim. Interestingly, when these two sites were interrogated together, the observed regulation was no different from miR-92 alone, suggesting a dominant role for miR-92 (28). Using our psiCheck2-Bim3′UTR vector, we determined that miR-17, miR-20, and miR-92 all participate in the posttranscriptional regulation of Bim (Fig. 6A). When combined, miR-17, miR-20a, and miR-92 had a greater effect on the 3′UTR than any one miRNA alone, suggesting coordinate regulation by these members of the miR-17∼92 cluster. Also, the combination of miR-17, miR-20a, and miR-92 induced Bim levels similar to the entire miR-17∼92 cluster (Fig. 6B). This implies that these three miRNAs are sufficient for the observed up-regulation of Bim by miR-17∼92.
The miR-17∼92 cluster of miRNAs plays an important role in both developmental processes and tumor progression (46). The discovery of the miR-17∼92 cluster followed observations that the chromosomal region containing its open reading frame was often amplified in lymphomas (31). Overexpression of miR-17∼92 increased tumor progression in the Eμ-Myc transgenic mouse model of lymphoma at least in part by inhibiting apoptosis (32). Other evidence suggests that miR-17∼92 can increase angiogenesis in a colon cancer model by repressing thrombospondin-1 (37). The miR-17∼92 cluster of miRNAs is essential for the development of B cells, because miR-17∼92−/− hematopoietic precursor cells cannot progress from pro-B cells to pre-B cells (27). The authors hypothesize that elevated Bim levels in the miR-17∼92−/− cells are responsible for the developmental block. In the present study, we demonstrate that dexamethasone partially functions through the repression of miR-17∼92 in lymphoma models, allowing for Bim up-regulation and subsequent apoptosis (Figs. 3 and 4). Based on our findings, it is reasonable to speculate that other processes, including negative selection of thymocytes and chemotherapeutic treatments, which lead to Bim induction and apoptosis in lymphocytes, may be regulated by the expression level of miR-17∼92.
Previously, there have been two reports using expression profiling methods to measure glucocorticoid-mediated alterations in miRNAs (47, 48). Rainer et al. (47) provided the first report detailing regulation of miRNAs by glucocorticoids using miRNA microarrays and qPCR. This report provides evidence for the up-regulation of miR-15b, miR-16, and miR-223, as well as the down-regulation of pri-miR-17∼92, but does not document the down-regulation of any mature miRNAs (47). During the final preparations of our manuscript, a second report used miRNA microarrays and deep sequencing to screen for miRNA changes during glucocorticoid-induced apoptosis of rat thymocytes. In this report, Smith et al. observe substantial down-regulation of miRNAs, including mature miRNAs from the miR-17∼92 cluster. Smith et al. (48) overexpress miR-17∼92 and observe a decrease in glucocorticoid-induced apoptosis, consistent with our findings in Fig. 4C. They also observe a decrease in the expression of the miRNA processing enzymes Dicer, Drosha, and Pasha and propose that the down-regulation of these enzymes is responsible for glucocorticoid-mediated global down-regulation of miRNAs (48).
Consistent with the findings of Rainer et al. (47), we observed up-regulation of miR-16 in dexamethasone-treated WEHI7.2 cells by RNase protection assay (Fig. 3B) and qPCR (data not shown), suggesting that down-regulation of Dicer/Drosha/Pasha, as suggested by Smith et al. (48), does not readily explain the ability of glucocorticoids to both down- and up-regulate miRNAs. There are several theories that could explain the differences observed among the three reports. First, although all three studies focus on well-established models of glucocorticoid-induced apoptosis, there may be differences in the processes of miRNA biogenesis, processing and turnover, and their response to glucocorticoids among the systems used. Second, differences could be explained by alternative methods of miRNA detection used in each study. And finally, the dexamethasone concentrations used in each study are different and could account for the differences observed. Thus, further investigation is needed to unravel the mechanisms by which glucocorticoids regulate miRNAs.
Because we see up- and down-regulation of different miRNAs in our model system, determining the precise mechanism through which glucocorticoids down-regulate miR-17∼92 will require further study. Using specific qPCR probes, we determined that glucocorticoids repress the pri-miR-17∼92 transcript, suggesting a transcriptional repression of the gene (Fig. 3, E and F). This agrees with previous findings showing down-regulation of pri-miR-17∼92 after glucocorticoid treatment (47). Several reports have identified that the transcriptional factors c-Myc (49) and the E2F family (50) transactivate the miR-17∼92 cluster of miRNAs. It also is well known that glucocorticoids repress the expression and activity of c-Myc in the same cells we have used (51), leading us to hypothesize that glucocorticoids transcriptionally repress miR-17∼92 through down-regulation of c-Myc.
In summary, our findings present a novel mechanism for the up-regulation of Bim after glucocorticoid treatment. These findings build on previous reports demonstrating the importance of Bim to glucocorticoid-induced apoptosis of immature lymphoblasts as well as previous reports documenting glucocorticoid-mediated regulation of miRNAs. Importantly, these findings are the first to directly link glucocorticoid-mediated alterations in miRNAs to the regulation of a protein involved in the induction of apoptosis. Also, our work further supports the role of miR-17∼92 in the development and progression of tumors, specifically by blocking apoptosis through repression of proapoptotic proteins; miRNAs currently are being evaluated as potential therapeutic targets (52), and our findings support targeting miR-17∼92 as a way to increase the effectiveness of glucocorticoid-induced apoptosis.
Materials and Methods
Cell culture
The WEHI7.2 murine T-cell lymphoma cell line was a gift of Diane Dowd and cultured in DMEM supplemented with 2 mm l-glutamine, 10% bovine calf serum (HyClone, Logan, UT), and 100 μm nonessential amino acids. The CEMC7 cell line was a gift of E. B. Thompson (7) and cultured in RPMI 1640 media supplemented with 2 mm l-glutamine, 10% fetal calf serum (HyClone), and 100 μm nonessential amino acids. WEHI7.2 cells were cultured at 1.5 × 105 cells/ml in a humidified 7% CO2 incubator. CEMC7 cells were cultured at 2 × 105 cells/ml in a humidified 5% CO2 incubator. Dexamethasone and RU-486 (mifepristone) were purchased from Sigma-Aldrich (St. Louis, MO).
RNA isolation, reverse transcription, qPCR, and RNase protection assay
When measuring mRNA transcripts, total RNA was isolated using Trizol reagent (Invitrogen, Carlsbad, CA). When also measuring miRNAs, total RNA was isolated using mirVANA miRNA isolation kit (Ambion, Austin, TX). RNA samples were resuspended in RNase-free water and quantified by measuring absorbance at 260 and 280 nm. When assaying mRNAs, total RNA was reverse transcribed using the TaqMan Gold RT-PCR kit (Applied Biosystems, Foster City, CA). The resulting cDNAs were amplified using Universal TaqMan master mix and primer/probe sets specific for the genes of interest (Applied Biosystems) on a Lightcycler 480 II (Roche, Indianapolis, IN). Relative quantification was determined using the ΔΔCt method using 18S rRNA and β-actin as reference genes. Bim, 18S rRNA, and β-actin primer/probe sets were commercially available (Applied Biosystems). Custom primer/probe sets were synthesized for pri-miR-17∼92 using primer express 3.0 software (Applied Biosystems). The primers used to detect pri-miR17∼92 are as follows: forward primer 5′-CCTAAGTGCTCCTTCTGGCATAA-3′, reverse primer 5′-CCCTACATGCTTGCTTGACTTG-3′, probe 6FAM-AAGTTATGTCCTCATCCAAT-MGBNFQ for mouse; forward primer 5′-GCCTGTCGCCCAATCAAA-3′, reverse primer 5′-ATGCAAACCTGCAAAACTAACCA-3′, probe 6FAM-TGTCCTGTTACTGAACACT-MGBNFQ for human). Deoxyribonuclease treatment (Ambion) was performed before reverse transcription in experiments quantifying pri-miR17∼92 levels.
TaqMan miRNA assays were used to measure miR-17∼92 miRNAs with qPCR (Applied Biosystems). Briefly, specific stem-loop primers were used to reverse transcribe mature miRNAs, and quantitative PCR was performed using specific primers and probes on a 7500 fast real-time PCR thermal cycler (Applied Biosystems). The reference RNA sno202 was used as an internal control, and relative quantification was performed using the ΔΔCt method.
RNase protection assays were carried out using the mirVana miRNA detection kit (Ambion). The primers used to detect miRNAs are as follows: miR-19a 5′-UCAGUUUUGCAUAGAUUUGCACACCAGAG-3′, miR-92 5′-ACAGGCCGGGACAAGUGCAAUACCAGAG-3′, miR-16 5′-CGCCAAUAUUUACGUGCUGCUACCAGAG-3′, miR-21 5′-UCAACAUCAGUCUGAUAAGCUACCAGAG-3′. Primers were end-labeled with γ32P-ATP and separated on a denaturing polyacrylamaide gel (Bio-Rad, Hercules, CA).
Immunoblotting
Whole cell lysates were obtained by suspending 5 million cells in radioimmunoprecipitation assay buffer [10 mm Tris-HCl (pH 7.6), 150 mm NaCl, 5 mm EDTA, 1% Triton X-100, 0.1% sodium dodecyl sulfate, 200 mm dithiothreitol, 0.5% sodium deoxycholate, and Complete Mini protease inhibitor cocktail (Roche)]. After protein concentration determination using Bradford Reagent (Bio-Rad), equal amounts of protein and sample buffer [50 nm Tris-HCl (pH 6.8), 2% sodium dodecyl sulfate, 100 mm dithiothreitol, 10% glycerol, and 0.05% bromophenol blue; final concentrations] were separated onto a 4–20% gradient SDS-PAGE gel, transferred to polyvinylidene fluoride membrane and blocked in Tris-buffered saline containing Tween 20 (0.1%) and milk or BSA (5%). The following primary antibodies were used: Bim (C34C5) (Cell Signaling Technology, Danvers, MA) and actin (AC-15) (Sigma-Aldrich). All immunoblots are representative of at least three independent experiments.
Apoptosis assays
Apoptotic nuclear morphology was evaluated on cells stained with Hoechst 33342 (10 μg/ml) for 15 min at 37 C and visualized by epifluorescence microscopy (Axiovert S100; Carl Zeiss AG, Oberkochen, Germany) using a ×40 oil objective (Carl Zeiss AG). Apoptosis was determined using Annexin V to measure extracellular expression of phosphatidylserine and propidium iodide to measure membrane permeability. Flow cytometric analysis for Annexin V positive cells was performed using an EPICS XL-MCL flow cytometer (Beckman Coulter, Pasadena, CA). Analysis of flow cytometric data was performed using WinList (Verity House Software, Topsham, ME). Similar levels of apoptosis are observed by the Hoechst nuclear morphology assay and the Annexin V flow cytometry assay.
Generation of the Bim 3′UTR luciferase plasmids
The mouse Bim 3′UTR was amplified using PCR from mouse genomic DNA with nested PCR primers. The following primer sequences were used: forward primer outside, 5′-CAGGATCTACATGCAGCCAGGATACGTGGC-3′; reverse primer outside, 5′-CTTCAGCAAGCGGTGTGTG-3′; forward primer inside, 5′-CCCTCGAGCAGGATCTACATGCAGCCA-3′ (underlined sequence is XhoI restriction site); reverse primer inside, 5′-GGGCGGCCGCTTTTGAAAGCTAGTCGCAAGT-3′ (underlined sequence is NotI restriction site). The resulting PCR amplified fragment was cloned into the XhoI/NotI sites in the 3′UTR of Renilla luciferase in psiCheck2 (Promega, Madison, WI) and verified by DNA sequencing. Due to spurious transcription of the psiCheck2 vector by dexamethasone, the Bim 3′UTR was excised from psiCheck2 and ligated into pmiRGLO for experiments with dexamethasone.
Bim 3′UTR luciferase assays
Dual-luciferase reporter assays were performed according to the recommendations provided with the dual-luciferase reporter kit (Promega). The 795-bp and 3.6-kb Bim promoters were generous gifts of Eric Lam (University of London, London, UK), and the 3.3-kb Bim promoter was a generous gift of Hamsa Puthalakath (Walter and Eliza Hall Institute of Medical Research, Parkville, Australia). Cells were nucleofected using an Amaxa Nucleofector II (WEHI7.2: solution L, program B-003; and CEMC7: solution C, program X-001; Lonza, Basel, Switzerland) with 2 μg of experimental plasmid and 100 ng of pGL4.74, a constitutively expressing Renilla luciferase vector (Promega). Sixteen hours after nucleofection, cells were treated with either vehicle or varying concentrations of dexamethasone, and a 0 h sample was taken. Fold induction was determined by dividing dexamethasone-treated cells by either 0 h or vehicle-treated cells.
To measure dexamethasone-mediated regulation of Bim through its 3′UTR, WEHI7.2 cells were nucleofected with 100 ng of pmiRGLO or pmiRGLO-Bim3′UTR. Sixteen hours after nucleofection, cells were treated with either VEH or varying concentrations of dexamethasone, and a 0 h sample was taken. After treatments, cells were lysed with passive lysis buffer solution, and luminescence of firefly and Renilla was measured using a PerkinElmer Victor3 plate reader (PerkinElmer, Waltham, MA). Background luminescence was determined using an equal volume of nontransfected cell lysate. Fold induction was quantified by dividing dexamethasone-mediated promoter activity by 0-h luminescence values. To remove spurious transcriptional changes by dexamethasone, pmiRGLO-Bim3′UTR samples were divided by pmiRGLO samples with identical treatments.
To measure direct regulation of the Bim 3′UTR by altering miR-17∼92 levels, psiCheck2-Bim3′UTR was nucleofected into WEHI7.2 cells. Sixteen hours after nucleofection, miRNA mimics for miR-17∼92 or inhibitors of miR-17∼92 were transfected as described above, and cells were assayed for firefly and Renilla luciferase 24 h later. Fold induction was quantified by dividing miR-17∼92 transfected cells by control transfected cells.
Overexpression and inhibition of miR-17∼92
Overexpression of miR-17∼92 was performed using miRNA mimics (QIAGEN, Valencia, CA) for individual miRNAs. For analysis of Bim levels and apoptosis, a mixture of all six miRNA mimics (200 nm each) or 1200 nm of a nontargeting control was electroporated (10 million cells, 0.2-cm cuvette subjected to a single 140 V, 10 millisecond square wave pulse) into WEHI7.2 cells. Four hours after electroporation, cells were treated with vehicle or dexamethasone for 30 h and assayed for Bim protein levels and apoptosis.
Inhibition of miR-17∼92 miRNAs was performed using 2′-O-methyl oligoribonucleotides with interspersed locked nucleic acids synthesized by Integrated DNA Technologies (Coralville, IA). These chemical modifications have been shown to display high specificity and potency for inhibiting miRNAs (53). The sequences used are as follows: anti-miR-17 5′-C*U*A*CCTGCACUGUAAGCACU*T*U*G-3′; anti-miR-18 5′-C*U*A*UCTGCACUAGATGCACC*T*U*A-3′; anti-miR-19a/b 5′-U*C*A*GUTUUGCATGGAUUTGC*A*C*A-3′; anti-miR-20a 5′-C*U*A*CCTGCACUAUAAGCACU*T*U*A-3′; anti-miR-92 5′-C*A*G*GCCGGGACAAGUGCA*A*T*A-3′, where underlined bases indicate locked nucleic acids positions and an asterisk denotes a phosphorothioate linkage. For analysis of Bim levels and apoptosis, a mixture of all five anti-miRNAs (200 nm each) or 1000 nm of mutated anti-miR-21 (5′-U*C*A*AGAUCAGTCUCAUAAG*G*U*A-3′) were nucleofected into WEHI7.2 cells with an Amaxa Nucleofector II (2 million cells, solution L, program B-003; Lonza). Four hours after nucleofection, cells were treated with vehicle or dexamethasone for 24 h and assayed for Bim protein levels and apoptosis.
Mice treatment
C57BL6 sex matched 6-wk-old mice were given ip injections of vehicle (PBS) or dexamethasone (250 μg in PBS); 18 h after injection, the mice were killed, and the thymocytes were isolated. miRNA levels were quantified as described above. Experiments using mice were approved by the Case Western Reserve University Institutional Animal Care and Use Committee.
Statistics
Prism software was used to perform ANOVA tests on group populations to determine the presence of significance. Two tailed t tests were used to determine significance between pairs of data. Error bars represent mean ± se of at least three independent experiments.
Acknowledgments
We thank Kim Lennox (Integrated DNA Technologies, Coralville, IA) for help cloning the Bim3′UTR and designing AMO's to miR-17∼92; Eric Lam (University of London, London, UK) and Hamsa Puthalakath (Walter and Eliza Hall Institute of Medical Research, Parkville, Australia) for their Bim promoter luciferase vectors; Shigemi Matsuyama for the mice used in this study; and Mark Jackson, Ed Stavnezer, George Dubyak, and Alex Huang for helpful suggestions regarding the manuscript.
This work was supported by National Institutes of Health Grants RO1 CA42755 (to C.W.D.), T32 HL007147 (to J.K.M.), and T32 GM007250 (to J.K.M.).
Disclosure Summary: The authors have nothing to disclose.
NURSA Molecule Pages:
Ligands: Dexamethasone | RU486.
Footnotes
- ALL
- Acute lymphoblastic leukemia
- AMO
- anti-miRNA oligonucleotide
- Bcl-2
- B-cell lymphoma 2
- BH
- Bcl-2 homology domain
- GR
- glucocorticoid receptor
- miRNA
- microRNA
- qPCR
- quantitative real-time PCR
- RNase
- ribonuclease
- shRNA
- short hairpin RNA
- siRNA
- small interfering RNA
- UTR
- untranslated region.
References
- 1. Rhen T, Cidlowski JA. 2005. Antiinflammatory action of glucocorticoids—new mechanisms for old drugs. N Engl J Med 353:1711–1723 [DOI] [PubMed] [Google Scholar]
- 2. Gaynon PS, Carrel AL. 1999. Glucocorticosteroid therapy in childhood acute lymphoblastic leukemia. Adv Exp Med Biol 457:593–605 [DOI] [PubMed] [Google Scholar]
- 3. Smets LA, Salomons G, van den Berg J. 1999. Glucocorticoid induced apoptosis in leukemia. Adv Exp Med Biol 457:607–614 [DOI] [PubMed] [Google Scholar]
- 4. Cidlowski JA, King KL, Evans-Storms RB, Montague JW, Bortner CD, Hughes FM., Jr 1996. The biochemistry and molecular biology of glucocorticoid-induced apoptosis in the immune system. Recent Prog Horm Res 51:457–490; discussion 490–451 [PubMed] [Google Scholar]
- 5. Schmidt S, Rainer J, Ploner C, Presul E, Riml S, Kofler R. 2004. Glucocorticoid-induced apoptosis and glucocorticoid resistance: molecular mechanisms and clinical relevance. Cell Death Differ 11(Suppl 1):S45–S55 [DOI] [PubMed] [Google Scholar]
- 6. Distelhorst CW. 2002. Recent insights into the mechanism of glucocorticosteroid-induced apoptosis. Cell Death Differ 9:6–19 [DOI] [PubMed] [Google Scholar]
- 7. Thompson EB, Johnson BH. 2003. Regulation of a distinctive set of genes in glucocorticoid-evoked apoptosis in CEM human lymphoid cells. Recent Prog Horm Res 58:175–197 [DOI] [PubMed] [Google Scholar]
- 8. Medh RD, Webb MS, Miller AL, Johnson BH, Fofanov Y, Li T, Wood TG, Luxon BA, Thompson EB. 2003. Gene expression profile of human lymphoid CEM cells sensitive and resistant to glucocorticoid-evoked apoptosis. Genomics 81:543–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang Z, Malone MH, He H, McColl KS, Distelhorst CW. 2003. Microarray analysis uncovers the induction of the proapoptotic BH3-only protein Bim in multiple models of glucocorticoid-induced apoptosis. J Biol Chem 278:23861–23867 [DOI] [PubMed] [Google Scholar]
- 10. Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. 2010. The BCL-2 family reunion. Mol Cell 37:299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Youle RJ, Strasser A. 2008. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol 9:47–59 [DOI] [PubMed] [Google Scholar]
- 12. O'Connor L, Strasser A, O'Reilly LA, Hausmann G, Adams JM, Cory S, Huang DC. 1998. Bim: a novel member of the Bcl-2 family that promotes apoptosis. EMBO J 17:384–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Strasser A. 2005. The role of BH3-only proteins in the immune system. Nat Rev Immunol 5:189–200 [DOI] [PubMed] [Google Scholar]
- 14. Letai A, Bassik MC, Walensky LD, Sorcinelli MD, Weiler S, Korsmeyer SJ. 2002. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell 2:183–192 [DOI] [PubMed] [Google Scholar]
- 15. Cory S, Huang DC, Adams JM. 2003. The Bcl-2 family: roles in cell survival and oncogenesis. Oncogene 22:8590–8607 [DOI] [PubMed] [Google Scholar]
- 16. Abrams MT, Robertson NM, Yoon K, Wickstrom E. 2004. Inhibition of glucocorticoid-induced apoptosis by targeting the major splice variants of BIM mRNA with small interfering RNA and short hairpin RNA. J Biol Chem 279:55809–55817 [DOI] [PubMed] [Google Scholar]
- 17. Bouillet P, Metcalf D, Huang DC, Tarlinton DM, Kay TW, Köntgen F, Adams JM, Strasser A. 1999. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science 286:1735–1738 [DOI] [PubMed] [Google Scholar]
- 18. Erlacher M, Michalak EM, Kelly PN, Labi V, Niederegger H, Coultas L, Adams JM, Strasser A, Villunger A. 2005. BH3-only proteins Puma and Bim are rate-limiting for γ-radiation- and glucocorticoid-induced apoptosis of lymphoid cells in vivo. Blood 106:4131–4138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bachmann PS, Gorman R, Mackenzie KL, Lutze-Mann L, Lock RB. 2005. Dexamethasone resistance in B-cell precursor childhood acute lymphoblastic leukemia occurs downstream of ligand-induced nuclear translocation of the glucocorticoid receptor. Blood 105:2519–2526 [DOI] [PubMed] [Google Scholar]
- 20. Dijkers PF, Medema RH, Lammers JW, Koenderman L, Coffer PJ. 2000. Expression of the pro-apoptotic Bcl-2 family member Bim is regulated by the forkhead transcription factor FKHR-L1. Curr Biol 10:1201–1204 [DOI] [PubMed] [Google Scholar]
- 21. Essafi A, Fernández de Mattos S, Hassen YA, Soeiro I, Mufti GJ, Thomas NS, Medema RH, Lam EW. 2005. Direct transcriptional regulation of Bim by FoxO3a mediates STI571-induced apoptosis in Bcr-Abl-expressing cells. Oncogene 24:2317–2329 [DOI] [PubMed] [Google Scholar]
- 22. Whitfield J, Neame SJ, Paquet L, Bernard O, Ham J. 2001. Dominant-negative c-Jun promotes neuronal survival by reducing BIM expression and inhibiting mitochondrial cytochrome c release. Neuron 29:629–643 [DOI] [PubMed] [Google Scholar]
- 23. Puthalakath H, O'Reilly LA, Gunn P, Lee L, Kelly PN, Huntington ND, Hughes PD, Michalak EM, McKimm-Breschkin J, Motoyama N, Gotoh T, Akira S, Bouillet P, Strasser A. 2007. ER stress triggers apoptosis by activating BH3-only protein Bim. Cell 129:1337–1349 [DOI] [PubMed] [Google Scholar]
- 24. Matsui H, Asou H, Inaba T. 2007. Cytokines direct the regulation of Bim mRNA stability by heat-shock cognate protein 70. Mol Cell 25:99–112 [DOI] [PubMed] [Google Scholar]
- 25. Ewings KE, Hadfield-Moorhouse K, Wiggins CM, Wickenden JA, Balmanno K, Gilley R, Degenhardt K, White E, Cook SJ. 2007. ERK1/2-dependent phosphorylation of BimEL promotes its rapid dissociation from Mcl-1 and Bcl-xL. EMBO J 26:2856–2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ley R, Balmanno K, Hadfield K, Weston C, Cook SJ. 2003. Activation of the ERK1/2 signaling pathway promotes phosphorylation and proteasome-dependent degradation of the BH3-only protein, Bim. J Biol Chem 278:18811–18816 [DOI] [PubMed] [Google Scholar]
- 27. Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, Jaenisch R, Sharp PA, Jacks T. 2008. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 132:875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. 2008. Lymphoproliferative disease and autoimmunity in mice with increased miR-17–92 expression in lymphocytes. Nat Immunol 9:405–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Carthew RW, Sontheimer EJ. 2009. Origins and Mechanisms of miRNAs and siRNAs. Cell 136:642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Filipowicz W, Bhattacharyya SN, Sonenberg N. 2008. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9:102–114 [DOI] [PubMed] [Google Scholar]
- 31. Ota A, Tagawa H, Karnan S, Tsuzuki S, Karpas A, Kira S, Yoshida Y, Seto M. 2004. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res 64:3087–3095 [DOI] [PubMed] [Google Scholar]
- 32. He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. 2005. A microRNA polycistron as a potential human oncogene. Nature 435:828–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sibley CH, Tomkins GM. 1974. Isolation of lymphoma cell variants resistant to killing by glucocorticoids. Cell 2:213–220 [DOI] [PubMed] [Google Scholar]
- 34. Dowd DR, MacDonald PN, Komm BS, Haussler MR, Miesfeld R. 1991. Evidence for early induction of calmodulin gene expression in lymphocytes undergoing glucocorticoid-mediated apoptosis. J Biol Chem 266:18423–18426 [PubMed] [Google Scholar]
- 35. Fontana L, Fiori ME, Albini S, Cifaldi L, Giovinazzi S, Forloni M, Boldrini R, Donfrancesco A, Federici V, Giacomini P, Peschle C, Fruci D. 2008. Antagomir-17-5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PLoS One 3:e2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mavrakis KJ, Wolfe AL, Oricchio E, Palomero T, de Keersmaecker K, McJunkin K, Zuber J, James T, Khan AA, Leslie CS, Parker JS, Paddison PJ, Tam W, Ferrando A, Wendel HG. 2010. Genome-wide RNA-mediated interference screen identifies miR-19 targets in Notch-induced T-cell acute lymphoblastic leukaemia. Nat Cell Biol 12:372–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT, Thomas-Tikhonenko A. 2006. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet 38:1060–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. 2005. Combinatorial microRNA target predictions. Nat Genet 37:495–500 [DOI] [PubMed] [Google Scholar]
- 39. Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. 2003. Prediction of mammalian microRNA targets. Cell 115:787–798 [DOI] [PubMed] [Google Scholar]
- 40. Hwang HW, Wentzel EA, Mendell JT. 2007. A hexanucleotide element directs microRNA nuclear import. Science 315:97–100 [DOI] [PubMed] [Google Scholar]
- 41. Zhang L, Insel PA. 2004. The pro-apoptotic protein Bim is a convergence point for cAMP/protein kinase A- and glucocorticoid-promoted apoptosis of lymphoid cells. J Biol Chem 279:20858–20865 [DOI] [PubMed] [Google Scholar]
- 42. Shinjyo T, Kuribara R, Inukai T, Hosoi H, Kinoshita T, Miyajima A, Houghton PJ, Look AT, Ozawa K, Inaba T. 2001. Downregulation of Bim, a proapoptotic relative of Bcl-2, is a pivotal step in cytokine-initiated survival signaling in murine hematopoietic progenitors. Mol Cell Biol 21:854–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sunters A, Fernández de Mattos S, Stahl M, Brosens JJ, Zoumpoulidou G, Saunders CA, Coffer PJ, Medema RH, Coombes RC, Lam EW. 2003. FoxO3a transcriptional regulation of Bim controls apoptosis in paclitaxel-treated breast cancer cell lines. J Biol Chem 278:49795–49805 [DOI] [PubMed] [Google Scholar]
- 44. Qi XJ, Wildey GM, Howe PH. 2006. Evidence that Ser87 of BimEL is phosphorylated by Akt and regulates BimEL apoptotic function. J Biol Chem 281:813–823 [DOI] [PubMed] [Google Scholar]
- 45. Dehan E, Bassermann F, Guardavaccaro D, Vasiliver-Shamis G, Cohen M, Lowes KN, Dustin M, Huang DC, Taunton J, Pagano M. 2009. βTrCP- and Rsk1/2-mediated degradation of BimEL inhibits apoptosis. Mol Cell 33:109–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mendell JT. 2008. miRiad roles for the miR-17–92 cluster in development and disease. Cell 133:217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rainer J, Ploner C, Jesacher S, Ploner A, Eduardoff M, Mansha M, Wasim M, Panzer-Grümayer R, Trajanoski Z, Niederegger H, Kofler R. 2009. Glucocorticoid-regulated microRNAs and mirtrons in acute lymphoblastic leukemia. Leukemia 23:746–752 [DOI] [PubMed] [Google Scholar]
- 48. Smith LK, Shah RR, Cidlowski JA. 2010. Glucocorticoids modulate microRNA expression and processing during lymphocyte apoptosis. J Biol Chem 285:36698–36708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. 2005. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435:839–843 [DOI] [PubMed] [Google Scholar]
- 50. Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, Major F, Ferbeyre G, Chartrand P. 2007. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem 282:2135–2143 [DOI] [PubMed] [Google Scholar]
- 51. Zhou F, Medh RD, Thompson EB. 2000. Glucocorticoid mediated transcriptional repression of c-myc in apoptotic human leukemic CEM cells. J Steroid Biochem Mol Biol 73:195–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kota J, Chivukula RR, O'Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, Mendell JR, Mendell JT. 2009. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 137:1005–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lennox KA, Behlke MA. 2010. A Direct Comparison of Anti-microRNA Oligonucleotide Potency. Pharm Res 27:1788–1799 [DOI] [PubMed] [Google Scholar]