Abstract
Trastuzumab (Herceptin®) is the first FDA-approved therapeutic targeting a HER-family receptor tyrosine kinase (HER2/ErbB2/neu). Although trastuzumab is effective in the treatment of HER2-positive breast cancer, a substantial proportion of patients will not respond to trastuzumab-based regimens (primary resistance), and those who do respond will often lose clinical benefit (i.e., secondary resistance). While multiple mechanisms underlying the development of secondary trastuzumab resistance have been identified, few studies have specifically examined the basis of primary trastuzumab resistance. Here, we review these studies, which, together, demonstrate that trastuzumab induces phenotypic changes in tumor cells, even when they are not growth inhibited by trastuzumab, including changes in gene expression. These changes have important clinical implications, including sensitization of malignant cells to other therapeutic drugs. In light of these observations, we propose that the conventional definition of “resistance” as it pertains to trastuzumab and, perhaps, to other targeted therapeutics, may require revision. The results of these studies will be useful in informing the direction of future basic and clinical research focused on overcoming primary trastuzumab resistance.
Keywords: EGFR/ErbB/HER, trastuzumab/herceptin, breast cancer, targeted therapeutics, primary resistance
Introduction
While the HER2-directed antibody trastuzumab is one of the early success stories among biologically targeted breast cancer therapeutics, some patients will not respond to this drug, and among responders, the majority will eventually relapse. This clinical scenario occurs in spite of steadfast efforts to select patients for trastuzumab treatment based on HER2-positive tumor status with expensive diagnostic tests [1]. Here, we review recent studies on the mechanistic basis of primary trastuzumab resistance. These studies suggest that multiple endpoints beyond tumor cell growth inhibition should be considered during the evaluation of trastuzumab efficacy, and perhaps the efficacy of other emerging biologically targeted therapeutics.
Proposed mechanisms of trastuzumab activity
Contrary to initial assumptions, trastuzumab does not appear to inhibit HER2 dimerization or ligand-dependent HER heterodimeric signaling [2]. Instead, three major mechanisms of trastuzumab inhibition of tumor growth and survival have gained some experimental support:
Targeting of immune cells to HER2-positive tumor cells. Immune cells, binding to trastuzumab Fc domains, effect antibody dependent cell-mediated cytotoxicity (ADCC) of HER2-positive tumor cells [3–5].
Inhibition of HER2 shedding. In certain breast cancer-derived cell lines, the extracellular domain (ECD) of HER2 is proteolytically shed from the cell surface [6–8], and the remaining truncated receptor (‘p95HER2’ or ‘t95HER2’; [9]), now freed from autoinhibition by the receptor’s ECD, exhibits constitutive tyrosine kinase activity [10, 11]. Trastuzumab binding to the HER2 receptor inhibits proteolytic shedding, perhaps via a steric or allosteric mechanism, resulting in decreased HER2 kinase activity [10]. This model is discussed in the context of recent studies demonstrating that p95HER2 also may be expressed from an alternate HER2 transcript (see section IV) [12].
Internalization and degradation of HER2. Trastuzumab binding to HER2 stimulates the recruitment of c-Cbl and resulting in subsequent HER2 ubiquitinlyation, internalization, and degradation [13–15].
Given the diversity of these proposed mechanisms of trastuzumab’s therapeutic activity, it is perhaps not surprising that our understanding of trastuzumab resistance, both primary and secondary, is incomplete. While the mechanistic basis for secondary resistance recently has been reviewed [16–18], the number of studies exploring the mechanistic basis of primary trastuzumab resistance is relatively limited. Here we review several recent studies on the mechanism of primary resistance to trastuzumab, and discuss unifying themes revealed by them.
Clinical studies reveal complex patterns of responsiveness to trastuzumab
Most patients with early stage breast cancer and a large proportion of patients with metastatic breast cancer have a measurable tumor response to trastuzumab-chemotherapy, as demonstrated by a reduction in tumor burden (i.e., partial or complete response). However, a fraction (~20%) of early stage breast cancer patients will not respond to trastuzumab, and ~70% of patients with metastatic disease who receive trastuzumab monotherapy are resistant to treatment [19, 20]. De novo or ‘primary’ resistance occurs when trastuzumab is ineffective for the treatment of breast cancer patients despite tumor expression of HER2. ‘Acquired’ or ‘secondary’ trastuzumab resistance occurs when patients who initially respond to trastuzumab experience trastuzumab-refractory relapse. Patients with HER2-positive breast cancer are typically treated with a combination of trastuzumab and chemotherapy, as exemplified in the pivotal National Surgical Adjuvant Breast and Bowel Project B31 and NCCTG N9831 trials. In both of these studies, while the addition of trastuzumab to chemotherapy reduced the chance of death among patients with early-stage HER2-positive breast cancer, survival among patients treated with chemotherapy alone was also high [21]. Since patients are not routinely treated with trastuzumab monotherapy, the relative contribution of each drug to reduced tumor burden, as well as the potential interactions among these drugs, can be difficult to assess, and both primary and secondary trastuzumab resistance must necessarily be associated with primary resistance to genotoxic therapies as well as to trastuzumab (except in the case of neoadjuvant trastuzumab monotherapy).
Putative mechanisms of primary trastuzumab resistance
Most studies on trastuzumab resistance have focused on the mechanisms underlying acquired or secondary trastuzumab resistance, using trastuzumab-sensitive cell lines such as SKBR3 and BT474 cultured with trastuzumab until a resistant phenotype emerges. Several recent studies, however, have examined in vitro models of primary trastuzumab resistance, or have studied the properties of trastuzumab resistant tumors, to explore the mechanistic basis for this phenomenon. The results of these studies are summarized schematically in Figure 1, and are discussed in more detail, below.
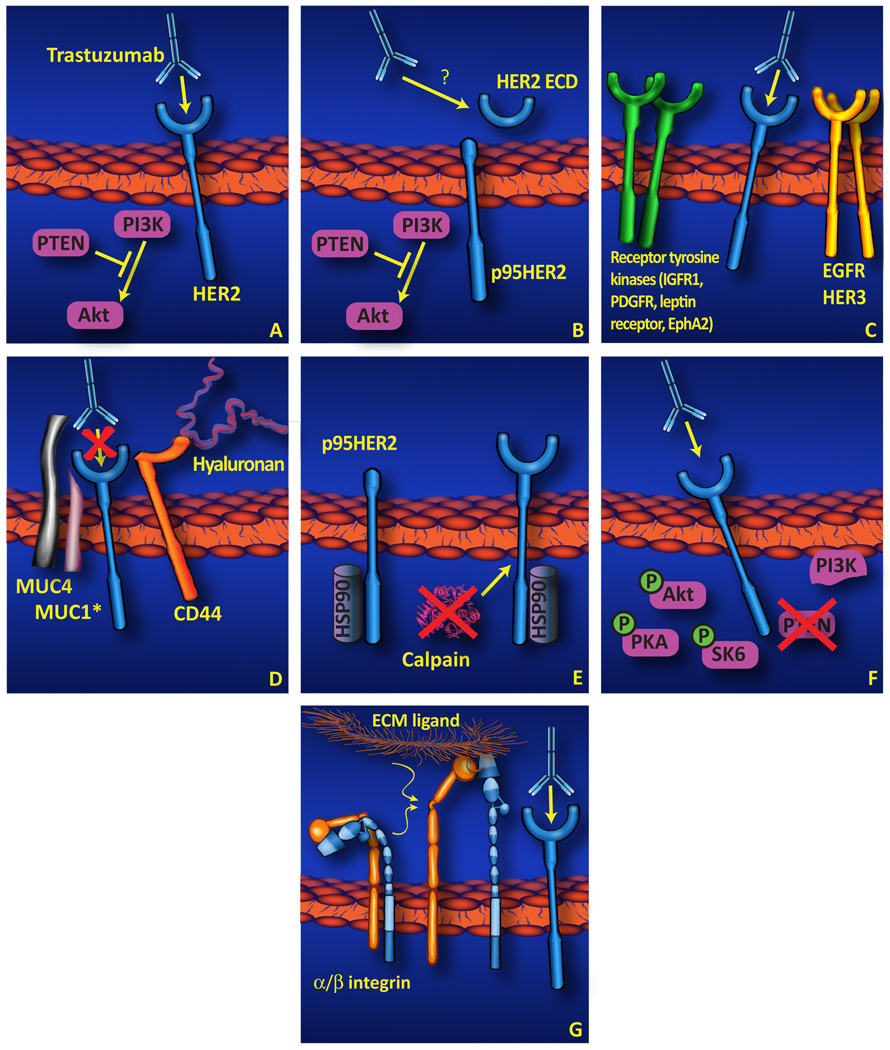
Figure 1. Potential mechanisms underlying primary trastuzumab resistance.
(A) While the proposed mechanisms of trastuzumab action are diverse (and are not mutually exclusive), one consensus viewpoint is that trastuzumab must be able to bind to the HER2 extracellular domain, and in doing so this antibody inhibits the association of PI3K and activated HER2, leading to decreased activation of Akt and subsequent inhibition of cell proliferation and survival. Even in the absence of a unifying hypothesis for the mechanism of trastuzumab inhibition of tumor cell growth, several alternate mechanisms to define the basis for primary trastuzumab resistance have been reported as summarized in this review, and are depicted schematically here, including: (B) Expression or proteolytic generation of p95HER2, a constitutively kinase-active HER2 isoform lacking the trastuzumab-binding site; (C) Compensatory signaling by other cell surface receptors including EGFR/HER3 and other receptor tyrosine kinases. (D) Physical blockade of trastuzumab/HER2 association by CD44/hyaluronin, MUC1*, or MUC4. (E) Increased HER2 stability in association with chaperone/HER2 interaction or downregulation of HER2-client proteases. (F) Constitutive activation of downstream effectors or cross-talk pathways. (G) Interaction of integrins and extracellular matrix components resulting in enhanced HER2-independent cell proliferation survival signaling. (H) Dispensability of HER2 (not displayed). In this model, HER2 expression may be stochastic and nonessential in some tumor cells.
N-terminal truncation of HER2
Alternate isoforms of all four members of the HER family have been described [9]. Soluble (s) HER2 isoforms arise from alternately-spliced transcripts of the HER2 gene resulting in 68 kDa [22] or 100 kDa [23] isoforms encompassing most of the HER2 ECD, or from proteolytic cleavage of full-length HER2 resulting in shed 105 kDa [7] or 110 kDa [6, 8] sHER2 fragments of the ECD. Proteolytic cleavage of full-length HER2 also yields a cell-surface associated fragment of ~95 kDa (termed ‘p95HER2’) encompassing a small fragment of the ECD, the transmembrane domain, and the intracellular domain (including the tyrosine kinase) [11].
While all of these isoforms may be relevant to clinical targeting of HER2 in cancer patients, it is the p95HER2 product that has received the most attention to date. Specifically, p95HER2, freed from ECD-mediated autoinhibition, is a constitutively active kinase and a potent oncogene [24]. Since p95HER2 lacks the trastuzumab binding domain of full-length HER2 (see Figure 1, panel B), it may also be an important mediator of primary trastuzumab resistance. Preclinical xenograft studies demonstrate that T47D [12] and MCF7 [25] cells stably expressing p95HER2 are insensitive to trastuzumab whereas cells stably expressing full-length HER2 are growth inhibited by trastuzumab. While retrospective studies have associated p95HER2 expression with an aggressive breast tumor phenotype and poor patient outcome [26, 27], only two studies have specifically examined the role of p95HER2 in primary trastuzumab resistance in breast cancer. Scaltriti et al. and Sperinde et al. both demonstrate an association between p95HER2 expression and failure to respond to primary trastuzumab regimens [25, 28].
Regulation of p95HER2 expression reveals additional complexities of the primary trastuzumab resistant phenotype. Initial studies demonstrated that full-length HER2 can be cleaved by a metalloprotease resulting in the generation of a p95HER2 membrane-associated fragment and a “shed” ECD fragment [6–8]; moreover, trastuzumab inhibits HER2 ECD shedding [10]. However, recent studies have identified an alternate transcript of HER2 which encodes a p95HER2 isoform lacking most or all of the ECD [12]. The relative contribution of these two distinct mechanisms to the synthesis of p95HER2 in tumor cells has not been determined, nor has the potential role of the shed ECD (or alternate sHER2 isoform[s]) been examined in the context of primary trastuzumab resistance.
Might the proteolytically shed sHER2 (ECD) isoform also play a role in primary trastuzumab resistance? Early studies by Brodowicz et al. demonstrated that sHER2 ECD, perhaps acting as an antibody sink, attenuated growth inhibitory effects of anti-HER2 antibodies on HER2-positive breast cancer cell lines [29] (see Figure 1, panel B); more recent studies by Ghedini et al. suggest that sHER2 may serve to bind to and deliver trastuzumab to the cell surface thereby synergistically inhibiting the growth of HER2-positive cells [30]. Clinical correlative studies on circulating sHER2 have demonstrated a trend or statistically significant correlation between decline in post-treatment vs. baseline serum sHER2 concentration and responsiveness to trastuzumab therapy [31, 32]. However, the limited biochemical characterization of this circulating isoform(s) of sHER2, as well as the limited number of functional studies on serum sHER2 prevent us from concluding that this isoform plays an active role in the development of primary trastuzumab resistance.
Dispensability and redundancy of HER2 signaling
Arnoud et al. demonstrated that primary trastuzumab resistance was inversely associated with breast cancer HER2 expression [33]. Complementary studies by Ginester et al. have identified basal phosphorylation of HER2 as a predictor of trastuzumab resistance in vitro; in these studies primary trastuzumab resistance was common among basal subtype cell lines and/or cell lines with low basal levels of phosphorylation of HER2 [34]. While the mechanism underlying reduced basal HER2 phosphorylation was not rigorously characterized in this study, the lack of constitutive HER2 phosphorylation in these cells suggests that HER2 may not an “addictive oncogene” under these conditions, in contrast to its role in BT-474 and SKBR-3 cells [35].
Complementary evidence in support of the dispensability or redundancy of HER2 signaling in certain breast carcinoma-derived cell lines has been reported by Narayan et al. [36]. In this study, several cell lines were selected that were not growth inhibited by trastuzumab or by EGFR inhibitors; long-term trastuzumab treatment sensitized these cells to EGFR inhibitor-mediated growth inhibition suggesting that HER2 and EGFR signaling may be compensatory in these cell lines, i.e., only one functional HER axis is required for cell proliferation. This concept is further supported by recent observations showing that trastuzumab-mediated growth inhibition can be overcome by ectopic expression of EGFR in SKBR3 cells [37], or by HB-EGF treatment of BT474 cells [38]. Similarly, an HB-EGF inhibitor renders MCF7-HER2 cells sensitive to trastuzumab [38]. High tumor EGFR expression levels also have been associated with primary resistance to trastuzumab neoadjuvant therapy [39].
Similarly, in two studies, mRNA profiles of pre-treatment breast tumor core biopsies were compared in patients with complete vs. non-complete pathologic response to trastuzumab in a neo-adjuvant setting. Harris et al. demonstrated that unresponsive patients (trastuzumab plus vinorelbine) were found to have elevated tumor expression levels of wnt family members, growth factors and growth factor receptors such as HGF, met, IGF-I, PDGF, leptin receptor, and pleitoropin; increased expression of IGF-1 in primary trastuzumab resistant breast tumors was confirmed by immunohistochemistry (see Figure 1, Panel C) [19]. More recent studies have demonstrated that IGFBP3, an inhibitor of IGF-1, synergizes with trastuzumab to inhibit the growth of primary trastuzumab resistant cells [40, 41]. By contrast, Vegran et al. did not identify growth factor receptors as part of a gene array profile upregulated in non-responsive (trastuzumab plus docetaxel) patient tumors, but did identify upregulation of certain cell signaling effectors in tumor samples from non-responsive patients (see below) [42].
In vitro studies also have identified the receptor tyrosine kinase EphA2 as commonly overexpressed in breast cancer cell lines resistant to trastuzumab but not in cell lines sensitive to trastuzumab [43], and EphA2 expression also has been correlated with poor survival in breast cancer patients [44, 45]. However, not all cell surface receptors cooperate to attenuate trastuzumab sensitivity. For example, granulocyte-colony stimulating factor in combination with trastuzumab can synergistically induce apoptosis in the primary trastuzumab resistant cell lines T47D and ZR-75-1 [46]. These studies illustrate the complex effect of cytokines on trastuzumab’s activity in tumor cells, and also the importance of considering the entire complement of tyrosine kinases in the context of the tumor cell’s resistance to trastuzumab.
Steric access to receptor and HER2 stability
As outlined above, several competing hypotheses have been proposed for the mechanism(s) of trastuzumab-mediated inhibition of tumor cell growth (see ‘II’, above). One unifying aspect among these mechanisms is that tumor cell growth inhibition is dependent on the binding of trastuzumab to the ECD of HER2. Therefore, processes or molecules which mask or prevent trastuzumab from binding to the HER2 receptor would be predicted to block trastuzumab activity. Experimental support for this hypothesis is summarized below.
Associations between HER2 and other cell surface signaling proteins, i.e., integrin β1 [47], and CD44 (hyaluronin receptor) [48], have been associated with primary trastuzumab resistance in the breast cancer-derived cell line JIMT-1 (see Figure 1, panel D). Studies using this cell line suggest that reduced trastuzumab binding to cell surface HER2 is mediated by steric hindrance associated with CD44 and/or MUC4 expression. This concept is supported by the observation that 4-methylumbelliferon, a hyaluronin synthesis inhibitor, increases trastuzumab binding to JIMT-1 cells, resulting in synergistic inhibition of JIMT-1 xenograft growth [48]. Similarly, MUC4, a high molecular mass proteoglycan, co-immunoprecipitates with HER2 from lysates of JIMT-1 cells, and also inhibits cell surface binding of trastuzumab [49]. In further support of this concept, siRNA’s directed against MUC4 increase trastuzumab binding to JIMT-1 cells [50]. Clearly, however, HER2 association with CD44 and/or MUC4 does not completely inhibit trastuzumab binding to JIMT-1 cells, nor does this association mask all HER2 epitopes, since pertuzumab, another HER2-directed monoclonal antibody, binds effectively to JIMT-1 cells [50].
The related proteoglycan MUC1 also has been implicated as a mediator of primary trastuzumab resistance. Proteolytic cleavage of full-length MUC1 yields a cell surface-associated fragment termed MUC1*. BT-474 cells with acquired trastuzumab resistance exhibit greatly increased concentrations of MUC1*, and siRNA knockdown of MUC1* or antibodies directed against MUC1* reverse acquired trastuzumab resistance in BT-474 cells [51]. MUC1* apparently plays a similar role in primary trastuzumab resistance: primary trastuzumab resistant ZR-75-30 and T47D cells express MUC1*, and either knockdown or antibody-mediated inhibition of MUC1* induce trastuzumab sensitivity in these cell lines. While MUC1* could potentially inhibit trastuzumab’s association with HER2 via steric hindrance, MUC1* expression has been independently correlated with induction of Erk phosphorylation and also with cell survival and proliferation [52, 53]. Therefore, the precise mechanism underlying MUC1*’s role in trastuzumab resistance remains unclear.
In addition to these associations with extracellular matrix and cell surface proteoglycans, cytosolic regulators of HER2 stability have been identified as critical mediators of primary trastuzumab resistance. HER2 was recently identified as a target of the cysteine protease calpain-1, and pharmacologic inhibition of calpain-1 desensitizes trastuzumab sensitive cells (see Figure 1, panel E) [54]. This finding is novel and unanticipated, as calpain is known to couple to EGFR and Src signaling to enhance motility and transformation [55], but in the case of HER2, calpain has been proposed to function as an signaling attenuator [54].
Molecular chaperones also have been implicated in the development of primary trastuzumab resistance. For example, HSP90 is well known to regulate the stability of client proteins such as HER2, decreasing receptor turnover thereby potentiating HER2 signaling (see Figure 1, panel E) [56]. Consequently, pharmacologic inhibition of HSP90 has become an area of intense study for overcoming trastuzumab resistance. In this regard, the cleaved tumorigenic HER2 fragment, p95HER2, has been identified as a client protein of the molecular chaperone HSP90, and trastuzumab resistant breast cancer cells expressing p95HER2 exhibit growth inhibition by the HSP90 inhibitors in vitro and in vivo [57, 58]. While these studies do not directly identify HSP90 as a mediator of primary trastuzumab resistance, they do validate HSP90 as a potential therapeutic target in trastuzumab resistant patients. Furthermore, expression of an HSP90 interacting protein, DARPP-32 and its truncated isoform t-Darpp, also have been shown to confer primary trastuzumab resistance in BT474 and SKBR-3 cells [59]. While DARPP-32 and tDarpp form a complex with HER2 and HSP90 in trastuzumab-resistant breast cancer cells [60], the function of DARPP-32 and tDarpp in this complex has not yet been examined.
Activation of downstream effectors and signal crosstalk
In a small cohort of stage II/III breast cancer patients, Yonemori et al. have found an association between responsiveness to neo-adjuvant therapy (trastuzumab plus chemotherapy) and tumor HER2 expression, but not with PTEN, p53, ER, PR or activated Akt expression [61]. In contrast, Nagata et al. have proposed that PTEN-deficiency in primary breast tumors from patients later treated with trastuzumab (for recurrent disease) is correlated with poor response to therapy [62]. In support of this finding, Migliaccio et al. have reported that patients treated with neo-adjuvant trastuzumab plus docetaxel therapy who exhibit low tumor PTEN expression or PIK3CA mutations are less likely to respond to neo-adjuvant therapy [63]. However, low PTEN expression or PIK3CA mutation status does not predict responsiveness to neoadjuvant lapatinib therapy, suggesting an involvement of EGFR signaling and/or differential responsiveness of tumors to HER2-directed tyrosine kinase inhibitors vs. therapeutic antibodies. Gori et al. further reported decreased survival in response to trastuzumab therapy among patients with elevated levels of phosphorylated MAPK [64]. Related in vitro studies using panels of breast cancer cell lines have identified PI3K expression [65], low PTEN expression [65], Akt phosphorylation [65], and S6K phosphorylation [66] as potential mediators of primary trastuzumab resistance in breast cancer; the role of PIK3CA mutation in primary trastuzumab resistance in vitro is supported by Katoaka et al. [66], but not by Koninki et al. [67].
Recent studies also have implicated protein kinase A (PKA) signaling as a mediator of trastuzumab resistance. Treatment with the adenylyl cyclase agonist forskolin confers partial resistance to trastuzumab-mediated survival signaling inhibition (i.e., Akt activation) in trastuzumab-sensitive cells, and siRNA-mediated knockdown of the PKA negative regulator PKA-RIIα also confers partial resistance to trastuzumab-mediated growth inhibition of BT474 cells [68]. Additionally, Vegran et al. have identified upregulation of PRKACA, the gene encoding the catalytic α-subunit of PKA, as an upregulated gene product in patients with trastuzumab resistant breast cancer [42]. Among the small cluster of upregulated gene products in patients with primary trastuzumab resistant breast cancer identified by Vegran and colleagues are several cell signaling and cell cycle regulators, including the WEE1 homologue, protein phosphatase 2A, and CDC14A [42].
Finally, Cheng et al. have demonstrated a role for the well-known protein synthesis regulator, eEF-2 kinase, in primary trastuzumab resistance. siRNA-mediated knockdown of eEF-2 kinase, a negative regulator of eukaryotic elongation factor-2, sensitizes MCF7 and MDA-MB-468 cells to trastuzumab [69].
Since numerous factors regulate PKA activity, cell cycle progression, and protein synthesis, these findings provide further evidence of the complexity of primary trastuzumab resistance (see Figure 1, panel F).
Cell: extracellular matrix interactions
Information from studies using carcinoma-derived cell lines is limited by the artificial and highly selected nature of these in vitro models. Nowhere is this limitation more apparent than in studies on the cell’s microenvironment, including those of cell/matrix and cell/tissue interactions. The vast majority of in vitro studies using carcinoma-derived cell lines use rigid tissue culture plastic as an adherence medium. One exception, however is a recent study by Weigelt et al., who studied the influence of the microenvironment on trastuzumab resistance (see Figure 1, panel G) [70] using matrix coated substrates. When grown on a laminin-rich matrix vs. untreated plastic, SKBR-3 cells are, remarkably, not growth inhibited by trastuzumab. This laminin matrix-mediated trastuzumab resistance was transduced through interactions between laminin and integrins, as evidenced by a reversal of trastuzumab resistance when SKBR-3 cells were treated with a β1-integrin inhibitory antibody [70]. In support of this concept, LAMA3, encoding laminin α3, has been identified as part of a gene profile upregulated among patients with primary trastuzumab resistant breast cancer [42].
In further support of this provocative finding, a recent retrospective study of patients with metastatic HER2-positive breast cancer has shown that β1-integrin expression is inversely correlated with survival among patients treated with trastuzumab but not with other regimens [71]. Moreover, transfection of β1 integrin into SKBR3 cells renders these cells resistant to trastuzumab-mediated growth inhibition [71]. Related results have been reported in another in vitro model (ovarian cancer) of long-term trastuzumab treatment in which tensin and RhoB regulation of tumor cell morphology has been proposed as a mediator of trastuzumab resistance [72].
Non-cytostatic effects of trastuzumab
Inhibition of cell proliferation in vitro, or of tumor growth in vivo are typically used as measures of efficacy for biologically targeted therapeutics, and with good reason. However, other more subtle phenotypic changes involving alternative biological endpoints may be manifest following trastuzumab treatment, as summarized in Figure 2. Critically, some of these latent phenotypes may be clinically relevant. Identification of these novel phenotypes may, therefore, allow us to more effectively treat breast cancer patients in the future.
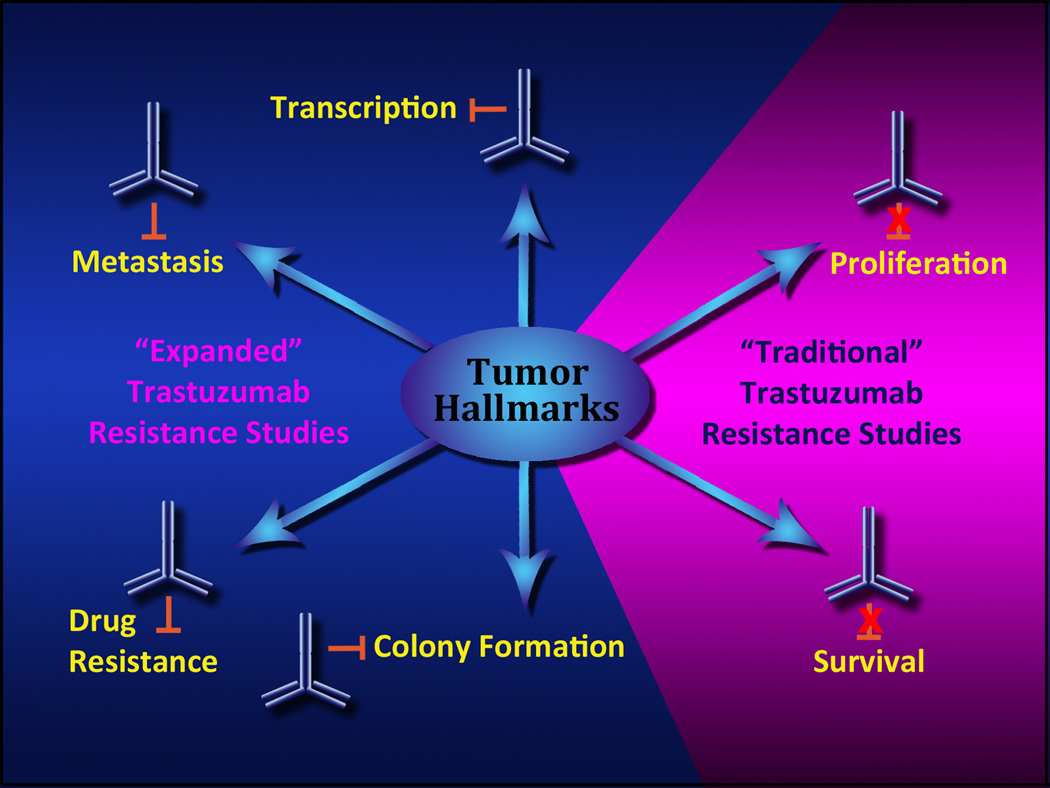
Figure 2. Models of primary trastuzumab resistance.
The majority of studies examining the action of trastuzumab (and subsequent acquired resistance to trastuzumab) have focused on studies of cell proliferation and survival (i.e., “traditional” trastuzumab resistance). As summarized in this review, emerging evidence suggests we need to consider a more expansive definition of trastuzumab action, by including diverse functional assays for biological endpoints such as migration and metastasis, anchorage dependence, patterns of gene expression, and the influence of trastuzumab on cell responses to other biologically targeted therapeutics. Multiple studies show that trastuzumab may influence all of these diverse cell phenotypes without directly influencing tumor cell proliferation or survival.
Trastuzumab mediates de novo sensitization to drugs, ligands, and radiation
Several reports have demonstrated that trastuzumab may render tumor cells sensitive to therapeutics, independent of tumor cell growth inhibition. We have demonstrated, using HER2-positive breast carcinoma cell lines not growth inhibited by trastuzumab, long-term trastuzumab treatment results in an increase in EGFR expression in three of five cell lines; all five cell lines also showed an increase in HER3 expression [73]. In this study, one cell line (MDA-MB-361) acquired sensitivity to the EGFR-targeted antibody cetuximab, and a second cell line (MDA-MB-453) acquired sensitivity to the EGFR small molecule inhibitor gefitinib. While such cell lines are not precise surrogates for the study of early stage HER2-positive breast cancer, this is the first study to demonstrate that long-term trastuzumab treatment of primary resistant cells has unambiguous effects on the tumor cell phenotype independent of this drug’s effect on cell proliferation. Subsequent studies demonstrating that primary trastuzumab resistant ovarian and lung cancer cells cultured with either trastuzumab or gefitinib (respectively), can sensitize these cells to other HER-directed therapeutics [74, 75], suggesting that this pattern of drug-induced HER-axis “reprogramming” may be a common theme in primary resistance to EGFR/HER2-directed therapeutics.
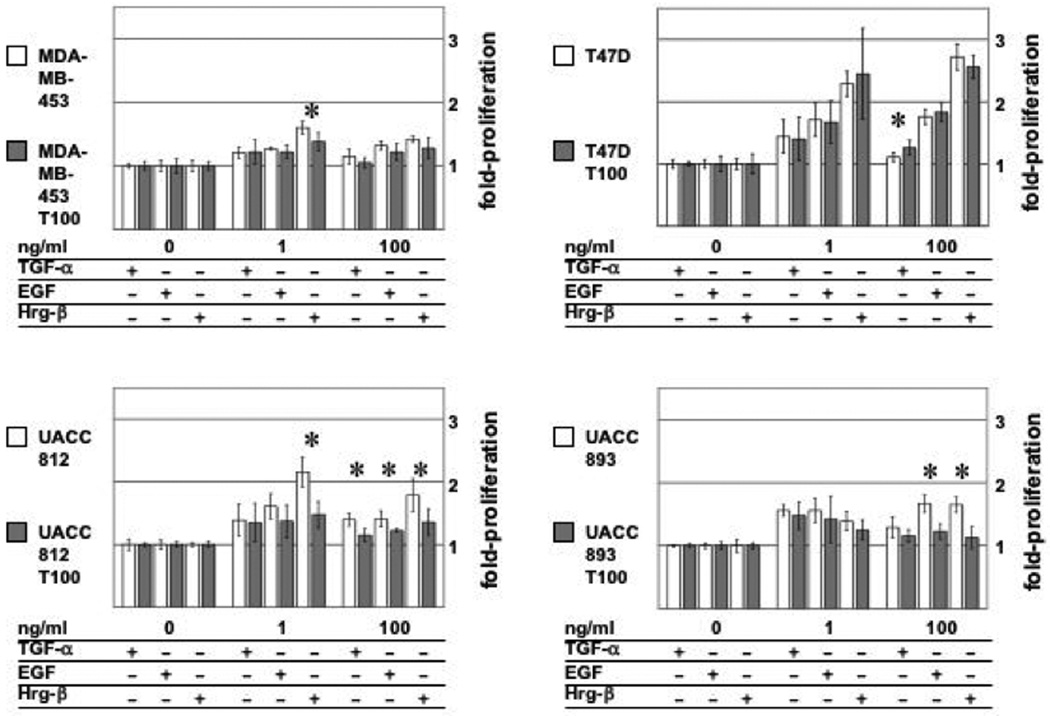
Concomitantly, long-term trastuzumab treatment also influences HER ligand-mediated cell proliferation. As shown in Figure 3, long-term treatment with trastuzumab alters both EGF and heregulin-mediated proliferative responses, despite the lack of trastuzumab cell growth inhibition in these cells. Similarly, patterns of HER ligand (EGF and heregulin)-stimulated phosphorylation of HER receptors and Akt are markedly different in gefitinib-resistant lung cancer cells cultured long-term with gefitinib [74]. This altered growth factor/ligand response phenotype may be related to changes in the expression pattern of HER receptors; the number of cell lines used in this initial study was limited, and so the results of these studies will need to be expanded and confirmed.
Figure 3. Long-term trastuzumab treatment alters proliferative response to HER family ligands in “trastuzumab-resistant” breast cancer cell lines.
The ability of HER ligands TGF-α, EGF, and Hrg-β to induce cell proliferation in long-term trastuzumab treated cells vs. parental cells was assayed using methods described previously [73]. Briefly, MDA-MB-453, T47D, UACC812, and UACC893 were cultured cells were cultured with (T100) or without 100 µg/ml trastuzumab for 12 weeks. Cells were plated in serum-free media overnight, then treated with TGF-α, EGF, or Hrg-β for 120 hours, and cell proliferation was measured by a WST-1-based colormetric assay. Fold increase in cell numbers is normalized against values determined for untreated cells. Statistical significance of cell proliferation variance between parental and T100 cells was determined by Student’s T-test. Asterisk denotes statistical variance (p<0.05) between parental and T100 cell line response to ligand.
Several reports indicate that trastuzumab induces de novo sensitization to other drugs, including sensitization of MDA-MB-453 cells to all-trans retinoic acid [76] and to the proteosome inhibitor bortezomib [77], sensitization of KPL-4 and JIMT-1 cells to an immunomodulatory oligonucleotide toll-like receptor 9 agonist [78], and as mentioned above, sensitization of JIMT-1 cells to antibodies directed against β1-integrin [71]. As some of these drugs are in various stages of clinical testing, again, the prospect of rapidly integrating this new information into the design of innovative clinical studies merits consideration.
One recent study has demonstrated synergy between trastuzumab and the anti-diabetic drug metformin. While trastuzumab alone had no effect on the formation of JIMT-1 cell mammospheres in vitro, metformin, alone or in synergy with trastuzumab, inhibited mammosphere formation [79]. Metformin has previously been demonstrated as protective for development of breast cancer [80], and a phase II trial recently has opened to evalaute the efficacy of trastuzumab plus metformin as neo-adjuvant therapy for patients with HER2-positive breast cancer [81]. As metformin exhibits pleiotropic effects on cells, the mechanism underlying metformin’s synergy with trastuzumab is unclear, but may include AMPK activation, cell cycle arrest, and/or metabolic alterations [80].
Finally, Liang et al. have shown that some trastuzumab-resistant breast cancer cells are sensitized to ionizing radiation following trastuzumab treatment [82]. Induction of apoptosis was not observed in MDA-MB-453 or MCF7-HER2 cells following trastuzumab treatment, but synergistic induction of apoptosis was noted with trastuzumab and ionizing radiation; this synergism could be duplicated using the inhibitor LY294002, suggesting that trastuzumab treatment inhibits PI3K activity in MCF7-HER2 cells. While the role of radiation therapy has been more limited in the treatment of breast cancer patients, the notion that drug priming of tumor cells prior to radiation treatment is one that has been exploited for the treatment of other adult solid tumors, and may be worth further consideration in breast cancer as well.
Trastuzumab mediates colony formation, dissemination and micrometastasis, and immune-mediated cytotoxicity
Barok et al. recently have described another prime example of a non-proliferative effect of trastuzumab treatment. Established JIMT-1 tumors in a xenograft model are not significantly growth inhibited by trastuzumab [83]. However, trastuzumab does effectively inhibit the level of circulating JIMT-1 tumor cells and distant micrometastases in this model [84], likely through an immune-mediated mechanism, which may explain the observation cited above (i.e., concurrent trastuzumab treatment inhibits both the seeding and proliferation of JIMT-1 cells a xenograft model) [83]. Similarly, KPL-4 cells, while not growth inhibited by trastuzumab in vitro, are growth inhibited by trastuzumab in a xenograft model only if trastuzumab has an intact FcγR [85]. UACC812 and UACC893 cells also are not growth inhibited by trastuzumab in vitro, but are sensitive to trastuzumab-mediated ADCC in vitro [67].
In related studies, trastuzumab has been shown to inhibit soft agar colony formation of SUM-190 and HCC-202 cells in vitro, despite the failure of this drug to inhibit the growth of these cells when assayed in two-dimensional growth conditions [65]. Conversely, at least one cell line (UACC-732) which is modestly growth inhibited by trastuzumab in two-dimensional growth conditions is not growth inhibited by trastuzumab when cultured in soft agar colony [65]. Trastuzumab also has been shown to inhibit colony formation (on tissue culture plastic) of freshly dispersed T47D cells [86], but not of cells already growing as a monolayer [73]. Remarkably, this inhibition of T47D colony formation can be reversed by induction of cyclin D1 [86]. While colony formation assays have long been used as a sensitive and stringent measure of cytotoxic activity, colony formation is also a measure of cell growth and plating (i.e., attachment) efficiency; therefore, interpretation of trastuzumab’s mechanism of action in these studies is challenging. These studies remind us of the importance of selecting in vitro assays that allow us to more precisely distinguish between the oncogenic vs. mitogenic aspects of HER mediated signal transduction [87] during all phases of drug development and testing, as well as the importance of such distinctions in defining the mechanisms of primary trastuzumab resistance.
Trastuzumab treatment induces changes in gene/protein expression and phosphorylation
One of the first indications of the complex relationship between trastuzumab sensitivity and changes in gene expression arose from studies on virally-infected tumor-derived cell lines [88]. In particular, the MCF7 cell line expresses low levels of HER2, which can be induced by treatment with estradiol, and trastuzumab can inhibit expression of estrogen receptor alpha in this cell line [89]. Epstein-Barr Virus (EBV) infected MCF7 cells express elevated levels of HER2 and HER3, and EBV-infected (or BARF0-transfected) cells acquire de novo sensitivity to trastuzumab [88]. Together, these results highlight the complex relationship between viral infection and changes in gene expression, and the impact of such changes on responsiveness to trastuzumab.
In contrast, a few studies have demonstrated that trastuzumab can induce changes in gene expression without inhibiting cell proliferation. For example, both the MDA-MB-436 and MCF7 cell lines exhibit reproducible changes in the pattern of trastuzumab-induced gene expression (as measured by cDNA microarray), including changes in expression patterns of the FK506-binding protein, interleukin 2 receptor β, MUC2, plasminogen activator, kit, and fos [90]. Similarly, in JIMT-1 cells, trastuzumab treatment does not inhibit Akt phosphorylation, which is contrary to trastuzumab’s effect on SKBR-3 cells; however, trastuzumab can down regulate Erk phosphorylation in both of these cell lines [91].
While the potential biological and clinical significance of these in vitro studies on gene expression is intriguing, there is not yet a direct link between any of these observations and their relationship to primary trastuzumab resistance. These observations do, however, further underscore the apparent disconnect between the failure of trastuzumab to inhibit tumor cell growth and the ability of this drug to induce many important phenotypic changes in the tumor cell.
Summary and conclusions
Trastuzumab is unquestionably effective as a therapeutic for some patients with “HER2-positive” breast cancer. However, the mechanisms of primary trastuzumab resistance have yet to be thoroughly characterized, and a simple constellation of the mediators of primary trastuzumab resistance has not been identified. The results of the early studies in this field, summarized here, while compelling in their demonstration that primary trastuzumab resistance can be correlated with certain patterns of gene expression, need to be interpreted with caution and also require further validation in prospective clinical trials. Patient heterogeneity, treatment of patients with combinations of trastuzumab and other drugs, small population/cohort sizes, (most studies included <50 patients), methodological differences in retrospective vs. prospective studies, tumor histological subtype (including primary vs. metastatic disease), and finally, the lack of external validation using independent patient populations, all limit our ability to extrapolate and generalize the findings from these studies across patient populations.
Despite these limitations, including the paucity of both clinically validated studies as well as adequate model systems, an important theme emerges from these studies: trastuzumab can induce important noncytostatic responses across model systems. Critically, some of these noncytostatic effects result in clinically significant phenotypic changes that may be useful in de novo sensitization of tumor cells to other drugs and/or treatment modalities. Moreover, these studies highlight the need to develop a better, comprehensive definition of resistance to trastuzumab, and perhaps to other biologically targeted drugs, that is consonant with the subtle but important phenotypic changes that may predictably be associated with exposure to biologically targeted therapeutics.
In conclusion, together, the unanticipated findings summarized here support the proposal that primary trastuzumab “resistance” should be defined using metrics beyond those restricted to conventional measures of drug inhibition of tumor cell proliferation. Recognition of these more subtle trastuzumab-induced biological phenotypes may be useful in the development of improved methods of cancer patient treatment, through the identification of common, targetable mediators of resistance that will allow us to better exploit all aspects of trastuzumab’s therapeutic potential.
Acknowledgements
Grant support: J.A. Wilken is supported by Susan G. Komen for the Cure. N.J. Maihle is supported by NIH CA 79808 and a “Senior Women in Medicine Professorship” from Yale University School of Medicine. We thank Ms. Tayf Badri for her assistance in preparation of this review, Drs. Andre T. Baron and Karin Rodland for editorial comments, and KJ Studios (www.kjstudios.com) for expert assistance with the artwork presented here.
References Cited
- 1.Allison M. The HER2 testing conundrum. Nat Biotechnol. 2010;28:117–119. doi: 10.1038/nbt0210-117. [DOI] [PubMed] [Google Scholar]
- 2.Nahta R, Hung MC, Esteva FJ. The HER-2-targeting antibodies trastuzumab and pertuzumab synergistically inhibit the survival of breast cancer cells. Cancer Res. 2004;64:2343–2346. doi: 10.1158/0008-5472.can-03-3856. [DOI] [PubMed] [Google Scholar]
- 3.Arnould L, et al. Trastuzumab-based treatment of HER2-positive breast cancer: an antibody-dependent cellular cytotoxicity mechanism? Br J Cancer. 2006;94:259–267. doi: 10.1038/sj.bjc.6602930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pegram M, et al. Inhibitory effects of combinations of HER-2/neu antibody and chemotherapeutic agents used for treatment of human breast cancers. Oncogene. 1999;18:2241–2251. doi: 10.1038/sj.onc.1202526. [DOI] [PubMed] [Google Scholar]
- 5.Slamon DJ, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 6.Pupa SM, et al. The extracellular domain of the c-erbB-2 oncoprotein is released from tumor cells by proteolytic cleavage. Oncogene. 1993;8:2917–2923. [PubMed] [Google Scholar]
- 7.Zabrecky JR, et al. The extracellular domain of p185/neu is released from the surface of human breast carcinoma cells, SK-BR-3. J Biol Chem. 1991;266:1716–1720. [PubMed] [Google Scholar]
- 8.Codony-Servat J, et al. Cleavage of the HER2 ectodomain is a pervanadate-activable process that is inhibited by the tissue inhibitor of metalloproteases-1 in breast cancer cells. Cancer Res. 1999;59:1196–1201. [PubMed] [Google Scholar]
- 9.Lafky JM, et al. Clinical implications of the ErbB/epidermal growth factor (EGF) receptor family and its ligands in ovarian cancer. Biochim Biophys Acta. 2008;1785:232–265. doi: 10.1016/j.bbcan.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Molina MA, et al. Trastuzumab (herceptin), a humanized anti-Her2 receptor monoclonal antibody, inhibits basal and activated Her2 ectodomain cleavage in breast cancer cells. Cancer research. 2001;61:4744–4749. [PubMed] [Google Scholar]
- 11.Christianson TA, et al. NH2-terminally truncated HER-2/neu protein: relationship with shedding of the extracellular domain and with prognostic factors in breast cancer. Cancer research. 1998;58:5123–5129. [PubMed] [Google Scholar]
- 12.Anido J, et al. Biosynthesis of tumorigenic HER2 C-terminal fragments by alternative initiation of translation. EMBO J. 2006;25:3234–3244. doi: 10.1038/sj.emboj.7601191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drebin JA, et al. Down-modulation of an oncogene protein product and reversion of the transformed phenotype by monoclonal antibodies. Cell. 1985;41:697–706. doi: 10.1016/s0092-8674(85)80050-7. [DOI] [PubMed] [Google Scholar]
- 14.Hurwitz E, et al. Suppression and promotion of tumor growth by monoclonal antibodies to ErbB-2 differentially correlate with cellular uptake. Proc Natl Acad Sci U S A. 1995;92:3353–3357. doi: 10.1073/pnas.92.8.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klapper LN, et al. Tumor-inhibitory antibodies to HER-2/ErbB-2 may act by recruiting c-Cbl and enhancing ubiquitination of HER-2. Cancer Res. 2000;60:3384–3388. [PubMed] [Google Scholar]
- 16.Baselga J, Swain SM. Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nat Rev Cancer. 2009;9:463–475. doi: 10.1038/nrc2656. [DOI] [PubMed] [Google Scholar]
- 17.Spector NL, Blackwell KL. Understanding the mechanisms behind trastuzumab therapy for human epidermal growth factor receptor 2-positive breast cancer. J Clin Oncol. 2009;27:5838–5847. doi: 10.1200/JCO.2009.22.1507. [DOI] [PubMed] [Google Scholar]
- 18.Pohlmann PR, Mayer IA, Mernaugh R. Resistance to Trastuzumab in Breast Cancer. Clin Cancer Res. 2009;15:7479–7491. doi: 10.1158/1078-0432.CCR-09-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris LN, et al. Predictors of resistance to preoperative trastuzumab and vinorelbine for HER2-positive early breast cancer. Clin Cancer Res. 2007;13:1198–1207. doi: 10.1158/1078-0432.CCR-06-1304. [DOI] [PubMed] [Google Scholar]
- 20.Wolff AC, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25:118–145. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 21.Romond EH, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. The New England journal of medicine. 2005;353:1673–1684. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 22.Doherty JK, et al. The HER-2/neu receptor tyrosine kinase gene encodes a secreted autoinhibitor. Proc Natl Acad Sci U S A. 1999;96:10869–10874. doi: 10.1073/pnas.96.19.10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scott GK, et al. A truncated intracellular HER2/neu receptor produced by alternative RNA processing affects growth of human carcinoma cells. Mol Cell Biol. 1993;13:2247–2257. doi: 10.1128/mcb.13.4.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pedersen K, et al. A naturally occurring HER2 carboxy-terminal fragment promotes mammary tumor growth and metastasis. Mol Cell Biol. 2009;29:3319–3331. doi: 10.1128/MCB.01803-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scaltriti M, et al. Expression of p95HER2, a truncated form of the HER2 receptor, and response to anti-HER2 therapies in breast cancer. J Natl Cancer Inst. 2007;99:628–638. doi: 10.1093/jnci/djk134. [DOI] [PubMed] [Google Scholar]
- 26.Molina MA, et al. NH(2)-terminal truncated HER-2 protein but not full-length receptor is associated with nodal metastasis in human breast cancer. Clin Cancer Res. 2002;8:347–353. [PubMed] [Google Scholar]
- 27.Saez R, et al. p95HER-2 predicts worse outcome in patients with HER-2-positive breast cancer. Clin Cancer Res. 2006;12:424–431. doi: 10.1158/1078-0432.CCR-05-1807. [DOI] [PubMed] [Google Scholar]
- 28.Sperinde J, Jin X, Banerjee J. Quantitation of p95HER2 in paraffin sections using a p95-specific antibody and correlation with outcome in a cohort of trastuzumab-treated breast cancer patients. Clin Cancer Res. 2010 doi: 10.1158/1078-0432.CCR-10-0410. In press. [DOI] [PubMed] [Google Scholar]
- 29.Brodowicz T, et al. Soluble HER-2/neu neutralizes biologic effects of anti-HER-2/neu antibody on breast cancer cells in vitro. Int J Cancer. 1997;73:875–879. doi: 10.1002/(sici)1097-0215(19971210)73:6<875::aid-ijc19>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 30.Ghedini GC, et al. Shed HER2 extracellular domain in HER2-mediated tumor growth and in Trastuzumab susceptibility. J Cell Physiol. 2010 doi: 10.1002/jcp.22257. [DOI] [PubMed] [Google Scholar]
- 31.Ali SM, et al. Serum HER-2/neu and relative resistance to trastuzumab-based therapy in patients with metastatic breast cancer. Cancer. 2008;113:1294–1301. doi: 10.1002/cncr.23689. [DOI] [PubMed] [Google Scholar]
- 32.Lennon S, et al. Utility of serum HER2 extracellular domain assessment in clinical decision making: pooled analysis of four trials of trastuzumab in metastatic breast cancer. J Clin Oncol. 2009;27:1685–1693. doi: 10.1200/JCO.2008.16.8351. [DOI] [PubMed] [Google Scholar]
- 33.Arnould L, et al. Pathologic complete response to trastuzumab-based neoadjuvant therapy is related to the level of HER-2 amplification. Clin Cancer Res. 2007;13:6404–6409. doi: 10.1158/1078-0432.CCR-06-3022. [DOI] [PubMed] [Google Scholar]
- 34.Ginestier C, et al. ERBB2 phosphorylation and trastuzumab sensitivity of breast cancer cell lines. Oncogene. 2007;26:7163–7169. doi: 10.1038/sj.onc.1210528. [DOI] [PubMed] [Google Scholar]
- 35.Weinstein IB. Cancer. Addiction to oncogenes--the Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 36.Narayan M, et al. Trastuzumab-induced HER reprogramming in "resistant" breast carcinoma cells. Cancer Res. 2009 doi: 10.1158/0008-5472.CAN-08-1056. In Press. [DOI] [PubMed] [Google Scholar]
- 37.Dua R, et al. EGFR over-expression and activation in high HER2, ER negative breast cancer cell line induces trastuzumab resistance. Breast cancer research and treatment. 2009 doi: 10.1007/s10549-009-0592-x. [DOI] [PubMed] [Google Scholar]
- 38.Yotsumoto F, et al. HB-EGF orchestrates the complex signals involved in triple-negative and trastuzumab-resistant breast cancer. Int J Cancer. 2010 doi: 10.1002/ijc.25472. [DOI] [PubMed] [Google Scholar]
- 39.Yonemori K, et al. Immunohistochemical expression of HER1, HER3, and HER4 in HER2-positive breast cancer patients treated with trastuzumab-containing neoadjuvant chemotherapy. J Surg Oncol. 2010;101:222–227. doi: 10.1002/jso.21486. [DOI] [PubMed] [Google Scholar]
- 40.Jerome L, et al. Recombinant human insulin-like growth factor binding protein 3 inhibits growth of human epidermal growth factor receptor-2-overexpressing breast tumors and potentiates herceptin activity in vivo. Cancer Res. 2006;66:7245–7252. doi: 10.1158/0008-5472.CAN-05-3555. [DOI] [PubMed] [Google Scholar]
- 41.Lu Y, et al. Insulin-like growth factor-I receptor signaling and resistance to trastuzumab (Herceptin) J Natl Cancer Inst. 2001;93:1852–1857. doi: 10.1093/jnci/93.24.1852. [DOI] [PubMed] [Google Scholar]
- 42.Vegran F, et al. Gene expression profile and response to trastuzumab-docetaxel-based treatment in breast carcinoma. Br J Cancer. 2009;101:1357–1364. doi: 10.1038/sj.bjc.6605310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhuang G, et al. Elevation of receptor tyrosine kinase EphA2 mediates resistance to trastuzumab therapy. Cancer research. 2010;70:299–308. doi: 10.1158/0008-5472.CAN-09-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin KJ, et al. Prognostic breast cancer signature identified from 3D culture model accurately predicts clinical outcome across independent datasets. PLoS One. 2008;3:e2994. doi: 10.1371/journal.pone.0002994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fournier MV, et al. Gene expression signature in organized and growth-arrested mammary acini predicts good outcome in breast cancer. Cancer Res. 2006;66:7095–7102. doi: 10.1158/0008-5472.CAN-06-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cavalloni G, et al. Granulocyte-colony stimulating factor upregulates ErbB2 expression on breast cancer cell lines and converts primary resistance to trastuzumab. Anti-cancer drugs. 2008;19:689–696. doi: 10.1097/CAD.0b013e3283050083. [DOI] [PubMed] [Google Scholar]
- 47.Mocanu MM, et al. Associations of ErbB2, beta1-integrin and lipid rafts on Herceptin (Trastuzumab) resistant and sensitive tumor cell lines. Cancer letters. 2005;227:201–212. doi: 10.1016/j.canlet.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 48.Palyi-Krekk Z, et al. Hyaluronan-induced masking of ErbB2 and CD44-enhanced trastuzumab internalisation in trastuzumab resistant breast cancer. Eur J Cancer. 2007;43:2423–2433. doi: 10.1016/j.ejca.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 49.Price-Schiavi SA, et al. Rat Muc4 (sialomucin complex) reduces binding of anti-ErbB2 antibodies to tumor cell surfaces, a potential mechanism for herceptin resistance. International journal of cancer. 2002;99:783–791. doi: 10.1002/ijc.10410. [DOI] [PubMed] [Google Scholar]
- 50.Nagy P, et al. Decreased accessibility and lack of activation of ErbB2 in JIMT-1, a herceptin-resistant, MUC4-expressing breast cancer cell line. Cancer research. 2005;65:473–482. [PubMed] [Google Scholar]
- 51.Fessler SP, et al. MUC1* is a determinant of trastuzumab (Herceptin) resistance in breast cancer cells. Breast cancer research and treatment. 2009;118:113–124. doi: 10.1007/s10549-009-0412-3. [DOI] [PubMed] [Google Scholar]
- 52.Hikita ST, et al. MUC1* mediates the growth of human pluripotent stem cells. PLoS ONE. 2008;3:e3312. doi: 10.1371/journal.pone.0003312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mahanta S, et al. A minimal fragment of MUC1 mediates growth of cancer cells. PLoS ONE. 2008;3:e2054. doi: 10.1371/journal.pone.0002054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kulkarni S, et al. Calpain regulates sensitivity to trastuzumab and survival in HER2-positive breast cancer. Oncogene. 2009 doi: 10.1038/onc.2009.422. [DOI] [PubMed] [Google Scholar]
- 55.Carragher NO, Frame MC. Calpain: a role in cell transformation and migration. Int J Biochem Cell Biol. 2002;34:1539–1543. doi: 10.1016/s1357-2725(02)00069-9. [DOI] [PubMed] [Google Scholar]
- 56.Citri A, Kochupurakkal BS, Yarden Y. The achilles heel of ErbB-2/HER2: regulation by the Hsp90 chaperone machine and potential for pharmacological intervention. Cell Cycle. 2004;3:51–60. [PubMed] [Google Scholar]
- 57.Chandarlapaty S, et al. Inhibitors of HSP90 block p95-HER2 signaling in Trastuzumab-resistant tumors and suppress their growth. Oncogene. 2010;29:325–334. doi: 10.1038/onc.2009.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leow CC, et al. Antitumor efficacy of IPI-504, a selective heat shock protein 90 inhibitor against human epidermal growth factor receptor 2-positive human xenograft models as a single agent and in combination with trastuzumab or lapatinib. Molecular cancer therapeutics. 2009;8:2131–2141. doi: 10.1158/1535-7163.MCT-08-1038. [DOI] [PubMed] [Google Scholar]
- 59.Hamel S, et al. Both t-Darpp and DARPP-32 can cause resistance to trastuzumab in breast cancer cells and are frequently expressed in primary breast cancers. Breast Cancer Res Treat. 2010;120:47–57. doi: 10.1007/s10549-009-0364-7. [DOI] [PubMed] [Google Scholar]
- 60.Belkhiri A, et al. Expression of t-DARPP mediates trastuzumab resistance in breast cancer cells. Clin Cancer Res. 2008;14:4564–4571. doi: 10.1158/1078-0432.CCR-08-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yonemori K, et al. Immunohistochemical expression of PTEN and phosphorylated Akt are not correlated with clinical outcome in breast cancer patients treated with trastuzumab-containing neo-adjuvant chemotherapy. Medical oncology (Northwood, London, England) 2008 doi: 10.1007/s12032-008-9127-2. [DOI] [PubMed] [Google Scholar]
- 62.Nagata Y, et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer cell. 2004;6:117–127. doi: 10.1016/j.ccr.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 63.Migliaccio I, et al. PI3 kinase activation and response to trastuzumab or lapatinib in HER-2 overexpressing locally advanced breast cancer (LABC). San Antonio Breast Cancer Symposium; San Antonio. 2008. Abstract 34. [Google Scholar]
- 64.Gori S, et al. EGFR, pMAPK, pAkt and PTEN status by immunohistochemistry: correlation with clinical outcome in HER2-positive metastatic breast cancer patients treated with trastuzumab. Ann Oncol. 2009;20:648–654. doi: 10.1093/annonc/mdn681. [DOI] [PubMed] [Google Scholar]
- 65.O'Brien NA, et al. Activated phosphoinositide 3-kinase/AKT signaling confers resistance to trastuzumab but not lapatinib. Mol Cancer Ther. 2010;9:1489–1502. doi: 10.1158/1535-7163.MCT-09-1171. [DOI] [PubMed] [Google Scholar]
- 66.Kataoka Y, et al. Association between gain-of-function mutations in PIK3CA and resistance to HER2-targeted agents in HER2-amplified breast cancer cell lines. Ann Oncol. 2010;21:255–262. doi: 10.1093/annonc/mdp304. [DOI] [PubMed] [Google Scholar]
- 67.Koninki K, et al. Multiple molecular mechanisms underlying trastuzumab and lapatinib resistance in JIMT-1 breast cancer cells. Cancer Lett. 2010;294:211–219. doi: 10.1016/j.canlet.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 68.Gu L, et al. Protein kinase A activation confers resistance to trastuzumab in human breast cancer cell lines. Clin Cancer Res. 2009;15:7196–7206. doi: 10.1158/1078-0432.CCR-09-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cheng Y, et al. Cytoprotective effect of the elongation factor-2 kinase-mediated autophagy in breast cancer cells subjected to growth factor inhibition. PLoS One. 2010;5:e9715. doi: 10.1371/journal.pone.0009715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weigelt B, et al. HER2 signaling pathway activation and response of breast cancer cells to HER2-targeting agents is dependent strongly on the 3D microenvironment. Breast cancer research and treatment. 2009 doi: 10.1007/s10549-009-0502-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lesniak D, et al. Beta1-integrin circumvents the antiproliferative effects of trastuzumab in human epidermal growth factor receptor-2-positive breast cancer. Cancer research. 2009;69:8620–8628. doi: 10.1158/0008-5472.CAN-09-1591. [DOI] [PubMed] [Google Scholar]
- 72.Delord JP, et al. Trastuzumab induced in vivo tissue remodelling associated in vitro with inhibition of the active forms of AKT and PTEN and RhoB induction in an ovarian carcinoma model. British journal of cancer. 2010;103:61–72. doi: 10.1038/sj.bjc.6605699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Narayan M, et al. Trastuzumab-induced HER reprogramming in "resistant" breast carcinoma cells. Cancer research. 2009;69:2191–2194. doi: 10.1158/0008-5472.CAN-08-1056. [DOI] [PubMed] [Google Scholar]
- 74.Jain A, et al. HER kinase axis receptor dimer partner switching occurs in response to EGFR tyrosine kinase inhibition despite failure to block cellular proliferation. Cancer Res. 2010;70:1989–1999. doi: 10.1158/0008-5472.CAN-09-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wilken JA, Webster KT, Maihle NJ. Trastuzumab Sensitizes Ovarian Cancer Cells to EGFR-targeted Therapeutics. J Ovarian Res. 2010;3:7. doi: 10.1186/1757-2215-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tari AM, et al. Her2/neu induces all-trans retinoic acid (ATRA) resistance in breast cancer cells. Oncogene. 2002;21:5224–5232. doi: 10.1038/sj.onc.1205660. [DOI] [PubMed] [Google Scholar]
- 77.Cardoso F, et al. Bortezomib (PS-341, Velcade) increases the efficacy of trastuzumab (Herceptin) in HER-2-positive breast cancer cells in a synergistic manner. Molecular cancer therapeutics. 2006;5:3042–3051. doi: 10.1158/1535-7163.MCT-06-0104. [DOI] [PubMed] [Google Scholar]
- 78.Damiano V, et al. A novel toll-like receptor 9 agonist cooperates with trastuzumab in trastuzumab-resistant breast tumors through multiple mechanisms of action. Clin Cancer Res. 2009;15:6921–6930. doi: 10.1158/1078-0432.CCR-09-1599. [DOI] [PubMed] [Google Scholar]
- 79.Vazquez-Martin A, et al. The anti-diabetic drug metformin suppresses self-renewal and proliferation of trastuzumab-resistant tumor-initiating breast cancer stem cells. Breast Cancer Res Treat. 2010 doi: 10.1007/s10549-010-0924-x. [DOI] [PubMed] [Google Scholar]
- 80.Papanas N, Maltezos E, Mikhailidis DP. Metformin and cancer: licence to heal? Expert Opin Investig Drugs. 2010 doi: 10.1517/13543784.2010.499122. [DOI] [PubMed] [Google Scholar]
- 81.Martin-Castillo B, et al. Incorporating the antidiabetic drug metformin in HER2-positive breast cancer treated with neo-adjuvant chemotherapy and trastuzumab: an ongoing clinical-translational research experience at the Catalan Institute of Oncology. Ann Oncol. 2010;21:187–189. doi: 10.1093/annonc/mdp494. [DOI] [PubMed] [Google Scholar]
- 82.Liang K, et al. Sensitization of breast cancer cells to radiation by trastuzumab. Molecular cancer therapeutics. 2003;2:1113–1120. [PubMed] [Google Scholar]
- 83.Barok M, et al. Trastuzumab causes antibody-dependent cellular cytotoxicity-mediated growth inhibition of submacroscopic JIMT-1 breast cancer xenografts despite intrinsic drug resistance. Molecular cancer therapeutics. 2007;6:2065–2072. doi: 10.1158/1535-7163.MCT-06-0766. [DOI] [PubMed] [Google Scholar]
- 84.Barok M, et al. Trastuzumab decreases the number of circulating and disseminated tumor cells despite trastuzumab resistance of the primary tumor. Cancer letters. 2008;260:198–208. doi: 10.1016/j.canlet.2007.10.043. [DOI] [PubMed] [Google Scholar]
- 85.Junttila TT, et al. Superior in vivo efficacy of afucosylated trastuzumab in the treatment of HER2-amplified breast cancer. Cancer Res. 2010;70:4481–4489. doi: 10.1158/0008-5472.CAN-09-3704. [DOI] [PubMed] [Google Scholar]
- 86.Wu K, et al. Flavopiridol and trastuzumab synergistically inhibit proliferation of breast cancer cells: association with selective cooperative inhibition of cyclin D1-dependent kinase and Akt signaling pathways. Molecular cancer therapeutics. 2002;1:695–706. [PubMed] [Google Scholar]
- 87.Boerner JL, Danielsen A, Maihle NJ. Ligand-independent oncogenic signaling by the epidermal growth factor receptor: v-ErbB as a paradigm. Experimental cell research. 2003;284:111–121. doi: 10.1016/s0014-4827(02)00096-4. [DOI] [PubMed] [Google Scholar]
- 88.Lin JH, et al. Dysregulation of HER2/HER3 signaling axis in Epstein-Barr virus-infected breast carcinoma cells. J Virol. 2007;81:5705–5713. doi: 10.1128/JVI.00076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chang CJ, et al. Modulation of HER2 expression by ferulic acid on human breast cancer MCF7 cells. Eur J Clin Invest. 2006;36:588–596. doi: 10.1111/j.1365-2362.2006.01676.x. [DOI] [PubMed] [Google Scholar]
- 90.Kauraniemi P, et al. Effects of Herceptin treatment on global gene expression patterns in HER2-amplified and nonamplified breast cancer cell lines. Oncogene. 2004;23:1010–1013. doi: 10.1038/sj.onc.1207200. [DOI] [PubMed] [Google Scholar]
- 91.Tanner M, et al. Characterization of a novel cell line established from a patient with Herceptin-resistant breast cancer. Molecular cancer therapeutics. 2004;3:1585–1592. [PubMed] [Google Scholar]