Abstract
The ALK kinase inhibitor crizotinib (PF-02341066) is clinically effective in patients with ALK-translocated cancers, but its efficacy will ultimately be limited by acquired drug resistance. Here we report the identification of a secondary mutation in ALK, F1174L, as one cause of crizotinib resistance in a patient with an inflammatory myofibroblastic tumor (IMT) harbouring a RANBP2-ALK translocation who progressed while crizotinib therapy. When present in cis with an ALK translocation, this mutation (also detected in neuroblastomas) causes an increase in ALK phosphorylation, cell growth and downstream signaling. Furthermore, the F1174L mutation inhibits crizotinib mediated downregulation of ALK signaling and blocks apoptosis in RANBP2-ALK Ba/F3 cells. A chemically distinct ALK inhibitor, TAE684, or the HSP90 inhibitor 17-AAG are both effective in models harbouring the F1174L ALK mutation. Our findings highlight the importance of studying drug resistance mechanisms in order to develop effective clinical treatments for patients with ALK-translocated cancers.
Keywords: Inflammatory myofibroblastic tumor, Anaplastic lymphoma kinase, kinase inhibitor, drug resistance
INTRODUCTION
Chromosomal translocations involving the anaplastic lymphoma kinase (ALK) have been detected in several human malignancies including in anaplastic large cell lymphomas (ALCLs), inflammatory myofibroblastic tumors (IMTs), and non-small cell lung carcinomas (NSCLCs) (1-3). ALK translocated fusion proteins include the entire ALK kinase domain, lead to constitutive ALK kinase activity and oncogenic transformation both in vitro and in vivo (3). Somatic mutations in the ALK kinase domain, including at position F1174L, have been detected in neuroblastomas and are also transforming in vitro and in vivo (4, 5). These findings have led to pre-clinical and clinical development of ALK kinase inhibitors including PF-2341066 (crizotinib) (6). ALK kinase inhibitors lead to apoptosis in vitro and tumor shrinkage in mouse models of EML4-ALK NSCLC (7, 8). To date, significant clinical activity, including tumor shrinkage in 60% of patients, has been observed in a phase I trial of crizotinib in EML4-ALK NSCLC (9). Furthermore, clinical efficacy of crizotinib has been observed in an IMT patient harbouring an ALK translocation (10). However, despite these dramatic effects, as with other kinase inhibitors, drug resistance (herein termed acquired resistance) to ALK kinase inhibitors is likely to emerge. An understanding of the acquired resistance mechanisms will be important for the development of additional ALK kinase inhibitors and/or combination therapeutic strategies.
METHODS
Patients
The patients were treated in a clinical trial that was sponsored by Pfizer, Inc. Tumor biopsies were obtained from under an IRB approved protocol. Both patients provided written informed consent. Total RNA was isolated using Trizol™ (Invitrogen, Carlsbad, CA) and purified using RNeasy™ minielute cleanup kit (Qiagen,Valencia, CA).
ALK genomic analyses
The ALK kinase domain was sequenced from all of the available specimens. Exon 23 of ALK was amplified from DNA using exon specific primers, PCR products cloned into a TOPOTA vector (Invitrogen, Carlsbad, CA), transformed into bacteria and individual clones sequenced. The PCR primers and conditions are available upon request. ALK fluorescence in situ hybridization (FISH) was performed using the break apart probes (Vysis LSI ALK Dual Color, Abbott Molecular, Des Plaines, IL) as previously described (11).
Expression Constructs and cell culture
The full length RANBP2-ALK cDNA from patient A and the EML4-ALK (Variant 1) cDNA from the H3122 cell line were cloned into pDNR-Dual (BD Biosciences). The F1174L mutation was introduced using site-directed mutagenesis (Stratagene) with mutant specific primers according to the manufacturer’s instructions and as previously described (12). All constructs were confirmed by DNA sequencing. Retroviral infection and culture of Ba/F3 cell were performed using previously described methods (12). Polyclonal cell lines were established by puromycin selection and subsequently cultured in the absence of interleukin-3 (IL-3). Uninfected Ba/F3 cells or those expressing EGFR delE746_A750 or the JP1536 empty vector were used as controls.
Antibodies and Western Blotting
Cell lysis, Western blotting and immunoblotting was performed as previously described (12). Anti-phospho-ALK (DF53), anti-phospho-Akt (Ser-473) and anti-total-Akt, were obtained from Cell Signaling Technology. Total ERK1/2 and phospho-ERK1/2 (pT185/pY187) antibodies were from Invitrogen (Carlsbad, CA). Immunoprecipitations were performed using anti-Flag-M2 agarose (Sigma-Aldrich Co.;St. Louis, MO). ALK immunohistochemistry (IHC) was performed using the mouse monoclonal anti-human CD246 (clone: ALK1, DAKO USA, Carpinteria, CA) as previously described (11).
Cell proliferation and growth assays
Crizotinib was provided by Pfizer. TAE684 was synthesized as previously described (7). Growth and inhibition of growth was assessed by MTS assay according to previously established methods (12). All experimental points were set up in six to twelve wells and all experiments were repeated at least three times.
RESULTS AND DISCUSSION
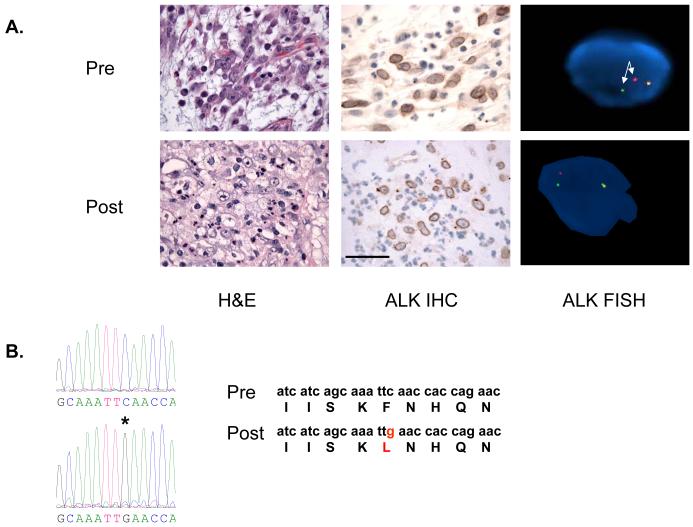
We identified 2 patients with ALK translocated cancers that developed clinical acquired resistance to crizotinib. Patient A, with IMT, achieved a partial response with crizotinib therapy lasting 8 months but subsequently developed re-growth of several tumor lesions and had these surgically removed (Table S1) (10). Both pre- and post-treatment tumor specimens had evidence of viable tumor, expressed ALK by IHC and contained an ALK translocation (Figure 1A). There was no evidence of ALK amplification in the post-treatment tumor (data not shown). This patient’s tumor was known to harbour the RANBP2-ALK translocation (10). Sequencing of the entire ALK kinase domain demonstrated that one of the clinically progressing tumor lesions contained a F1174L mutation (Figure 1B). This was not detected by direct sequencing, or by cloning and sequencing of individual clones, in the 2 other progressing lesions, or by direct sequencing in 2 other tumours that were clinically stable at the time of surgery (Table S1). Furthermore it was not detected in the pre-treatment tumor specimen even by cloning and sequencing of individual clones (Table S1). Patient B, with EML4-ALK NSCLC, achieved a partial response with crizotinib treatment but developed acquired resistance following 5 months of therapy. At the time of progression a liver biopsy of a growing lesion was performed (Table S1). EML4-ALK variant 1 was identified by RT-PCR but no secondary mutations in ALK were detected in the acquired resistance tumor specimen.
Figure 1. Tumor from crizotinib resistant patient contains a secondary ALK mutation.
A. Comparison of pre-treatment and post-treatment biopsy specimens. Both specimens contain viable tumor and both tumors express ALK by IHC which is localized to the nuclear membrane. FISH analyses demonstrate an ALK translocation (split red and green signals; arrows). Scale bar 50 μM. B. Sequence tracing from pre and post-treatment tumor specimens. There is a C to G mutation (asterix) in codon 3522 in exon 23 resulting in the F1174L mutation. This is not detected in the pre-treatment tumor.
We next evaluated the biologic impact of the F1174L mutation. Both RANPB2-ALK and RANBP-ALK F1174L led to IL-3 independent growth of Ba/F3 cells (Figure 2A) but the growth was faster in the presence of the F1174L mutation. This increased growth rate was mirrored by a greater baseline ALK phosphorylation of RANBP2-ALK F1174L compared to RANBP2-ALK (Figure 2B) and by increased downstream AKT and ERK 1/2 phosphorylation (Figure 2C). The RANB2-ALK F1174L cells were significantly more resistant to crizotinib (Figure 2D) and the F1174L mutation diminished crizotinib mediated inhibition of ALK signalling and blocked apoptosis (Figures S1A and B). We also introduced the F1174L mutation into the background of EML4-ALK found in NSCLC(3). Similar to RANPB2-ALK, the EML4-ALK F1174L Ba/F3 cells grew faster (Figure 3A), had a greater baseline ALK phosphorylation (Figure 3B) and were more resistant to crizotinib growth inhibition than EML4-ALK Ba/F3 cells (Figure 3C). Consistent with these findings on growth, greater concentrations of crizotinib were required to inhibit ALK phosphorylation in the EML4-ALK F1174L cells compared to those with EML4-ALK alone (Figure 3B). Collectively, our studies demonstrate that the F1174L mutation imparts both biologic and drug resistance properties to cancers harbouring ALK translocations. Furthermore, patients with neuroblastoma harbouring the F1174L ALK mutation may have a transient or no clinical benefit from crizotinib treatment using the current dosing schedules (13).
Figure 2. Impact of F1174L on growth and signaling in Ba/F3 cells harboring RANBP2-ALK.
A. IL-3 independent Ba/F3 cells expressing RANBP2-ALK F1174L proliferate faster compared to cells expressing RANBP2-ALK. * p < 0.05; ** p < 0.001 B. Ba/F3 cells with indicated genotypes were treated with increasing concentrations of crizotinib for 6 hours. Cell extracts were immunoprecipitated with an anti-FLAG antibody followed immunoblotting to detect the indicated proteins. C. Presence of F1174L mutation in the background of an ALK translocation leads to enhanced AKT and ERK 1/2 signaling. Cell extracts were immunoblotted to detect the indicated proteins. D. Ba/F3 cells were treated with crizotinib at the indicated concentrations, and viable cells were measured after 72 hours of treatment and plotted relative to untreated controls. There is a significant effect of the F1174L mutation at 300 nM (p < 0.001).
Figure 3. Impact of the F1174L mutation on growth and signaling in Ba/F3 cells expressing EML4-ALK.
A. IL-3 independent Ba/F3 cells expressing EML4-ALK F1174L proliferate faster compared to cells expressing EML4-ALK. ** p < 0.001 B. EML4-ALK and EML4-ALK F1174L Ba/F3 cells were treated with increasing concentrations of PF-02341066 for 6 hours. Cell extracts were immunoprecipitated with an anti-FLAG antibody followed immunoblotting to detect the indicated proteins. C. Ba/F3 cells were treated with crizotinib at the indicated concentrations, and viable cells were measured after 72 hours of treatment and plotted relative to untreated controls. There is a significant effect of the F1174L mutation at 300 nM (p < 0.001).
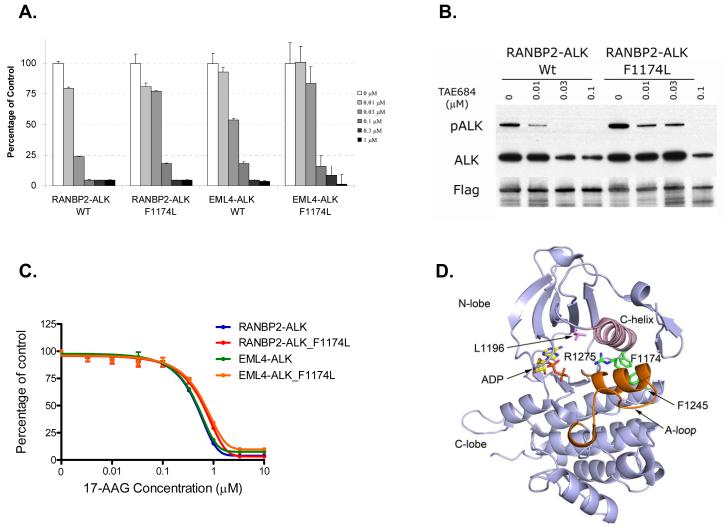
Crizotinib is administered continuously daily (250 mg BID), reaching a median through plasma concentration of 57 nM of free drug, and is clinically effective at this dosing in ALK rearranged IMT and NSCLC (9, 10, 13). Our preclinical studies suggest that higher doses of crizotinib could be used to overcome the F1174L mediated resistance mechanism (Figure 2D). This could potentially be achieved using intermittent administration of higher doses of crizotinib, to achieve a higher Cmax, sufficient to inhibit ALK phosphorylation in the presence of F1174L. Similarly, some imatinib resistance mutations (including F359V, M244V, Q252H and E355G), many of which effect the conformational change in ABL, cause a relative drug resistance which can be overcome by higher drug doses in vitro and in some cases clinically by using higher doses of imatinib (14, 15). In order to develop additional therapeutic strategies we evaluated both a structurally unrelated ALK kinase inhibitor, TAE684, and the heat shock protein (HSP) 90 inhibitor 17-AAG in the crizotinib resistant models (7, 16). Although the F1174L mutation slightly increased the IC50 for TAE684 against RANBP-ALK Ba/F3 cells (59 nM with F1174L; 22 nM without), the IC50 was still substantially below the concentrations of crizotinib (IC50 200 nM) required to inhibit growth and ALK phosphorylation in the parental RANBP-ALK Ba/F3 cells (Figures 2B, 2D and 4A). Similar findings were observed for EML4-ALK Ba/F3 cells (Figure 4A and data not shown). The effects of TAE684 on growth were also mirrored at the level of ALK phosphorylation (Figure 4B). Recent clinical studies have identified anti-tumor activity of the HSP90 inhibitor IPI-504 in ALK translocated NSCLC (17). We thus evaluated the effects of the HSP90 inhibitor 17-AAG in models harbouring the F1174L mutation (Figure 4C). Ba/F3 cells with and without F1174L were equally sensitive to 17-AAG in vitro (Figure 4C). As many ALK inhibitors are in pre-clinical development and several HSP90 inhibitors currently undergoing clinical development, our findings provide direct therapeutic strategies for patients that develop crizotinib resistance.
Figure 4. Therapeutic strategies against cancers harboring the F1174L crizotinib resistance mutation.
A. Ba/F3 cells were treated with TAE684 at the indicated concentrations, and viable cells were measured after 72 hours of treatment and plotted relative to untreated controls. B. RANBP2-ALK and RANBP2-ALK F1174L Ba/F3 cells were treated with increasing concentrations of TAE684 for 6 hours. Cell extracts were immunoprecipitated with an anti-FLAG antibody followed immunoblotting to detect the indicated proteins. C. IL-3 independent Ba/F3 cells harboring ALK translocations with or without the F1174L mutation are equally sensitive to the HSP90 inhibitor 17-AAG. Viable cells were measured after 72 hours of treatment and plotted relative to untreated controls. D. Ribbon diagram depicting the crystal structure of ALK kinase in the inactive conformation in complex with ADP. The sidechains of F1174 and selected other neuroblastoma mutations are shown in green, and the gatekeeper residue L1196 is shown in magenta. Note that F1174 is not in contact with the ATP-binding cleft. Interestingly, these neuroblastoma mutations cluster along a helix formed in the activation loop (A-loop, colored orange) in this inactive structure. The mutations may destabilize this helix to promote the active conformation. They may also affect the position of the C-helix (pink), which is known to play a key regulatory role in some kinases. Figure is drawn from PDB ID 3LCT (18).
The structural basis for the crizotinib resistance of the F1174L mutation is not entirely clear. Examination of the recently published crystal structure of ALK in an inactive conformation reveals that the F1174L mutation is not in direct contact with the ATP-binding pocket, where both crizotinib and TAE684 are expected to bind (Figure 4D) (18, 19). Thus the F1174L mutation is unlikely to confer resistance via direct steric interactions. Crizotinib is known to bind an inactive conformation of MET (20) and a recently released crystal structure in complex with ALK indicates that it binds a distinct inactive conformation in ALK (PDB ID 2XP2). The activating F1174L mutation must promote the active conformation of the kinase, and therefore may disfavor binding of crizotinib analogous to some imatinib resistance mutations in ABL (14). A more definitive understanding of the mechanism of resistance, and the differential effect of the mutation on crizotinib versus TAE684, will require detailed binding and structural studies of these inhibitors with the F1174L mutant.
The current study highlights the need to study drug resistance mechanisms from cancer patients treated with kinase inhibitors. The molecular, cellular and structural understanding of drug resistance mechanisms will continue to reveal therapeutic insights for the development of future anti-cancer therapies.
Supplementary Material
Acknowledgements
This study was supported by NIH R01CA136851 (N.S.G. and P.A.J.), R01CA135257 (P.A.J.) and by the NCI Lung SPORE P50CA090578 (P.A.J. and G.I.S.).
References
- 1.Morris SW, Kirstein MN, Valentine MB, et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science. 1994;263:1281–4. doi: 10.1126/science.8122112. [DOI] [PubMed] [Google Scholar]
- 2.Lawrence B, Perez-Atayde A, Hibbard MK, et al. TPM3-ALK and TPM4-ALK oncogenes in inflammatory myofibroblastic tumors. Am J Pathol. 2000;157:377–84. doi: 10.1016/S0002-9440(10)64550-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–67. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 4.Chen Y, Takita J, Choi YL, et al. Oncogenic mutations of ALK kinase in neuroblastoma. Nature. 2008;455:971–4. doi: 10.1038/nature07399. [DOI] [PubMed] [Google Scholar]
- 5.George RE, Sanda T, Hanna M, et al. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature. 2008;455:975–8. doi: 10.1038/nature07397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen JG, Zou HY, Arango ME, et al. Cytoreductive antitumor activity of PF-2341066, a novel inhibitor of anaplastic lymphoma kinase and c-Met, in experimental models of anaplastic large-cell lymphoma. Mol Cancer Ther. 2007;6:3314–22. doi: 10.1158/1535-7163.MCT-07-0365. [DOI] [PubMed] [Google Scholar]
- 7.Koivunen JP, Mermel C, Zejnullahu K, et al. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res. 2008;14:4275–83. doi: 10.1158/1078-0432.CCR-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soda M, Takada S, Takeuchi K, et al. A mouse model for EML4-ALK-positive lung cancer. Proc Natl Acad Sci U S A. 2008;105:19893–7. doi: 10.1073/pnas.0805381105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bang Y, Kwak EL, Shaw AT, et al. Clinical activity of the oral ALK inhibitor PF-02341066 in ALK-positive patients with non-small cell lung cancer (NSCLC) Journal of Clinical Oncology. 2010;28 abstract 3. [Google Scholar]
- 10.Butrynski JE, D’Adamo DR, Horick JL, et al. Crizotinib in ALK rearranged Infammatory Myofibroblastic Tumor. 2010 doi: 10.1056/NEJMoa1007056. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mino-Kenudson M, Chirieac LR, Law K, et al. A novel, highly sensitive antibody allows for the routine detection of ALK-rearranged lung adenocarcinomas by standard immunohistochemistry. Clin Cancer Res. 2010;16:1561–71. doi: 10.1158/1078-0432.CCR-09-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou W, Ercan D, Chen L, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462:1070–4. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwak EL, Camidge DR, Clark J, et al. Clinical activity observed in a phase I dose escalation trial of an oral c-met and ALK inhibitor, PF-02341066. Journal of Clinical Oncology. 2009;27:15s. abstract 3509. [Google Scholar]
- 14.Shah NP, Nicoll JM, Nagar B, et al. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–25. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 15.Kantarjian H, Pasquini R, Hamerschlak N, et al. Dasatinib or high-dose imatinib for chronic-phase chronic myeloid leukemia after failure of first-line imatinib: a randomized phase 2 trial. Blood. 2007;109:5143–50. doi: 10.1182/blood-2006-11-056028. [DOI] [PubMed] [Google Scholar]
- 16.Galkin AV, Melnick JS, Kim S, et al. Identification of NVP-TAE684, a potent, selective, and efficacious inhibitor of NPM-ALK. Proc Natl Acad Sci U S A. 2007;104:270–5. doi: 10.1073/pnas.0609412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sequist LV, Natale RB, Senzer NN, et al. Association between anaplastic lymphoma kinase rearrangements (rALK) and the clinical activity of IPI-504 (retaspimycin hydrochloride), a novel Hsp90 inhibitor, in patients with non-small cell lung cancer (NSCLC) J Clin Oncol. 2010;28:15s. doi: 10.1200/JCO.2010.30.8338. Abstract 7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee CC, Jia Y, Li N, et al. Crystal structure of the anaplastic lymphoma kinase (ALK) catalytic domain. Biochem J. 2010 doi: 10.1042/BJ20100609. [DOI] [PubMed] [Google Scholar]
- 19.Bossi RT, Saccardo MB, Ardini E, et al. Crystal structures of anaplastic lymphoma kinase in complex with ATP competitive inhibitors. Biochemistry. 2010;49:6813–25. doi: 10.1021/bi1005514. [DOI] [PubMed] [Google Scholar]
- 20.Timofeevski SL, McTigue MA, Ryan K, et al. Enzymatic characterization of c-Met receptor tyrosine kinase oncogenic mutants and kinetic studies with aminopyridine and triazolopyrazine inhibitors. Biochemistry. 2009;48:5339–49. doi: 10.1021/bi900438w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.