The evolution of dengue viruses has had a major impact on their virulence for humans and on the epidemiology of dengue disease around the world. Although antigenic and genetic differences in virus strains had become evident, it is mainly due to the lack of animal models of disease that has made it difficult to detect differences in virulence of dengue viruses. However, phylogenetic studies of many different dengue virus samples have led to the association between specific genotypes (within serotypes) and the presentation of more or less severe disease. Currently, dengue viruses can be classified as being of epidemiologically low, medium, or high impact; i.e., some viruses may remain in sylvatic cycles of little or low transmissibility to humans, others produce dengue fever (DF) only, and some genotypes have been associated with the potential to cause the more severe dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS) in addition to DF. Although the factors that contribute to dengue virus epidemiology are complex, studies have suggested that specific viral structures may contribute to increased replication in human target cells and to increased transmission by the mosquito vector; however, the immune status and possibly the genetic background of the host are also determinants of virulence or disease presentation. As to the question of whether dengue viruses are evolving toward virulence as they continue to spread throughout the world, phylogenetic and epidemiological analyses suggest that the more virulent genotypes are now displacing those that have lower epidemiological impact; there is no evidence for the transmission of antigenically aberrant, new strains.
I. Introduction
Understanding dengue virus variation is especially important because we still know little about the disease; what has become evident over the many years of dengue research is that dengue epidemiology is determined by many factors, including those in the host, vector, and environment. Dengue virus evolution is also determined by many complex interactions, from the cellular to the populational level, in humans and mosquitoes. Some of the genetic changes that occur during the natural transmission cycles of dengue viruses have ultimately affected virulence or the potential to cause disease in humans, and it would benefit us greatly if we could narrow these down for inclusion in the development of control measures. The understanding of these virus-specified determinants of virulence has been difficult to achieve because of the lack of in vivo and in vitro markers to correlate with disease severity in humans. Therefore, what little we understand today has been acquired indirectly by the association between evolutionary groupings of virus strains and their epidemiological and/or disease clinical presentations. These associations are still being revealed, as in the case of dengue viruses of serotypes 1 and 4, about which we have little information, and as more dengue virus samples are being analyzed. It is also evident that we have a limited window in time from which to derive our information—virus fossils do not exist, and virus strains have only recently begun to be acquired and stored properly, with documentation of the associated epidemiological and clinical associations. Therefore, it behooves us to understand the mechanisms of dengue virus evolution, but we are only at the beginning stages of being able to associate specific viral sequences or structures with virulence for humans. The study of laboratory-prepared, attenuated dengue viruses is the subject of other reviews in this volume, and virulence in that context refers to the propensity of candidate vaccine viruses to cause any disease at all. This review attempts to summarize our current interpretations of dengue virus sequence analyses and how we can use this information to follow, understand, and possibly predict the evolution of dengue viruses around the globe.
II. Genetic Variation within Dengue Serotypes
As with other viruses, the evidence for strain differences among dengue viruses was first detected serologically using antibodies made by inoculating laboratory animals (Sabin, 1952). However, none of these animals showed signs of disease comparable to humans. Nonhu-man primates develop transient viremias, but even the apes, our most closely related primates, do not develop the rash and hemorrhagic symptoms consistent with DF or DHF. Some investigators have resorted to using mouse neurovirulence (after intracranial inoculation) as a surrogate model of virulence for humans. The first observations concerning differences in strain virulence came from clinical and epidemiological associations, where more severe disease was associated with specific outbreaks and the virus strains were isolated from patients with hemorrhagic disease (Barnes and Rosen, 1974; Gubler et al., 1978; Rosen, 1977, 1986). Meanwhile, others observed an increase in disease severity in patients who had been infected by more than one dengue virus serotype; this became known as the immune enhancement phenomenon (Halstead, 1970, 1988). Thus, two opposing theories of dengue pathogenesis came into existence, with one describing virulence as a viral characteristic and the other relating the severity of disease to the immune status of the host. Controversies surrounding both of these theories still exist, but it is most probable that both the virus and the host immune system play a role in dengue pathogenesis.
The first genetic evidence for differences between dengue viruses of the same serotype came from RNA fingerprinting studies (Repik et al., 1983; Vezza et al., 1980). This method uses an enzyme to digest viral RNA into strands, with the number and size of strands varying according to the entire viral sequence. This is a relatively crude surveying technique that does not give results that are directly comparable across strains. The resulting groupings of viral samples were called “topotypes,” reflecting the two-dimensional topology of the RNA strand electrophoresis patterns. Another method, using a similar approach of digesting a cDNA copy of the viral RNA template with endonucleases, gave somewhat more resolution, but it also failed to identify the regions of the genome in which the sequences differed among viruses (Kerschner et al., 1986). It was not until the mid-1980s that direct sequencing of specific genome regions became amenable to the study of many different viruses. Because primer-extension sequencing off of the viral RNA required very large quantities of relatively pure virus preparations to obtain clear sequence information, the first analyses of dengue virus variation used relatively short sequences for comparison or used very few strains (Blok et al., 1989; Chu et al., 1989; Rico-Hesse, 1990). What has become clear is that with more sequence information, i.e., either full genomic sequences for one virus or entire genes for many different viruses, our understanding of dengue evolution and virulence has increased dramatically. Current controversies center on how to interpret this information rather than whether dengue viruses differ in their potential to cause disease. The classification of viruses into genetic groups (“genotypes”) within serotypes is constantly changing, as methods for sequencing and evolutionary analysis improve and the available database expands.
Current methods for obtaining dengue virus sequences no longer require a viable virus isolate; in fact, the entire genome sequence can be obtained by enzymatic amplification of the viral RNA template in the patient’s blood sample (Leitmeyer et al., 1999). Thus, most of the sequences available from the 1990s to date were generated with the reverse transcriptase-polymerase chain reaction (RT-PCR), which has substantially improved the quality or fidelity of the sequences available from many laboratories around the world. Unfortunately, there is no uniform approach to determining which sequences will be compared among strains; this has led to a vast quantity of information available on the sequence database (GenBank) for which there has been no systematic interpretation. In addition, there are numerous sequences that contain errors (sequencing or editing artifacts), which have led to serious mistakes in interpretation, especially regarding the possibility of intramolecular recombination (see later). Therefore, it is wise to obtain virus isolates for further genetic characterization if in fact the initial sequence information obtained proves to be unusual.
III. Phylogenetics of Dengue Viruses
As sequencing methods have become more accessible technically and financially, there has been a concomitant increase in the number of dengue virus strains that have been analyzed. This has led to a refinement in the analyses of dengue evolution (thus we now speak of “microevolution” of dengue viruses), but this advance has also been dependent on the development of statistical methods for determining sequence relationships, known as “phylogenetics.” In the last decade, the computer algorithms used in the generation of phylogenetic or evolutionary trees of virus relationships have increased in number and complexity; this is a field of research that is evolving on its own, and most virologists tend to apply the algorithms that are handy, compatible with their computer, or are used by nearby colleagues. Therefore, it is difficult to say which phylogenetic method is best unless one does several comparisons under specific dataset assumptions (number of taxa, character weighing, rooting options, etc.). Currently, the consensus among “phylogeneticists” is that a large number of taxa (virus strains), with long sequence strings (i.e., one gene or more) analyzed by the maximum likelihood (ML) method, with its incorporated transition/transversion rate calculation, and with bootstrap (statistical) support for the branching patterns, are probably sufficient to yield an accurate phylogenetic tree for viral genotype classification (Hillis, 1998; Lemmon and Milinkovitch, 2002). Calculations of molecular clocks (rates of evolution over time), theoretical ancestors, and selective pressure estimations require many assumptions for which we have no hard evidence; these interpretations may be misleading when based solely on laboratory or in vitro observations. It is clear that we also suffer from a taxa inclusion bias because we obtain samples from humans who are ill (and historically analyze those from patients with more severe disease) and do not have enough samples from vertebrates involved in sylvatic cycles or from mosquitoes. Thus, quantitations of natural dengue virus diversity (Holmes, 1998; Zanotto et al., 1996) are premature. Currently, the accuracy of phylogenetic trees is usually limited by the number and type of taxa analyzed and the investigator’s access to high-speed computing resources.
The selection of domains or genome regions for sequence comparison is very important with regard to analysis outcome and limits of interpretation. When using the maximum parsimony method of phylogenetic analysis, it is common to state how many parsimony informative sites were included in the dataset; however, this approach is not available in ML and we are left with bootstrap values to support the monophyletic or genotype groupings after estimating numerous trees. It has become clear that different areas of the dengue virus genome evolve or fix mutations at different rates and will sometimes exhibit “hot spots” of higher mutation rates within a region (e.g., the E gene or the 3′-untranslated region). In general, when long enough sequences are used, the trees generated from different genome regions usually correspond or overlap, and discrepancies occur only when trying to interpret the minor branches of the trees (i.e., genotype groupings usually remain the same). It is this characteristic of varying mutation rates across the genome that prevents us from establishing a uniform cutoff rate of divergence for different genotypic groups; thus, it is important to state that arbitrary cutoffs for genotypes apply only to the region or sequences being compared (Rico-Hesse, 1990). Until more complete genome sequences of dengue viruses are available, we will not be able to establish the natural, full range of nucleotide or amino acid variability within serotypes and genotypes; in addition, viruses or templates should be of low passage from their original sources to avoid artificial selection of mutants (Lee et al., 1997). This is a difficult but not impossible task given the large number of mutations that can potentially occur in all areas of the dengue genome. At this point in time the actual mechanisms governing the natural selection of dengue mutants have not been determined, and we are limited to statistical inferences of positive or negative selection on specific genome regions or amino acids (Twiddy et al., 2002a, 2002b). Suffice it to say that these mechanisms appear to influence codon usage (Jenkins et al., 2001), RNA folding (Brinton and Dispoto, 1988; Leitmeyer et al., 1999; Shi et al., 1996), or protein structures of dengue and other flaviviruses; i.e., there are structural and functional limitations to the plasticity of dengue viruses in addition to the dogma of immune selection by host antibodies.
The possibility of intramolecular recombination among dengue viruses has received considerable attention (Gould et al., 2001; Holmes and Burch, 2000; Holmes et al., 1999; Uzcategui et al., 2001; Worobey et al., 1999); however, no virus isolates meeting stringent criteria for recombination have yet been described. In one instance, most of the purported recombinant sequences acquired from the GenBank database were shown to contain sequencing artifacts, which made them behave as recombinants in statistical analyses (Worobey et al., 1999). In other examples, the virus isolate can no longer be obtained for independent verification (Holmes et al., 1999) or the investigators were not able to meet virological criteria for recombinant classification (Tolou et al., 2001; Uzcategui et al., 2001). Such criteria include obtaining a virus isolate whose purity is confirmed by plaquing, probe hybridization, and/or neutralization with serotype-specific antisera, ruling out mixed templates, and direct sequencing of multiple ampli-cons from several RNA template preparations, as has been done with polioviruses (Cuervo et al., 2001; Liu et al., 2000). Although the possibility of recombination among dengue viruses may not be remote because dual (serotype) infection of humans has been demonstrated (Gubler et al., 1985; Laille et al., 1991; Lorono-Pino et al., 1999), the probability of simultaneous infection of cells in human or vector hosts may be low because of replication interference (Dittmar et al., 1982). However, this phenomenon has not kept other single-stranded, nonsegmcntcd RNA viruses, such as poliovirus, from often producing inter- and intraserotype recombinants, which has occurred in vaccine (live-attenuated, trivalent Sabin strain) recipients. It remains to be determined whether and at what frequency dengue viruses do undergo recombination in nature, where the main concern would be the creation of interserotype hybrids, which might be capable of escaping immunity to the four known serotypes. So far, the aforementioned phylogenetic or statistical methods for detecting recombinants have possibly demonstrated evidence for dengue virus hybrids within but not across serotypes.
The ultimate application of understanding virus evolution is to derive information that could be helpful in disease control. Thus, it is important that all of the information used for phylogenetic tree interpretation meet certain standards so that our conclusions are not erroneous. Both the clinical and epidemiological information we use to relate samples or genotypes to disease potential should therefore clearly meet dengue case definitions for disease classification (i.e., DF or DHF/DSS). Countries reporting outbreaks or epidemics should use the resources of reference laboratories to confirm their findings. In fact, the best samples for full genome sequencing of dengue viruses have come from prospective studies, where patients are enrolled and sampled when febrile and their clinical progress documented along with other immunological and epidemiological parameters (Rico-Hesse et al., 1998; Vaughn et al., 2000). Dengue virus phylogenies cannot be interpreted without clear information about their phenotypic or biological properties, and the trees will only be as accurate as the tests used to address our hypotheses. Therefore, different trees can be generated with differing sequences or discrete genome regions, depending on the question posed. For most dengue serotypes, we are still attempting to determine which genotypes are associated with higher virulence, severe disease, or larger epidemics. However, for dengue serotype 2 and 3 viruses, we appear to have identified genotypes that have undergone greater spread than the other genotypes and have the potential to cause DHF. The transmission of these genotypes is being monitored in several countries and the ministries of health have understood the urgency to reduce transmission of these strains, albeit by vector reduction.
In an attempt to clarify or unify the current classification of dengue genotypes within serotypes, phylogenies of all four serotypes are described using nucleotides from the entire E gene region. All sequences were aligned with representatives of the other serotypes (using the Clustal W program) (Aiyar, 2000; Higgins et al., 1996) and were compared by the computing-intensive ML method, with inherent estimations of transitions/transversions, and with bootstrap support for branching patterns (Swofford, 2002). Only the latest versions of sequences deposited in the GenBank database were used for comparisons to avoid the inclusion of laboratory artifacts or errors. These results and interpretations should not be construed to represent actuality but rather the best approximations of dengue virus evolutionary relationships we have at this time.
A. Phytogeny of Dengue Serotype 1
The first genetic comparison of dengue type 1 strains was reported in 1983 using RNA fingerprinting (Repik et al., 1983); these investigators were able to distinguish three geographic groupings (Caribbean, Pacific/Southeast Asian, and African) among 12 different strains isolated at different times. A review published in 1990 mentioned the possibility of up to eight topotypes of serotype 1 viruses, but the original data were not published (Trent et al., 1990). Subsequent E gene sequencing studies confirmed the original groupings and also noted a failure to detect specific virulence markers (Chu et al., 1989); that is, there was no correlation of specific amino acid sequences in this gene with dengue disease of greater or lesser clinical severity. Another sequencing study, using only 240 nucleotides from the E/NS1 gene junction, showed that 40 serotype-1 strains could be classified into five genotypic groups when using an arbitrary cutoff point of 6% divergence for groupings, but because more strains were analyzed, the global routes of transmission of these viruses could be followed (Rico-Hesse, 1990). A large number of strains were analyzed in another report (Chungue et al., 1995), but this study used a different region of the genome (180 nucleotides from E gene) for comparisons and the results were not directly comparable; however, these investigators used the 6% divergence cutoff mentioned earlier to distinguish three genotypes. Only recently has a more complete study of serotype 1 strains been reported (Goncalvez et al., 2002), where the full E gene sequences were determined, allowing for the correction of some of those reported earlier. The trees reported in that study differ from the analysis shown here only in that representatives of the other serotypes were included as an outgroup in the current study. Also, genotypes are denoted here by their apparent geographical origins (and not necessarily to where they have since spread) and not by numbers (Roman numerals).
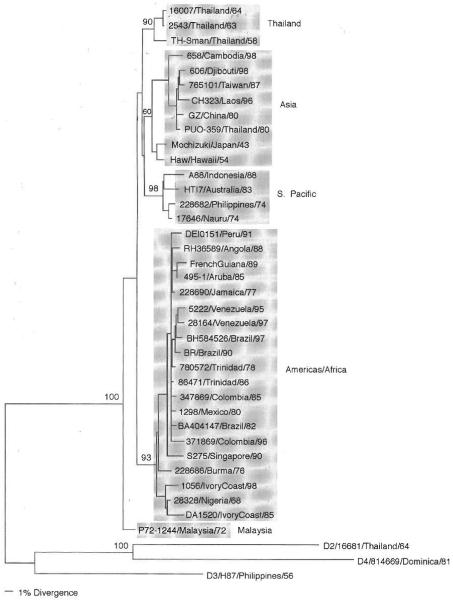
The phylogeny of 36 dengue serotype 1 viruses is shown in Fig. 1. The statistical analyses of validity of branching patterns (bootstrap) continue to support the classification of these viruses into five genotypes: sylvatic/Malaysia, Americas/Africa, South Pacific, Asia, and Thailand. The tree also supports the previous hypothesis (Wang et al., 2000) that sylvatic strains, one from Malaysia in this case, evolved earlier from a hypothetical ancestor shared by all dengue viruses, which is why most sylvatic viruses are basal (branch off first) in all ML-estimated trees. However, only one dengue type 1 virus has been isolated under these ecological conditions (from a sentinel monkey) (Rudnick, 1965); it is also unclear if this virus cycle still exists, as field work has ceased and other isolates from Malaysia are now from urban epidemics and involve other genotypes (Chow et al., 1994). It is clear, however, that this sylvatic genotype, albeit represented by one isolate, is of low epidemiological importance to humans, as it is not causing detectable disease in humans and there is no evidence that reintroduc-tion from a sylvatic cycle is required for persistence of dengue transmission year-round. Some of the other four genotypes may no longer exist either, as one of them is represented only by older Thai strains (from 1954 to 1964) and the Japan/Hawaii strains that are basal to the Asian genotype (including a Thai 1980 strain) have no other closely related neighbors. Only further sampling will allow clarification of these observations.
Fig 1.
Phylogeny of 36 serotype 1 dengue viruses using 1485 nucleotides from the E gene. Representatives of the other serotypes were used as an outgroup. Strains are denoted by number, country of isolation, and year of isolation; genotypes are shaded and names given to the right. Bootstrap values of statistical support for major branches are shown as percentage equivalents.
Because the clinical classifications of the patients from which some of these older viruses were obtained are unclear and because sequences for serotype 1 viruses, which have caused more severe dengue, are lacking in most analyses (e.g., recent Thai viruses), it is not possible to find an association between some of these genotypes and increased virulence. Only from the most recent study do we know that the American/African genotype has the potential to cause DHF because of the inclusion of two samples from patients in Venezuela (Goncalvez et al., 2002). In addition, this genotype had been spreading to many other countries or geographic regions during the past decade (see Brazil, 1997; Colombia, 1996; Peru, 1991; and Venezuela, 1997). Therefore, this genotype would be considered of higher epidemiological impact than the sylvatic genotype. It is evident from these results that a more complete and systematic survey of serotype 1 samples is necessary before a link can be established between specific genotypes and virulence of these viruses.
B. Phytogeny of Dengue Serotype 2
Dengue viruses of serotype 2 have traditionally been studied in more detail than those belonging to the other serotypes because of their association with more frequent and severe epidemics. In southeast Asia, the first detailed descriptions of disease and epidemics were those caused by viruses belonging to serotype 2 (Burke et al., 1988; Sangkawibha et al., 1984); in the Americas, the appearance of serotype 2 virus was associated with the first epidemics of DHF in this region (Kouri et al., 1983). It was with viruses of this serotype that numerous methods for detecting genetic differences among strains were first attempted: antigen signature analysis compared specific epitopes using monoclonal antibodies to the E glycoprotein (Monath et al., 1986), restriction enzyme mapping of cDNA and probing detected some nucleotide differences (Kerschner et al., 1986), and RNA fingerprinting of many different patient samples from the same location gave an estimate of the large number of variants circulating during 1 year in Thailand (Walker et al., 1988). One large fingerprinting study concluded that numerous variants circulated in southeast Asia over a 25-year period (Trent et al., 1989), and in one summary, up to 10 distinct topotypes of serotype 2 viruses could be identified (Trent et al., 1990). It was not until the first sequencing studies that an attempt was made to identify the genetic differences between strains from DF patients and viruses isolated from DHF patients. Comparisons of E gene sequences from 12 serotype 2 viruses showed no correlation between disease severity and specific nucleotides or amino acids (Blok et al., 1989), and a similar conclusion was drawn after comparison of the NS1 gene from eight virus strains (Blok et al., 1991). Subsequent full genome analyses of viruses from patients in southeast Asia also failed to identify specific sites that might determine virulence (Mangada and Igarashi, 1998; Pandey and Igarashi, 2000).
Sequence analysis of many more strains of serotype 2 first gave rise to the idea that specific genotypes could show differences in virulence potential, resulting in epidemics of DHF. A study of 40 strains from different areas of the tropical world, collected over a 45-year period, showed that dengue serotype 2 viruses could be classified into five genotypes by comparison of the E/NS1 gene junction sequences (240 nucleotides) (Rico-Hesse, 1990). Subsequent comparisons of E gene sequences from 16 strains (Lewis et al., 1993), then more sylva-tic strains (Wang et al., 2000), and, more recently, the comparison of only the 3′-untranslated region of serotype 2 strains continued to support these groupings (Shurtleff et al., 2001). Subsequently, after the analysis of many other samples, especially those from southeast Asia, a region with a long history of DHF epidemics and numerous cocirculat-ing virus variants, the groupings were broken down into four genotypes (Rico-Hesse et al., 1998): sylvatic/west Africa, Americas, southeast Asia, and Malaysia/Indian subcontinent (Fig. 2). It became clear that the introduction of the southeast Asian genotype into the Americas in 1981, specifically into Cuba, and its subsequent spread to other parts of the Caribbean were directly associated with the appearance of DHF in the western hemisphere (Rico-Hesse, 1990). Prior to 1981, there had been sporadic reports of cases of dengue with hemorrhagic manifestations, but it was unclear if the DHF/DSS case definitions had been met. It took a much more detailed analysis of the viruses circulating in the Americas to directly link more severe dengue disease with the southeast Asian genotype virus. The availability of samples from patients with well-documented clinical records from three countries with DHF cases (Brazil, Mexico, and Venezuela) helped prove this point (Rico-Hesse et al., 1997). Others have continued to corroborate this hypothesis (Gubler, 1998) and, to this date, all dengue serotype 2 viruses from DHF/DSS patients have been shown to belong to the southeast Asian genotype. The American and Malaysia/Indian subcontinent genotypes have so far been isolated from patients with DF only and are therefore considered to be of intermediate epidemiological impact.
Fig 2.
Phylogeny of 50 serotype 2 dengue viruses using 1485 nucleotides from the E gene. Representatives of the other serotypes were used as an outgroup. Strains are denoted by number, country of isolation, and year of isolation; genotypes are shaded and names given to the right. Bootstrap values of statistical support for major branches are shown as percentage equivalents.
An anomaly that currently has no explanation is the repeated, independent isolation of viruses genetically similar to the original New Guinea C (NGC) prototype (isolated in 1944) virus by several laboratories around the world: Cuba (Guzman et al., 1995), Venezuela (Rico-Hesse et al., 1997), Mexico (P. A. Armstrong, unpublished), China (GenBank, AF204177), and Vietnam (Twiddy et al, 2002a). Because these contemporary, independently isolated and sequenced samples vary (in general, <2%) from the old, high-passage NGC prototype virus, it is possible that these isolates represent contamination with the NGC laboratory strain, which had been passaged to fix some mutations. The NGC virus has been used for many decades in most national laboratories as the reference strain for serotype 2. It could have potentially been maintained under different in vitro conditions, which could have made the resulting sequences different for these countries. The only other explanation seems to be that NGC is, in fact, being transmitted in these countries. However, there is very little evidence to support this, as these isolations are very rare. The disease associated with these samples is not unusual in that there are a mixture of DF and DHF cases from which these were purportedly isolated; this is the only type of isolate that remains from the Cuban 1981 epidemic (Guzman et al., 1995).
As for serotype 1 viruses, serotype 2 sylvatic viruses do not seem to spread to other urban areas even within west Africa (the Malaysia/ Indian subcontinent genotype has been imported) and could be classified as being of low epidemiological impact. Furthermore, because of their basal or ancestral position on the phylogenetic tree (Fig. 2), it seems likely that they were the first to evolve from the progenitor shared by all four serotypes. Because of their great genetic distance from the other genotypes, it is hard to determine if they are the origin of all other serotype 2 viruses. Further research is required to explain the low transmissibility or virulence of these viruses to humans and whether this is a consequence of their adaptation to canopy-dwelling mosquito vectors, which do not bite or infect large numbers of humans.
Serotype 2 viruses have also shown the potential to establish transmission in very distant areas of the world. It has been documented previously that the southeast Asian and the Malaysia/Indian subcontinent genotypes have spread from one continent to another in a matter of years (Rico-Hesse, 1990). Classical epidemiological investigations have helped trace the pathways of these introductions and the establishment of endemic or hyperendemic cycles of transmission in some countries (e.g., Venezuela, Brazil). Currently, the main concern is that the southeast Asian genotype, of high epidemiological impact, continues to spread through several countries, including Peru and Mexico. In the case of Peru, good epidemiological evidence shows that the native, American genotype did not cause DHF in patients, even upon secondary infection (Watts et al., 1999). It remains to be seen if and how severe the epidemics will become upon the establishment of transmission of the southeast Asian genotype (Rocha et al., 2002). In Mexico, the introduction of the southeast Asian genotype has been gradual, with the first documented viruses from cases in south and central Mexico in 1995 (Rico-Hesse et al., 1997), but with some areas of northern Mexico (close to the Texas border) still occupied by the American genotype (P. A. Armstrong and R. Rico-Hesse, personal communication). The availability of a very large database of serotype 2 sequences (several hundred strains) will facilitate the documentation of movement of dengue 2 around the world.
C. Phylogeny of Dengue Serotype 3
The first published report of the analysis of genetic variation in serotype 3 viruses occurred in 1972 (Russell and McCown, 1972), but used serologic tests to show this difference; the geographical distinction of strains (Puerto Rico and Tahiti, separate from Asian strains) still holds true. Fingerprinting studies identified five topotypes for this serotype (versus 8 and 10 for dengues 1 and 2, respectively) (Trent et al., 1990); this lower number of distinct groups is evident in serotype 4 viruses also. Thus, the interpretation of the phylogeny of serotype 3 viruses is somewhat more complex because the rates of fixation seem to be lower, i.e., there is less distance or divergence between the genotypic groups.
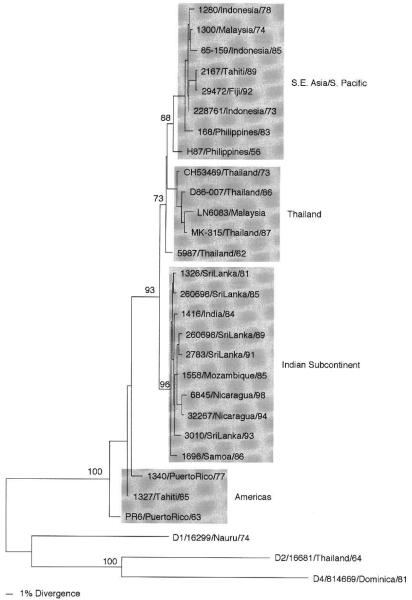
The first phylogenetic analysis of these viruses required the comparison of the entire E gene sequences (Lanciotti et al., 1994), and four genotypes were distinguished. This classification has remained unchanged with the study and inclusion of new isolates (Fig. 3), although the lines demarcating genotypic groups are somewhat blurred mainly by older strains (see Philippines 1956 and Thailand 1962). Again, most of the genotypes can be identified by geographical origin, but there is no evidence for a sylvatic group: Americas (most basal), Indian subcontinent, Thailand, southeast Asia/South Pacific.
Fig 3.
Phytogeny of 26 serotype 3 dengue viruses using 1479 nucleotides from the E gene. Representatives of the other serotypes were used as an outgroup. Strains are denoted by number, country of isolation, and year of isolation; genotypes are shaded and names given to the right. Bootstrap values of statistical support for major branches are given as percentage equivalents.
In this case, the genotype with low epidemiological potential seems to be the one that circulated in the Americas prior to 1989; isolates of this type came from patients with DF only. In the 1980s through the 1990s, in both the Americas and the South Pacific, there were two independent introductions of new genotypes: the southeast Asian genotype was associated with large epidemics with DHF in Tahiti and Fiji (Chungue et al., 1993) and the Indian subcontinent genotype was introduced to Central America in the mid-1990s (Balmaseda et al., 1999; Harris et al., 1998, 2000; Usuku et al, 2001), displacing the American genotype in both areas.
Evidence shows that the Indian subcontinent genotype has evolved, since 1989, to produce more DHF in Sri Lanka (Lanciotti et al., 1994), a country that maintains good epidemiological and virological records. Current studies are focused on documenting the mutations responsible for this phenotypic variation, as was done for viruses of serotype 2 (Messer et al., 2002). In prospective studies in Thailand, the direct correlation between viremia caused by dengue serotype 3 viruses and the severity of disease (DHF) has also been shown (Libraty et al., 2002). Thus, it is important to determine if some of these genotypes also replicate to higher levels in human cells, which are targets involved in the pathogenesis of dengue disease. The more virulent Indian subcontinent genotype has also been associated with epidemics currently occurring in Mexico and Central and South America, and viruses from these outbreaks need to be studied more closely, especially with the new assay systems described later. It remains to be seen if viruses of serotype 3 have consistent genetic differences in regions of the viral genome that encode determinants of more severe disease.
D. Phytogeny of Dengue Serotype 4
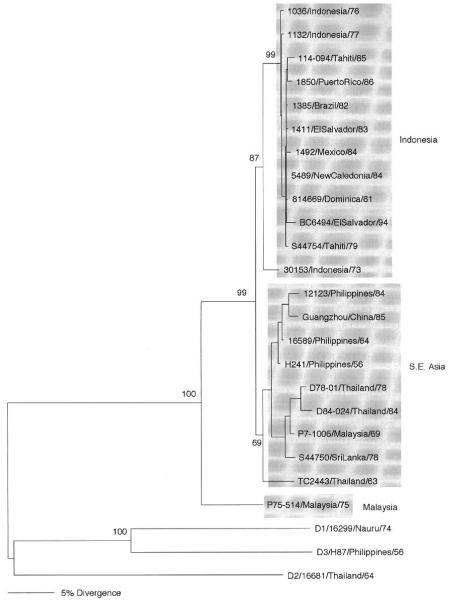
Dengue viruses of serotype 4 were shown to differ antigenically and genetically in 1986 (Henchal et al., 1986), and five topotypes were identified in 1990 (Trent et al., 1990). However, comparison of short (180-nucleotide) sequences from the E gene failed to classify 28 isolates into genotypic groups (Chungue et al., 1995). Only after the comparison of full E gene sequences was it possible to distinguish two genotypes (Lanciotti et al., 1997) and the rates of variation among 20 strains were shown to reach a maximum of 6%. The inclusion of longer nucleotide sequences (preM-E genes) has failed to add more resolution to the phylogenetic trees, and only two genotypes could be resolved (Fig. 4); the inclusion of only E gene sequences from one sylvatic virus (Wang et al., 2000) adds one extra group, as for serotype 1 viruses. Thus, there are currently the Malaysia, southeast Asia, and Indonesia genotypes. As with the other serotypes, it is not known if the sylvatic cycle in Malaysia still exists, as that country had imported the urban cycle virus, apparently from southeast Asia (see Malaysia 1969). At this point in time, it seems that the genotype that includes viruses from Indonesia, the South Pacific, and the Americas is the one of greater epidemiological impact, if only because it continues to spread to other countries, mainly in the Americas (Lanciotti et al., 1997). It is obvious that we know much less about the genetic variation of viruses of this serotype other than that their rates of mutation in nature are lower and that there is less transmission of this virus, as evidenced by the lack of epidemics associated strictly with these viruses.
Fig 4.
Phylogeny of 22 serotype 4 dengue viruses using 1635 nucleotides from preM-E genes, except for two strains from Malaysia, for which only E gene sequences were available (1485 nucleotides). Representatives of the other serotypes were used as an outgroup. Strains are denoted by number, country of isolation, and year of isolation; genotypes are shaded and names given to the right. Bootstrap values of statistical support for major branches are shown as percentage equivalents.
IV. Viral Determinants of Disease
As mentioned earlier, dengue virus virulence has been difficult to measure because of the lack of in vivo and in vitro models of disease. One approach to determining the viral sequences or structures involved in virulence has been to compare the entire genome amplified directly from samples from patients with clearly differing disease presentations, after having the virus isolates identified as to serotype and genotype, usually from the same blood sample. This requires samples of serum or plasma containing high titers of virus from patients in the acute phase of disease (before the onset of severe symptoms, if they develop) and ultralow temperature storage (−70 °C) until processing and sequencing with RT-PCR methods. So far, these conditions have been met only for dengue viruses belonging to serotype 2, with a detailed comparison between sequences belonging to the American (low virulence) and southeast Asian (high virulence) genotypes (Leitmeyer et al., 1999). The results pointed to consistent differences between these genotypes in certain nucleotides and folding patterns of the 5′- and 3′-untranslated regions of the viral genome, along with amino acid charge differences in the prM, E, NS4b, and NS5 genes. Others had shown that mutations in the untranslated regions of dengue serotype 4 infectious clones yielded viruses with altered mouse neurovirulence (Blaney et al., 2001) and that differences in amino acid position 390 of the E glycoprotein of serotype 2 viruses also have such an effect (Sanchez and Ruiz, 1996). The working hypothesis is that substitutions at these positions affect RNA structures and properties of the E protein and contribute in some way to alter the pathogenesis of DHF. Support for this hypothesis has come from complex, prospective studies showing that severe disease correlates with higher titers of virus in the bloodstream when associated with southeast Asian genotype virus infection (Vaughn et al., 2000).
Current studies of viral determinants of disease involve the modification of infectious clones (Kinney et al., 1997) and the measurement of the effects of mutations in new, target cell assay systems. The discovery of dendritic cells as primary targets for dengue virus replication (Wu et al., 2000; in contrast to many earlier studies focusing on monocytes as target cells) has led to attempts to use this characteristic as a correlate of differences in virulence. For instance, it has been shown (Cologna and Rico-Hesse, 2003) that the replacement of southeast Asian genotype 5′- and 3′-untranslated regions and one amino acid (E-390) with American genotype structures significantly reduce the level of replication of infectious clone-derived viruses in human, primary dendritic cell cultures. The virus yield (measured by quantitative RT-PCR) of the triple mutant (three regions substituted) was identical to that of a wild-type American genotype virus in the same assays, whereas that of the unmodified southeast Asian virus was 10-fold higher. Others have shown that the modification of amino acid residue 390 alone of the E protein reduces the rate of replication of dengue virus infectious clones in human monocytes (Pryor et al., 2001). It is anticipated that dendritic cell assay systems may serve as an in vitro surrogate for dengue virus virulence, but many more detailed investigations are necessary to determine the molecular mechanisms involved in the differences in virus yield. However, a comparison of 12 different wild-type, low-passage, dengue serotype 2 viruses belonging to the southeast Asian and American genotypes almost uniformly showed lower virus output by the latter in primary human dendritic cells. It remains to be seen whether these dengue viruses behave similarly in other human target cells, but it is tempting to speculate that the differences in the behavior of these viruses could explain the increase in cytokine release (i.e., as a result of higher virus burdens in infected monocytes and dendritic cells) and the subsequent plasma leakage by endothelial cells seen in DHF (Bosch et al., 2002). Another assay system under development involves the use of dengue virus-infected, primary lymphocyte cell cultures in chambers below human, primary endothelial cell cultures to measure the effects of cytokine secretion by lymphocytes on the latter cells as a surrogate for plasma leakage in vivo (C. Kubelka, R. Cologna, and R. Rico-Hesse, personal communication). Thus, the availability of in vitro systems to measure the effect of virus infection on isolated human target cells might also help us understand the mechanisms behind immune enhancement of disease. Other desirable models of disease might also include immuno-deficient mice that have been reconstituted with human cells or factors that may be involved in the pathogenic cascade that leads to more severe dengue disease. However, no one has yet reported successful en-graftment of dendritic cells or the reproduction of severe dengue symptoms in such an animal model (An et al., 1999; Lin et al., 1998; Wu et al., 1995).
V. Viral Determinants of Transmission
Another way of measuring dengue virus epidemic potential comes from studies of the ability of different viruses to infect and be transmitted to other humans by the mosquito vector, Aedes aegypti (Gubler et al., 1979). It follows that if some dengue viruses are capable of producing higher and longer viremias in their human hosts, these viruses could possibly infect more vectors and thus have a higher chance for transmission. This hypothesis was tested by infecting low-passage (F2–F4 generation) A. aegypti mosquitoes collected in McAllen, Texas, and Iquitos, Peru, with the same 12 wild-type dengue virus preparations used for dendritic cell infections described earlier and measuring time for virus dissemination to the salivary glands as a surrogate for transmission potential (Armstrong and Rico-Hesse, 2001). These mosquitoes were infected per os with the same amount of virus as measured by quantitative RT-PCR and mosquito infectious dose. Results suggested that the transmission of southeast Asian genotype viruses is more robust, with more efficient infection and dissemination in mosquitoes from different geographical areas. However, others have shown that mosquito populations also vary in their ability to become infected by, and to disseminate, a single dengue virus, albeit a very high passage strain (Bosio et al., 1998, 2000). It is clear that mosquitoes vary in their ability to get infected and reproduce enough virus to become infectious to humans by injection of their saliva. Laboratory-maintained strains of A. aegypti seem to have lost their potential to replicate dengue viruses differentially (southeast Asian versus American genotype), and there is also some variability of this transmission efficiency within field-derived populations (Armstrong and Rico-Hesse, 2001). Additional studies are needed to identify the factors that influence mosquito vectorial capacity, including the genetics of high and low transmitters (Miller and Mitchell, 1991).
VI. Viral Displacement and Epidemiology
Although the efficiency of dengue virus transmission is a complex phenomenon, the displacement of one genotype by another more “virulent” type has been documented in the past and is currently occurring in some countries. To understand this process, it is important to have good continuous epidemiological surveillance systems in place to detect the spread of different dengue virus genotypes. Such systems have allowed for the detection of the displacement of the American genotype of dengue serotype 2 by the southeast Asian genotype in the Americas in at least four countries in the past (Brazil, Colombia, Mexico, and Venezuela) (Rico-Hesse et al., 1997), and presently in one country (Peru) (Rocha et al., 2002). The failure to detect DHF in Peru has been associated with the lack of transmission of the southeast Asian genotype, although the American genotype virus was being transmitted at high rates (Watts et al., 1999). Unfortunately for the people of Peru, the southeast Asian genotype virus has been detected in mosquitoes in Iquitos, and this regional population has therefore become a natural test bed for the virus virulence hypothesis.
The same phenomenon of displacement seems to be occurring for dengue serotype 3, where the more virulent Indian subcontinent genotype had appeared after a nearly 20-year hiatus by the native American genotype in Nicaragua and Guatemala (Balmaseda et al., 1999; Usuku et al., 2001). The transmission of this virus has been associated with massive outbreaks of DF and DHF in Brazil, Ecuador, el Salvador, and Honduras (World Health Organization, 2002). These introductions and/or displacements have been clearly correlated with the appearance of DHF. Thus, by monitoring the transmission of specific dengue genotypes, we can predict the appearance of more severe epidemics. This phenomenon seems to occur uniformly because virus transmission is so high that invariably those with the propensity to present with severe disease become infected. However, more studies are needed to identify the viral determinants that impart to the virus an increased potential to replicate in certain hosts and vectors and a subsequent role in the epidemiology and pathogenesis of dengue disease.
Acknowledgments
Many thanks to Ferdinando Liprandi, William Messer, and Aravinda de Silva for providing sequences before publication. Financial support was provided by the National Institutes of Health (Grant AI50123) and the Kleberg Foundation.
References
- Aiyar A. The use of CLUSTAL W and CLUSTAL X for multiple sequence alignment. Methods Mol. Biol. 2000;132:221–241. doi: 10.1385/1-59259-192-2:221. [DOI] [PubMed] [Google Scholar]
- An J, Kimura-Kuroda J, Hirabayashi Y, Yasui K. Development of a novel mouse model for dengue virus infection. Virology. 1999;263:70–77. doi: 10.1006/viro.1999.9887. [DOI] [PubMed] [Google Scholar]
- Armstrong PM, Rico-Hesse R. Differential susceptibility of Aedes aegypti to infection by the American and southeast Asian genotypes of dengue type 2 virus. Vector Borne Zoonotic Dis. 2001;1:159–168. doi: 10.1089/153036601316977769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmaseda A, Sandoval E, Perez L, Gutierrez CM, Harris E. Application of molecular typing techniques in the 1998 dengue epidemic in Nicaragua. Am. J. Trop. Med Hyg. 1999;61:893–897. doi: 10.4269/ajtmh.1999.61.893. [DOI] [PubMed] [Google Scholar]
- Barnes WSJ, Rosen L. Fatal hemorrhagic disease and shock associated with primary dengue infection on a Pacific island. Am. J. Trop. Med Hyg. 1974;23:495–506. doi: 10.4269/ajtmh.1974.23.495. [DOI] [PubMed] [Google Scholar]
- Blaney JE, Jr., Johnson DH, Firestone CY,, Hanson CT, Murphy BR, Whitehead SS. Chemical mutagenesis of dengue virus type 4 yields mutant viruses which are temperature sensitive in Vero cells or human liver cells and attenuated in mice. J. Virol. 2001;75:9731–9740. doi: 10.1128/JVI.75.20.9731-9740.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blok J, Gibbs AJ, McWilliam SM, Vitarana UT. NS1 gene sequences from eight dengue-2 viruses and their evolutionary relationships with other dengue-2 viruses. Arch. Virol. 1991;118:209–223. doi: 10.1007/BF01314031. [DOI] [PubMed] [Google Scholar]
- Blok J, Samuel S, Gibbs AJ, Vitarana UT. Variation of the nucleotide and encoded amino acid sequences of the envelope gene from eight dengue-2 viruses. Arch. Virol. 1989;105:39–53. doi: 10.1007/BF01311115. [DOI] [PubMed] [Google Scholar]
- Bosch I, Xhaja K, Estevez L, Raines G, Melichar H, Warke RV, Fournier MV, Ennis FA, Rothman AL. Increased production of interleukin-8 in primary human monocytes and in human epithelial and endothelial cell lines after dengue virus challenge. J. Virol. 2002;76:5588–5597. doi: 10.1128/JVI.76.11.5588-5597.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosio CF, Beaty BJ, Black WCI. Quantitative genetics of vector competence for dengue-2 virus in Aedes aegypti. Am. J. Trop. Med. Hyg. 1998;59:965–970. doi: 10.4269/ajtmh.1998.59.965. [DOI] [PubMed] [Google Scholar]
- Bosio CF, Fulton RE, Salasek ML, Beaty BJ, Black WCT. Quantitative trait loci that control vector competence for dengue-2 virus in the mosquito. Aedes aegypti. Genetics. 2000;156:687–698. doi: 10.1093/genetics/156.2.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinton MA, Dispoto JH. Sequence and secondary structure analysis of the 5′-terminal region of flavivirus genome RNA. Virology. 1988;162:290–299. doi: 10.1016/0042-6822(88)90468-0. [DOI] [PubMed] [Google Scholar]
- Burke DS, Nisalak A, Johnson DE, Scott RM. A prospective study of dengue infections in Bangkok. Am. J. Trop. Med. Hyg. 1988;38:172–180. doi: 10.4269/ajtmh.1988.38.172. [DOI] [PubMed] [Google Scholar]
- Chow VT, Seah CL, Chan YC. Comparative analysis of NS3 sequences of temporally separated dengue 3 virus strains isolated from southeast Asia. Intervirology. 1994;37:252–258. doi: 10.1159/000150386. [DOI] [PubMed] [Google Scholar]
- Chu MC, O’Rourke EJ, Trent DW. Genetic relatedness among structural protein genes of dengue 1 virus strains. J. Gen. Virol. 1989;70:1701–1712. doi: 10.1099/0022-1317-70-7-1701. [DOI] [PubMed] [Google Scholar]
- Chungue E, Cassar O, Drouet MT, Guzman MG, Laille M, Rosen L, Deubel V. Molecular epidemiology of dengue-1 and dengue-4 viruses. J. Gen. Virol. 1995;76:1877–1884. doi: 10.1099/0022-1317-76-7-1877. [DOI] [PubMed] [Google Scholar]
- Chungue E, Deubel V, Cassar O, Laille M, Martin PM. Molecular epidemiology of dengue 3 viruses and genetic relatedness among dengue 3 strains isolated from patients with mild or severe form of dengue fever in French Polynesia. J. Gen. Virol. 1993;74:2765–2770. doi: 10.1099/0022-1317-74-12-2765. [DOI] [PubMed] [Google Scholar]
- Cologna R, Rico-Hesse R. American genotype structures decrease dengue virus output from human monocytes and dendritic cells. J. Virol. 2003;77:3929–3938. doi: 10.1128/JVI.77.7.3929-3938.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo NS, Guillot S, Romanenkova N, Combiescu M, Aubert-Combiescu A, Seghier M, Caro V, Crainic R, Delpeyroux F. Genomic features of intertypic recombinant Sabin poliovirus strains excreted by primary vaccinees. J. Virol. 2001;75:5740–5751. doi: 10.1128/JVI.75.13.5740-5751.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar D, Castro A, Haines H. Demonstration of interference between dengue virus types in cultured mosquito cells using monoclonal antibody probes. J. Gen. Virol. 1982;59:273–282. doi: 10.1099/0022-1317-59-2-273. [DOI] [PubMed] [Google Scholar]
- Goncalvez AP, Escalante AA, Pujol FH, Ludert JE, Tovar D, Salas R, Liprandi F. Diversity and evolution of the envelope gene of dengue virus type 1. Virology. 2002;303:110–119. doi: 10.1006/viro.2002.1686. [DOI] [PubMed] [Google Scholar]
- Gould EA, de Lamballerie X, Zanotto PM, Holmes EC. Evolution, epidemiology, and dispersal of flaviviruses revealed by molecular phylogenies. Adv. Virus Res. 2001;57:71–103. doi: 10.1016/s0065-3527(01)57001-3. [DOI] [PubMed] [Google Scholar]
- Gubler DJ. Dengue and dengue hemorrhagic fever. Clin. Microbiol. Rev. 1998;11:480–496. doi: 10.1128/cmr.11.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler DJ, Kuno G, Sather GE, Waterman SH. A case of natural concurrent human infection with two dengue viruses. Am. J. Trop. Med. Hyg. 1985;34:170–173. doi: 10.4269/ajtmh.1985.34.170. [DOI] [PubMed] [Google Scholar]
- Gubler DJ, Nalim S, Tan R, Saipan H, Sulianti Saroso J. Variation in susceptibility to oral infection with dengue viruses among geographic strains of Aedes aegypti. Am. J. Trop. Med. Hyg. 1979;28:1045–1052. doi: 10.4269/ajtmh.1979.28.1045. [DOI] [PubMed] [Google Scholar]
- Gubler DJ, Reed D, Rosen L, Hitchcock JR., Jr. Epidemiologic, clinical, and virologic observations on dengue in the Kingdom of Tonga. Am. J. Trop. Med. Hyg. 1978;27:581–589. doi: 10.4269/ajtmh.1978.27.581. [DOI] [PubMed] [Google Scholar]
- Guzman MG, Deubel V, Pelegrino JL, Rosario D, Marrero M, Sariol C, Kouri G. Partial nucleotide and amino acid sequences of the envelope and the envelope/nonstructural protein-1 gene junction of four dengue-2 virus strains isolated during the 1981 Cuban epidemic. Am. J. Trop. Med. Hyg. 1995;52:241–246. doi: 10.4269/ajtmh.1995.52.241. [DOI] [PubMed] [Google Scholar]
- Halstead SB. Observations related to pathogenesis of dengue hemorrhagic fever: VI. Hypotheses and discussion. Yale J. Biol. Med. 1970;42:350–362. [PMC free article] [PubMed] [Google Scholar]
- Halstead SB. Pathogenesis of dengue: Challenges to molecular biology. Science. 1988;239:476–481. doi: 10.1126/science.3277268. [DOI] [PubMed] [Google Scholar]
- Harris E, Roberts TG, Smith L, Selle J, Kramer LD, Valle S, Sandoval E, Balmaseda A. Typing of dengue viruses in clinical specimens and mosquitoes by single-tube multiplex reverse transcriptase PCR. J. Clin. Microbiol. 1998;36:2634–2639. doi: 10.1128/jcm.36.9.2634-2639.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E, Videa E, Perez L, Sandoval E, Tellez Y, Perez ML, Cuadra R, Rocha J, Idiaquez W, Alonso RE, Delgado MA, Campo LA, Acevedo F, Gonzalez A, Amador JJ, Balmaseda A. Clinical, epidemiologic, and virologic features of dengue in the 1998 epidemic in Nicaragua. Am. J. Trop. Med. Hyg. 2000;63:5–11. doi: 10.4269/ajtmh.2000.63.5. [DOI] [PubMed] [Google Scholar]
- Henchal EA, Repik PM, McCown JM, Brandt WE. Identification of an antigenic and genetic variant of dengue-4 virus from the Caribbean. Am. J. Trop. Med. Hyg. 1986;35:393–400. doi: 10.4269/ajtmh.1986.35.393. [DOI] [PubMed] [Google Scholar]
- Higgins DG, Thompson JD, Gibson TJ. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- Hillis DM. Taxonomic sampling, phylogenetic accuracy, and investigator bias. Syst. Biol. 1998;47:3–8. doi: 10.1080/106351598260987. [DOI] [PubMed] [Google Scholar]
- Holmes EC. Molecular epidemiology and evolution of emerging infectious diseases. Br. Med. Bull. 1998;54:533–543. doi: 10.1093/oxfordjournals.bmb.a011708. [DOI] [PubMed] [Google Scholar]
- Holmes EC, Burch SS. The causes and consequences of genetic variation in dengue virus. Trends Microbiol. 2000;8:74–77. doi: 10.1016/s0966-842x(99)01669-8. [DOI] [PubMed] [Google Scholar]
- Holmes EC, Worobey M, Rambaut A. Phylogenetic evidence for recombination in dengue virus. Mol. Biol. Evol. 1999;16:405–409. doi: 10.1093/oxfordjournals.molbev.a026121. [DOI] [PubMed] [Google Scholar]
- Jenkins GM, Pagel M, Gould EA, de A. Zanotto PM, Holmes EC. Evolution of base composition and codon usage bias in the genus Flavivirus. J. Mol. Evol. 2001;52:383–390. doi: 10.1007/s002390010168. [DOI] [PubMed] [Google Scholar]
- Kerschner JH, Vorndam AV, Monath TP, Trent DW. Genetic and epidemiological studies of dengue type 2 viruses by hybridization using synthetic deoxyoligonucleotides as probes. J. Gen. Virol. 1986;67:2645–2661. doi: 10.1099/0022-1317-67-12-2645. [DOI] [PubMed] [Google Scholar]
- Kinney RM, Butrapet S, Chang GJ, Tsuchiya KR, Roehrig JT, Bhamarapra-vati N, Gubler DJ. Construction of infectious cDNA clones for dengue 2 virus: Strain 16681 and its attenuated vaccine derivative, strain PDK-53. Virology. 1997;230:300–308. doi: 10.1006/viro.1997.8500. [DOI] [PubMed] [Google Scholar]
- Kouri G, Mas P, Guzman MG, Soler M, Goyenechea A, Morier L. Dengue hemorrhagic fever in Cuba, 1981: Rapid diagnosis of the etiologic agent. Bull. Pan. Am. Health Organ. 1983;17:126–132. [PubMed] [Google Scholar]
- Laille M, Deubel V, Sainte-Marie FF. Demonstration of concurrent dengue 1 and dengue 3 infection in six patients by the polymerase chain reaction. J. Med. Virol. 1991;34:51–54. doi: 10.1002/jmv.1890340109. [DOI] [PubMed] [Google Scholar]
- Lanciotti RS, Gubler DJ, Trent DW. Molecular evolution and phylogeny of dengue-4 viruses. J. Gen. Virol. 1997;78:2279–2284. doi: 10.1099/0022-1317-78-9-2279. [DOI] [PubMed] [Google Scholar]
- Lanciotti RS, Lewis JG, Gubler D, Trent DW. Molecular evolution and epidemiology of dengue-3 viruses. J. Gen. Virol. 1994;75:65–75. doi: 10.1099/0022-1317-75-1-65. [DOI] [PubMed] [Google Scholar]
- Lee E, Weir RC, Dalgarno L. Changes in the dengue virus major envelope protein on passaging and their localization on the three-dimensional structure of the protein. Virology. 1997;232:281–290. doi: 10.1006/viro.1997.8570. [DOI] [PubMed] [Google Scholar]
- Leitmeyer KC, Vaughn DW, Watts DM, Salas R, Villalobos I, de Chacon, Ramos C, Rico-Hesse R. Dengue virus structural differences that correlate with pathogenesis. J. Virol. 1999;73:4738–4747. doi: 10.1128/jvi.73.6.4738-4747.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon AR, Milinkovitch MC. The metapopulation genetic algorithm: An efficient solution for the problem of large phylogeny estimation. Proc. Natl. Acad. Sci. USA. 2002;99:10516–10521. doi: 10.1073/pnas.162224399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JA, Chang GJ, Lanciotti RS, Kinney RM, Mayer LW, Trent DW. Phylogenetic relationships of dengue-2 viruses. Virology. 1993;197:216–224. doi: 10.1006/viro.1993.1582. [DOI] [PubMed] [Google Scholar]
- Libraty DH, Endy TP, Houng HS, Green S, Kalayanarooj S, Suntayakorn S, Chansiriwongs W, Vaughn DW, Nisalak A, Ennis FA, Rothman AL. Differing influences of virus burden and immune activation on disease severity in secondary dengue-3 virus infections. J. Infect. Dis. 2002;185:1213–1221. doi: 10.1086/340365. [DOI] [PubMed] [Google Scholar]
- Lin YL, Liao CL, Chen LK, Yeh CT, Liu CI, Ma SH, Huang YY, Huang YL, Kao CL, King CC. Study of dengue virus infection in SCID mice engrafted with human K562 cells. J. Virol. 1998;72:9729–9737. doi: 10.1128/jvi.72.12.9729-9737.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HM, Zheng DP, Zhang LB, Oberste MS, Pallansch MA, Kew OM. Molecular evolution of a type 1 wild-vaccine poliovirus recombinant during widespread circulation in China. J. Virol. 2000;74:11153–11161. doi: 10.1128/jvi.74.23.11153-11161.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorono-Pino MA, Cropp CB, Farfan JA, Vorndam AV, Rodriguez-Angulo EM, Rosado-Paredes EP, Flores-Flores LF, Beaty BJ, Gubler DJ. Common occurrence of concurrent infections by multiple dengue virus serotypes. Am. J. Trop. Med. Hyg. 1999;61:725–730. doi: 10.4269/ajtmh.1999.61.725. [DOI] [PubMed] [Google Scholar]
- Mangada MN, Igarashi A. Molecular and in vitro analysis of eight dengue type 2 viruses isolated from patients exhibiting different disease severities. Virology. 1998;244:458–466. doi: 10.1006/viro.1998.9093. [DOI] [PubMed] [Google Scholar]
- Messer WB, Vitarana UT, Sivananthan K, Elvtgala J, Preethimala LD, Ramesh R, Withana N, Gubler DJ, De Silva AM. Epidemiology of dengue in Sri Lanka before and after the emergence of epidemic dengue hemorrhagic fever. Am. J. Trop. Med. Hyg. 2002;66:765–773. doi: 10.4269/ajtmh.2002.66.765. [DOI] [PubMed] [Google Scholar]
- Miller BR, Mitchell CJ. Genetic selection of a flavivirus-refractory strain of the yellow fever mosquito Aedes aegypti. Am. J. Trop. Med. Hyg. 1991;45:399–407. doi: 10.4269/ajtmh.1991.45.399. [DOI] [PubMed] [Google Scholar]
- Monath TP, Wands JR, Hill LJ, Brown NV, Marciniak RA, Wong MA, Gentry MK, Burke DS, Grant JA, Trent DW. Geographic classification of dengue-2 virus strains by antigen signature analysis. Virology. 1986;154:313–324. doi: 10.1016/0042-6822(86)90457-5. [DOI] [PubMed] [Google Scholar]
- Pandey BD, Igarashi A. Severity-related molecular differences among nineteen strains of dengue type 2 viruses. Microbiol. Immunol. 2000;44:179–188. doi: 10.1111/j.1348-0421.2000.tb02480.x. [DOI] [PubMed] [Google Scholar]
- Pryor MJ, Carr JM, Hocking H, Davidson AD, Li P, Wright PJ. Replication of dengue virus type 2 in human monocyte-derived macrophages: Comparisons of isolates and recombinant viruses with substitutions at amino acid 390 in the envelope glycoprotein. Am. J. Trop. Med. Hyg. 2001;65:427–434. doi: 10.4269/ajtmh.2001.65.427. [DOI] [PubMed] [Google Scholar]
- Repik PM, Dalrymple JM, Brandt WE, McCown JM, Russell PK. RNA fingerprinting as a method for distinguishing dengue 1 virus strains. Am. J. Trop. Med. Hyg. 1983;32:577–589. doi: 10.4269/ajtmh.1983.32.577. [DOI] [PubMed] [Google Scholar]
- Rico-Hesse R. Molecular evolution and distribution of dengue viruses type 1 and 2 in nature. Virology. 1990;174:479–493. doi: 10.1016/0042-6822(90)90102-w. [DOI] [PubMed] [Google Scholar]
- Rico-Hesse R, Harrison LM, Nisalak A, Vaughn DW, Kalayanarooj S, Green S, Rothraan AL, Ennis FA. Molecular evolution of dengue type 2 virus in Thailand. Am. J. Trop. Med. Hyg. 1998;58:96–101. doi: 10.4269/ajtmh.1998.58.96. [DOI] [PubMed] [Google Scholar]
- Rico-Hesse R, Harrison LM, Salas RA, Tovar D, Nisalak A, Ramos C, Boshell J, de Mesa MT, Nogueira RM, da Rosa AT. Origins of dengue type 2 viruses associated with increased pathogenicity in the Americas. Virology. 1997;230:244–251. doi: 10.1006/viro.1997.8504. [DOI] [PubMed] [Google Scholar]
- Rocha CF, Morrison AC, Sihuincha M, Scott TW, Kochel TJ. Am. Soc. Trop. Med. Hyg. Denver. CO. 2002 Meeting abstracts. [Google Scholar]
- Rosen L. “The Emperor’s New Clothes” revisited, or reflections on the pathogenesis of dengue hemorrhagic fever. Am. J. Trop. Med. Hyg. 1977;26:337–343. doi: 10.4269/ajtmh.1977.26.337. [DOI] [PubMed] [Google Scholar]
- Rosen L. The pathogenesis of dengue haemorrhagic fever. Bylae Tot Samt. 1986:40–42. [PubMed] [Google Scholar]
- Rudnick A. Studies of the ecology of dengue in Malaysia: A preliminary report. J. Med. Entomol. 1965;2:203–208. doi: 10.1093/jmedent/2.2.203. [DOI] [PubMed] [Google Scholar]
- Russell PK, McCown JM. Comparison of dengue-2 and dengue-3 virus strains by neutralization tests and identification of a subtype of dengue-3. Am. J. Trop. Med. Hyg. 1972;21:97–99. doi: 10.4269/ajtmh.1972.21.97. [DOI] [PubMed] [Google Scholar]
- Sabin AB. Research on dengue during World War II. Am. J. Trop. Med. Hyg. 1952;1:30–50. doi: 10.4269/ajtmh.1952.1.30. [DOI] [PubMed] [Google Scholar]
- Sanchez IJ, Ruiz BH. A single nucleotide change in the E protein gene of dengue virus 2 Mexican strain affects neurovirulence in mice. J. Gen. Virol. 1996;77:2541–2545. doi: 10.1099/0022-1317-77-10-2541. [DOI] [PubMed] [Google Scholar]
- Sangkawibha N, Rojanasuphot S, Ahandrik S, Viriyapongse S, Jatanasen S, Salitul V, Phanthumachinda B, Halstead SB. Risk factors in dengue shock syndrome: A prospective epidemiologic study in Rayong, Thailand. I. The 1980 outbreak. Am. J. Epidemiol. 1984;120:653–669. doi: 10.1093/oxfordjournals.aje.a113932. [DOI] [PubMed] [Google Scholar]
- Shi PY, Brinton MA, Veal JM, Zhong YY, Wilson WD. Evidence for the existence of a pseudoknot structure at the 3′ terminus of the flavivirus genomic RNA. Biochemistry. 1996;35:4222–4230. doi: 10.1021/bi952398v. [DOI] [PubMed] [Google Scholar]
- Shurtleff AC, Beasley DW, Chen JJ, Ni H, Suderman MT, Wang H, Xu R, Wang E, Weaver SC, Watts DM, Russell KL, Barrett AD. Genetic variation in the 3′ non-coding region of dengue viruses. Virology. 2001;281:75–87. doi: 10.1006/viro.2000.0748. [DOI] [PubMed] [Google Scholar]
- Swofford D. PAUP*, Phylogenetic Analysis Using Parsimony (*and Other Methods) 4.0bl0. Sinauer; Sunderland, MA: 2002. [Google Scholar]
- Tolou HJ, Couissinier-Paris P, Durand JP, Mercier V, de Pina JJ, de Micco P, Billoir F, Charrel RN, de Lamballerie X. Evidence for recombination in natural populations of dengue virus type 1 based on the analysis of complete genome sequences. J. Gen. Virol. 2001;82:1283–1290. doi: 10.1099/0022-1317-82-6-1283. [DOI] [PubMed] [Google Scholar]
- Trent DW, Grant JA, Monath TP, Manske CL, Corina M, Fox GE. Genetic variation and microevolution of dengue 2 virus in southeast Asia. Virology. 1989;172:523–535. doi: 10.1016/0042-6822(89)90195-5. [DOI] [PubMed] [Google Scholar]
- Trent DW, Manske CL, Fox GE, Chu MC, Kliks S, Monath TP. The molecular epidemiology of dengue viruses: Genetic variation and microevolution. Appl. Virol. Res. 1990;2:293–315. [Google Scholar]
- Twiddy SS, Farrar JJ, Vinh Chau N, Wills B, Gould EA, Gritsun T, Lloyd G, Holmes EC. Phylogenetic relationships and differential selection pressures among genotypes of dengue-2 virus. Virology. 2002a;298:63–72. doi: 10.1006/viro.2002.1447. [DOI] [PubMed] [Google Scholar]
- Twiddy SS, Woelk CH, Holmes EC. Phylogenetic evidence for adaptive evolution of dengue viruses in nature. J. Gen. Virol. 2002b;83:1679–1689. doi: 10.1099/0022-1317-83-7-1679. [DOI] [PubMed] [Google Scholar]
- Usuku S, Castillo L, Sugimoto C, Noguchi Y, Yogo Y, Kobayashi N. Phylogenetic analysis of dengue-3 viruses prevalent in Guatemala during 1996–1998. Arch. Virol. 2001;146:1381–1390. doi: 10.1007/s007050170098. [DOI] [PubMed] [Google Scholar]
- Uzcategui NY, Camacho D, Comach G, Cuello de Uzcategui R, Holmes EC, Gould EA. Molecular epidemiology of dengue type 2 virus in Venezuela: Evidence for in situ virus evolution and recombination. J. Gen. Virol. 2001;82:2945–2953. doi: 10.1099/0022-1317-82-12-2945. [DOI] [PubMed] [Google Scholar]
- Vaughn DW, Green S, Kalayanarooj S, Innis BL, Nimmannitya S, Suntayakorn S, Endy TP, Raengsakulrach B, Rothman AL, Ennis FA, Nisalak A. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J. Infect. Dis. 2000;181:2–9. doi: 10.1086/315215. [DOI] [PubMed] [Google Scholar]
- Vezza AC, Rosen L, Repik P, Dalrymple J, Bishop DH. Characterization of the viral RNA species of prototype dengue viruses. Am. J. Trop. Med. Hyg. 1980;29:643–652. doi: 10.4269/ajtmh.1980.29.643. [DOI] [PubMed] [Google Scholar]
- Walker RJ, Henchal EA, Blok J, Repik P, Henchal LS, Burke DS, Robbins SJ, Gorman BM. Variation in dengue type 2 viruses isolated in Bangkok during 1980. J. Gen. Virol. 1988;69:591–602. doi: 10.1099/0022-1317-69-3-591. [DOI] [PubMed] [Google Scholar]
- Wang E, Ni H, Xu R, Barrett AD, Watowich SJ, Gubler DJ, Weaver SC. Evolutionary relationships of endemic/epidemic and sylvatic dengue viruses. J. Virol. 2000;74:3227–3234. doi: 10.1128/jvi.74.7.3227-3234.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts DM, Porter KR, Putvatana P, Vasquez B, Calampa C, Hayes CG, Halstead SB. Failure of secondary infection with American genotype dengue 2 to cause dengue haemorrhagic fever. Lancet. 1999;354:1431–1434. doi: 10.1016/S0140-6736(99)04015-5. [DOI] [PubMed] [Google Scholar]
- World Health Organization Disease outbreak news. 2002 Accessible at http://www.who.int/csr/don/en.
- Worobey M, Rambaut A, Holmes EC. Widespread intra-serotype recombination in natural populations of dengue virus. Proc. Natl. Acad. Sci. USA. 1999;96:7352–7357. doi: 10.1073/pnas.96.13.7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SJ, Grouard-Vogel G, Sun W, Mascola JR, Brachtel E, Putvatana R, Louder MK, Filgueira L, Marovich MA, Wong HK, Blauvelt A, Murphy GS, Robb ML, Innes BL, Birx DL, Hayes CG, Frankel SS. Human skin Langerhans cells are targets of dengue virus infection. Nature Med. 2000;6:816–820. doi: 10.1038/77553. [DOI] [PubMed] [Google Scholar]
- Wu SJ, Hayes CG, Dubois DR, Windheuser MG, Kang YH, Watts DM, Sieckmann DG. Evaluation of the severe combined immunodeficient (SCID) mouse as an animal model for dengue viral infection. Am. J. Trop. Med. Hyg. 1995;52:468–476. doi: 10.4269/ajtmh.1995.52.468. [DOI] [PubMed] [Google Scholar]
- Zanotto PM, Gould EA, Gao GF, Harvey PH, Holmes EC. Population dynamics of flaviviruses revealed by molecular phylogenies. Proc. Natl. Acad. Sci. USA. 1996;93:548–553. doi: 10.1073/pnas.93.2.548. [DOI] [PMC free article] [PubMed] [Google Scholar]