Abstract
Background
The current criteria for recreational water quality evaluation are primarily based on measurements of fecal indicator bacteria growth. However, these criteria often fail to predict the presence of waterborne human pathogenic viruses. To explore the possibility of direct use of human enteric viruses as improved human fecal contamination indicators, human adenovirus (HAdV) was tested as a model in this study.
Findings
In order to establish a highly sensitive protocol for effective detection of HAdV in aquatic environments, sixteen published PCR primer sets were re-optimized and comparatively evaluated. Primer sets nehex3deg/nehex4deg, ADV-F/ADV-R, and nested PCR primer sets hex1deg/hex2deg and nehex3deg/nehex4deg were identified to be the most sensitive ones, with up to 1,000 fold higher detection sensitivity compared to other published assays. These three PCR protocols were successfully employed to detect HAdV in both treated and untreated urban wastewaters, and also in 6 of 16 recreational water samples collected around the island of Oahu, Hawaii.
Conclusions
Findings from this study support the possible use of enteric viruses for aquatic environmental monitoring, specifically for the essential routine monitoring of Hawaiian beach waters using the optimized PCR protocol to detect HAdV at certain water sites to ensure a safe use of recreational waters.
Findings
Occurrence of enteric virus contamination in recreational waters has been a major health concern worldwide in recent years [1-5]. However, the current recreational water quality criteria based on the concentration of fecal bacterial indicators (total coliforms, fecal coliforms, and enterococci) often fail to predict the presence of human pathogenic enteric viruses [6-9]. Therefore, enteric viruses have been suggested as alternative indicators of fecal contamination in aquatic environments [6,10,11] due to their low infectious dose [12,13], long survival period in the environment [6,14,15], high resistance to several wastewater treatments [16-18], and the stringent host specificity that makes them free of environmental multiplicity [11]. In addition, detection of human enteric viruses using the library-independent molecular methods (i.e. PCR) is much less laborious and time-consuming compared to the traditional growth-based assessment [11].
The European union regulation has already listed enteroviruses as a parameter governing water quality [11]. However, many studies have suggested that human adenovirus (HAdV), the only enteric virus with double-stranded DNA instead of RNA genome, would make a better candidate as a fecal pollution indicator because of its known stability and persistence in aquatic environments compared to other enteric viruses [11,18,19].
It is well known that monitoring the presence of enteric viruses could be challenging due to the relatively low level of viral particles existing in environmental waters. However, this limitation can be overcome by the use of improved methods for aquatic sample concentration, viral nucleic acids extraction, and more sensitive viral detection techniques [11]. There are currently a number of PCR protocols reported in literature for HAdV detection. However, little is known about their sensitivity or specificity for HAdV detection, particularly for detecting HAdV in environmental waters.
In this study, a total of 16 sets of published HAdV PCR primers, including 3 primer sets for nested PCR, were tested in a side-by-side comparison using a single source of viral DNA for determining detection sensitivity. The selected primer sets were summarized in Table 1. All primer sets were initially tested under standard laboratory PCR conditions with single source HAdV DNA extracted from an in vitro cultured cell sample using QIAamp DNA mini kit (Qiagen, CA) according to the manufacturer's instructions. As shown in Table 2, among all 16 sets tested, only 8 primer sets including hex1deg/hex2deg, nehex3deg/negex4deg, ADV-F/ADV-R, XuHex1/XuHex2, hexDEGF/hexDEGR, AdF/AdR, AdV1/AdV2, and AdV3ne/AdV4ne were able to generate PCR products of the expected sizes in a 25 μL volume reaction containing 1X Taq reaction buffer (Mg2+ free) (New England Biolabs, NEB, MA), 1.5 mM MgCl2 solution (NEB, MA), 200 nM of each dNTP (Sigma-Aldrich, MO), 0.1 μg/μL of BSA (NEB, MA), 400 nM of forward and reverse primers (Integrated DNA technologies, IA), and 2 units of Taq DNA polymerase (provided by Dr. Tung Hoang, University of Hawaii at Manoa), with a Master Cycler Gradient (Eppendorf, Germany). Amplification started with an initial denaturation at 94°C for 5 min, followed by 40 cycles of denaturation at 94°C for 30 sec, annealing at 56°C for 30 sec, extension at 72°C for 30 sec, and a final extension at 72°C for 5 min. All PCR products were subjected to 2% agarose gel electrophoreis alongside a 50-bp DNA marker (NEB, MA), stained with Ethidium Bromide (Sigma-Aldrich, MO) and viewed with the Molecular Imager Gel Doc XR+ system (BioRad Laboratories Inc., CA).
Table 1.
Oligonucleotide sequences used for detection of HAdV
| Primer | Sequence (5'→ 3')a | +/-b | Target | Ampliconsize (bp) | References |
|---|---|---|---|---|---|
| Q-Padv-F | AACGGCCGCTACTGCAAG | + | Swine AdV hexon | 68 | Hundesa et al., 2009 [24] |
| Q-Padv-R | AGCAGCAGGCTCTTGAGG | - | |||
| hex1deg (outer) | GCCSCARTGGKCWTACATGCACATC | + | Hexon | 301 | Allard et al., 2001 [25] |
| hex2deg (outer) | CAGCACSCCICGRATGTCAAA | - | |||
| nehex3deg (inner) | GCCCGYGCMACIGAIACSTACTTC | + | 171 | ||
| nehex4deg (inner) | CCYACRGCCAGIGTRWAICGMRCYTTGTA | - | |||
| ADV-F | GCCACGGTGGGGTTTCTAAACTT | + | Hexon | 131 | Gunson et al., 2009 [26] |
| ADV-R | GCCCCAGTGGTCTTACATGCACATC | - | |||
| XuHex1 | TTCCCCATGGCICAYAACAC | + | Hexon | 482 | Xu et al. 2000 [27] |
| XuHex2 | CCCTGGTAKCCRATRTTGTA | - | |||
| hexDEGF | CAGGACGCCTCGGRGTAYCTSAG | + | Hexon | 103 | Damen et al., 2008 [28] |
| hexDEGR | GGAGCCACVGTGGGRTT | - | |||
| AdE1 | TCCCTACGATGCAGACAACG | + | Fiber | 967 | Xu et al. 2000 [27] |
| AdE2 | AGTGCCATCTATGCTATCTCC | - | |||
| AdF1 | ACTTAATGCTGACACGGGCAC | + | 541-586 | ||
| AdF2 | TAATGTTTGTGTTACTCCGCTC | - | |||
| AdF | CWTACATGCACATCKCSGG | + | Hexon | ~75 | Hernroth et al., 2002 [29] |
| AdR | CRCGGGCRAAYTGCACCAG | - | |||
| HAdV-ABCDEF-hexon25fc | CARTGGKCDTACATGCACATC | + | Hexon | Kuo et al., 2009 [30] | |
| HAdV-E-hexon373r | CCAGRCTGTTGTAGGCAGTG | - | 349 | ||
| HAdV-F-hexon265r | CCACGGCCAGCGTAAAGC | - | 241 | ||
| hexAA1885 (outer) | GCCGCAGTGGTCTTACATGCACAGC | + | Hexon | 300 | Allard et al., 1990 [31] |
| hexAA1913 (outer) | CAGCACGCCGCGGATGTCAAAGT | - | |||
| nehexAA1893 (inner) | GCCACCGAGACGTACTTCAGCCTG | + | Hexon | 142 | Allard et al., 1992 [32] |
| nehexAA1905 (inner) | TTGTACGAGTACGCGGTATCCTCGCGGTC | - | |||
| JTVFF | AACTTTCTCTCTTAATAGACGCC | + | Fiber | 117 | Jothikumar et al., 2005 [33] |
| JTVFR | AGGGGGCTAGAAAACAAAA | - | |||
| AdV1 (outer) | CAAGATGGCCACCCCCTCG | + | hexon | 329 | Oh et al., 2003 [34] |
| AdV2 (outer) | CGATCCAGCACGCCGCGGATGTC | - | |||
| AdV3 (inner) | AATGGTCTTACATGCACAT | + | 253 | ||
| AdV4 (inner) | ACCCGGTTGTCGCCCACGGCCAG | - | |||
a. R = A+G; Y = C+T;S = C+G;W = A+T;H = A+C+T;B = C+G+T;V = A+C+G;D = A+G+T;N = all
b. Polarity
c. Forward primer for both HAdV-E-hexon373r and HAdV-F-hexon265r
Table 2.
Optimized PCR conditions and detection limits for each primer set
| Primer | Stda | Optimized condition | Detection limit | ||
|---|---|---|---|---|---|
| Tannel | [MgCl2] | [primer] | |||
| hex1deg/hex2deg | ✓ | 54-60 | 1.5 mM | 600-800 nM | 10-5 X |
| nehex3deg/nehex4deg | ✓ | 58-60 | 2.0 mM | 600-800 nM | 10-6-7 X |
| ADV-F/ADV-R | ✓ | 51.6-55.4 | 1.5 mM | 600-1000 nM | 10-7 X |
| XuHex1/XuHex2 | ✓ | 54-60 | 1.5 mM | 1000 nM | 10-5 X |
| hexDEGF/hexDEGR | ✓ | 59.2 | 3.0 mM | 600-800 nM | 10-5 X |
| AdF/AdR | ✓ | 60 | 1.5 mM | 600 nM | 10-6-7 X |
| AdV1/AdV2 | ✓ | 51.6-55.4 | 2.0 mM | 600-800 nM | 10-4X |
| AdV3ne/AdV4ne | ✓ | 55.4 | 1.5 mM | 400-800 nM | 10-4 X |
| Hex1deg/hex2deg; nehex3deg/nehex4degb | 10-7 X | ||||
| AdV1/AdV2; AdV3ne/AdV4neb | 10-5 X | ||||
a. Standard condition as described in text.
b. Nested PCR.
The 8 primer sets that successfully generated products of respective sizes (Table 1) were subjected to PCR condition optimization. Different annealing temperatures, MgCl2 concentration and amounts of primers were combined and adjusted to improve the sensitivity of individual detection protocols. The sensitivity of the detection assay was first evaluated at 6 annealing temperatures ranging from 50°C to 60°C. Using the obtained optimal annealing temperature, each set then was tested at 4 selected MgCl2 concentrations ranging from 1.5 to 4.0 mM. The last optimization step was to evaluate the effect of selected primer concentrations from 0.2 μM to 0.8 μM on detection sensitivity. The final optimized PCR conditions for each primer set are listed in Table 2. Detection sensitivity of each primer set was evaluated under the final optimized condition using a serial of 10-fold dilution of a single-source HAdV DNA as template. The detection limits were based on the highest dilution that gave a clear positive signal after PCR amplification. As shown in Table 2, the sensitivities among all tested primers ranged from 10-4 X to 10-7 X dilution, indicating a detection difference of 1,000 fold. Among all, sets nehex3deg/nehex4deg, ADV-F/ADV-R, and AdF/AdR were identified to be the most sensitive ones for ADV detection. Nested PCR using hex1deg/hex2deg and nehex3deg/nehex4deg exhibited similar sensitivity by showing the positive results with stronger detection signals when the same dilution was used. Due to the small size of the final product generated by AdF/AdR (less than 100 bp), only nehex3deg/nehex4deg, ADV-F/ADV-R, and nested PCR sets hex1deg/hex2deg and nehex3deg/nehex4deg were employed in a survey study directed to testing sewage and environmental water samples for naturally occurring HAdV and the detection protocol validation.
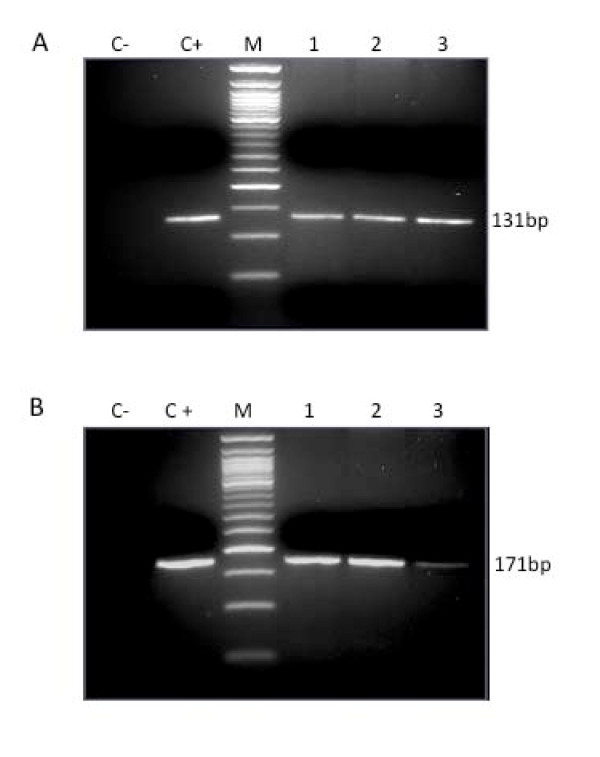
From previous experience examining human norovirus contamination, we learned that PCR conditions optimized using a single clinical source of viral nucleic acids might not act as specifically on total nucleic acids extracted from a complex microbial community, especially when the target viruses only exist in a very small amount compared to other organisms, which is often the case for naturally occurring enteric viruses in environmental waters. Urban wastewaters of human origin are known to contain a large volume of enteric viruses due to characteristic shedding in high numbers in the feces of infected individuals [11,20]. Therefore, the three selected protocols were first validated for their potential application in HAdV detection in environmental samples by testing treated and untreated urban wastewater samples collected from Sand Island Wastewater Treatment Plant (SIWWTP, Hawaii) during May 2010. The SIWWTP processes around 60 million gallons of wastewater daily, accounting for approximately 85% of Oahu's wastewater. Samples were collected from 3 different treatment stages, including untreated raw influence, post-primary clarifying/pre-disinfection stage, and post-disinfection/effluence. Water samples were concentrated according to the filtration-based method described by Katayama et al. [21] with modifications. Briefly, negatively charged type HA filter membranes (Millipore Corporation, MA) with a 0.45-μm pore size and 90-mm diameter were used with a vacuum pump system. MgCl2 was added to the sewage samples at a final concentration of 25 mM before filtration was performed. One hundred milliliters of wastewater sample was filtered through the membranes for viral absorption. The recovered membranes were subjected to nucleic acid extraction using the PowerWater DNA isolation kit (MoBio Laboratories, CA) according to the manufacturer's instructions. Five microliters of the total DNA was used as the template for HAdV detection using the optimized PCR protocols with primer sets ADV-F/ADV-R and nehex3deg/nehex4deg. As expected, HAdV was detected consistently by the two detection methods in all three stages of the collected urban wastewater samples with clean, single bands as shown in Figure 1.
Figure 1.
Agarose gel electrophoresis of PCR detection of HAdV in urban wastewaters of three different treatments stages. (A) Amplified with primer set ADV-F/ADV-R. (B) amplified with primer set nehex3deg/nehex4deg. HAdV were detected from 100 mL of untreated raw influenced (lane 1), pre-disinfection (lane 2), and post-disinfection/effluence (lane 3) stages. Lane M = 50-bp DNA marker, lane C+ = positive control using HAdV DNA, and lane C- = no template control.
Following validation with sewage samples, we evaluated the three protocols for potential use in detecting naturally occurring HAdV in environmental water samples. Sixteen surface water samples were collected from different recreational water bodies composed of both marine and fresh waters around the island of Oahu during June 2010. Sample sites, conditions, and filtration volumes are summarized in Table 3. To ensure the reliability of water filtration and nucleic acid extraction processes, a field blank sample comprised of 2 L distilled water as a negative control, and a spiked sample made by adding 50 mL of HAdV-positive untreated wastewater sample into 2 L of seawater collected from Diamond Head Beach Park as a positive control were also carried out using the same filtration and DNA extraction procedures for all environmental samples. Surface waters collected from fresh water bodies were subjected to initial treatment with 25 mM MgCl2 for 5 min at room temperature prior to filtration.
Table 3.
HAdV prevalence in Hawaiian urban wastewater and environmental waters
| Sample | PCR | Nested PCR | ||||
|---|---|---|---|---|---|---|
| primary | nested | |||||
| Site | Condition | Volume | ADV-F ADV-R | nehex3deg nehex4deg | hex1deg hex2deg | hex1deg/hex2deg; nehex3deg/nehex4deg |
| SIWWTP influence tank | Sewage | 100 mL | + | + | + | ND |
| SIWWTP clarifying tank | Sewage | 100 mL | + | + | + | ND |
| SIWWTP effluence tank | Sewage | 100 mL | + | + | + | ND |
| Sand Island State Recreational Area | Seawater | 2.00 L | - | - | - | + |
| Kailua Bay | Seawater | 2.00 L | + | - | - | + |
| Waikiki Beach | Seawater | 2.00 L | - | + | - | + |
| Ala Wai Canal | Freshwater | 2.00 L | - | - | - | + |
| Wahiawa freshwater | Freshwater | 2.00 L | - | - | - | + |
| Manoa stream | Freshwater | 2.00 L | + | + | - | + |
| Ala Moan Park/Magic Island | Seawater | 2.00 L | - | - | - | - |
| Diamond Head Beach Park | Seawater | 2.00 L | - | - | - | - |
| Maili Beach Park | Seawater | 2.00 L | - | - | - | - |
| Waianae/Pokai Bay | Seawater | 2.00 L | - | - | - | - |
| Waialae Beach Park | Seawater | 2.00 L | - | - | - | - |
| Maunalua Bay Beach Park | Seawater | 2.00 L | - | - | - | - |
| Kalaka Bay Beach Park | Seawater | 2.00 L | - | - | - | - |
| Bellows Field Beach Park | Seawater | 0.80 La | - | - | - | - |
| West Loch Community Shoreline Park | Seawater | 0.80 La | - | - | - | - |
| Kailua Stream | Freshwater | 0.50 La | - | - | - | - |
| Field Blank | dH2O | 2.00 L | - | - | - | - |
| Spike control | S+Sb | 2 +0.05 L | + | + | + | + |
a. 2L sample water was initially prepared but actual volume passed through the filter membrane.
b. 2 L of seawater + 50 mL of sewage water.
c. ND = not done
As shown in Table 3, six sites including Sand Island State Recreational Area, Kailua Bay, Waikiki Beach, Ala Wai Canal, Wahiawa freshwater, and Manoa stream were all positive for HAdV by using the nested PCR protocol with hex1deg/hex2deg and nehex3deg/nehex4deg. As a comparison, Kailua Bay and Manoa stream were HAdV positive using the ADV-F/ADV-R PCR protocol, while HAdV was found in the surface waters of Waikiki Beach and Manoa Stream when nehex3deg/nehex4deg PCR protocol was employed. Manoa Stream was the only site at which all three protocols confirmed HAdV presence. All HAdV positive PCR products were recovered after electrophoresis from 2% agarose gel using QIAquick Gel Extraction kit (Qiagen, CA) by following the manufacture's instruction. The recovered DNAs were sent to the Advanced Studies in Genomics, Proteomics and Bioinformatics (ASGPB, University of Hawaii at Manoa) for sequencing to confirm the positive detection of HAdV.
The resulting positive detection of HAdV from environmental water by employing highly sensitive detection methods clearly indicates that low concentration is not an impossible obstacle to overcome when detecting human enteric viruses from environmental waters. In this study, the comparative analysis of several PCR assays currently available for HAdV detection has led to the identification of three highly sensitive PCR protocols, which were successfully employed for effective detection of HAdV in different types of aquatic environments in Hawaii. In addition, the nested PCR appeared to be superior to the other two protocols for detecting HAdV in environmental waters, suggesting this protocol should be a priority for use in facilitating early detection of HAdV contamination in future.
As the only enteric virus containing a double-stranded DNA genome, HAdV has been shown to be up to 60 times more resistant to UV irradiation than its RNA enteric virus counterparts [17,22] and is able to persist and remain infectious in the environment for a long period of time [6]. In addition, HAdV occurrence was reported to have great correlation with other human enteric viruses, especially hepatitis A virus [19,23]. With several well-established stable cell lines available, HAdV could also be subjected to infectious studies and future exposure risk assessment. Overall, HAdV would make an ideal candidate as a potential molecular index for enteric viral contamination of recreational waters.
It should be noted that this is the first report of HAdV detection in Hawaiian environmental waters using PCR methods to the best of our knowledge. The high prevalence of HAdV in Oahu waters revealed from this study should raise public awareness of a more serious beach contamination issue than previously expected. These new findings strongly argue the importance and necessity of including these established, sensitive HAdV detection protocols into routine water quality monitoring for better protection of the public from recreational waterborne illness associated with enteric viruses. Current research in this laboratory is directed to the establishment of in vitro infectivity assays for water samples collected from the HAdV positive sites and to the determination of a possible correlation between the PCR detection and actual viral infectivity, thus providing baseline information important for the interpretation and assessment of PCR-based HAdV detection and actual health risk to the public.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
HT carried out all experimental testing, data analysis, and drafted the manuscript; and YL conceived of the study, experimental design and data analysis, and manuscript revision. Both authors read and approved the final manuscript
Contributor Information
Hsin-I Tong, Email: hsini@hawaii.edu.
Yuanan Lu, Email: ylu@pbrc.hawaii.edu.
Acknowledgements
The authors would like to thank Rey-Ian Transfiguracion for his assistance on water samples collection and Mary Margaret Byron for reviewing the manuscript. This work was supported in part by grants from the Centers for Oceans and Human Health (COHH) program, the National Institutes of Environmental Health Sciences (P50ES012740) and the National Science Foundation (OCE04-32479 and OCE09-11000).
References
- Papapetropoulou M, Vantarakis AC. Detection of adenovirus outbreak at a municipal swimming pool by nested PCR amplification. J Infect. 1998;36:101–103. doi: 10.1016/S0163-4453(98)93414-4. [DOI] [PubMed] [Google Scholar]
- Lee S, Kim S. Detection of infectious enteroviruses and adenoviruse in tap water in urban areas in Korea. Water Res. 2002;36:248–256. doi: 10.1016/S0043-1354(01)00199-3. [DOI] [PubMed] [Google Scholar]
- Albinana-Gimenez N, Clemente-Casares P, Bofill-Mas S, Hundesa A, Ribas F, Girones R. Distribution of human polyoma-viruses, adenoviruses, and hepatitis E virus in the environment and in a drinking-water treatment plant. Environ Sci Technol. 2006;40:7416–7422. doi: 10.1021/es060343i. [DOI] [PubMed] [Google Scholar]
- Xagoraraki I, Kuo DHW, Wong K, Wong M, Rose JB. Occurrence of human adenoviruses at two recreational beaches of the great lakes. Appl Environ Microbiol. 2007;73:7874–7881. doi: 10.1128/AEM.01239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair RG, Jones EL, Gerba CP. Viruses in recreational water-borne disease outbreaks: a review. J Appl Microbiol. 2009;107:1769–1780. doi: 10.1111/j.1365-2672.2009.04367.x. [DOI] [PubMed] [Google Scholar]
- Jiang S, Noble R, Chu W. Human adenoviruses and coliphages in urban runoff-impacted coastal waters of Southern California. Appl Environ Microbiol. 2001;67:179–184. doi: 10.1128/AEM.67.1.179-184.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble RT, Fuhrman JA. Enteroviruses detected by reverse transcriptase polymerase chain reaction from the coastal waters of Santa Monica Bay, California: low correlation to bacterial indicator levels. Hydrobiologia. 2001;460:175–184. doi: 10.1023/A:1013121416891. [DOI] [Google Scholar]
- Fong TT, Griffin DW, Lipp EK. Molecular assays for targeting human and bovine enteric viruses in coastal waters and their application for library-independent source tracking. Appl Environ Microbiol. 2005;71:2070–2078. doi: 10.1128/AEM.71.4.2070-2078.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa AC, Arias CF, Sánchez-Colón S, Mazari-hiriart M. Comparative study of enteric viruses, coliphages and indicator bacteria for evaluating water quality in a tropical high-altitude system. Environ Health. 2009;8:49–58. doi: 10.1186/1476-069X-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Environmental Protection Agency. Drinking Water contaminant candidate list. EPA. 1998. p. 815-F-98-002.
- Fong TT, Lipp EK. Enteric viruses of humans and animals in aquatic environments: health risks, detection, and potential water quality assessment tools. Microbiol Mol Biol Rev. 2005;69:357–371. doi: 10.1128/MMBR.69.2.357-371.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas CN, Rose JB, Gerba CP, Regli R. Risk assessment of viruses in drinking water. Risk Anal. 1993;13:545–552. doi: 10.1111/j.1539-6924.1993.tb00013.x. [DOI] [PubMed] [Google Scholar]
- Teunis PFM, Moe CL, Lui P, Miller SE, Lindesmith L, Baric RS, Le Pendu J, Calderon RL. Norwalk virus: how infectious is it? J Med Virol. 2008;80:1468–1476. doi: 10.1002/jmv.21237. [DOI] [PubMed] [Google Scholar]
- Wetz JJ, Lipp EK, Griffin DW, Lukasik J, Wait D, Sobsey MD, Scott TM, Rose JB. Presence, infectivity, and stability of enteric viruses in seawater: relationship to marine water quality in the Florida Keys. MaMar Pollut Bull. 2004;48:698–704. doi: 10.1016/j.marpolbul.2003.09.008. [DOI] [PubMed] [Google Scholar]
- Enriquez CE, Hurst CJ, Gerba CP. Survival of the enteric adenoviruses 40 and 41 in tap, sea, and waste water. Wat Res. 1995;29:2548–2553. doi: 10.1016/0043-1354(95)00070-2. [DOI] [Google Scholar]
- Fujioka RS, Yoneyama BS. Sunight inactivation of human enteric viruses and fecal bacteria. Water Sci Technol. 2002;46:291–295. [PubMed] [Google Scholar]
- Gerba CP, Gramos DM, Nwachuku N. Comparative inactivation of enteroviruses and adenovirus 2 by UV light. Appl Environ Microbiol. 2002;68:5167–5169. doi: 10.1128/AEM.68.10.5167-5169.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston-Enriquez JA, Haas CN, Jacangelo J, Riley K, Gerba CP. Inactivation of feline calicivirus and adenovirus type 40 by UV radiation. Appl Environ Microbiol. 2003;69:577–582. doi: 10.1128/AEM.69.1.577-582.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina S, Puig M, Lucena F, Jofre J, Girones R. Viral pollution in the environment and in shellfish: human adenovirus detection by PCR as an index of human viruses. Appl Environ Microbiol. 1998;64:3376–3382. doi: 10.1128/aem.64.9.3376-3382.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree KD, Gerba CP, Rose JB, Haas CN. Waterborne adenovirus: a risk assessment. Wat Sci Technol. 1997;35:1–6. [Google Scholar]
- Katayama H, Shimasaki A, Ohgaki S. Development of a virus concentration metods and its application to detection of enterovirus and Norwalk virus from coastal sweater. Appl Environ Microbiol. 2002;68:1033–1039. doi: 10.1128/AEM.68.3.1033-1039.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng QS, Gerba CP. Comparative inactivation of enteric adenoviruses, poliovirus and coliphages by ultraviolet irradiation. Water Res. 1996;30:2665–2668. doi: 10.1016/S0043-1354(96)00179-0. [DOI] [Google Scholar]
- Muniain-Mujika I, Calvo M, Lucena F, Girones R. Comparative analysis of viral pathogens and potential indicators in shellfish. Int J Food Microbiol. 2003;83:75–85. doi: 10.1016/S0168-1605(02)00324-0. [DOI] [PubMed] [Google Scholar]
- Hundesa A, Maluquer de Motes C, Albinana-Gimenez N, Rodriguez-Manzano J, Bofill-Mas S, Suñen E, Rosina Girones R. Development of a qPCR assay for the quantification of porcine adenoviruses as an MST tool for swine fecal contamination in the environment. J Virol Methods. 2009;158:130–135. doi: 10.1016/j.jviromet.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Allard A, Albinsson B, Wadell G. Rapid typing of human adenoviruses by a general PCR combined with restriction endonuclease analysis. J clin microl. 2001;39:498–505. doi: 10.1128/JCM.39.2.498-505.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunson RN, Maclean AR, Shepherd SJ, Carman WF. Simultaneous detection and quantitation of cytomegalovirus, Epstein-Barr virus, and adenovirus by use of real-time PCR and pooled standards. J Clin Microbiol. 2009;47:765–770. doi: 10.1128/JCM.01721-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, McDonough MC, Erdman DD. Species-specific identification of human adenoviruses by a multiplex PCR assay. J Clin Microbiol. 2000;38:4114–4120. doi: 10.1128/jcm.38.11.4114-4120.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damen M, Minnaar R, Glasius P, van der Ham A, Koen G, Wertheim P, Beld M. Real-time PCR with an internal control for detection of all known human adenovirus serotypes. J Clin Microbiol. 2008;46:3997–4003. doi: 10.1128/JCM.00563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernroth BE, Conden-Hansson A, Rehnstam-Holm A, Girones R, Allard AK. Environmental factors influencing human viral pathogens and their potential indicator organisms in the blue mussel, Mytilu eduli: the first Scandinavian report. Appl Environ Microbiol. 2002;68:4523–4533. doi: 10.1128/AEM.68.9.4523-4533.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo DHW, Simmons F, Xagoraraki I. A new set of PCR assays for the identification of multiple human adenovirus species in environmental samples. J Appl Microbiol. 2009;107:1219–29. doi: 10.1111/j.1365-2672.2009.04300.x. [DOI] [PubMed] [Google Scholar]
- Allard A, Girones R, Juto P, Wadell G. Polymerase chain reaction for detection of adenoviruses in stool samples. J Clin Microbiol. 1990;28:2659–2667. doi: 10.1128/jcm.28.12.2659-2667.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard A, Albinsson B, Wadell G. Detection of adenoviruses in stools from healthy persons and patients with diarrhea by two-step polymerase chain reaction. J Med Virol. 1992;37:149–157. doi: 10.1002/jmv.1890370214. [DOI] [PubMed] [Google Scholar]
- Jothikumar N, Hill VR, Lu X, Sobsey MD, Erdman DD. Quantitative real-time PCR assays for detection of human adenoviruses and identification of serotypes 40 and 41. Appl Envrion Microbiol. 2005;71:3131–3136. doi: 10.1128/AEM.71.6.3131-3136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh D, Gaedicke G, Schreier E. Viral agents of acute gastroenteritis in German children: prevalence and molecular diversity. J Med Virol. 2003;71:82–93. doi: 10.1002/jmv.10449. [DOI] [PubMed] [Google Scholar]