Abstract
The DNA-dependent protein kinase catalytic subunit (DNA-PKcs) and Ku heterodimer together form the biologically critical DNA-PK complex that plays key roles in the repair of ionizing radiation-induced DNA double-strand breaks through the non-homologous end-joining (NHEJ) pathway. Despite elegant and informative electron microscopy studies, the mechanism by which DNA-PK co-ordinates the initiation of NHEJ has been enigmatic due to limited structural information. Here, we discuss how the recently described small angle X-ray scattering structures of full-length Ku heterodimer and DNA-PKcs in solution, combined with a breakthrough DNA-PKcs crystal structure, provide significant insights into the early stages of NHEJ. Dynamic structural changes associated with a functionally important cluster of autophosphorylation sites play a significant role in regulating the dissociation of DNA-PKcs from Ku and DNA. These new structural insights have implications for understanding the formation and control of the DNA-PK synaptic complex, DNA-PKcs activation and initiation of NHEJ. More generally, they provide prototypic information for the phosphatidylinositol-3 kinase-like (PIKK) family of serine/threonine protein kinases that includes Ataxia Telangiectasia-Mutated (ATM) and ATM-, Rad3-related (ATR) as well as DNA-PKcs.
Keywords: DNA-PKcs, non-homologous end joining, DNA double strand break repair, SAXS, phosphorylation
Introduction
Cellular DNA is continually exposed to endogenous and exogenous agents that cause multiple forms of DNA damage. This damage must be faithfully and efficiently repaired in order to maintain the integrity of the genome and to ensure reliable duplication and inheritance of genetic material. One of the most deleterious forms of DNA damage is the double-strand break (DSB), which can arise through replication fork collapse or exposure to free radicals, reactive oxygen species, chemical agents such as chemotherapeutic drugs, UV and ionizing radiation (IR) [1].
IR-induced DSBs are formed when two single strand (ss) breaks occur in close proximity on opposite strands of the DNA. IR-induced DSBs are characterized by the presence of additional DNA damage, including base lesions and abasic sites within one to two helical turns of the DNA (so called clustered DNA damage), and frequently contain non-ligatable end groups such as 3′ phosphate or 3′-phosphoglycolate groups that must be removed prior to ligation [2–5]. In mammalian cells, the majority of IR-induced DSBs are repaired by the non-homologous end-joining (NHEJ) pathway. NHEJ is also responsible for the repair of DSBs produced during V(D)J recombination and, to a lesser extent, class switch recombination, thus defects in NHEJ can lead to defects in DSB repair, increased radiation sensitivity and immune deficiencies [6–10]. Accordingly, the core NHEJ factors have largely been identified based on their requirement for cellular survival after IR and by their function in V(D)J recombination. These core factors include the Ku70/80 heterodimer, the DNA-dependent protein kinase catalytic subunit (DNA-PKcs), Artemis, XRCC4, DNA ligase IV and XLF (also called Cernunnos).
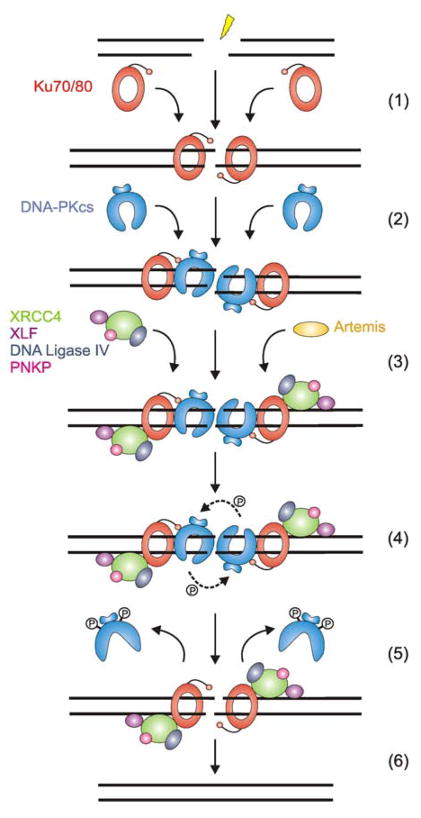
NHEJ is thought to proceed through three key steps: recognition of the break, DNA processing to remove non-ligatable ends or other forms of damage at the termini, and finally ligation of the DNA ends (Figure 1). Recognition of the DSB is carried out by the Ku heterodimer (Step 1, Figure 1), which is also required for recruitment of DNA-PKcs, the XRCC4/DNA ligase IV complex and XLF, reflecting its essential role in NHEJ [11–16]. In current models, binding of Ku to DNA is followed by recruitment of DNA-PKcs, which causes the inward translocation of Ku, positioning DNA-PKcs at the extreme DNA termini [15,17] (Step 2, Figure 1). Formation of the DNA-PK holoenzyme is dependent upon DNA binding as in the absence of DNA the complex does not form [18].
Figure 1. A simple model for NHEJ.
IR induces DNA ss breaks on opposite DNA strands resulting in a DSB with overhanging ends. In step 1 of NHEJ, the DSB is detected and bound by the Ku70/80 heterodimer (red). Once bound to the DSB, the flexible CTR of Ku80 recruits DNA-PKcs (blue) which induces inward translocation of Ku and positions DNA-PKcs at the extremity of the DSB (step 2). In step 3, the DNA ends are processed by one or more possible enzymes that include Artemis (yellow), PNKP (pink), DNA polymerases (not shown), or MRN (not shown). Either before or after end processing, DNA-PKcs undergoes autophosphorylation (step 4, indicated by arrows/dashed lines), resulting in a conformational change that opens the central DNA binding cavity, resulting in the release of autophosphorylated DNA-PKcs from DNA (step 5). In the final step (step 6), the XRCC4/DNA ligase IV complex (green/grey) ligates the DNA ends in a reaction that is stimulated by XLF (purple).
The interaction of two DNA-PKcs molecules on adjacent sides of the DSB (a configuration often referred to as a synaptic complex) (Step 2, Figure 1) stimulates the protein kinase activity of DNA-PKcs, leading to DNA-PKcs autophosphorylation and dissociation (Steps 4 and 5, Figure 1). Depending on the complexity of the DSB and the nature of the break termini, different processing factors may also be recruited (Step 3, Figure 1). Potential processing factors include Artemis, an endonuclease which interacts with, and is activated by, DNA-PKcs [19–26], as well as the Mre11-Rad50-Nbs1 (MRN) exo/endonuclease complex [27–30]. DNA polymerases μ and λ are reported to fill-in missing nucleotides [31,32], and polynucleotide kinase/phosphatase (PNKP), which interacts with XRCC4, removes 3′ phosphate groups and/or adds 5′-phosphate groups to DNA termini prior to ligation [33–35]. The final step in NHEJ is DNA ligation, which is carried out by the XRCC4/DNA ligase IV complex (Step 6, Figure 1). In addition, XLF interacts with XRCC4 and the XRCC4/ligase IV complex to stimulate end joining [36–39] by promoting re-adenylation of ligase IV [40]. In this review, we focus on the early events of NHEJ, in particular the key role DNA-PKcs and Ku play in coordinating the cellular response to IR-induced DSBs.
DNA-PKcs protein kinase activity is critical for NHEJ
DNA-PKcs is a member of the phosphatidylinositol-3 kinase-like (PIKK) family of serine/threonine protein kinases that also includes Ataxia Telangiectasia-Mutated (ATM) and ATM-, Rad3–related (ATR) [41]. DNA-PKcs is a large polypeptide of over 4000 amino acids and, like other PIKKs, is composed of a large N-terminal domain, predicted to be largely a-helical, and a C-terminal kinase domain flanked by FAT (FRAP, ATM, TRRAP) and FAT-C terminal (FAT-C) domains (Figure 2). DNA-PKcs is recruited to DNA ends through interaction with the Ku heterodimer [42]. Ku is composed of 70 and ~80 kDa subunits that together form a basket shaped structure which encircles a molecule of dsDNA [43]. The asymmetrical ring allows the Ku70 subunit to bind in the major groove proximal to the DSB and the Ku80 subunit in the minor groove distal to the break site [15]. In addition, both Ku70 and Ku80 contain unique C-terminal regions. The C-terminal region of Ku70 contains a SAP (SAF-A/B, Acinus and PIAS) domain that has been proposed to interact with chromatin [44,45], while the C-terminal region (CTR) of Ku80 contains a globular helical domain suggested to be involved in protein-protein interactions [46,47] in addition to a region at the extreme C terminus that is required for interaction with DNA-PKcs [48–50].
Figure 2. Domain map of DNA-PKcs showing identified post-translational modifications.
Amino acid numbers in DNA-PKcs (accession number P78527) corresponding to approximate domain boundaries (determined from the UniProt protein database [114]) are shown in black. The N-terminal alpha-helical domain (amino acids 1–2882) is shown in green. Positions of the L-rich (Leucine-rich), FAT (pink), kinase (yellow) and FATC (magenta) domains were determined by UniProt and confirmed in NCBI [115]. Identified phosphorylation sites are indicated in blue and acetylation sites are shown in red with the approximate positions indicated by either a circle for individual sites or a line for clustered sites. In vitro sites are shown in italics and in vivo phosphorylation sites are highlighted in bold with DNA damage-induced phosphorylation sites indicated by asterisks. Phosphorylation sites within the ABCDE/Thr-2609 cluster are boxed. References for phosphorylation sites are provided in Supplementary Table 1. The acetylation site at amino acid 2 is from [99]; other acetylation sites are from [98]. Caspase cleavage sites (Asp2713 and Asp2983) are from [112] and are indicated by dashed lines.
Small angle X-ray scattering (SAXS) analysis [51] of the full length Ku heterodimer in solution reveals that the Ku80 CTR forms a flexible arm consistent with its ability to interact with and recruit DNA-PKcs to DSBs [52]. Furthermore, the dimensions of the Ku80 CTR are such that in addition to recruiting and positioning the DNA-PKcs molecule proximal to the Ku heterodimer, it may also help to stabilize the adjacent DNA-PKcs molecule on the opposite side of the DSB, hence stabilizing the overall synaptic complex [52]. Indeed, such tethering of the DNA-PK molecules across the break may promote trans-autophosphorylation, which facilitates dissociation of DNA-PKcs from the DSB (discussed below).
The protein kinase activity of DNA-PKcs is essential for NHEJ as small molecule inhibitors or mutations that inactivate DNA-PKcs kinase activity result in radiation sensitivity and defects in DSB repair [53–55]. Identification of the physiological substrates of DNA-PKcs is therefore critical for understanding its role in NHEJ. Like other PIKK family members, DNA-PKcs phosphorylates many substrates, including itself (Supplementary Table 1), Artemis and XLF on serines or threonines that are followed by glutamines, i.e. SQ or TQ motifs [26,56–61]. However, an increasing number of in vitro substrates of DNA-PKcs are phosphorylated on serines or threonines that are followed by other amino acids, frequently leucine or tyrosine. For example, non-SQ/TQ in vitro phosphorylation sites have been identified in DNA-PKcs itself [59] (Supplementary Table 1); Ku70 and Ku80 [62]; XRCC4 [63,64]; XLF [60]; Artemis [26,61], DNA ligase IV [65], the Werner syndrome helicase, WRN [66,67] and heterogenous nuclear ribonuclear protein, hnRNP-U (also known as scaffold attachment factor-A or SAF-A) [68,69]. Moreover, phosphorylation of non-SQ/TQ sites in DNA-PKcs [70,71], XLF [60] and hnRNP-U [68,69] are DNA-damage induced and DNA-PK-dependent in vivo suggesting that these motifs are bona fide DNA-PK targets.
Surprisingly, although DNA-PKcs phosphorylates multiple proteins in vitro, relatively few phosphorylation events have been shown to be required for NHEJ in vivo. For example, although DNA-PKcs phosphorylates Ku, XRCC4, XLF and DNA ligase IV in vitro, phosphorylation at these sites does not seem to be essential for DSB repair in vivo [60,64,65,72]. Similarly, although Artemis is highly phosphorylated by DNA-PK in vitro [26,61,73], phosphorylation is predominantly ATM-dependent in vivo [74]. Indeed, to date, the most established substrate of DNA-PKcs is DNA-PKcs itself. DNA-PKcs is highly phosphorylated in vitro [59,75] and is phosphorylated at many of the same sites in vivo in response to DNA damage [56,59,70,71,75–77] (Figure 2 and Supplementary Table 1). DNA damage-induced phosphorylation of DNA-PKcs has been reported to be DNA-PKcs-dependent, i.e. consistent with autophosphorylation [16,70,71,75,78] although DNA-PKcs can also be phosphorylated by the related PIKKs, ATM and ATR [76,77].
Several lines of evidence suggest that phosphorylation of the Thr2609/ABCDE cluster of phosphorylation sites (see Figure 2) plays a significant role in regulating the dissociation of DNA-PKcs from Ku and DNA. Purified DNA-PKcs and Ku proteins co-immunoprecipitate in the absence of ATP but DNA-PKcs dissociates from Ku when ATP is included in the reaction [79–81] and phosphorylation-induced dissociation is reversed by protein phosphatases [82]. In contrast, purified DNA-PKcs containing serine/threonine to alanine mutations at six phosphorylation sites within the Thr2609 cluster is considerably less efficient at dissociation from Ku in vitro than is wild type DNA-PKcs [52,70,83]. Together, these studies support a model whereby DNA-PKcs is recruited to DNA-bound Ku, which stimulates the protein kinase activity of DNA-PKcs, resulting in autophosphorylation in trans across the DSB (Step 4, Figure 1) [71] and release of autophosphorylated DNA-PKcs from DNA-bound Ku (Step 5, Figure 1). Further support for this model comes from in vivo studies which have shown that wild type DNA-PKcs is released from sites of laser-induced damage more rapidly than either kinase-dead DNA-PKcs or DNA-PKcs containing alanine substitutions at Ser2056 and the Thr2609/ABCDE cluster [16,52]. Significantly, cells expressing DNA-PKcs in which the Thr2609/ABCDE cluster has been mutated to alanine are more radiation sensitive that cells lacking DNA-PKcs altogether [84]. Moreover, in these cells, the major alternative DSB repair pathway, homologous recombination, is suppressed [85], suggesting that autophosphorylation of DNA-PKcs may play a role in regulating pathway choice as well as NHEJ progression.
Although these studies point to a critical role for Thr2609/ABCDE phosphorylation in the regulation of NHEJ, the in vitro DNA-PKcs autophosphorylation sites identified to date (shown in Figure 2 and Supplementary Table 1) represent only a fraction of the total sites autophosphorylated [83] and the effects of additional autophosphorylation events on DNA-PKcs function in vitro and in vivo have yet to be determined. Moreover, phosphorylation at different sites within DNA-PKcs can have different biological outcomes [85,86], therefore the regulation of DNA-PKcs activity in vivo may be complex. Adding to this complexity, several recent proteomics studies have identified multiple additional in vivo phosphorylation [87–97] and acetylation sites [98,99] in DNA-PKcs (Figure 2 and Supplementary Table 1), and it will be interesting to determine both the enzymes responsible for these post-translational modifications as well as their effects on DNA-PK function.
Autophosphorylation-induced conformational plasticity in DNA-PKcs
Due in part to its large size (>4000 amino acids), characterization of DNA-PKcs has been difficult, however, elegant cryo-electron microscopy (cryo-EM) and negative stain EM studies have provided considerable information on the overall structure and dimensions of DNA-PKcs, albeit at low resolution [18,100–106]. These image-based structures consistently reveal a globular-shaped monomeric molecule with overall dimensions of approximately 70–120 × 130 × 150–160 Å [101,106–108]. The protein is composed of several recognizable regions: a head or crown domain and a palm or base as well as arms and a brow/forehead that surround a central intramolecular cavity or channel with dimensions suitable for binding single stranded and/or double-stranded (ds) DNA [101,102,106]. (Figure 3A). Indeed, dsDNA has been modeled into this large channel [106,107], whilst ssDNA has been modeled into a smaller channel within the head domain [102,106]. However, precise localization of the different domains of DNA-PKcs within these structures has proved challenging, with different models placing the kinase domain at opposite locations in the overall structure [103,104,106,109].
Figure 3. Structural models of DNA-PKcs.
(A) Cryo-EM structure of DNA-PKcs at ~7 Å showing the crown/head (red), arms (yellow), forehead/brow (purple), and base (blue), with the DNA binding cavity in the centre of the molecule. Reprinted with permission from [106]; (B) X-ray structure of DNA-PKcs in complex with the Ku80CTR at 6.6 Å from [110] (PDB ID 3KGV) showing the N-terminal HEAT/α-helical domains or arms (green), the FAT and FATC domains (magenta), and the kinase domain in yellow. A putative DNA binding domain within the central cavity is shown in blue and the forehead in light green. The regions where the HEAT repeat/α-helical arms abut the FAT/kinase/FATC domains (indicated by blue arrows) are predicted to exhibit conformational flexibility [110]. Phosphorylation within or close to this flexible region may induce conformational changes that regulate the interaction of DNA-PKcs with DNA (see text for details); (C) left - Single SAXS envelope for non-phosphorylated DNA-PKcs. Centre, superimposition of the DNA-PKcs crystal structure (coloured as in Fig 3B) on the average SAXS envelope. The enclosed cavity in the head region, observed in the EM structures, is observed in the SAXS data as dark shading (see text for details). Right, centre panel rotated by 90°; (D) left – Single SAXS envelope for autophosphorylated DNA-PKcs. Centre, average SAXS envelopes for autophosphorylated DNA-PKcs superimposed with the X-ray structure. Right, average SAXS envelope for autophosphorylated DNA-PKcs, rotated by 90° as in panel C.
The recently reported X-ray crystallographic structure of DNA-PKcs (in complex with the Ku80 CTR) at 6.6 Å has helped resolve some of these important questions, and reveals additional clues as to the function of DNA-PKcs [110]. In keeping with the cryo-EM structures, the X-ray structure of DNA-PKcs has a head or crown domain atop a ring-shaped palm or base region composed of two arms (shown in green in Figure 3B) that encircle a central open cavity [110]. These arms (which correspond to the N-terminal ~2880 residues of the protein, shown in green in Figure 2) are composed of multiple anti-parallel HEAT repeats, and other a-helical structures, which fold back on themselves and reverse direction creating a gap at the base of the molecule (Figure 3B). The arms thus form a pincer-like structure that encircles the putative dsDNA binding channel. Importantly, this structural arrangement places the FAT, kinase and FAT-C domains at the apex of the pincer arms [110] (magenta and yellow in Figure 3B). The regions at the top of the arms, abutting the FAT-kinase-FAT-C domains, are predicted to have considerable flexibility (indicated by blue arrows in Figure 3B), and may represent hinge regions that allow DNA-PKcs to undergo dynamic conformational changes [110].
The solution structure of DNA-PKcs obtained by SAXS reveals a similarly shaped molecule, with a recognizable head/crown and a large ring structure in the palm/base, corresponding to the central dsDNA binding channel (Figure 3C) that compares well with previous cryo-EM and low-resolution X-ray structures [52,111]. The SAXS structure also shows an enclosed cavity in the head region (indicated by the dark shading in Figure 3C) which may be analogous to the ssDNA binding channel proposed from EM structures [106].
Significantly, the solution structure of phosphorylated DNA-PKcs reveals that autophosphorylation produces a large conformational change and an increase in the D-max from 155Å to 180Å [52] (Figures 3C,D). As discussed above, the hinge regions identified in the crystal structure [110] (indicated by the blue arrows in Figure 3B) are in close proximity to the C-terminal FAT-kinase-FAT-C domains located at the apex of the ring (coloured magenta and yellow in Figure 3B). Since the FAT domain begins at residue 2883 (Figure 2), it seems likely that the cluster of in vivo phosphorylation sites, including the functionally significant Thr2609/ABCDE cluster, is located in close proximity to the hinge regions. Taken together the SAXS and crystallography data are consistent with autophosphorylation inducing a conformational change that increases the gap at the base of the pincers, opening the central DNA binding cavity and releasing DNA-PKcs from dsDNA [52,110,111]. Interestingly, DNA-PKcs also contains two caspase cleavage sites (Asp2713 and Asp2983) [112], suggesting that the hinge region is also proteolytically sensitive, allowing apoptotic cleavage of DNA-PKcs to separate the C-terminal FAT/kinase/FATC domain from the N-terminal DNA binding domain (Figure 2).
Conclusions and Perspectives
Recent structural data from crystallography and SAXS solution analyses have contributed significantly to our understanding of the initiating events of NHEJ and the dynamics of both assembly and disassembly of the DNA-PK complex. In particular, the recent X-ray structure provides the most detailed glimpse to date of the overall structure of DNA-PKcs [110]. This breakthrough crystal structure, in concert with SAXS structures of non-phosphorylated and autophosphorylated DNA-PKcs [52], provides a probable structural basis for the autophosphorylation-induced release of DNA-PKcs from DNA, which may be important not only for completion of NHEJ but also for regulation of pathway choice. Moreover, since other PIKK family members are also composed of an α-helical N-terminal domain and C-terminal FAT-kinase-FATC domains [41], the related protein kinases, ATM and ATR, will likely share a similar structure to that of DNA-PKcs. Indeed, the similarity in cryo-EM structures of DNA-PKcs and ATM, both of which exhibit a “pincer-arm” structure, supports this notion [113].
Together, these results provide a foundation for a more complete understanding of the structural dynamics of the DNA-bound DNA-PKcs-Ku complex, as well as the later steps in the repair of DSBs by NHEJ. However, the available structures are still of low resolution, preventing the assignment of individual residues, and further work is required to fully characterize the effects of phosphorylation on DNA-PKcs structure and function. Similarly, although low-resolution structures have been generated for the DNA-bound DNA-PKcs-Ku complex [52,105], the precise positioning of both the DNA and Ku within these complexes has made interpretation difficult and further work will provide more detailed information on the precise geometry of the putative DNA-PK synaptic complex. Nevertheless, taken together these studies illustrate how biochemical, cellular and structural approaches integrate to further our understanding of the functional roles of DNA repair machines in the DNA damage response.
Supplementary Material
Acknowledgments
We thank Michal Hammel for help with SAXS analyses and figures and Alberta Innovates-Health Solutions, the Canadian Institutes for Health Research and National Cancer Institute Structural Cell Biology of DNA Repair Machines grant CA92584 for support.
Abbreviations
- DNA-PK
DNA-dependent protein kinase
- DNA-PKcs
DNA-PK catalytic subunit
- ds
double-stranded
- DSB
DNA double strand break
- IR
ionizing radiation
- NHEJ
non-homologous end joining
- ss
single-stranded
- SAXS
small angle X-ray scattering
- XLF
XRCC4 like factor
- XRCC4
X-ray cross complementing gene 4
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA repair and mutagenesis. American Society for Microbiology; 2006. [Google Scholar]
- 2.Curtis SB. Lethal and potentially lethal lesions induced by radiation--a unified repair model. Radiat Res. 1986;106:252–270. [PubMed] [Google Scholar]
- 3.O’Neill P, Fielden EM. Primary free radical processes in DNA. Adv Radiat Biol. 1993;17:53–120. [Google Scholar]
- 4.Regulus P, Duroux B, Bayle PA, Favier A, Cadet J, Ravanat JL. Oxidation of the sugar moiety of DNA by ionizing radiation or bleomycin could induce the formation of a cluster DNA lesion. Proc Natl Acad Sci U S A. 2007;104:14032–14037. doi: 10.1073/pnas.0706044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.von Sonntag C. Free-Radical-Induced DNA Damage and Its Repair: A Chemical Perspective. Springer-Verlag; New York: 2006. [Google Scholar]
- 6.Christmann M, Tomicic MT, Roos WP, Kaina B. Mechanisms of human DNA repair: an update. Toxicology. 2003;193:3–34. doi: 10.1016/s0300-483x(03)00287-7. [DOI] [PubMed] [Google Scholar]
- 7.Jeggo PA. Identification of genes involved in repair of DNA double-strand breaks in mammalian cells. Radiat Res. 1998;150:S80–91. [PubMed] [Google Scholar]
- 8.Karagiannis TC, El-Osta A. Double-strand breaks: signaling pathways and repair mechanisms. Cell Mol Life Sci. 2004;61:2137–2147. doi: 10.1007/s00018-004-4174-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verkaik NS, Esveldt-van Lange RE, van Heemst D, Bruggenwirth HT, Hoeijmakers JH, Zdzienicka MZ, van Gent DC. Different types of V(D)J recombination and end-joining defects in DNA double-strand break repair mutant mammalian cells. Eur J Immunol. 2002;32:701–709. doi: 10.1002/1521-4141(200203)32:3<701::AID-IMMU701>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 10.Lieber MR, Grawunder U, Wu X, Yaneva M. Tying loose ends: roles of Ku and DNA-dependent protein kinase in the repair of double-strand breaks. Curr Opin Genet Dev. 1997;7:99–104. doi: 10.1016/s0959-437x(97)80116-5. [DOI] [PubMed] [Google Scholar]
- 11.Calsou P, Delteil C, Frit P, Drouet J, Salles B. Coordinated assembly of Ku and p460 subunits of the DNA-dependent protein kinase on DNA ends is necessary for XRCC4-ligase IV recruitment. J Mol Biol. 2003;326:93–103. doi: 10.1016/s0022-2836(02)01328-1. [DOI] [PubMed] [Google Scholar]
- 12.Chen L, Trujillo K, Sung P, Tomkinson AE. Interactions of the DNA ligase IV-XRCC4 complex with DNA ends and the DNA-dependent protein kinase. J Biol Chem. 2000;275:26196–26205. doi: 10.1074/jbc.M000491200. [DOI] [PubMed] [Google Scholar]
- 13.Nick McElhinny SA, Snowden CM, McCarville J, Ramsden DA. Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol Cell Biol. 2000;20:2996–3003. doi: 10.1128/mcb.20.9.2996-3003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yano K, Morotomi-Yano K, Wang SY, Uematsu N, Lee KJ, Asaithamby A, Weterings E, Chen DJ. Ku recruits XLF to DNA double-strand breaks. EMBO Rep. 2008;9:91–96. doi: 10.1038/sj.embor.7401137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoo S, Dynan WS. Geometry of a complex formed by double strand break repair proteins at a single DNA end: recruitment of DNA-PKcs induces inward translocation of Ku protein. Nucleic Acids Res. 1999;27:4679–4686. doi: 10.1093/nar/27.24.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uematsu N, Weterings E, Yano K, Morotomi-Yano K, Jakob B, Taucher-Scholz G, Mari PO, van Gent DC, Chen BP, Chen DJ. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J Cell Biol. 2007;177:219–229. doi: 10.1083/jcb.200608077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calsou P, Frit P, Humbert O, Muller C, Chen DJ, Salles B. The DNA-dependent protein kinase catalytic activity regulates DNA end processing by means of Ku entry into DNA. J Biol Chem. 1999;274:7848–7856. doi: 10.1074/jbc.274.12.7848. [DOI] [PubMed] [Google Scholar]
- 18.Yaneva M, Kowalewski T, Lieber MR. Interaction of DNA-dependent protein kinase with DNA and with Ku: biochemical and atomic-force microscopy studies. Embo J. 1997;16:5098–5112. doi: 10.1093/emboj/16.16.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeggo P, O’Neill P. The Greek Goddess, Artemis, reveals the secrets of her cleavage. DNA Repair (Amst) 2002;1:771–777. doi: 10.1016/s1568-7864(02)00096-4. [DOI] [PubMed] [Google Scholar]
- 20.Drouet J, Frit P, Delteil C, de Villartay JP, Salles B, Calsou P. Interplay between Ku, Artemis, and the DNA-dependent protein kinase catalytic subunit at DNA ends. J Biol Chem. 2006;281:27784–27793. doi: 10.1074/jbc.M603047200. [DOI] [PubMed] [Google Scholar]
- 21.Gu J, Li S, Zhang X, Wang LC, Niewolik D, Schwarz K, Legerski RJ, Zandi E, Lieber MR. DNA-PKcs regulates a single-stranded DNA endonuclease activity of Artemis. DNA Repair (Amst) 2010;9:429–437. doi: 10.1016/j.dnarep.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niewolik D, Pannicke U, Lu H, Ma Y, Wang LC, Kulesza P, Zandi E, Lieber MR, Schwarz K. DNA-PKcs dependence of Artemis endonucleolytic activity, differences between hairpins and 5′ or 3′ overhangs. J Biol Chem. 2006;281:33900–33909. doi: 10.1074/jbc.M606023200. [DOI] [PubMed] [Google Scholar]
- 23.Touvrey C, Couedel C, Soulas P, Couderc R, Jasin M, de Villartay JP, Marche PN, Jouvin-Marche E, Candeias SM. Distinct effects of DNA-PKcs and Artemis inactivation on signal joint formation in vivo. Mol Immunol. 2008;45:3383–3391. doi: 10.1016/j.molimm.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 24.van der Burg M, Ijspeert H, Verkaik NS, Turul T, Wiegant WW, Morotomi-Yano K, Mari PO, Tezcan I, Chen DJ, Zdzienicka MZ, van Dongen JJ, van Gent DC. A DNA-PKcs mutation in a radiosensitive T-B- SCID patient inhibits Artemis activation and nonhomologous end-joining. J Clin Invest. 2009;119:91–98. doi: 10.1172/JCI37141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma Y, Schwarz K, Lieber MR. The Artemis:DNA-PKcs endonuclease cleaves DNA loops, flaps, and gaps. DNA Repair (Amst) 2005;4:845–851. doi: 10.1016/j.dnarep.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 26.Goodarzi AA, Yu Y, Riballo E, Douglas P, Walker SA, Ye R, Harer C, Marchetti C, Morrice N, Jeggo PA, Lees-Miller SP. DNA-PK autophosphorylation facilitates Artemis endonuclease activity. Embo J. 2006;25:3880–3889. doi: 10.1038/sj.emboj.7601255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lloyd J, Chapman JR, Clapperton JA, Haire LF, Hartsuiker E, Li J, Carr AM, Jackson SP, Smerdon SJ. A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage. Cell. 2009;139:100–111. doi: 10.1016/j.cell.2009.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams RS, Dodson GE, Limbo O, Yamada Y, Williams JS, Guenther G, Classen S, Glover JN, Iwasaki H, Russell P, Tainer JA. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell. 2009;139:87–99. doi: 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie A, Kwok A, Scully R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat Struct Mol Biol. 2009;16:814–818. doi: 10.1038/nsmb.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhuang J, Jiang G, Willers H, Xia F. Exonuclease function of human Mre11 promotes deletional nonhomologous end joining. J Biol Chem. 2009;284:30565–30573. doi: 10.1074/jbc.M109.059444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andrade P, Martin MJ, Juarez R, Lopez de Saro F, Blanco L. Limited terminal transferase in human DNA polymerase mu defines the required balance between accuracy and efficiency in NHEJ. Proc Natl Acad Sci U S A. 2009;106:16203–16208. doi: 10.1073/pnas.0908492106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Capp JP, Boudsocq F, Bertrand P, Laroche-Clary A, Pourquier P, Lopez BS, Cazaux C, Hoffmann JS, Canitrot Y. The DNA polymerase lambda is required for the repair of non-compatible DNA double strand breaks by NHEJ in mammalian cells. Nucleic Acids Res. 2006;34:2998–3007. doi: 10.1093/nar/gkl380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chappell C, Hanakahi LA, Karimi-Busheri F, Weinfeld M, West SC. Involvement of human polynucleotide kinase in double-strand break repair by non-homologous end joining. Embo J. 2002;21:2827–2832. doi: 10.1093/emboj/21.11.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karimi-Busheri F, Daly G, Robins P, Canas B, Pappin DJ, Sgouros J, Miller GG, Fakhrai H, Davis EM, Le Beau MM, Weinfeld M. Molecular characterization of a human DNA kinase. J Biol Chem. 1999;274:24187–24194. doi: 10.1074/jbc.274.34.24187. [DOI] [PubMed] [Google Scholar]
- 35.Koch CA, Agyei R, Galicia S, Metalnikov P, O’Donnell P, Starostine A, Weinfeld M, Durocher D. Xrcc4 physically links DNA end processing by polynucleotide kinase to DNA ligation by DNA ligase IV. Embo J. 2004;23:3874–3885. doi: 10.1038/sj.emboj.7600375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 37.Buck D, Malivert L, de Chasseval R, Barraud A, Fondaneche MC, Sanal O, Plebani A, Stephan JL, Hufnagel M, le Deist F, Fischer A, Durandy A, de Villartay JP, Revy P. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 38.Gu J, Lu H, Tsai AG, Schwarz K, Lieber MR. Single-stranded DNA ligation and XLF-stimulated incompatible DNA end ligation by the XRCC4-DNA ligase IV complex: influence of terminal DNA sequence. Nucleic Acids Res. 2007;35:5755–5762. doi: 10.1093/nar/gkm579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai CJ, Kim SA, Chu G. Cernunnos/XLF promotes the ligation of mismatched and noncohesive DNA ends. Proc Natl Acad Sci U S A. 2007;104:7851–7856. doi: 10.1073/pnas.0702620104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Riballo E, Woodbine L, Stiff T, Walker SA, Goodarzi AA, Jeggo PA. XLF-Cernunnos promotes DNA ligase IV-XRCC4 re-adenylation following ligation. Nucleic Acids Res. 2009;37:482–492. doi: 10.1093/nar/gkn957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lempiainen H, Halazonetis TD. Emerging common themes in regulation of PIKKs and PI3Ks. Embo J. 2009;28:3067–3073. doi: 10.1038/emboj.2009.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gottlieb TM, Jackson SP. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- 43.Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 44.Lehman JA, Hoelz DJ, Turchi JJ. DNA-dependent conformational changes in the Ku heterodimer. Biochemistry. 2008;47:4359–4368. doi: 10.1021/bi702284c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Z, Zhu L, Lin D, Chen F, Chen DJ, Chen Y. The three-dimensional structure of the C-terminal DNA-binding domain of human Ku70. J Biol Chem. 2001;276:38231–38236. doi: 10.1074/jbc.M105238200. [DOI] [PubMed] [Google Scholar]
- 46.Harris R, Esposito D, Sankar A, Maman JD, Hinks JA, Pearl LH, Driscoll PC. The 3D solution structure of the C-terminal region of Ku86 (Ku86CTR) J Mol Biol. 2004;335:573–582. doi: 10.1016/j.jmb.2003.10.047. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Z, Hu W, Cano L, Lee TD, Chen DJ, Chen Y. Solution structure of the C-terminal domain of Ku80 suggests important sites for protein-protein interactions. Structure (Camb) 2004;12:495–502. doi: 10.1016/j.str.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 48.Gell D, Jackson SP. Mapping of protein-protein interactions within the DNA-dependent protein kinase complex. Nucleic Acids Res. 1999;27:3494–3502. doi: 10.1093/nar/27.17.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singleton BK, Torres-Arzayus MI, Rottinghaus ST, Taccioli GE, Jeggo PA. The C terminus of Ku80 activates the DNA-dependent protein kinase catalytic subunit. Mol Cell Biol. 1999;19:3267–3277. doi: 10.1128/mcb.19.5.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 51.Rambo RP, Tainer JA. Bridging the solution divide: comprehensive structural analyses of dynamic RNA, DNA, and protein assemblies by small-angle X-ray scattering. Current Opinion in Structural Biology. 2010;20:128–137. doi: 10.1016/j.sbi.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hammel M, Yu Y, Mahaney BL, Cai B, Ye R, Phipps BM, Rambo RP, Hura GL, Pelikan M, So S, Abolfath RM, Chen DJ, Lees-Miller SP, Tainer JA. Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. J Biol Chem. 2010;285:1414–1423. doi: 10.1074/jbc.M109.065615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao Y, Thomas HD, Batey MA, Cowell IG, Richardson CJ, Griffin RJ, Calvert AH, Newell DR, Smith GC, Curtin NJ. Preclinical evaluation of a potent novel DNA-dependent protein kinase inhibitor NU7441. Cancer Res. 2006;66:5354–5362. doi: 10.1158/0008-5472.CAN-05-4275. [DOI] [PubMed] [Google Scholar]
- 54.Hosoi Y, Miyachi H, Matsumoto Y, Ikehata H, Komura J, Ishii K, Zhao HJ, Yoshida M, Takai Y, Yamada S, Suzuki N, Ono T. A phosphatidylinositol 3-kinase inhibitor wortmannin induces radioresistant DNA synthesis and sensitizes cells to bleomycin and ionizing radiation. Int J Cancer. 1998;78:642–647. doi: 10.1002/(sici)1097-0215(19981123)78:5<642::aid-ijc19>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 55.Kurimasa A, Kumano S, Boubnov NV, Story MD, Tung CS, Peterson SR, Chen DJ. Requirement for the kinase activity of human DNA-dependent protein kinase catalytic subunit in DNA strand break rejoining. Mol Cell Biol. 1999;19:3877–3884. doi: 10.1128/mcb.19.5.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soubeyrand S, Pope L, Pakuts B, Hache RJ. Threonines 2638/2647 in DNA-PK are essential for cellular resistance to ionizing radiation. Cancer Res. 2003;63:1198–1201. [PubMed] [Google Scholar]
- 57.O’Neill T, Dwyer AJ, Ziv Y, Chan DW, Lees-Miller SP, Abraham RH, Lai JH, Hill D, Shiloh Y, Cantley LC, Rathbun GA. Utilization of oriented peptide libraries to identify substrate motifs selected by ATM. J Biol Chem. 2000;275:22719–22727. doi: 10.1074/jbc.M001002200. [DOI] [PubMed] [Google Scholar]
- 58.Lees-Miller SP, Sakaguchi K, Ullrich SJ, Appella E, Anderson CW. Human DNA-activated protein kinase phosphorylates serines 15 and 37 in the amino-terminal transactivation domain of human p53. Mol Cell Biol. 1992;12:5041–5049. doi: 10.1128/mcb.12.11.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Douglas P, Sapkota GP, Morrice N, Yu Y, Goodarzi AA, Merkle D, Meek K, Alessi DR, Lees-Miller SP. Identification of in vitro and in vivo phosphorylation sites in the catalytic subunit of the DNA-dependent protein kinase. Biochem J. 2002;368:243–251. doi: 10.1042/BJ20020973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu Y, Mahaney BL, Yano K, Ye R, Fang S, Douglas P, Chen DJ, Lees-Miller SP. DNA-PK and ATM phosphorylation sites in XLF/Cernunnos are not required for repair of DNA double strand breaks. DNA Repair (Amst) 2008;7:1680–1692. doi: 10.1016/j.dnarep.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ma Y, Pannicke U, Lu H, Niewolik D, Schwarz K, Lieber MR. The DNA-dependent protein kinase catalytic subunit phosphorylation sites in human Artemis. J Biol Chem. 2005;280:33839–33846. doi: 10.1074/jbc.M507113200. [DOI] [PubMed] [Google Scholar]
- 62.Chan DW, Ye R, Veillette CJ, Lees-Miller SP. DNA-dependent protein kinase phosphorylation sites in Ku 70/80 heterodimer. Biochemistry. 1999;38:1819–1828. doi: 10.1021/bi982584b. [DOI] [PubMed] [Google Scholar]
- 63.Lee KJ, Jovanovic M, Udayakumar D, Bladen CL, Dynan WS. Identification of DNA-PKcs phosphorylation sites in XRCC4 and effects of mutations at these sites on DNA end joining in a cell-free system. DNA Repair (Amst) 2004;3:267–276. doi: 10.1016/j.dnarep.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 64.Yu Y, Wang W, Ding Q, Ye R, Chen D, Merkle D, Schriemer D, Meek K, Lees-Miller SP. DNA-PK phosphorylation sites in XRCC4 are not required for survival after radiation or for V(D)J recombination. DNA Repair (Amst) 2003;2:1239–1252. doi: 10.1016/s1568-7864(03)00143-5. [DOI] [PubMed] [Google Scholar]
- 65.Wang YG, Nnakwe C, Lane WS, Modesti M, Frank KM. Phosphorylation and regulation of DNA ligase IV stability by DNA-dependent protein kinase. J Biol Chem. 2004;279:37282–37290. doi: 10.1074/jbc.M401217200. [DOI] [PubMed] [Google Scholar]
- 66.Perry JJ, Asaithamby A, Barnebey A, Kiamanesch F, Chen DJ, Han S, Tainer JA, Yannone SM. Identification of a coiled-coil in WRN that facilitates multimerization and promotes exonuclease processivity. J Biol Chem. 2010 doi: 10.1074/jbc.M110.124941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yannone SM, Roy S, Chan DW, Murphy MB, Huang S, Campisi J, Chen DJ. Werner syndrome protein is regulated and phosphorylated by DNA-dependent protein kinase. J Biol Chem. 2001;276:38242–38248. doi: 10.1074/jbc.M101913200. [DOI] [PubMed] [Google Scholar]
- 68.Berglund FM, Clarke PR. hnRNP-U is a specific DNA-dependent protein kinase substrate phosphorylated in response to DNA double-strand breaks. Biochem Biophys Res Commun. 2009;381:59–64. doi: 10.1016/j.bbrc.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 69.Britton S, Froment C, Frit P, Monsarrat B, Salles B, Calsou P. Cell nonhomologous end joining capacity controls SAF-A phosphorylation by DNA-PK in response to DNA double-strand breaks inducers. Cell Cycle. 2009;8:3717–3722. doi: 10.4161/cc.8.22.10025. [DOI] [PubMed] [Google Scholar]
- 70.Douglas P, Cui X, Block WD, Yu Y, Gupta S, Ding Q, Ye R, Morrice N, Lees-Miller SP, Meek K. The DNA-dependent protein kinase catalytic subunit is phosphorylated in vivo on threonine 3950, a highly conserved amino acid in the protein kinase domain. Mol Cell Biol. 2007;27:1581–1591. doi: 10.1128/MCB.01962-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Meek K, Douglas P, Cui X, Ding Q, Lees-Miller SP. trans Autophosphorylation at DNA-dependent protein kinase’s two major autophosphorylation site clusters facilitates end processing but not end joining. Mol Cell Biol. 2007;27:3881–3890. doi: 10.1128/MCB.02366-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Douglas P, Gupta S, Morrice N, Meek K, Lees-Miller SP. DNA-PK-dependent phosphorylation of Ku70/80 is not required for non-homologous end joining. DNA Repair (Amst) 2005;4:1006–1018. doi: 10.1016/j.dnarep.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 73.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 74.Zhang X, Succi J, Feng Z, Prithivirajsingh S, Story MD, Legerski RJ. Artemis is a phosphorylation target of ATM and ATR and is involved in the G2/M DNA damage checkpoint response. Mol Cell Biol. 2004;24:9207–9220. doi: 10.1128/MCB.24.20.9207-9220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chan DW, Chen BP, Prithivirajsingh S, Kurimasa A, Story MD, Qin J, Chen DJ. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 2002;16:2333–2338. doi: 10.1101/gad.1015202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen BP, Uematsu N, Kobayashi J, Lerenthal Y, Krempler A, Yajima H, Lobrich M, Shiloh Y, Chen DJ. Ataxia telangiectasia mutated (ATM) is essential for DNA-PKcs phosphorylations at the Thr-2609 cluster upon DNA double strand break. J Biol Chem. 2007;282:6582–6587. doi: 10.1074/jbc.M611605200. [DOI] [PubMed] [Google Scholar]
- 77.Yajima H, Lee KJ, Zhang S, Kobayashi J, Chen BP. DNA double-strand break formation upon UV-induced replication stress activates ATM and DNA-PKcs kinases. J Mol Biol. 2009;385:800–810. doi: 10.1016/j.jmb.2008.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen BP, Chan DW, Kobayashi J, Burma S, Asaithamby A, Morotomi-Yano K, Botvinick E, Qin J, Chen DJ. Cell cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double strand breaks. J Biol Chem. 2005;280:14709–14715. doi: 10.1074/jbc.M408827200. [DOI] [PubMed] [Google Scholar]
- 79.Lees-Miller SP, Chen YR, Anderson CW. Human cells contain a DNA-activated protein kinase that phosphorylates simian virus 40 T antigen, mouse p53, and the human Ku autoantigen. Mol Cell Biol. 1990;10:6472–6481. doi: 10.1128/mcb.10.12.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chan DW, Lees-Miller SP. The DNA-dependent protein kinase is inactivated by autophosphorylation of the catalytic subunit. J Biol Chem. 1996;271:8936–8941. doi: 10.1074/jbc.271.15.8936. [DOI] [PubMed] [Google Scholar]
- 81.Merkle D, Douglas P, Moorhead GB, Leonenko Z, Yu Y, Cramb D, Bazett-Jones DP, Lees-Miller SP. The DNA-dependent protein kinase interacts with DNA to form a protein-DNA complex that is disrupted by phosphorylation. Biochemistry. 2002;41:12706–12714. doi: 10.1021/bi0263558. [DOI] [PubMed] [Google Scholar]
- 82.Douglas P, Moorhead GB, Ye R, Lees-Miller SP. Protein phosphatases regulate DNA-dependent protein kinase activity. J Biol Chem. 2001;276:18992–18998. doi: 10.1074/jbc.M011703200. [DOI] [PubMed] [Google Scholar]
- 83.Block WD, Yu Y, Merkle D, Gifford JL, Ding Q, Meek K, Lees-Miller SP. Autophosphorylation-dependent remodeling of the DNA-dependent protein kinase catalytic subunit regulates ligation of DNA ends. Nucleic Acids Res. 2004;32:4351–4357. doi: 10.1093/nar/gkh761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ding Q, Reddy YV, Wang W, Woods T, Douglas P, Ramsden DA, Lees-Miller SP, Meek K. Autophosphorylation of the catalytic subunit of the DNA-dependent protein kinase Is required for efficient end processing during DNA double-strand break repair. Mol Cell Biol. 2003;23:5836–5848. doi: 10.1128/MCB.23.16.5836-5848.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cui X, Yu Y, Gupta S, Cho YM, Lees-Miller SP, Meek K. Autophosphorylation of DNA-dependent protein kinase regulates DNA end processing and may also alter double-strand break repair pathway choice. Mol Cell Biol. 2005;25:10842–10852. doi: 10.1128/MCB.25.24.10842-10852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Meek K, Dang V, Lees-Miller SP. DNA-PK: the means to justify the ends? Adv Immunol. 2008;99:33–58. doi: 10.1016/S0065-2776(08)00602-0. [DOI] [PubMed] [Google Scholar]
- 87.Stokes MP, Rush J, Macneill J, Ren JM, Sprott K, Nardone J, Yang V, Beausoleil SA, Gygi SP, Livingstone M, Zhang H, Polakiewicz RD, Comb MJ. Profiling of UV-induced ATM/ATR signaling pathways. Proc Natl Acad Sci U S A. 2007;104:19855–19860. doi: 10.1073/pnas.0707579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 89.Mayya V, Lundgren DH, Hwang SI, Rezaul K, Wu L, Eng JK, Rodionov V, Han DK. Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Sci Signal. 2009;2:ra46. doi: 10.1126/scisignal.2000007. [DOI] [PubMed] [Google Scholar]
- 90.Nousiainen M, Sillje HH, Sauer G, Nigg EA, Korner R. Phosphoproteome analysis of the human mitotic spindle. Proc Natl Acad Sci U S A. 2006;103:5391–5396. doi: 10.1073/pnas.0507066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 92.Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, Brunak S, Mann M. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 93.Oppermann FS, Gnad F, Olsen JV, Hornberger R, Greff Z, Keri G, Mann M, Daub H. Large-scale proteomics analysis of the human kinome. Mol Cell Proteomics. 2009;8:1751–1764. doi: 10.1074/mcp.M800588-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y, Hu Y, Tan Z, Stokes M, Sullivan L, Mitchell J, Wetzel R, Macneill J, Ren JM, Yuan J, Bakalarski CE, Villen J, Kornhauser JM, Smith B, Li D, Zhou X, Gygi SP, Gu TL, Polakiewicz RD, Rush J, Comb MJ. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 95.Daub H, Olsen JV, Bairlein M, Gnad F, Oppermann FS, Korner R, Greff Z, Keri G, Stemmann O, Mann M. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol Cell. 2008;31:438–448. doi: 10.1016/j.molcel.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 96.Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bennetzen MV, Larsen DH, Bunkenborg J, Bartek J, Lukas J, Andersen JS. Site-specific phosphorylation dynamics of the nuclear proteome during the DNA damage response. Mol Cell Proteomics. 2010;9:1314–1323. doi: 10.1074/mcp.M900616-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 99.Gauci S, Helbig AO, Slijper M, Krijgsveld J, Heck AJ, Mohammed S. Lys-N and trypsin cover complementary parts of the phosphoproteome in a refined SCX-based approach. Anal Chem. 2009;81:4493–4501. doi: 10.1021/ac9004309. [DOI] [PubMed] [Google Scholar]
- 100.Cary RB, Peterson SR, Wang J, Bear DG, Bradbury EM, Chen DJ. DNA looping by Ku and the DNA-dependent protein kinase. Proc Natl Acad Sci U S A. 1997;94:4267–4272. doi: 10.1073/pnas.94.9.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chiu CY, Cary RB, Chen DJ, Peterson SR, Stewart PL. Cryo-EM imaging of the catalytic subunit of the DNA-dependent protein kinase. J Mol Biol. 1998;284:1075–1081. doi: 10.1006/jmbi.1998.2212. [DOI] [PubMed] [Google Scholar]
- 102.Leuther KK, Hammarsten O, Kornberg RD, Chu G. Structure of DNA-dependent protein kinase: implications for its regulation by DNA. Embo J. 1999;18:1114–1123. doi: 10.1093/emboj/18.5.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rivera-Calzada A, Maman JD, Spagnolo L, Pearl LH, Llorca O. Three-dimensional structure and regulation of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) Structure. 2005;13:243–255. doi: 10.1016/j.str.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 104.Rivera-Calzada A, Spagnolo L, Pearl LH, Llorca O. Structural model of full-length human Ku70-Ku80 heterodimer and its recognition of DNA and DNA-PKcs. EMBO Rep. 2007;8:56–62. doi: 10.1038/sj.embor.7400847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Spagnolo L, Rivera-Calzada A, Pearl LH, Llorca O. Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol Cell. 2006;22:511–519. doi: 10.1016/j.molcel.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 106.Williams DR, Lee KJ, Shi J, Chen DJ, Stewart PL. Cryo-EM structure of the DNA-dependent protein kinase catalytic subunit at subnanometer resolution reveals alpha helices and insight into DNA binding. Structure. 2008;16:468–477. doi: 10.1016/j.str.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Boskovic J, Rivera-Calzada A, Maman JD, Chacon P, Willison KR, Pearl LH, Llorca O. Visualization of DNA-induced conformational changes in the DNA repair kinase DNA-PKcs. Embo J. 2003;22:5875–5882. doi: 10.1093/emboj/cdg555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Llorca O, Pearl LH. Electron microscopy studies on DNA recognition by DNA-PK. Micron. 2004;35:625–633. doi: 10.1016/j.micron.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 109.Brewerton SC, Dore AS, Drake AC, Leuther KK, Blundell TL. Structural analysis of DNA-PKcs: modelling of the repeat units and insights into the detailed molecular architecture. J Struct Biol. 2004;145:295–306. doi: 10.1016/j.jsb.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 110.Sibanda BL, Chirgadze DY, Blundell TL. Crystal structure of DNA-PKcs reveals a large open-ring cradle comprised of HEAT repeats. Nature. 2010;463:118–121. doi: 10.1038/nature08648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Perry JJ, Cotner-Gohara E, Ellenberger T, Tainer JA. Structural dynamics in DNA damage signaling and repair. Current Opinion in Structural Biology. 2010;20:283–294. doi: 10.1016/j.sbi.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Song Q, Lees-Miller SP, Kumar S, Zhang Z, Chan DW, Smith GC, Jackson SP, Alnemri ES, Litwack G, Khanna KK, Lavin MF. DNA-dependent protein kinase catalytic subunit: a target for an ICE-like protease in apoptosis. Embo J. 1996;15:3238–3246. [PMC free article] [PubMed] [Google Scholar]
- 113.Llorca O, Rivera-Calzada A, Grantham J, Willison KR. Electron microscopy and 3D reconstructions reveal that human ATM kinase uses an arm-like domain to clamp around double-stranded DNA. Oncogene. 2003;22:3867–3874. doi: 10.1038/sj.onc.1206649. [DOI] [PubMed] [Google Scholar]
- 114.The Universal Protein Resource (UniProt) [date accessed June 15th 2010];2010 doi: 10.1093/nar/gkp846. available at http://www.uniprot.org/uniprot/P78527. [DOI] [PMC free article] [PubMed]
- 115.NCBI protein database. [date accessed June 15th 2010];2010 available at http://www.ncbi.nlm.nih.gov/protein/P78527.3.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.