Abstract
Excess fatty acids are closely associated with metabolic dysfunction. The deleterious effects of fatty acids relate, in part, to their ability to up-regulate proinflammatory cytokines and propagate a state of systemic inflammation. CREBh is a recently identified transcription factor that appears to be required for hepatic synthesis of C-reactive protein (CRP). Recent data suggest that fatty acids can up-regulate CREBh, thus establishing a potential molecular link between fatty acids and inflammation. The aim of the current study was to examine the nature and mechanisms of fatty acid-mediated regulation of CREBh. H4IIE liver cells were incubated in the absence or presence of varying concentrations (50–500 μM) of albumin-bound, long-chain saturated (palmitate, stearate) or unsaturated (oleate, linoleate) fatty acids (1–16 hours). All fatty acids significantly increased CREBh gene expression via transcriptional mechanisms, at concentrations as low as 50 μM. Palmitate- or oleate-mediated upregulation of CREBh was not inhibited by triacsin C, an inhibitor of long-chain fatty acyl CoA synthetase, or by the PPARα antagonist, MK886. Inhibition of proteasome activity with MG132 or lactacystin, or inclusion of insulin reduced palmitate- and oleate-mediated increases in CREBh mRNA. Finally, we examined fatty acid regulation of CREBh in vivo. Male Wistar rats were exposed to a 4-hour pancreatic clamp combined with infusion of saline or a mixed lipid emulsion. CREBh mRNA and protein were significantly increased in rats exposed to the lipid infusion compared to the saline group. Collectively, these results may have important implications for metabolic diseases characterized by excess fatty acids, insulin resistance and inflammation.
Keywords: Endoplasmic reticulum, inflammation, obesity, liver
INTRODUCTION
CREBh is a recently identified, liver-specific, endoplasmic reticulum-localized transcription factor [1]. Mobilization of CREBh from the endoplasmic reticulum (ER) to the nucleus can be activated by pro-inflammatory cytokines, and CREBh is required for the hepatic synthesis of C-reactive protein and serum amyloid P-component [2], suggesting that CREBh is critical to the acute inflammatory response.
Excess free fatty acids are closely associated with metabolic dysfunction and represent a putative link between obesity and chronic diseases such as non-alcoholic fatty liver disease and cardiovascular disease [3–4]. At least some of the deleterious effects of excess fatty acids relate to their ability to up-regulate proinflammatory cytokines and propagate a state of systemic inflammation. Recent data suggest that fatty acids can up-regulate CREBh in HepG2 cells [5], thus establishing a putative molecular link between fatty acids and inflammation. However, several questions remain regarding how and under what conditions fatty acids regulate CREBh. The aim of the present study was to examine these questions by identifying potential molecular and cellular mediators of fatty acid-mediated regulation of CREBh, and to determine if this regulation occurs in vivo. Our results demonstrate that up-regulation of CREBh occurs in vitro and in vivo at fatty acid concentrations within the physiological range (i.e. 50 μM). This regulation occurs via transcriptional mechanisms that involve proteasome activity and insulin.
METHODS
Experimental agents
Fatty acids (Sigma Chemical Company, St. Louis, MO) were complexed to bovine serum albumin (BSA) at a 2:1 molar ratio [6]. Briefly, a 20 mM solution of fatty acid in 0.01 M NaOH was incubated at 70°C for 30 min. Addition of 1 N NaOH was used to promote solubilization. Fatty acid soaps were then complexed with fatty acid-free BSA in phosphate-buffered saline to achieve the appropriate 2:1 (fatty acid-to-albumin) molar ratio. Studies were performed in the absence or presence of the following compounds (all purchased from Sigma unless otherwise noted): Actinomycin-D (10 μg/ml), an inhibitor of transcription; triacsin C (10 μM), an inhibitor of long-chain fatty acyl CoA synthetase; MK886 (10 μM), a PPARα antagonist; WY14644 (10 μM), a PPARα agonist; lipopolysaccharide (LPS) (100 ng/ml), a toll-like receptor-4 (TLR4) agonist; zymosan (40μg/ml), a toll-like receptor-2 (TLR2) agonist; the proteasome inhibitors, MG132 (20 μM) and lactacystin (10 μM); insulin (100 nM); and the phosphatidylinositol 3 (PI3) kinase inhibitors wortmannin (1 μM) (Calbiochem, San Diego, CA) and LY294002 (50 μM) (Millipore, Bellerica, MA).
Cell culture
H4IIE liver cells (American Type Culture Collection, Manassas, VA), a rat liver hepatoma cell line, were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM), and supplemented with 10% fetal bovine serum, penicillin and streptomycin sulfate. Experiments were performed at 80–100% cell confluence.
Animals
Male Wistar Crl(WI)BR rats (Charles River, Wilmington, MA) weighing ~180 g upon arrival (7–8 wks of age) were provided free access to a purified high starch diet (Research Diets, Inc., New Brunswick, NJ) [7] and water for 1 wk. Rats were housed in pairs in a temperature- and humidity-controlled environment with a 12-hr light:dark cycle. All procedures were reviewed and approved by the Colorado State University institutional animal care and use committee.
Pancreatic clamp with lipid infusion
Following the acclimatization period, 10 rats underwent a pancreatic clamp to examine the acute effects of fatty acids on CREBh gene and protein expression. Catheters were placed in the carotid artery and jugular vein under general anesthesia as previously described [7–8]. Experiments were performed following 4–5 days of recovery, by which time body weights had recovered to at least 95% of their pre-surgery levels. Experiments were performed on conscious rats that were allowed unrestricted movement.
On the day of the experiment, extensions were added to the catheters and rats were allowed to rest for 15 minutes. Following a baseline blood sample (−10 min), a priming infusion of somatostatin release-inhibiting hormone (SRIF) and insulin was provided for 5 minutes (−10 to −5 min). Following the priming infusion, SRIF and insulin were infused at constant rates of 1.2 μg·kg−1·min−1 and 1 mU·kg−1·min−1, respectively, and a variable glucose infusion (20% dextrose) was used to maintain basal glucose levels. After five minutes, an infusion of the mixed lipid emulsion, Liposyn II (Hospira, Lake Forest, IL), sufficient to increase free fatty acid levels ~3-fold (LIPID, n=5), or saline containing glycerol (CON, n=5) was initiated and maintained, along with the SRIF, insulin and glucose, for 4 hrs (0 to 240 min). Liposyn II is a 20% triglyceride emulsion containing 10% safflower oil, 10% soybean oil, 1.2% egg phosphatides and 2.5% glycerin, with the following fatty acid composition: 65.8% linoleic, 17.7% oleic, 8.8% palmitic, 3.4% stearic, and 4.2% linolenic acid. Blood samples were obtained from the arterial catheter frequently during the first hour and then at ~10–15 min intervals during the final 3 hours. At the end of the experiment, rats were deeply anesthetized with sodium pentobarbital and liver samples were processed for RNA analysis and biochemical assays as described below.
Plasma measures
Blood samples were centrifuged, and plasma was either used immediately for glucose analysis or stored at −80° C. Glucose was measured with an automated analyzer (Beckman Instruments, Fullerton, CA); insulin was analyzed by ELISA (Linco Research, St. Charles, MO), and free fatty acid levels were determined using the WAKO NEFA-C kit (Richmond, VA).
RNA isolation and analysis
Total RNA was extracted with TRIzol reagent using the manufacturer’s protocol (Invitrogen, Carlsbad, CA). For Real Time PCR, reverse transcription was performed using 0.5 μg of DNase-treated RNA, Superscript II RnaseH- and random hexamers. PCR reactions were performed in 96 well plates using transcribed cDNA and IQ-SYBR green master mix (Bio Rad, Hecula, CA) using the following primer sets designed by the Beacon designer program version 3.1: 1) CREBh, forward: CGGTCCTTCTGCTGTCCTTC; reverse: CGTGGTTGTGGAGGGTTCTG; 2) CCAAT/enhancer-binding protein homologous protein (CHOP), forward: CCAGCAGAGGTCACAAGCAC; reverse: CGCACTGACCACTCTGTTTC; 3) growth arrest and DNA damage-inducible protein 34 (GADD34), forward: CTTCCTCTGTCGTCCTCGTCTC; reverse: CCCGCCTTCCTCCCAAGTC; 4) β2-microglobulin(control gene), forward: GGTGACCGTGATCTTTCTGGTG; reverse: GGATGGCGAGAGTACACTTGAATT; and 5) cyclophilin (control gene), forward: GTCAACCCCACCGTGTTCTTC; reverse: ACTTTGTCTGCAAACAGCTCGAA.
PCR efficiency was between 90 and 105% for primer and probe sets and linear over five orders of magnitude. Melting curve analysis and gel electrophoresis was used to examine the specificity of products generated for each primer set. Reactions were run in duplicate and the data recorded as the change in cycle threshold (ΔCT) for the target gene compared to the ΔCT for β2-microglobulin [9]. When cyclophilin was used in place of β2- microglobulin identical results were observed. The ΔCT ratio was averaged for control samples and the fold change calculated as the ratio of individual experimental group samples to this average.
Western blot analysis
Tissue or cells were harvested using a lysis buffer containing 20 mM HEPES, pH 7.4, 1% Triton X-100, 10% glycerol, 2 mM EGTA, 1 mM sodium vanadate, 2 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 50 mM β-glycerophosphate, 3 mM benzamidine, 10 μM leupeptin, 5 μM pepstatin, and 10 μg/ml aprotinin. Equivalent amounts of protein (50–100 μg) were subjected to SDS-PAGE, transferred to Hybond-P membranes (Amersham Pharmacia Biotech, Piscataway, NJ), and the membranes incubated with antiserum raised in two rabbits against a peptide consisting of the N-terminal sequence IAVLLLSFALIILPSISPFNSNKVDSPGDFVPVRVFSRTLHNH of mouse CREBh (Pacific Immunology, Ramona, CA) and actin (Sigma). Proteins were detected using horseradish peroxidase-conjugated secondary antibodies and an enhanced chemiluminescence reagent (Pierce, Rockford, IL). Density was quantified using a UVP Bioimaging system (Upland, CA).
Tracer studies
Liver cells were incubated with [3H]-oleate or [3H]-palmitate in the presence or absence of triacsin C. Lipids were extracted as described previously [10]. Oxidation of oleate or palmitate was estimated as [3H]-H2O production in medium as previously described [11]. Radioactivity was determined in a Liquid Scintillation Counter (Hewlett Packard).
CREBh mRNA stability and transcription
Stability of CREBh mRNA was determined in the absence or presence of Actinomycin-D (10 μg/ml). Liver cells were harvested for RNA analysis at 3, 5, 7, and 9 hrs. CREBh transcription was measured by nuclear run-on assay as described previously [12].
Data analysis
For in vitro experiments, data were analyzed using analysis of variance or independent t-tests. For the clamp experiment, group differences (i.e. control vs lipid) in the dependent variables were determined by one-or two-way analysis of variance (ANOVA). Changes in plasma-derived substances (e.g. glucose, insulin) from 0 to 240 min of the clamp were determined using repeated measures ANOVA. Post-hoc comparisons among means were made using the Tukey’s test. Statistical significance was set at p<0.05. All data are reported as the means ± SDEV for 3–8 independent experiments. The specific number of individual experiments are listed in each figure legend.
RESULTS
Fatty acids increase CREBh expression in vitro at physiologic concentrations
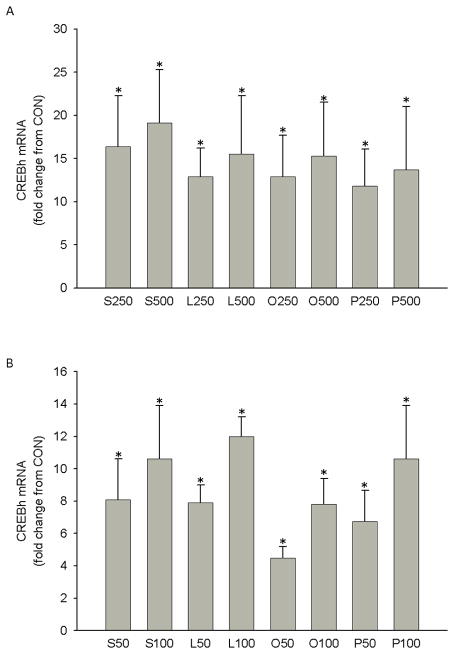
Danno et al. recently demonstrated that palmitate or oleate at concentrations of 200 or 500 μM, and eicosapentanoic acid at concentrations of 100 and 250 μM, increased CREBh mRNA (100–300%) and CREBh promoter activity (50–100%) in HepG2 cells [5]. To corroborate and extend these findings, we incubated H4IIE liver cells with varying concentrations of long chain-saturated (palmitate or stearate) or -unsaturated (linoleate or oleate) fatty acids for 6 hrs. We found that all fatty acids significantly increased CREBh mRNA at concentrations as low as 50 μM (Fig. 1a & 1b).
Figure 1.
Effect of fatty acids on CREBh mRNA in H4IIE liver cells. a H4IIE liver cells were incubated with stearate, linoleate, oleate, palmitate or control media containing 8 mM glucose (CON) for 6 hr. Data are expressed relative to CON which was set to 1. S250, stearate at 250 μM (n=4); S500, stearate at 500 μM (n=4); L250, linoleate at 250 μM (n=4); L500, linoleate at 500 μM (n=4); O250, oleate at 250 μM (n=13); O500, oleate at 500 μM, (n=32); P250, palmitate at 250 μM (n=16); P500, palmitate at 500 μM (n=32); *, significantly different from CON. b Effect of physiologic doses of fatty acids (50 and 100 μM) on CREBh gene mRNA in H4IIE liver cells. S50, stearate at 50 μM (n=8); S100, stearate at 100 μM (n=8); L50, linoleate at 50 μM (n=4); L100, linoleate at 100 μM (n=4); O50, oleate at 50 μM (n=4); O100, oleate at 100 μM, (n=4); P50, palmitate at 50 μM (n= 8); p100, palmitate at 100 μM (n= 8); *, significantly different from CON; all data are reported as the means ± SDEV.
Fatty acids increase CREBh transcription
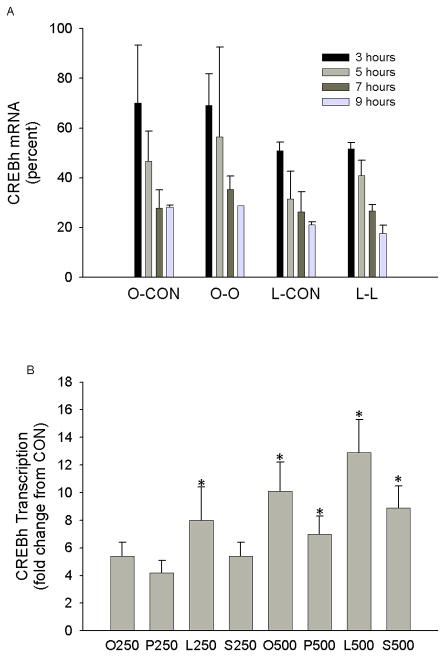
We next examined whether fatty acid-mediated induction of CREBh mRNA in H4IIE liver cells involves changes in mRNA stability and/or transcription. Although oleate or linoleate did not enhance CREBh stability (Fig. 2a), all fatty acids (oleate, linoleate, palmitate and stearate) increased CREBh transcription by4- to 12-fold at concentrations of 250 and 500 μM (Fig. 2b). These data demonstrate that the observed up-regulation of CREBh mRNA by fatty acids in H4IIE liver cells occurs primarily through activation of gene transcription. These experiments also indicate that fatty acids do not differ in their ability to up-regulate CREBh mRNA; thus subsequent experiments primarily utilized oleate and palmitate.
Figure 2.
Effect of fatty acids on CREBh mRNA stability and transcription in H4IIE liver cells. a Effect of fatty acids on CREBh mRNA stability in H4IIE liver cells. Cells were cultured in the presence of oleate (O; 250 μM) or linoleate (L; 250 μM). Transcription was then halted with the addition of actinomycin D (10 μg/ml), and incubations were continued for 3–9 hours in the presence of oleate (250 μM), linoleate (250 μM) or control media (CON). Data are expressed relative to the 0 time point (immediately before addition of actinomycin D), which was set to 100%; n=2–4 for all conditions. b The effect of fatty acids (3 hr) on CREBh gene transcription, as measured by nuclear run-on assay, in H4IIE liver cells. Data are expressed relative to control (CON), which was set to 1. O250, oleate at 250 μM; O500, oleate at 500 μM; P250, palmitate at 250 μM; P500, palmitate at 500 μM; L250, linoleate at 250 μM; L500, linoleate at 500 μM; S250, stearate at 250 μM; S2=500, stearate at 500 μM *, significantly different from CON; n=3 for all conditions; all data are reported as the means ± SDEV.
Fatty acid induction of CREBh does not require fatty acid metabolism
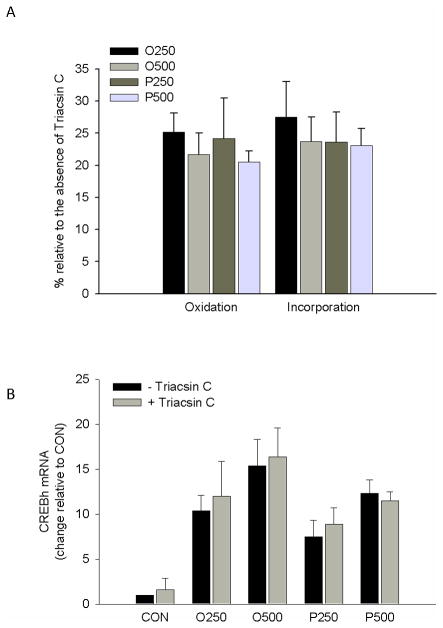
To examine if the fatty acid-mediated increase in CREBh gene expression required fatty acid metabolism, H4IIE liver cells were incubated (6 hrs) with [3H]-oleate and albumin-bound oleate (250 or 500 μM) or [3H]-palmitate and albumin-bound palmitate (250 or 500 μM) in the absence or presence of triacsin C, an inhibitor of long chain acyl CoA synthetase. Triacsin C reduced tracer estimated fatty acid oxidation and fatty acid incorporation into total lipid by approximately 75% (Fig. 3a), but did not reduce oleate or palmitate-mediated up-regulation of CREBh mRNA (Fig. 3b).
Figure 3.
The effect of inhibitors of fatty acid metabolism on fatty acid induction of CREBh mRNA. a Effectiveness of triacsin C (10μM, 6hr) on tracer estimated fatty acid oxidation and labeled fatty acid incorporation into lipid. O250, [3H]-oleate + albumin-bound oleate at 250 μM; O500, [3H]-oleate + albumin-bound oleate at 500 μM; P250, [3H]-palmitate + albumin-bound palmitate at 250 μM; P500, [3H]-palmitate + albumin-bound palmitate at 500 μM; *, significantly different than without Triacsin C; n=3 for all conditions. b Effect of triacsin C (10μM, 6hr) on fatty acid-mediated up-regulation of CREBh mRNA. O250, albumin-bound oleate at 250 μM; O500, albumin-bound oleate at 500 μM; P250, albumin-bound palmitate at 250 μM; P500, albumin-bound palmitate at 500 μM; n=4 for all conditions; all data are reported as the means ± SDEV.
PPARα and fatty acid-mediated CREBh gene expression
Recent data suggest that the PPARα agonist, fenofibrate, increases CREBh mRNA in HepG2 cells [5]. Therefore, we next examined whether PPARα was involved in fatty acid-mediated regulation of CREBh mRNA. Similar to previously published data., CREBh mRNA was significantly increased by the PPARα agonist, WY14644, and this increase was prevented by the PPARα antagonist, MK886 (Fig. 4a). However, despite the well know ability of fatty acids to activate PPARα [13–14], MK886 did not reduce oleate-mediated activation of CREBh mRNA (Fig 4b; similar results were found for linoleate and palmitate (data not shown)). These data suggest that fatty acid regulation of CREBh mRNA does not involve PPARα signaling.
Figure 4.
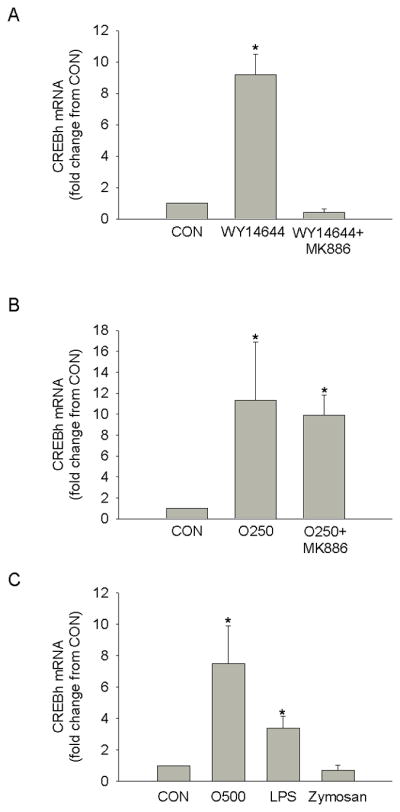
The effect of PPARα on fatty acid-mediated CREBh gene expression. a Effect of the PPARα agonist, WY14644 (10μM, 6hr), with and without a 16 hr pre-incubation with the PPARα antagonist, MK886 (10μM), on CREBh mRNA in H4IIE liver cells. b Effect of 250 μM of oleate (O250, 6hr) with and without a 16 hr pre-incubation with the PPARα antagonist, MK886 (10μM), on CREBh mRNA in H4IIE liver cells. c Effect of 6hr incubation with the TLR4 agonist, LPS (100 ng/ml), the TLR2 agonist, zymosan (40 ug/ml) or oleate (500 μM; O500) on CREBh mRNA in H4IIE liver cells. *, significantly different from control (CON); n=3 for all conditions; all data are reported as the means ± SDEV.
Like PPARα, toll-like receptors (TLRs) have emerged as important mediators of the biologic effects of fatty acids. In particular, TLR4 has been shown to mediate the pro-inflammatory effects of fatty acids in various cell types [15–16]. To begin to elucidate the role of TLRs in CREBh regulation, H4IIE liver cells were incubated with LPS, an activator of TLR4, or zymosan, an activator of TLR2. LPS, but not zymosan, significantly increased CREBh mRNA. These data suggest that TLR4-mediated signaling can regulate CREBh mRNA (Fig. 4c).
Fatty acid- mediated increase in CREBh gene expression is blocked by inhibitors of proteasome activity
Previous studies have demonstrated that fatty acid activation of proinflammatory cytokines and propagation of a systemic inflammatory state involves proteasome activation [17–20]. Therefore, we next examined the effects of inhibitors of proteasome activity, MG132 or lactacystin, on fatty acid-mediated up-regulation of CREBh mRNA. MG132 or lactacystin prevented the increase in CREBh mRNA in response to palmitate or oleate (Fig. 5). Importantly, upregulation of the ER stress related genes, CHOP and GADD34, were not affected (supplemental Fig. 1), suggesting that these inhibitors did not indiscriminately reduce overall gene expression. These data suggest that the fatty acid up-regulation of CREBh mRNA requires proteasome activity.
Figure 5.
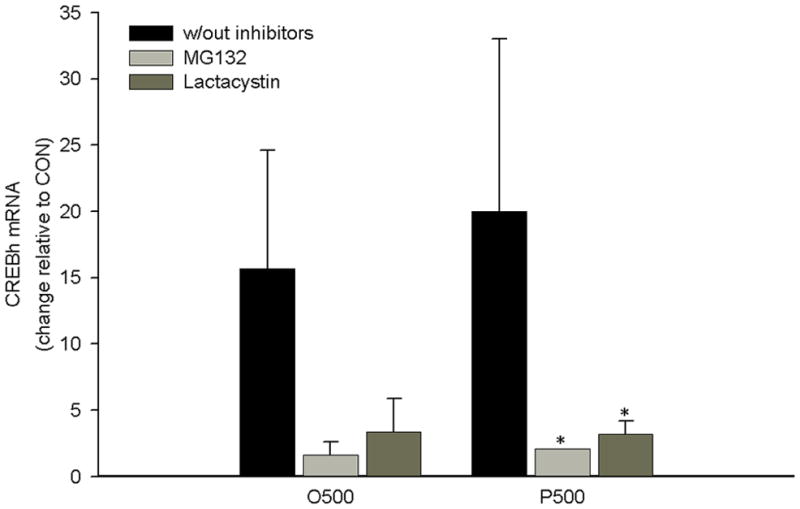
The effect of the proteasome inhibitors MG132 (20 μM, 6 hrs) and lactacystin (10 μM, 6 hrs) on fatty acid-mediated up-regulation of CREBh mRNA. O500, oleate at 500 μM; P500, palmitate at 500 μM; *, significantly different from same condition without inhibitors; n=3 for all conditions; all data are reported as the means ± SDEV.
Fatty acid- mediated increase in CREBh gene expression is blocked by insulin via PI3-kinase
Recent data indicate that CREBh mRNA is suppressed in the fed state [5]. We hypothesized that this suppression may be due to postprandial hyperinsulinemia. Indeed, insulin (100 nM) abolished palmitate- and oleate-mediated up-regulation of CREBh mRNA (Fig. 6a–6b) and the PI 3-kinase inhibitors, wortmannin (Fig. 6a) or LY294002 (Fig. 6b), interfered with insulin-mediated suppression of fatty acid up-regulation of CREBh. These data suggest that insulin-mediated signaling through PI3K is an important determinant of fatty acid-mediated regulation of CREBh mRNA in liver cells.
Figure 6.
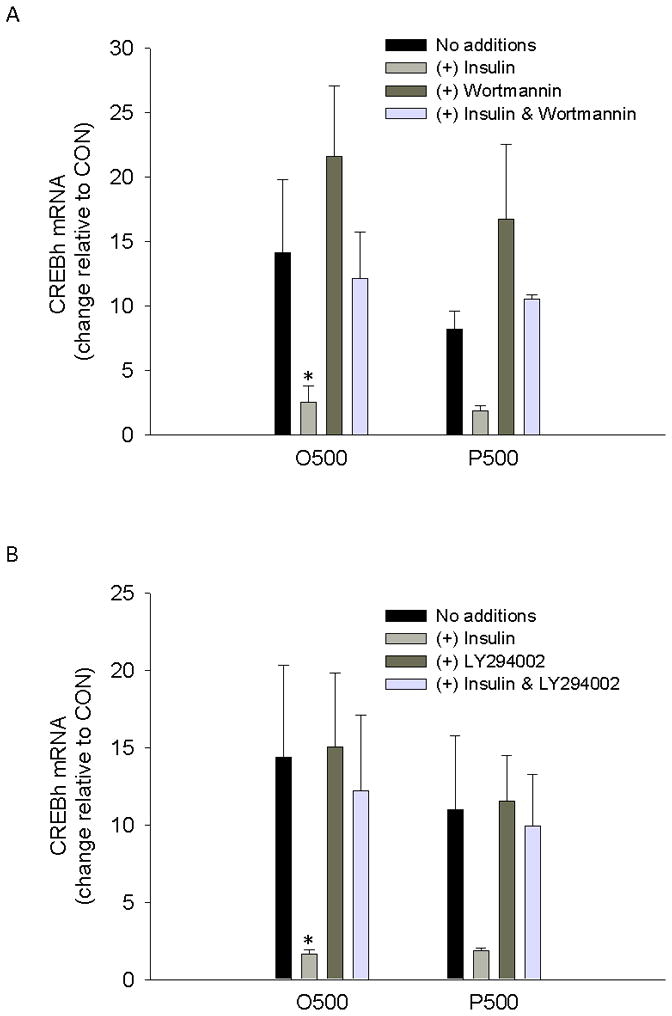
The effect of insulin and/or inhibitors of PI3K on fatty acid induction of CREBh gene expression. a Effect of insulin (100nM) and/or the PI3K inhibitor, wortmannin (1 μM), on fatty acid mediated up-regulation of CREBh gene expression; O500, oleate at 500 μM; P500, palmitate at 500 μM; *, significantly different from same condition with no additions; n=3 for all conditions b Effect of insulin (100nM) and/or the PI3K inhibitor, LY294002 (50 μM), on fatty acid mediated up-regulation of CREBh gene expression; All incubations were 6 hr in duration and data are presented relative to the low control (CON); O500, oleate at 500 μM; P500, palmitate at 500 μM; *, significantly different from same condition with no additions; n=3 for all conditions; all data are reported as the means ± SDEV.
Fatty acids increase CREBh expression in vivo
Our in vitro results indicated that long chain saturated and unsaturated fatty acids increase CREBh gene expression. To examine fatty acid regulation of CREBh in vivo, a mixed lipid emulsion (LIPID), containing both saturated and unsaturated fatty acids, or saline + glycerol control (CON) was infused into rats for 4 hrs. Body weights on the day of the study were not significantly different between LIPID and CON (242 ± 15 vs. 282 ± 49 g). Pre-infusion glucose levels were lower in LIPID compared to CON (113 ± 4 mg/dl vs. 131 ± 9 mg/dl; p<0.05), whereas insulin (0.7 ± 0.2 vs 0.6 ± 0.1 ng/ml) and free fatty acids (0.5 ± 0.4 vs. 0.5 ± 0.4 mM) were not significantly different between groups. Insulin values averaged over the 4-hr clamp did not differ between LIPID and CON (0.63 ± 0.10 vs. 0.57 ± 0.05 ng/ml) and no differences were observed between groups at any time point. Similarly, glucose values did not differ at any timepoint or when averaged over the duration of the clamp (131.4 ± 7.7 vs. 134.9 ± 8.1 mg/dl). Free fatty acids increased approximately 4-fold in LIPID compared to CON (p<0.05) (Fig. 7a). CREBh mRNA was undetectable in the heart or kidney (data not shown), but was increased ~3.5-fold in livers taken from LIPID compared to CON (Fig. 7b). In agreement with previous reports [2], the newly raised CREBh antiserum detected a single 75 kD protein in whole liver lysates but not in lysates from skeletal muscle or kidney (data not shown). Importantly, hepatic CREBh protein was significantly increased in LIPID compared to CON (Fig. 7c). These data suggest that fatty acid regulation of CREBh gene and protein expression occurs in vivo.
Figure 7.
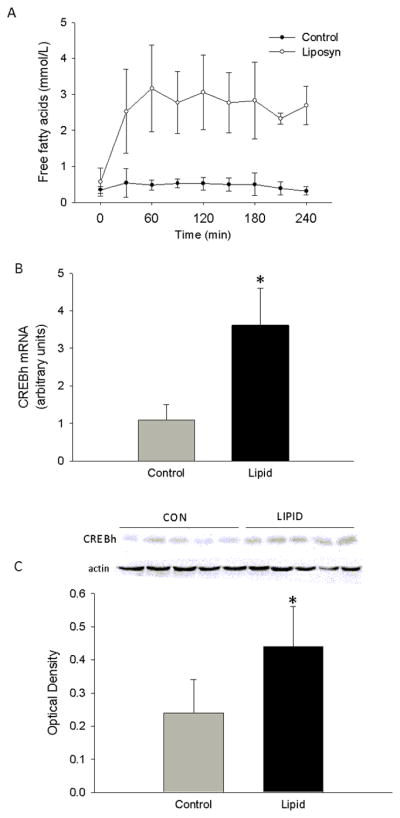
Effect of a 4-hr infusion of either a lipid emulsion (LIPID, n=5) or saline + glycerol (CON, n=5) on a plasma free fatty acids, b hepatic CREBh mRNA and (c) hepatic CREBh protein, in Male Wistar rats. Baseline blood samples were drawn at −10 min (fasting), after which time priming infusions of somatostatin and insulin were initiatedfor 5 min (−10 to −5 min). At −5 min, the infusion rates were reduced to1.2 μg·kg−1·min−1 and 1 mU·kg−1·min−1 for somatostatin and insulin, respectively, and a variable glucose infusion was initiated. A mixed lipid emulsion (Liposyn II) or saline infusion was then initiated and maintained, along with the somatostatin, insulin and glucose, for 4 hours (0 to 240 min); c optical density represents the ratio of CREBh:actin *, significantly different from control at corresponding time point; all data are reported as the means ± SDEV.
DISCUSSION
Elevated fatty acids are closely linked to metabolic disorders, including obesity, type 2 diabetes, and cardiovascular disease. One putative mechanism by which fatty acids promote metabolic dysfunction is via propagation of a pro-inflammatory state, although it is presently not clear how fatty acids promote inflammation. CREBh is a recently identified liver-specific transcription factor belonging to the CREB/ATF family. Zhang et al., found that genetically altered mice lacking CREBh have scarcely detectable levels of C-reactive protein in the basal state and following stimulation with proinflammatory cytokines or bacterial LPS, suggesting that CREBh is required for initiation of the acute phase response and systemic inflammation [2]. The aim of the present study was to extend recent findings linking fatty acids to the regulation of CREBh. Our primary results demonstrate that: (1) long-chain saturated (palmitate and stearate) and unsaturated (oleate and linoleate) fatty acids increase CREBh transcription at physiological concentrations; (2) oleate and palmitate-mediated regulation of CREBh appears to occur independently of fatty acid metabolism and PPARα signaling; (3) oleate and palmitate regulation of CREBh appears to be dependent upon proteasome activity; (4) oleate- and palmitate-mediated regulation of CREBh expression is repressed by PI3 kinase-dependent insulin signaling and (4) fatty acid-mediated regulation of CREBh occurs in vivo.
Regulated intramembrane proteolysis (RIP) of endoplasmic reticulum membrane-anchored transcription factors is critical to several cellular processes, including sterol homeostasis [21]. CREBh is a RIP-regulated liver-specific transcription factor that is required for the hepatic synthesis of acute phase response genes [2]. Thus, CREBh-mediated outcomes will be dependent on both the amount of CREBh produced and its cleavage and entry into the nucleus. Danno et al. recently demonstrated that palmitate or oleate at concentrations of 200 or 500 μM, and eicosapentanoic acid at concentrations of 100 and 250 μM increased CREBh mRNA (100–300%) and CREBh promoter activity (50–100%) in HepG2 cells [5]. Data in the present study confirm and extend these data by demonstrating that physiologic concentrations of long chain saturated or unsaturated fatty acids increase CREBh mRNA and endogenous transcription. Thus, the ability of fatty acids to increase CREBh transcription may be an important molecular link to the development of inflammation.
Very little is known about the regulation of CREBh mRNA. The proteasome is the major site of cellular protein degradation, and thus, plays an important role in numerous cellular processes [22]. Recent data suggest that the proteasome inhibitor MG132 prevents fatty acid induction of the pro-inflammatory enzyme cyclooxygenase-2 in endometrial stromal cells [23]. In the present study, the role of the proteasome in fatty acid-mediated regulation of CREBh mRNA was examined using two distinct proteasome inhibitors, MG132 and lactacystin. Both inhibitors prevented palmitate- and oleate-mediated increase in CREBh mRNA. Thus, we have identified an integral role for the proteasome in fatty acid-mediated transcriptional regulation of CREBh.
Recent data indicate that CREBh is suppressed in the fed state, but that this suppression is mitigated in ob/ob mice [5], suggesting that insulin and insulin action are important determinants of CREBh expression. To address this issue, we examined the effects of insulin and PI3-kinase inhibitors on palmitate and oleate regulation of CREBh mRNA in H4IIE liver cells. Insulin dramatically reduced the fatty acid-mediated increase in CREBh, and this effect was blocked by two different PI3-kinase inhibitors. These data led to the hypothesis that the combination of excess lipid delivery and reduced insulin action, as seen in many obese individuals, may be an environment that promotes elevated CREBh levels, and thus the capacity to propagate a pro-inflammatory state. This hypothesis predicts that selective elevation of free fatty acids in vivo should increase CREBh in the liver. Indeed, infusion of a mixed lipid emulsion that selectively increased free fatty acid levels increased CREBh mRNA and protein in the liver of rats. It is unclear whether this increase in CREBh expression was due to elevated fatty acids, per se, or to factors activated consequent to the lipid infusion. For example, Zhang et al., found that proinflammatory cytokines TNFα and IL6, which are commonly elevated following lipid infusion, increased CREBh mRNA in hepatoma cells, and that CREBh cleavage was induced following intraperitoneal injections of IL6 plus IL1β in wild type mice [2]. Our data in isolated liver cells suggest that fatty acids can directly regulated the expression of CREBh mRNA, but future studies are needed to discern the respective effects of fatty acids, cytokines and other factors common to the obese mileau in the regulation of CREBh in vivo.
Although the present study provides relevant information regarding the mechanisms by which fatty acids regulate CREBh mRNA, future studies could provide an additional layer of mechanistic insight. For example, given our results that an intact proteasome was necessary for CREBh activation, it seems plausible that a transcription factor that requires the proteasome for transcriptional activity is involved in fatty acid-mediated CREBh regulation. One such protein is NFκB, since it is sequestered in the cytoplasm by its inhibitory subunit IκB, and requires proteasome-dependent degradation of IκB for its activation [24]. Given its importance in regulating inflammatory pathways, the role of NFκB in fatty acid regulation of CREBh warrants examination in future studies. We also found that fatty acid metabolism was not required for CREBh induction; thus the proximal target of fatty acids may include a cell surface factor(s) that acts independently of cellular metabolism. Recent data suggest that TLRs, in particular TLR4, play a critical role in innate immunity, and are involved in fatty acid-mediated activation of the inflammatory response in adipose tissue [16,15]. To begin to examine the role of TLRs in fatty acid-mediated activation of CREBh, we incubated liver cells with LPS, an activator of TLR4, or zymosan, an activator of TLR2. We found that LPS, but not zymosan, significantly increased CREBh mRNA. Thus, future studies will examine the role of TLR4 in fatty acid-mediated regulation of CREBh.
In summary, data from the current study indicate that fatty acids up-regulate CREBh in vitro and in vivo. This up-regulation occurs at physiologically relevant fatty acid concentrations via activation of gene transcription and is dependent on proteasome activity and PI3-kinase-mediated insulin signaling. These results may have implications for the development and/or exacerbation of fatty acid-mediated inflammation in insulin deficient/resistant states.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Diabetes and Digestive and Kidney Diseases (NIDDK) grants DK-072017 and DK-47416, and the Lillian Fountain Smith Foundation Endowment. CLG was supported by an individual postdoctoral fellowship, NIDDK F32DK082166-01.
References
- 1.Omori Y, Imai J, Watanabe M, Komatsu T, Suzuki Y, Kataoka K, Watanabe S, Tanigami A, Sugano S. Creb-h: A novel mammalian transcription factor belonging to the creb/atf family and functioning via the box-b element with a liver-specific expression. Nucleic Acids Res. 2001;29 (10):2154–2162. doi: 10.1093/nar/29.10.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang K, Shen X, Wu J, Sakaki K, Saunders T, Rutkowski DT, Back SH, Kaufman RJ. Endoplasmic reticulum stress activates cleavage of crebh to induce a systemic inflammatory response. Cell. 2006;124 (3):587–599. doi: 10.1016/j.cell.2005.11.040. [DOI] [PubMed] [Google Scholar]
- 3.Semenkovich CF. Fatty acid metabolism and vascular disease. Trends Cardiovasc Med. 2004;14 (2):72–76. doi: 10.1016/j.tcm.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Rader DJ. Effect of insulin resistance, dyslipidemia, and intra-abdominal adiposity on the development of cardiovascular disease and diabetes mellitus. Am J Med. 2007;120 (3 Suppl 1):S12–18. doi: 10.1016/j.amjmed.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Danno H, Ishii KA, Nakagawa Y, Mikami M, Yamamoto T, Yabe S, Furusawa M, Kumadaki S, Watanabe K, Shimizu H, Matsuzaka T, Kobayashi K, Takahashi A, Yatoh S, Suzuki H, Yamada N, Shimano H. The liver-enriched transcription factor crebh is nutritionally regulated and activated by fatty acids and pparalpha. Biochem Biophys Res Commun. 2009;391(2):1222–7. doi: 10.1016/j.bbrc.2009.12.046. [DOI] [PubMed] [Google Scholar]
- 6.Listenberger LL, Ory DS, Schaffer JE. Palmitate-induced apoptosis can occur through a ceramide-independent pathway. J Biol Chem. 2001;276 (18):14890–14895. doi: 10.1074/jbc.M010286200. [DOI] [PubMed] [Google Scholar]
- 7.Pagliassotti MJ, Shahrokhi KA, Moscarello M. Involvement of liver and skeletal muscle in sucrose-induced insulin resistance: Dose-response studies. Am J Physiol. 1994;266 (5 Pt 2):R1637–1644. doi: 10.1152/ajpregu.1994.266.5.R1637. [DOI] [PubMed] [Google Scholar]
- 8.Popovic V, Popovic P. Permanent cannulation of aorta and vena cava in rats and ground squirrels. J Appl Physiol. 1960;15:727–728. doi: 10.1152/jappl.1960.15.4.727. [DOI] [PubMed] [Google Scholar]
- 9.Muller PY, Janovjak H, Miserez AR, Dobbie Z. Processing of gene expression data generated by quantitative real-time rt-pcr. Biotechniques. 2002;32(6):1372–1374. 1376, 1378–1379. [PubMed] [Google Scholar]
- 10.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37 (8):911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 11.Milburn JL, Jr, Hirose H, Lee YH, Nagasawa Y, Ogawa A, Ohneda M, BeltrandelRio H, Newgard CB, Johnson JH, Unger RH. Pancreatic beta-cells in obesity. Evidence for induction of functional, morphologic, and metabolic abnormalities by increased long chain fatty acids. J Biol Chem. 1995;270 (3):1295–1299. doi: 10.1074/jbc.270.3.1295. [DOI] [PubMed] [Google Scholar]
- 12.Wang D, Wei Y, Schmoll D, Maclean KN, Pagliassotti MJ. Endoplasmic reticulum stress increases glucose-6-phosphatase and glucose cycling in liver cells. Endocrinology. 2006;147 (1):350–358. doi: 10.1210/en.2005-1014. [DOI] [PubMed] [Google Scholar]
- 13.Jump DB, Botolin D, Wang Y, Xu J, Christian B, Demeure O. Fatty acid regulation of hepatic gene transcription. J Nutr. 2005;135(11):2503–2506. doi: 10.1093/jn/135.11.2503. 135/11/2503 [pii] [DOI] [PubMed] [Google Scholar]
- 14.Pawar A, Jump DB. Unsaturated fatty acid regulation of peroxisome proliferator-activated receptor alpha activity in rat primary hepatocytes. J Biol Chem. 2003;278(38):35931–35939. doi: 10.1074/jbc.M306238200. M306238200 [pii] [DOI] [PubMed] [Google Scholar]
- 15.Wong SW, Kwon MJ, Choi AM, Kim HP, Nakahira K, Hwang DH. Fatty acids modulate toll-like receptor 4 activation through regulation of receptor dimerization and recruitment into lipid rafts in a reactive oxygen species-dependent manner. J Biol Chem. 2009;284(40):27384–27392. doi: 10.1074/jbc.M109.044065. M109.044065 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaeffler A, Gross P, Buettner R, Bollheimer C, Buechler C, Neumeier M, Kopp A, Schoelmerich J, Falk W. Fatty acid-induced induction of toll-like receptor-4/nuclear factor-kappab pathway in adipocytes links nutritional signalling with innate immunity. Immunology. 2009;126(2):233–245. doi: 10.1111/j.1365–2567.2008.02892.x. IMM2892 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Staiger K, Staiger H, Weigert C, Haas C, Haring HU, Kellerer M. Saturated, but not unsaturated, fatty acids induce apoptosis of human coronary artery endothelial cells via nuclear factor-kappab activation. Diabetes. 2006;55 (11):3121–3126. doi: 10.2337/db06-0188. [DOI] [PubMed] [Google Scholar]
- 18.Coll T, Jove M, Rodriguez-Calvo R, Eyre E, Palomer X, Sanchez RM, Merlos M, Laguna JC, Vazquez-Carrera M. Palmitate-mediated downregulation of peroxisome proliferator-activated receptor-gamma coactivator 1alpha in skeletal muscle cells involves mek1/2 and nuclear factor-kappab activation. Diabetes. 2006;55 (10):2779–2787. doi: 10.2337/db05-1494. [DOI] [PubMed] [Google Scholar]
- 19.Weigert C, Brodbeck K, Staiger H, Kausch C, Machicao F, Haring HU, Schleicher ED. Palmitate, but not unsaturated fatty acids, induces the expression of interleukin-6 in human myotubes through proteasome-dependent activation of nuclear factor-kappab. J Biol Chem. 2004;279 (23):23942–23952. doi: 10.1074/jbc.M312692200. [DOI] [PubMed] [Google Scholar]
- 20.Kharroubi I, Ladriere L, Cardozo AK, Dogusan Z, Cnop M, Eizirik DL. Free fatty acids and cytokines induce pancreatic beta-cell apoptosis by different mechanisms: Role of nuclear factor-kappab and endoplasmic reticulum stress. Endocrinology. 2004;145 (11):5087–5096. doi: 10.1210/en.2004-0478. [DOI] [PubMed] [Google Scholar]
- 21.Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: A control mechanism conserved from bacteria to humans. Cell. 2000;100 (4):391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 22.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 23.Derecka K, Sheldrick EL, Wathes DC, Abayasekara DR, Flint AP. A ppar-independent pathway to pufa-induced cox-2 expression. Mol Cell Endocrinol. 2008;287 (1–2):65–71. doi: 10.1016/j.mce.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 24.Perkins ND. Integrating cell-signalling pathways with nf-kappab and ikk function. Nat Rev Mol Cell Biol. 2007;8 (1):49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.