Abstract
Background
The horn fly, Haematobia irritans (Linnaeus, 1758) (Diptera: Muscidae) is one of the most important ectoparasites of pastured cattle. Horn flies infestations reduce cattle weight gain and milk production. Additionally, horn flies are mechanical vectors of different pathogens that cause disease in cattle. The aim of this study was to conduct a functional genomics study in female horn flies using Expressed Sequence Tags (EST) analysis and RNA interference (RNAi).
Results
A cDNA library was made from whole abdominal tissues collected from partially fed adult female horn flies. High quality horn fly ESTs (2,160) were sequenced and assembled into 992 unigenes (178 contigs and 814 singlets) representing molecular functions such as serine proteases, cell metabolism, mitochondrial function, transcription and translation, transport, chromatin structure, vitellogenesis, cytoskeleton, DNA replication, cell response to stress and infection, cell proliferation and cell-cell interactions, intracellular trafficking and secretion, and development. Functional analyses were conducted using RNAi for the first time in horn flies. Gene knockdown by RNAi resulted in higher horn fly mortality (protease inhibitor functional group), reduced oviposition (vitellogenin, ferritin and vATPase groups) or both (immune response and 5'-NUC groups) when compared to controls. Silencing of ubiquitination ESTs did not affect horn fly mortality and ovisposition while gene knockdown in the ferritin and vATPse functional groups reduced mortality when compared to controls.
Conclusions
These results advanced the molecular characterization of this important ectoparasite and suggested candidate protective antigens for the development of vaccines for the control of horn fly infestations.
Background
The horn fly, Haematobia irritans (Linnaeus, 1758) (Diptera: Muscidae) is one of the most important ectoparasites of pastured cattle [1]. This fly was originally introduced from Europe and currently represents a tremendous health problem for cattle in the Americas from Southern Canada to Argentina [2]. Although horn flies parasitize mainly cattle, occasionally they feed on horses, sheep and dogs [3].
The developmental cycle of H. irritans is very short, taking from 10 to 14 days to complete. Larvae and pupae develop on dung and once the flies emerge from pupae, immediately start and remain feeding on cattle during their whole life. Flies leave the host only to move to others or to lay eggs on fresh manure [1]. Both males and females feed 24 to 38 times per day ingesting an average of 14.3 mg blood per fly [4].
Horn flies infestations interfere with animal feeding, thus producing significant reductions in weight gain and milk production [5,6]. The economic impact of H. irritans on livestock in the United States was estimated in approximately US$1 billion annually [7,8]. In dairy cattle, infestations higher than 200 flies per animal produce a loss of 520 ml milk and 28 kg weight daily [6]. In beef cattle, H. irritans infestations can cause a reduction of 8.1 kg weight daily [5]. Moreover, the skin lesions caused by the intermittent feeding of horn flies produce significant hide damages, affecting considerably the leather industry [9]. Additionally, horn flies are mechanical vectors of different pathogens that cause disease in cattle [10-14].
The control of horn flies has been primarily based on the use of chemical insecticides [15,16]. This control strategy has been partially successful but has resulted in the selection of flies resistant to most commercially available insecticides [15-17]. In addition to resistance, chemical insecticides affect other living organisms, contribute to environmental pollution and contaminate cattle products for human consumption.
Recently, research has been conducted to develop new horn fly control strategies that are cost-effective and environmentally friendly. The efficacy of the entomopathogenic fungi, Metarhizium anisopalinae, against horn fly larvae was very high in vitro [18]. However, field application of entomopathogenic fungi for biological control of horn flies is difficult. The use of female-specific conditional lethality systems has been also considered but not yet developed [19].
The immunological control of ectoparasite infestations was demonstrated through cattle vaccination against tick infestations [20,21]. The effect of anti-tick vaccines on the reduction of cattle tick infestations and the transmission of some tick-borne pathogens [21-23] and preliminary results obtained in insect vector species [24-32] have provided evidence that protective antigens may be used for development of vaccines with the dual target control of both arthropod infestations and reduction of vector capacity to transmit pathogens that impact human and animal health. Recently, Cupp et al. [33] demonstrated that horn flies fed on cattle immunized with the anti-clotting factor thrombostasin, took smaller blood meals and the egg development was delayed. Although other molecules have been proposed as vaccine candidates against horn flies [16,34,35], further research is needed to identify new vaccine candidates for effective control of horn fly infestations.
Recently, RNA interference (RNAi) was proposed as a method to identify candidate tick protective antigens [36] and was used for the screening of tick genes with potential applications in vaccine development [37-39].
The aim of this study was to conduct a functional genomics study in female horn flies using Expressed Sequence Tags (EST) analysis and RNAi. The results of this study will advance the molecular characterization of this important ectoparasite and suggested candidate protective antigens for the development of vaccines for the control of horn fly infestations.
Results
Assembly and annotation of female horn fly Expressed Sequence Tags (ESTs)
A cDNA library was made from whole abdominal tissues collected from partially fed adult female horn flies. From 2,462 sequenced ESTs, 302 and 2,160 were low and high quality ESTs, respectively (Table 1). Empty or vector ESTs were not obtained.
Table 1.
Statistics of horn fly EST assembly.
| Number of sequences | 2,462 |
|---|---|
| Mean length ± S.D. before vector stripping | 877 ± 44 bp |
| Mean length ± S.D. after vector stripping | 653 ± 32 bp |
| High quality EST reads | 2,160 |
| Assembled ESTs | 2,160 |
| No. unigenes | 992 |
| Mean length ± S.D. | 758 ± 46 bp |
| No. unigenes with more than 20 ESTs | 13 |
| No. unigenes with 5-20 ESTs | 44 |
| No. unigenes with less than 5 ESTs | 935 |
| No. contigs | 178 |
| No. singlets | 814 |
| Novelty (unigenes/assembled ESTs) | 46% |
| Redundancy (1-Novelty) | 54% |
Since the female horn fly cDNA library was not normalized, the EST distribution per contig was quantified to determine the redundancy level of our EST dataset. High quality ESTs were assembled into 992 unigenes (178 contigs and 814 singlets) (Table 1; Additional file 1: Table S1), representing 46% novelty (unigenes/assembled ESTs) in our dataset. ESTs (814) present as singleton sequences represented 82% of all unigenes, while 72 unigenes (7%) contained only two ESTs. On average, the number of ESTs per unigene was 2.2, which suggested a low diversity in our dataset.
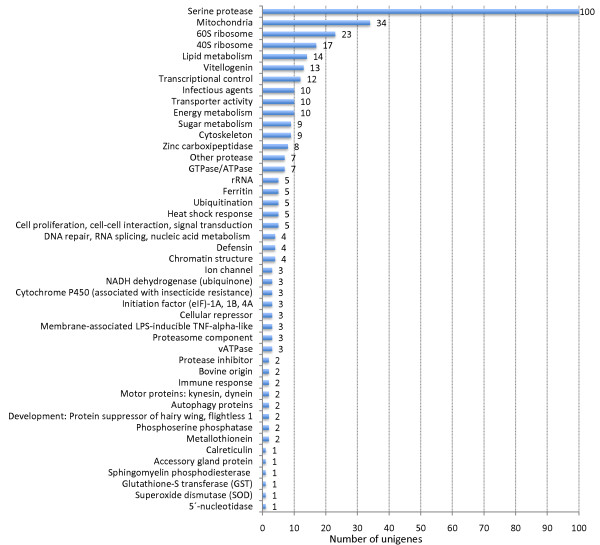
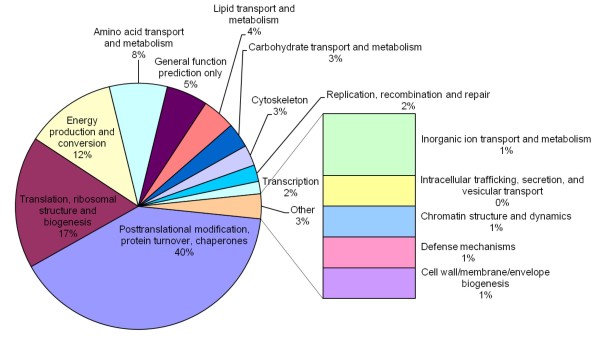
BLAST searches to TrEMBL and Swiss-Prot databases assigned 367 proteins to molecular function Gene Ontology (GO) terms (Figure 1). One hundred unigenes (10%) containing 535 ESTs (25%) corresponded to serine proteases. Other molecular functions represented in the unigenes included those involved in cell metabolism, mitochondrial function, transcription and translation, transport, chromatin structure, vitellogenesis, cytoskeleton, DNA replication, cell response to stress and infection, cell proliferation and cell-cell interactions, intracellular trafficking and secretion, and development (Figure 1). Of the 367 unigenes with molecular function GO assignments, 184 could be assigned to Clusters of Orthologous Groups of proteins (COG) (Figure 2). The COG comprising posttranslational modification, protein turnover and chaperones contained 40% of proteins with COG assignments, followed by translation, ribosomal structure and biogenesis (17%) and energy production and conversion (12%) (Figure 2).
Figure 1.
Functional grouping of horn fly unigenes based on Gene Ontology (GO) molecular function assignments.
Figure 2.
Horn fly unigene assignment to Clusters of Orthologous Groups of proteins (COG; http://www.ncbi.nlm.nih.gov/COG).
A relatively large set of 449 unigenes (45%) lacked any significant sequence similarity (Blast E values > 10-5) to any sequence available in the public databases. Of all the 543 unigenes with significant sequence similarity to previously published sequences, 88.8% were most similar to Diptera (Drosophila spp., H. irritans, Musca domestica, Lucilia cuprina, Glossina spp., Stomoxys calcitrans, Aedes aepgypti, Sarcophaga spp., Phlebotomus papatasi, Chrysomyia bezziana, Anopheles gambiae, Ceratitis stricta, Trichopalpus fraterna, Automola atomaria, Nanna tibiella, Bactrocera dorsalis, Lutzomyia longipalpis, Eristalinus punctulatus, and Ophiomyia sp.), 1.5% to other insect species (Spodoptera frugiperda, Tribolium castaneum, Gryllus bimaculatus, Lonomia obliqua, Nasonia vitripennis, and Lymantria dispar), 5.5% to other eukaryotic organisms, and 4.2% to microorganisms (Table 2).
Table 2.
Distribution of annotated unigenes to different species.
| Species | Total No. of unigenes | Ratio (%) |
|---|---|---|
| All organisms | 543 | 100 |
| Insects (Diptera) | 482 | 88.8 |
| Drosophila spp. | 289 | 53.2 |
| H. irritans | 89 | 16.4 |
| Musca domestica | 22 | 4.0 |
| Lucilia cuprina | 16 | 2.9 |
| Glossina spp. | 15 | 2.8 |
| Stomoxys calcitrans | 14 | 2.6 |
| Aedes aepgypti | 11 | 2.0 |
| Sarcophaga spp. | 9 | 1.7 |
| Phlebotomus papatasi | 3 | 0.6 |
| Chrysomyia bezziana | 3 | 0.6 |
| Anopheles gambiae | 3 | 0.6 |
| Ceratitis stricta | 1 | 0.2 |
| Trichopalpus fraterna | 1 | 0.2 |
| Automola atomaria | 1 | 0.2 |
| Nanna tibiella | 1 | 0.2 |
| Bactrocera dorsalis | 1 | 0.2 |
| Lutzomyia longipalpis | 1 | 0.2 |
| Eristalinus punctulatus | 1 | 0.2 |
| Ophiomyia sp. | 1 | 0.2 |
| Insects (Other) | 8 | 1.5 |
| Tribolium castaneum | 2 | 0.4 |
| Gryllus bimaculatus | 2 | 0.4 |
| Spodoptera frugiperda | 1 | 0.2 |
| Lonomia obliqua | 1 | 0.2 |
| Nasonia vitripennis | 1 | 0.2 |
| Lymantria dispar | 1 | 0.2 |
| Other eukaryotes | 30 | 5.5 |
| Microorganisms | 23 | 4.2 |
Over 90% of best hits matched to insects. Only less than 10% of all best hits matched to other eukaryotes and microorganisms.
Thirteen unigenes assembled from 505 sequence reads, contained more than 20 ESTs, most probably representing transcripts with highest abundance in abdominal tissues of partially fed female horn flies (Table 3). As expected from the results of the annotation of the entire EST dataset, 10 (77%) of these unigenes corresponded to serine proteases (Table 3). The second largest group of ESTs was derived from mitochondrial transcripts (Table 3). The analysis of serine protease unigene sequences showed that although some of them may be paralogs (for example unigenes 1-2 and 3-5; Table 3), other probably reflect sequence polymorphisms within the horn fly population because they had 97%-98% nucleotide sequence identity (for example between unigenes 3-5; Table 3).
Table 3.
Transcripts with highest abundance in abdominal tissues of partially fed female horn flies.
| Unigene annotation | ||||
|---|---|---|---|---|
| Unigene No. [GenBank accession number] | Number of clustered ESTs | Sequence identity | Clusters of Orthologous Groups of proteins (COG) | Molecular function Gene Ontology (GO) term |
| 1 [HO004732] | 21 | L. cuprina clone sbsp9 serine proteinase | Posttranslational modification, protein turnover, chaperones | Secreted trypsin-like serine protease |
| 2 [HO004733] | 21 | L. cuprina clone sbsp9 serine proteinase | Posttranslational modification, protein turnover, chaperones | Secreted trypsin-like serine protease |
| 3 [HO004734] | 22 | H. irritans serine protease | Posttranslational modification, protein turnover, chaperones | Secreted trypsin-like serine protease |
| 4 [HO004735] | 25 | H. irritans serine protease | Posttranslational modification, protein turnover, chaperones | Secreted trypsin-like serine protease |
| 5 [HO004736] | 25 | H. irritans serine protease | Posttranslational modification, protein turnover, chaperones | Secreted trypsin-like serine protease |
| 6 [HO004737] | 26 | D. pseudoobscura GA21163-PA (Dpse\GA21163) | Amino acid transport and metabolism | Zinc carboxypeptidase |
| 7 [HO004738] | 28 | H. irritans serine protease mRNA | Posttranslational modification, protein turnover, chaperones | Secreted trypsin-like serine protease |
| 8 [HO004739] | 30 | L. cuprina clone sbsp9 serine proteinase | Posttranslational modification, protein turnover, chaperones | Secreted trypsin-like serine protease |
| 9 [HO004740] | 33 | H. irritans serine protease | Posttranslational modification, protein turnover, chaperones | Secreted trypsin-like serine protease |
| 10 [HO004741] | 37 | H. irritans mitochondrion, complete genome | Mitochondria | |
| 11 [HO004742] | 40 | H. irritans serine protease | Posttranslational modification, protein turnover, chaperones | Secreted trypsin-like serine protease |
| 12 [HO004743] | 90 | H. irritans serine protease | Posttranslational modification, protein turnover, chaperones | Secreted trypsin-like serine protease |
| 13 [HO004744] | 107 | H. irritans mitochondrion, complete genome | Mitochondria | |
Functional characterization of horn fly ESTs by RNAi
For functional genomics studies, selected unigene functional groups were used in RNAi experiments in female horn flies (Table 4). These groups included serine protease, protease inhibitor, vitellogenin (VTG), ubiquitination, ferritin (FER), vacuolar (H+)-ATPase (vATPase), proteasome component, immune response and 5'-nucleotidase (5'-NUC) ESTs and were selected based on their putative function in insect biology and previous results of RNAi experiments in other arthropods (see Discussion). As controls, ESTs with sequence identity to Nora virus and Wolbachia endosymbionts were selected (Table 4). The injection of these control dsRNAs did not affect horn fly mortality (β = -0.01, Wald Chi2 = 0.01, P = 0.91) and oviposition (P > 0.05) when compared to buffer-injected flies in 14 independent RNAi experiments (Table 5), thus supporting their use as controls. Significant gene knockdown was obtained for at least one targeted unigene sequence on each group except for the serine protease group 1 in which significant gene expression silencing was not obtained for any of the unigenes included in the analysis (Table 5). For some sequences, gene knockdown was observed as early as 6 h post-injection (hpi) and lasted at least until 36 hpi (Table 5). For other sequences in groups 8 and 9, gene knockdown was not detected until after 12 hpi (Table 5). In most cases, gene expression silencing was higher than 70% when compared to the control group (Table 5).
Table 4.
Horn fly unigene functional groups selected for RNAi.
| Group N° | Functional group | N° of unigenesa | Unigenes selected for RNAib |
|---|---|---|---|
| 1 | Serine protease | 100 | 5, 10, 14, 19, 42, 90, 224, 230 |
| 2 | Protease inhibitor | 2 | 2_B12, 24_H02 |
| 3 | Vitellogenin | 13 | 7, 20, 37, 76, 89, 145, 176, 7_D07 |
| 4 | Ubiquitination | 5 | 84, 146, 4_E04, 5_G03, 7_B08 |
| 5 | Ferritin | 5 | 26, 39, 154, 156, 10_A09 |
| 6 | vATPase | 3 | 7_F08, 9_A08, 17_H03 |
| 7 | Proteasome component | 3 | 6_G04, 7_A04, 12_H09 |
| 8 | Immune response | 2 | 6_F11, 10_G05 |
| 9 | 5'-nucleotidase | 1 | 13_D07 |
| 10 (negative control) | Infectious agents: Nora virus Wolbachia endosymbionts |
3 3 |
191 2_E12 |
aNumber of unigenes grouped into this category.
bWhen the number of unigenes in the functional group was greater than 5, unigenes were selected for RNAi to include all different genes present in this category. Unigenes GenBank accession numbers are shown in Table 6.
Table 5.
Results of RNAi experiments in female horn flies.
| Group No.a | Cumulative percent mortality | Oviposition (eggs per survived fly)b | Expression silencing (average% ± SD) with respect to group 10 controlc | dsRNA injectedd | |||||
|---|---|---|---|---|---|---|---|---|---|
| 12 hpi | 24 hpi | 36 hpi | 6 hpi | 12 hpi | 24 hpi | 36 hpi | |||
| 1 (Serine protease) |
57 ± 6 | 69 ± 11 | 78 ± 2 | 0.88 ± 0.23 | ND | ND | 57 ± 50 0 ± 0 |
ND | 90 230 |
| 2* (Protease inhibitor) |
73 ± 11 | 84 ± 7 | 94 ± 1 | 2.56 ± 1.80 | ND | ND | 83 ± 3** | ND | 2_B12 |
| 3 (Vitellogenin) |
55 ± 11 | 67 ± 8 | 74 ± 8 | 0.31 ± 0.9** | ND | ND | 0 ± 0 0 ± 0 100 ± 2* |
ND | 7 20 76 |
| 4 (Ubiquitination) |
42 ± 12 | 67 ± 11 | 83 ± 16 | 0.64 ± 0.63 | ND | ND | 92 ± 6* 70 ± 24 46 ± 43 |
ND | 84 146 4_E04 |
| 5* (Ferritin) |
24 ± 23 | 39 ± 37 | 47 ± 41 | 0.22 ± 0.17** | ND | ND | 68 ± 13* 86 ± 86 |
ND | 26 154 |
| 6* (vATPase) |
17 ± 13 | 24 ± 11 | 34 ± 3 | 0.08 ± 0.05** | ND | ND | 78 ± 6* 100 ± 1* 99 ± 8* |
ND | 7_F08 9_A08 17_H03 |
| 7 (Proteasome component) |
64 ± 3 | 76 ± 8 | 84 ± 11 | 0.38 ± 0.03** | 98 ± 2* 100 ± 6* |
100 ± 2* 67 ± 9* |
ND ND | 0 ± 0 ND | 7_A04 12_H09 |
| 8* (Immune response) |
66 ± 7 | ND | 99 ± 8 | 0.23 ± 0.09** | 100 ± 8* 0 ± 0 |
92 ± 5 0 ± 0 |
ND ND | 96 ± 9* 98 ± 8* |
6_F11 10_G05 |
| 9* (5'-nucleotidase) |
50 ± 11 | ND | 91 ± 22 | 0.12 ± 0.09** | 0 ± 0 | 98 ± 9* | ND | 70 ± 7* | 13_D07 |
| 10 (negative control) |
45 ± 24 | 57 ± 30 | 64 ± 27 | 1.26 ± 0.90 | --- | --- | --- | --- | 191 2_E12 |
| Injection buffer | 46 ± 28 | 57 ± 30 | 65 ± 29 | 1.50 ± 1.01 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | None |
aThe average ± S.D. of two independent RNAi experiments for each of the test groups 1-9 and 14 experiments for the negative control and injection buffer groups is shown. One hundred flies were used on each RNAi experiment. Cumulative percent mortality was evaluated in female horn flies at 12, 24 and 36 hpi. Survival curves (temporal rates of mortality) were compared between different treatments and the control group 10 using Cox Proportional Hazards Survival Regression analysis (*P < 0.05). Abbreviation: ND, not determined.
bOviposition data from test groups 1-9 and the injection buffer control were compared with the unrelated dsRNA-injected control group 10 by Student's t-test (**P < 0.005).
cFor analysis of expression silencing, the mRNA levels of each knockdown gene were determined by real-time RT-PCR in 4 individual flies each at 6, 12 and 24 or 36 hpi. Percent expression silencing shown at different hpi corresponds to each individual gene targeted by dsRNA injection. The mRNA levels were normalized against horn fly 16S rRNA and the mean of the duplicate normalyzed Ct values from test dsRNA-injected groups 1-9 and in the injection buffer control were compared with the unrelated dsRNA-injected control group 10 by Student's t-test (*P < 0.05, **P < 0.005).
dUnigenes GenBank accession numbers are shown in Table 6.
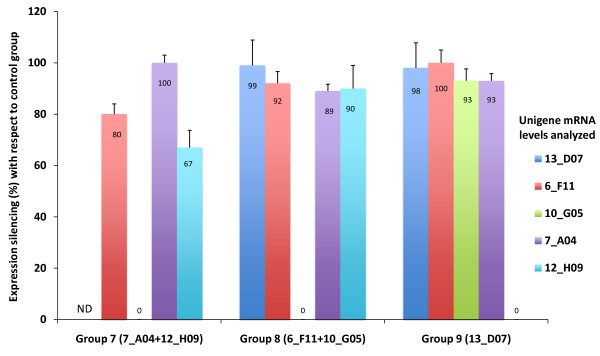
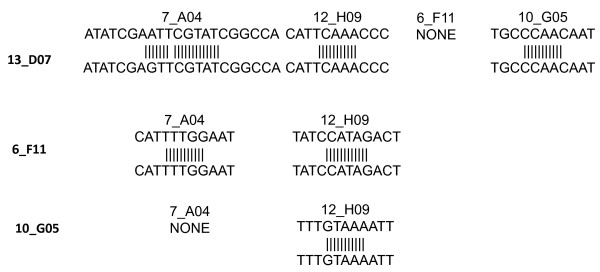
To analyze RNAi off-target effects, the expression of genes not targeted by the injected dsRNA was analyzed at 12 hpi in functional groups 7-9 (Figure 3). The results showed that the expression of genes not targeted by the injected dsRNA was silenced in all three groups analyzed (Figure 3), thus suggesting RNAi off-target effects in horn flies. Pairwise sequence alignments identified regions with homology ≥ 11 bp in some sequences (Figure 4). However, only one region had 21 bp homology between unigene sequences 13_D07 and 7_A04 (Figure 4).
Figure 3.
Analysis of RNAi off-target effects in female horn flies. Gene expression silencing (average % + SD) with respect to the control group was determined by real-time RT-PCR at 12 hpi in horn flies (N = 4) injected with different dsRNAs in three of the functional groups characterized by RNAi.
Figure 4.
Pairwise sequence alignment of dsRNA sequences showing homology regions ≥ 11 nucleotides. Sequence alignments were done between the sequences used in the experiment described in figure 3. Abbreviation: ND, not determined.
Injection of dsRNAs in the serine protease and ubiquitination functional groups did not affect fly mortality (β = 0.09, Wald Chi2 = 2.68, P = 0.10 and β = 0.07, Wald Chi2 = 3.60, P = 0.08, respectively) or oviposition (P > 0.05) when compared to controls (Table 5). The knockdown of a protease inhibitor gene (elastase) resulted in higher fly mortality (β = 0.60, Wald Chi2 = 13.35, P = 0.0002) but did not affect oviposition (P > 0.05) when compared to controls (Table 5). VTG-2 and proteasome component genes knockdown did not affect fly mortality (β = 0.03, Wald Chi2 = 1.42, P = 0.22 and β = 0.13, Wald Chi2 = 2.59, P = 0.107, respectively) but significantly (P < 0.005) reduced oviposition (Table 5). When the expression of immune response and 5'-NUC genes was silenced, higher fly mortality (β = -0.46, Wald Chi2 = 7.39, P = 0.006 and β = 0.35, Wald Chi2 = 4.65, P = 0.03, respectively) and reduced oviposition (P < 0.005) were obtained when compared to controls (Table 5). Interestingly, knockdown of FER light chain and vATPase genes resulted in lower fly mortality (β = 0.21, Wald Chi2 = 5.12, P = 0.02 and β=-0.16, Wald Chi2 = 14.70, P = 0.0001, respectively), and 6 and 16-fold decrease in oviposition (P < 0.005) when compared to control dsRNA-injected flies (Table 5).
Discussion
The effective control of horn fly infestations requires the design of new control strategies. Genomics and functional genomics studies are important to understand basic biological questions and to identify new targets for improved control strategies. Recently, gene expression analysis was reported in horn fly embryos, larvae and adult females [19,35]. However, this is the first report of functional genomics studies in this species.
ESTs sequenced and assembled in this study provided new sequence information for horn fly. The assembled unigenes without sequence similarity to sequences in public databases probably represented unique transcripts for horn fly or corresponded to proteins that have not yet been identified in related organisms due to incomplete genomic information. However, it cannot be excluded that the identified ESTs represent parts of known proteins whose similarities are located in parts of the sequence that are not covered by the analyzed ESTs.
The number of ESTs assembled into a certain unigene roughly reflected the relative abundance of corresponding mRNAs since the cDNA library from female horn flies used in this study was not normalized. We found that 100 unigenes, containing 25% of the ESTs, corresponded to serine proteases, indicating that this group represented the most abundantly expressed genes in abdominal tissues of partially fed female horn flies. In fact, the unigenes with the largest number of ESTs represented members of the serine protease family, thus suggesting that posttranslational modification and protein turnover were highly active in partially fed female flies.
The high proportion of ESTs present as singleton sequences when compared to contigs reflected a low diversity in our dataset, probably due to the presence of paralogs and sequence polymorphisms for some unigenes. In fact, sequence analysis of serine protease unigenes makes at this point difficult to discriminate between paralogs and ESTs representing sequence polymorphisms within the horn fly population.
RNAi was used to functionally characterize selected horn fly genes in adult female flies. To our knowledge, this is the first report of RNAi in horn flies. RNAi has been used to study gene function in insects and other arthropods [37,40-49] and to screen for candidate protective antigens in ticks [36-39]. Although with some fly mortality probably due to dsRNA injection with a Hamilton syringe, the RNAi method used here produced reproducible results in female horn flies. The failure to demonstrate gene knockdown for some sequences in serine protease and other functional groups studied could be due to unknown factors affecting RNAi in horn flies, because gene expression silencing did not occur until after 24-36 hpi for these genes or due to the existence of paralogs in these groups that affected the efficacy of gene knockdown. Gene silencing mediated by RNAi depends on short interfering RNAs (siRNAs) and micro RNAs (miRNAs). These RNAs have unique features, namely a defined size of 19-21 pb, and characteristic two-nucleotide single-stranded 3' overhangs and 5' monophosphate groups [50]. Although RNAi off-target effects were shown in horn flies, most sequence alignments resulted in homology regions of 11 bp only and in some cases no homology ≥ 11 bp was found. These results suggested differences in RNAi specificity and sensitivity, a fact that needs to be fully characterized to understand and efficiently use RNAi in horn flies and other organisms [37,48,51].
The aim of this study was to conduct a functional genomics study in female horn flies combining EST analysis with RNAi. Therefore, we will focus the discussion on unigene functional groups characterized by RNAi.
Serine proteases
Serine proteases are a group of endopeptidases involved in several processes such as digestion, immune response, blood clotting and inflammation. In female horn flies, 10% of the assembled unigenes, containing more than 500 ESTs, were identified as serine proteases. In agreement with these results, Guerrero et al. [19] recently showed that serine proteases are differentially expressed in fly adult stages when compared to larvae. Significant gene knockdown was not obtained for any of the genes targeted by dsRNA injection in this group. Consequently, RNAi did not affect fly mortality or oviposition. In other arthropods, silencing of serine proteases expression by RNAi showed that these proteins are involved in blood digestion, oocyte maturation, development and immune response [40,42-44,46,52-55].
Protease inhibitors
The protease inhibitor genes identified in female horn flies corresponded to serpins, inhibitors of serine proteases and thus involved in the same biological processes discussed before for serine proteases. A horn fly serine protease inhibitor gene was previously cloned and characterized, suggesting that these genes may be involved in the control of fly endogenous and pathogen proteases [56,57]. In mosquitoes, serpin RNAi affected insect immune response [58]. The elastase inhibitor gene knockdown significantly increased horn fly mortality at 12, 24 and 36 hpi. Thus, the effect of elastase inhibitor RNAi described here in horn flies may be the result of impaired fly protease control and/or the effect of increased susceptibility to persistent pathogen infections resulting from diminished immune response.
Vitellogenin
VTGs constitute a multigene superfamily encoding for egg yolk precursor proteins expressed in the females of arthropods and other oviparous organisms [59]. In cockroaches, ants and ticks, knockdown of VTG receptor (VTG-R), essential for VTG uptake into developing oocytes, disrupts egg formation [41,60-62]. In honeybees, silencing of VTG expression by RNAi affects honeybee workers developmental behavior [63,64]. Similar to results of VTG-R knockdown in other arthropods, silencing of VTG-2 expression in horn flies reduced oviposition in 4-fold when compared to controls.
Ubiquitination
Ubiquitination is a post-translational modification carried out by a set of enzymes that affect protein proteasomal degradation, stability, function, and intracellular localization [65]. In this functional group, horn fly genes involved in the ubiquitination pathway such as ubiquitin-1 (UBQ-1), UBQ-protein ligase, and UBQ hydrolase were included. In this group, only the UBQ-protein ligase expression was significantly silenced after RNAi. Although UBQ-protein ligase has been shown to regulate apoptosis in Drosophila [66], knockdown of this gene did not affect horn fly mortality or oviposition. Thus, it may be possible that the phenotype resulting from silencing the UBQ-protein ligase expression was not evident in horn flies under our experimental conditions. Additionally, knockdown of other ubiquitination genes may be required in order to have a significant effect on horn fly mortality and oviposition. For example, UBQ knockdown in ticks causes mortality and reduces oviposition and Anaplasma marginale infection/multiplication in the guts [39,67].
Ferritin
FER is the main protein for intracellular iron storage and consists of 2 types of subunits, a heavy (ferroxidase sites) and a light chain (nucleation sites) [68]. FER light (3 unigens containing 5 ESTs) and heavy (2 unigenes containing 4 ESTs) chains were not among the most abundant ESTs identified in female horn flies. However, Guerrero et al. [35] found FER light chain as one of the most abundant transcripts in horn fly larvae. These results suggested differences in the FER expression between horn fly larvae and adult females. FER light chain knockdown in horn flies significantly reduced oviposition (6-fold with respect to controls), but surprisingly fly mortality was reduced when compared to controls. In ticks, FER RNAi reduces not only oviposition but also feeding and A. marginale infection levels in IDE8 cells [67,68].
vATPase
vATPase is a multisubunit enzyme that mediates acidification of eukaryotic intracellular organelles and has been shown to be required for the normal function of the Golgi complex, endoplasmic reticulum, vacuoles and endocytotic and exocytotic vesicles [69]. vATPase was also implicated in immunity [70]. Guerrero et al. [35] identified vATPase as one of the most abundant transcripts in horn fly larvae. However, in adult females, only 3 ATPase unigenes were assembled with one EST each, those suggesting like previously for FER, differences in the vATPase expression between horn fly larvae and adult females. Genetic knockout of vATPase subunits resulted in lethal phenotypes in fruit flies (Drosophila melanogaster), flour beetles (T. castaneum), pea aphids (Acyrthosiphon pisum), and tobacco hornworms (Manduca sexta) [69,71] and reduced influenza virus replication in Drosophila cells [72]. RNAi of vATPase expression in ticks resulted in testis and salivary gland degeneration, suggesting a role for this molecule in the function of these organs [73] and reduced A. marginale infection in Dermacentor variabilis tick guts but not pathogen multiplication in IDE8 tick cells [67]. vATPase knockdown in horn flies resulted in 16-fold reduction in oviposition but, as with FER light chain, fly mortality was reduced when compared to controls. These results suggested that despite the important function of vATPase in all arthropods, developmental stage-specific and species-specific differences might exist that could explain the results obtained after gene knockdown in horn flies.
Proteasome component
Proteasomes are large protein complexes involved in protein proteolysis that are functionally related to ubiquitination and thus essential for eukaryotic cells [74]. Experiments in D. melanogaster showed that knockdown of proteasome subunits leads to increased levels of ubiquitin conjugates, cell cycle defects, DNA overreplication, and apoptosis [74,75]. In tick cells, 26S proteasome knockdown resulted in lower A. marginale infection levels when compared to controls but did not affect tick survival, feeding and reproduction [67]. However, based on the essential proteasome function in eukaryotic cells, it was not surprising to observe a decrease in oviposition in horn flies injected with proteasome components dsRNAs targeting proteasome subunit beta (two unigenes) and proteasome maturation protein (one unigene). As previously shown in D. melanogaster [74,75], proteasome subunits knockdown in horn flies may affect cell cycle and DNA replication thus resulting in reduced oviposition.
Immune response
Innate immune response is essential for insect survival. Only two unigens were assembled into this category and knockdown in female horn flies. Assembled unigenes encoded for putative T-cell immunomodulatory protein and RNAse L inhibitor. Silencing of these genes resulted in higher horn fly mortality and lower oviposition when compared to controls. These RNAi results may be due to an effect of gene knockdown on increased susceptibility to persistent pathogen infections resulting from impaired immune response in horn flies. Knockdown of immune response genes may affect the mechanisms involved in the control of persistent infections such as those caused by Nora virus and Wolbachia spp. [76-78], which could affect horn fly mortality and ovisposition. RNAi knockdown of immune response genes in other arthropods results in increased mortality and higher pathogen infection levels [79-81].
5'-nucleotidase
5'-NUC and other ectonucleotidases control the levels of extracellular nucleotides and nucleosides that act as signaling molecules involved in a wide spectrum of biological effects [82]. 5'-NUC is commonly expressed in the salivary glands of blood-sucking ectoparasites [83-88]. Herein, as previously shown in ticks [36], 5'-NUC knockdown resulted in higher fly mortality and lower oviposition when compared to controls. As in other organisms, these results suggested an essential function for 5'-NUC in horn fly females.
Conclusions
In summary, a cDNA library was constructed from whole abdominal tissues collected from partially fed adult female horn flies and 2,160 high quality ESTs were sequenced and assembled into 992 unigenes (178 contigs and 814 singlets) representing molecular functions such as serine proteases, cell metabolism, mitochondrial function, transcription and translation, transport, chromatin structure, vitellogenesis, cytoskeleton, DNA replication, cell response to stress and infection, cell proliferation and cell-cell interactions, intracellular trafficking and secretion, and development. A method was developed for RNAi that produced reproducible results in horn flies. Functional analyses by RNAi showed the effect of some genes on horn fly mortality and oviposition. These results advanced the molecular characterization of this important ectoparasite and suggested candidate protective antigens for the development of vaccines for the control of horn fly infestations. Based on RNAi results, some of the candidate antigens to be considered for cattle vaccination experiments against horn flies include those within VTG, immune response and 5'-NUC functional groups.
Methods
Rearing of horn flies
H. irritans were reared under laboratory conditions as reported by Schmidt et al. [89]. A horn fly colony was established with flies originally collected in a cattle farm close to Ciudad Victoria, Tamaulipas, Mexico. About 2,000 flies were collected from 2 infested animals and transported in a 20 × 30 cm mosquito netting aluminum cage. Flies were allowed to lay eggs over a water container during 12 h. Eggs were collected and incubated into fresh bovine feces during 5 days. Pupae were collected and placed in Petri dishes located inside mosquito netting aluminum cages for molting into adult flies. After molting, flies were fed twice a day using pieces of cotton impregnated with fresh defibrinated bovine blood obtained from a naive cow. All the horn fly developmental phases were kept under a photoperiod of 12 h light: 12 h darkness at 28-32°C and 70-80% relative humidity [89].
Analysis of expressed sequence tags (ESTs) in adult female horn flies
Total RNA was isolated from whole abdominal tissues collected from 1,500 partially fed adult female horn flies using Trizol (Sigma, St. Louis, MO, EUA). The cDNA library was synthesized using the SMART™ cDNA Library Construction Kit (Clontech, Mountain View, CA, USA) at Creative Biolabs (Port Jefferson Station, NY, USA; http://www.creativebiolabs.com). cDNAs were cloned into the pBluescript II SK vector (Agilent Technologies, Inc., Santa Clara, CA, USA). The library had more than 1×106 primary clones, with >90% recombinants with inserts >500 bp (average cDNA length >1,000 bp). A total of 2,462 ESTs were 5' sequenced (Creative Biolabs). The cDNA Annotation System software (CAS; Bioinformatics and Scientific IT Program (BSIP), Office of Technology Information Systems (OTIS), National Institute of Allergy and Infectious Diseases (NIAID), Bethesda, MD, USA) http://exon.niaid.nih.gov was used for automated sequence clean up, assembly, blasting against multiple sequence databases (ncbi non-redundant nucleotide and protein sequence databases, H. irritans EST sequences [35] and databases of mosquito- and tick-specific sequences http://www.ncbi.nlm.nih.gov/; http://www.vectorbase.org/index.php) and Gene Ontology (GO) assignments. Comparison with the ncbi Clusters of Orthologous Groups of proteins (COG; http://www.ncbi.nlm.nih.gov/COG) was also performed. Nucleotide sequences were aligned using the program AlignX (Vector NTI Suite V 5.5, InforMax, North Bethesda, MD, USA). Gene sequences were deposited in the GenBank with accession numbers HO000420-HO001165 and HO004499-HO004744.
RNAi in adult female horn flies
Oligonucleotide primers (pBLUET75: 5'-TAATACGACTCACTATAGGGTACTTCGAGGTCGACGGTATCGAT-3' and pBLUET73: 5'-TAATACGACTCACTATAGGGTACTCAATTAACCCTCACTAAAGGGA-3') were synthesized specific for vector DNA sequences flanking the horn fly cDNA insert and containing T7 promoter sequences (in italics) for in vitro transcription and synthesis of dsRNA. PCR reactions were performed from individual or pooled cDNA clones (when more than one unigene was included in the functional group analyzed; Table 2) using the Access RT-PCR system (Promega, Madison, WI, USA) in a 50 μl reaction mixture. The resultant amplicons were purified using the Wizard 96-well PCR purification system (Promega). In vitro transcription and purification of dsRNA was done using the Megascript RNAi kit (Ambion, Austin, TX, USA). The dsRNA was quantified by spectrometry. Adult partially fed female flies were injected with approximately 0.1 μl of dsRNA (1 × 109-1 × 1011 molecules per μl) in the abdominal segment. The injections were done with a Hamilton syringe with a 1 inch, 33 gauge needle. Control flies were injected with unrelated dsRNA or injection buffer (10 mM Tris-HCl, pH 7, 1 mM EDTA) (Table 5). One hundred flies were used in each group. After injection with dsRNA, female flies were kept in petri dishes for one hour and then transferred to wired 20 × 30 cm boxes. Flies were fed using impregnated cotton with fresh defibrinated blood obtained from a naive cow and reared as described before. Fly mortality was evaluated at 12, 24 and 36 hpi. Survival curves (temporal rates of mortality) were compared between different treatments and controls using Cox Proportional Hazards Survival Regression analysis (SPSS Inc., Chicago, IL, USA). Oviposition (number of eggs per survived fly) was also evaluated and the results in test dsRNA-injected groups and in the injection buffer control were compared with the unrelated dsRNA-injected control group by Student's t-test (P = 0.05).
Gene expression silencing was evaluated in 4 individual flies each at 6, 12 and 36 or 24 hpi. The mRNA levels of each knockdown gene were determined using sequence-specific oligonucleotide primers (Table 6) and the iScript One-Step RT-PCR Kit with SYBR Green and the iQ5 thermal cycler (Bio-Rad, Hercules, CA, USA) following manufacturer's recommendations. A dissociation curve was run at the end of the reaction to ensure that only one amplicon was formed and that the amplicon denatured consistently in the same temperature range for every sample [90]. The mRNA levels were normalized against horn fly 16S rRNA (Table 4) using the genNorm method (ddCT method as implemented by Bio-Rad iQ5 Standard Edition, Version 2.0) [91]. In all cases, the mean of the duplicate values was used and normalyzed Ct values from test dsRNA-injected groups and in the injection buffer control were compared with the unrelated dsRNA-injected control group by Student's t-test (P = 0.05).
Table 6.
Primer sets and real-time RT-PCR conditions used for analysis of selected H. irritans ESTs.
| Unigene ID (Genbank accession number) | Gene description | Upstream/downstream primer sequences (5'-3') | PCR annealing conditions |
|---|---|---|---|
| 13_D07 (HO000820) |
5'-nucleotidase | AGTGGACAAATGTCCCGAAG AGCATTGGGGTTTGAATGAG | 55°C, 30 s |
| 6_F11 (HO000609) |
T-cell immunomodulatory protein | CCGGTGACTTTGATGGAGAT GATAATGGCTCCCCTTTGGT | 55°C, 30 s |
| 10_G05 (HO000738) |
RNase L inhibitor | GCCGATCGTGTTATTGTCCT CCGGATCGTTTTTGTTCAGT | 55°C, 30 s |
| 7_A04 (HO000619) |
Proteasome subunit beta | CAGGCGAGGTCCATTATTGT AGTGCGCGACCTCAAGTAGT | 55°C, 30 s |
| 12_H09 (HO000808) |
Proteasome maturation protein | GAGGAATCGTGAGGGTTTGA ACATGGGGTTGTCGGATAAA | 55°C, 30 s |
| 7_F08 (HO000644) |
vATPase subunit d | TGTTTTTCCGTCACCAGTCA GGCACAAACCCTCCAAGTAA | 60°C, 30 s |
| 9_A08 (HO000679) |
vATPase subunit f | TGTTGGATTCTTGCTTGGTG GGCACTGGTGATGTATGTGC | 60°C, 30 s |
| 17_H03 (HO001053) |
vATPase proteolipid subunit | GTCCAGCCAGACTGTGATGA AATCAATCGCGGACAAAAAC | 60°C, 30 s |
| 26 (HO004524) |
Ferritin light chain | TGATCATGTTGAACCCGAGA CGGCTGGTCAATTTCTTGAT | 60°C, 30 s |
| 154 (HO004652) |
Ferritin heavy chain | GTTGTTGCCCCTGCTGTATT TGAAAAGTGGGCTCCCATAG | 60°C, 30 s |
| 84 (HO004582) |
Ubiquitin-protein ligase | TCGCATCTGTTTGGATGTGT CGGGAAAACTTTTGAGTCCA | 60°C, 30 s |
| 146 (HO004644) |
Ubiquitin family (UBQ-1) | CCCGACCAACAACGTTTAAT CGACGAAGACGGTGAATTTT | 60°C, 30 s |
| 4_E04 (HO000538) |
Ubiquitin carboxyl-terminal hydrolase | AGCCAGAGATGTTGGAATGG TCGATGTAAATTGCCGCATA | 60°C, 30 s |
| 7 (HO004505) |
Vitellogenin 3 | GAGCTTTTTGCGTTGTAGCC ACAAAAGTGGGAGCAACACC | 60°C, 30 s |
| 20 (HO004518) |
Vitellogenin 1 | GAGCTTTTTGCGTTGTAGCC ACAAAAGTGGGAGCAACACC | 60°C, 30 s |
| 76 (HO004574) |
Vitellogenin 2 | ACGGCCGGTTGTGAGATTAT AGCATCTTTTTCGGTCTTGC | 60°C, 30 s |
| 2_B12 (HO000479) |
Elastase inhibitor | CAAGGGTGAATGGGAAAAGA TAAAGGCCTTCACGTTCCTG | 60°C, 30 s |
| 90 (HO004588) |
Serine protease, midgut specific trypsin, secreted | TGCGTTATATTCCGTTGGTG CTTTGTCAACGGCATAAGCA | 60°C, 30 s |
| 230 (HO004728) |
Serine protease | TGGCTACAATGAATGCAAGC GGTTAGCACCAGGGAACGTA | 60°C, 30 s |
| (FJ025436) | H. irritans 16S rRNA | TTTAAATGGCCGCAGTATCC GATTTATAGGGTCTTCTCGTCTTTT | 60°C, 30 s |
Authors' contributions
LT reared horn flies, prepared RNA samples, did the RNAi experiments and helped with drafting the manuscript. CA participated in coordination of the study, RNAi experiments and drafting of the manuscript. NA, RCG and CA conducted the real-time RT-PCR analyses. RCG helped with drafting the manuscript. JF participated in design and coordination of the study, analyzed EST and RNAi data and drafted the manuscript. RRC and HQR participated in coordination of the study. All authors read and approved the final manuscript.
Supplementary Material
Table S1. Assembling and analysis of H. irritans high quality ESTs.
Contributor Information
Lorena Torres, Email: lorenatorresrdz@gmail.com.
Consuelo Almazán, Email: consuelo_almazan@yahoo.com.mx.
Nieves Ayllón, Email: neve_8659@hotmail.com.
Ruth C Galindo, Email: ruthcecilia.galindo@uclm.es.
Rodrigo Rosario-Cruz, Email: rockdrig@yahoo.com.mx.
Héctor Quiroz-Romero, Email: hquiroz@servidor.unam.mx.
José de la Fuente, Email: jose_delafuente@yahoo.com.
Acknowledgements
Joaquín Vicente (IREC, Spain) is acknowledged for conducting statistical analysis of fly survival curves. This research was supported by SAGARPA-CONACYT (project 12260) and SEP-CONACYT (project 25772), Mexico to CA, the INIA (project FAU2008-00014-00-00) and the Unversity of Castilla La Mancha (project 00030072 "Ayudas para acciones de cooperación al desarrollo"), Spain to JF. Lorena Torres was supported by CONACYT, grant 42883 and Promep-UAT, Mexico. R.C. Galindo was funded by MEC, Spain. Publication of this paper was possible through the grant 151779, FOMIX, Tamaulipas, Mexico.
References
- Harwood RF, James MT. Entomología Médica y Veterinaria. Uteha, Mexico, DF. 1993.
- Barros AT, Guglielmone AA, Martins JR. Mosca de los cuernos: Control sustentable y resistencia a los insecticidas. Red Ectopar. 2002. pp. 1–10.
- Quiroz RH. Parasitología y Enfermedades Parasitarias de los Animales Domésticos. 3. Limusa. México, DF; 2005. [Google Scholar]
- Cupp EW, Cupp MS, Ribeiro JM, Kunz SE. Blood-feeding strategy of Haematobia irritans (Diptera:Muscidae) J Med Entomol. 1998;35:591–595. doi: 10.1093/jmedent/35.4.591. [DOI] [PubMed] [Google Scholar]
- Steelman CD, Brown MA, Gbur EE, Tolley G. The effects of hair density of beef cattle on Haematobia irritans horn fly populations. Med Vet Entomol. 1997;11:257–264. doi: 10.1111/j.1365-2915.1997.tb00404.x. [DOI] [PubMed] [Google Scholar]
- Johnson NN, Mayer DG. Estimation of the effects of buffalo fly (Haematobia irritans exigua) on the estimation of dairy cattle based on a meta-analysis of literature data. Med Vet Entomol. 1999;13:372–376. doi: 10.1046/j.1365-2915.1999.00179.x. [DOI] [PubMed] [Google Scholar]
- Kunz SE, Kemp DH. Insecticides and acaricides: resistance and environmental impact. Rev Sci Tech. 1994;13:1249–1286. doi: 10.20506/rst.13.4.816. [DOI] [PubMed] [Google Scholar]
- Cupp EW, Cupp MS, Ribeiro JM, Kunz SE. Blood-feeding strategy of Haematobia irritans (Díptera: Muscidae) J Med Entomol. 1998;35:591–595. doi: 10.1093/jmedent/35.4.591. [DOI] [PubMed] [Google Scholar]
- Guglielmone AA, Gimeno E, Idiart J, Fisher WF, Volpogni MM, Quaino O, Anziani OS, Flores SG, Warnke O. Skin lesions and cattle hide damage from Haematobia irritans infestations. Med Vet Entomol. 1999;13:324–329. doi: 10.1046/j.1365-2915.1999.00167.x. [DOI] [PubMed] [Google Scholar]
- Sinshaw A, Abebe G, Desquesnes M, Yoni W. Biting flies and Tripanosoma vivax infection in three highland districts bordering lake Tana, Ethiopia. Vet Parasitol. 2006;142:35–46. doi: 10.1016/j.vetpar.2006.06.032. [DOI] [PubMed] [Google Scholar]
- Abril C, Nimmervoll H, Pilo P, Brodard I, Korczak B, Markus S. Rapid diagnosis and quantification of Francisella tularensis in organs of naturally infected common squirrel monkeys (Saimiri sciureus) Vet Microbiol. 2007;127:203–208. doi: 10.1016/j.vetmic.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Gillespie BE, Owens WE, Nickerson SC, Oliver SP. Deoxyribonucleic acid fingerprinting of Staphylococcus aureus from heifer mammary secretions from horn flies. J Dairy Sci. 1999;82:1581–1585. doi: 10.3168/jds.S0022-0302(99)75386-5. [DOI] [PubMed] [Google Scholar]
- Spier SJ, Leutenegger CM, Carroll SP, Loye JE, Pusteria JB, Carpenter TE, Mihalyi JE, Madigan JE. Use of real-time polymerase chain reaction-based fluorogenic 5' nuclease assay to evaluate insect vectors of Corynebacterium pseudotuberculosis infections in horses. Amer J Vet Res. 2004;65:829–834. doi: 10.2460/ajvr.2004.65.829. [DOI] [PubMed] [Google Scholar]
- Rodríguez SD, García Ortiz MA, Jiménez Ocampo R, Vega y Murguía C. Molecular epidemiology of bovine anaplasmosis with a particular focus in Mexico. Infect Genet Evol. 2009;9:1092–1101. doi: 10.1016/j.meegid.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Kunz SE, Schmidt CD. The pyretroid resistance problem in the horn fly. J Agric Entomol. 1985;2:358–363. [Google Scholar]
- Oyarzún MP, Quiroz A, Birkett MA. Insecticide resistance in the horn fly: alternative control strategies. Med Vet Entomol. 2008;22:188–202. doi: 10.1111/j.1365-2915.2008.00733.x. [DOI] [PubMed] [Google Scholar]
- Kunz SE, Kemp DH. Insecticides and acaricides: Resistance and environmental impact. Rev Sci Tech. 1994;13:1249–1286. doi: 10.20506/rst.13.4.816. [DOI] [PubMed] [Google Scholar]
- Mochi DA, Monteiro AC, Ribeiro Machado AC, Yoshida L. Efficiency of entomopathogenic fungi in the control of eggs and larvae of the horn fly Haematobia irritans (Diptera: Muscidae) Vet Parasitol. 2009;167:62–66. doi: 10.1016/j.vetpar.2009.09.046. [DOI] [PubMed] [Google Scholar]
- Guerrero FD, Dowd SE, Sun Y, Saldivar L, Wiley GB, Macmil SL, Najar F, Roe BA, Foil LD. Microarray analysis of female- and larval-specific gene expression in the horn fly (Diptera: Muscidae) J Med Entomol. 2009;46:257–270. doi: 10.1603/033.046.0210. [DOI] [PubMed] [Google Scholar]
- Willadsen P, Riding GA, McKenna RV, Kemp DH, Tellam RL, Nielsen JN, Lahstein J, Cobon GS, Gough JM. Immunological control of a parasitic arthropod: identification of a protective antigen from Boophilus microplus. J Immunol. 1989;143:1346–1351. [PubMed] [Google Scholar]
- de la Fuente J, Almazán C, Canales M, Pérez de la Lastra JM, Kocan KM, Willadsen P. A ten-year review of commercial vaccine performance for control of tick infestations on cattle. Anim Health Res Rev. 2007;8:23–28. doi: 10.1017/S1466252307001193. [DOI] [PubMed] [Google Scholar]
- de la Fuente J, Rodríguez M, Redondo M, Montero C, García-García JC, Méndez L, Serrano E, Valdés M, Enríquez A, Canales M, Ramos E, de Armas CA, Rey S, Rodríguez JL, Artiles M, García L. Field studies and cost-effectiveness analysis of vaccination with Gavac™ against the cattle tick Boophilus microplus. Vaccine. 1998;16:366–373. doi: 10.1016/S0264-410X(97)00208-9. [DOI] [PubMed] [Google Scholar]
- de la Fuente J, Kocan KM. Advances in the identification and characterization of protective antigens for development of recombinant vaccines against tick infestations. Expert Rev Vaccines. 2003;2:583–593. doi: 10.1586/14760584.2.4.583. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Belkaid Y, Garfield MK, Mendez S, Kamhawi S, Rowton ED, Sacks DL, Ribeiro JM. Toward a defined anti-Leishmania vaccine targeting vector antigens: characterization of a protective salivary protein. J Exp Med. 2001;194:331–342. doi: 10.1084/jem.194.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal AA, Patterson PS, Sacci JB, Vaughan JA, Paul C, Collins WE, Wirtz RA, Azad AF. Anti-mosquito midgut antibodies block development of Plasmodium falciparum and Plasmodium vivax in multiple species of Anopheles mosquitoes and reduce vector fecundity and survivorship. Proc Natl Acad Sci USA. 2001;98:5228–5233. doi: 10.1073/pnas.091447398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida AP, Billingsley PF. Induced immunity against the mosquito Anopheles stephensi (Diptera: Culicidae): effects of cell fraction antigens on survival, fecundity, and Plasmodium berghei (Eucoccidiida: Plasmodiidae) transmission. J Med Entomol. 2002;39:207–214. doi: 10.1603/0022-2585-39.1.207. [DOI] [PubMed] [Google Scholar]
- Suneja A, Gulia M, Gakhar SK. Blocking of malaria parasite development in mosquito and fecundity reduction by midgut antibodies in Anopheles stephensi (Diptera: Culicidae) Arch Insect Biochem Physiol. 2003;52:63–70. doi: 10.1002/arch.10068. [DOI] [PubMed] [Google Scholar]
- Milleron RS, Ribeiro JM, Elnaime D, Soong L, Lanzaro G. Negative effect of antibodies against maxadilan on the fitness of the sand fly vector of American visceral leishmaniasis. Am J Trop Med Hyg. 2004;70:278–285. [PubMed] [Google Scholar]
- Titus RG, Bishop JV, Mejia JS. The immunomodulatory factors of arthropod saliva and the potential for these factors to serve as vaccine targets to prevent pathogen transmission. Parasite Immunol. 2006;4:131–141. doi: 10.1111/j.1365-3024.2006.00807.x. [DOI] [PubMed] [Google Scholar]
- Kedzierski L, Zhu Y, Handman E. Leishmania vaccines: progress and problems. Parasitol. 2006;133:S87–S112. doi: 10.1017/S0031182006001831. [DOI] [PubMed] [Google Scholar]
- Saul A. Mosquito stage, transmission blocking vaccines for malaria. Curr Opin Infect Dis. 2007;5:476–481. doi: 10.1097/QCO.0b013e3282a95e12. [DOI] [PubMed] [Google Scholar]
- Canales M, Naranjo V, Almazán C, Molina R, Tsuruta SA, Szabó MPJ, Manzano-Roman R, Pérez de la Lastra JM, Kocan KM, Jiménez MI, Lucientes J, Villar M, de la Fuente J. Conservation and immunogenicity of the mosquito ortholog of the tick protective antigen, subolesin. Parasitol Res. 2009;105:97–111. doi: 10.1007/s00436-009-1368-2. [DOI] [PubMed] [Google Scholar]
- Cupp MS, Cupp EW, Navarre C, Wisnewski N, Brandt KS, Silver GM, Zhang D, Panangala V. Evaluation of a recombinant salivary gland protein (thrombostasin) as a vaccine candidate to disrupt blood-feeding by horn flies. Vaccine. 2004;22:2285–2297. doi: 10.1016/j.vaccine.2003.11.024. [DOI] [PubMed] [Google Scholar]
- Wijffels G, Hughes S, Gough J, Allen J, Don A, Marshall K, Kay B, Kemp D. Peritrophins of adult dipteran ectoparasites and their evaluation as vaccine antigens. Int J Parasitol. 1999;29:1363–77. doi: 10.1016/S0020-7519(99)00070-3. [DOI] [PubMed] [Google Scholar]
- Guerrero FD, Dowd SE, Nene VM, Foil LD. Expressed cDNAS from embryonic and larval stages of the horn fly (Diptera: Muscidae) J Med Entomol. 2008;45:686–692. doi: 10.1603/0022-2585(2008)45[686:ECFEAL]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- de la Fuente J, Almazán C, Blouin EF, Naranjo V, Kocan KM. RNA interference screening in ticks for identification of protective antigens. Parasitol Res. 2005;96:137–141. doi: 10.1007/s00436-005-1351-5. [DOI] [PubMed] [Google Scholar]
- de la Fuente J, Kocan KM, Almazán C, Blouin EF. RNA interference for the study and genetic manipulation of ticks. Trends Parasitol. 2007;23:427–433. doi: 10.1016/j.pt.2007.07.002. [DOI] [PubMed] [Google Scholar]
- de la Fuente J, Manzano-Roman R, Naranjo V, Kocan KM, Zivkovic Z, Blouin EF, Canales M, Almazán C, Galindo RC, Step DL, Villar M. Identification of protective antigens by RNA interference for control of the lone star tick, Amblyomma americanum. Vaccine. 2010;28:1786–1795. doi: 10.1016/j.vaccine.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Almazán C, Lagunes R, Villar M, Canales M, Rosario-Cruz R, Jongejan F, de la Fuente J. Identification and characterization of Rhipicephalus (Boophilus) microplus candidate protective antigens for the control of cattle tick infestations. Parasitol Res. 2010;106:471–479. doi: 10.1007/s00436-009-1689-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T, Tsuji N, Islam MK, Kamio T, Fujisaki K. Gene silencing of a cubilin-related serine proteinase from the hard tick Haemaphysalis longicornis by RNA interference. J Vet Med Sci. 2004;66:1471–3. doi: 10.1292/jvms.66.1471. [DOI] [PubMed] [Google Scholar]
- Ciudad L, Piulachs MD, Bellés X. Systemic RNAi of the cockroach vitellogenin receptor results in a phenotype similar to that of the Drosophila yolkless mutant. FEBS J. 2006;273:325–35. doi: 10.1111/j.1742-4658.2005.05066.x. Erratum in: FEBS J. 2006 Feb;273(3):671. [DOI] [PubMed] [Google Scholar]
- Kambris Z, Brun S, Jang IH, Nam HJ, Romeo Y, Takahashi K, Lee WJ, Ueda R, Lemaitre B. Drosophila immunity: a large-scale in vivo RNAi screen identifies five serine proteases required for Toll activation. Curr Biol. 2006;16:808–13. doi: 10.1016/j.cub.2006.03.020. [DOI] [PubMed] [Google Scholar]
- Araujo RN, Campos IT, Tanaka AS, Santos A, Gontijo NF, Lehane MJ, Pereira MH. Brasiliensin: A novel intestinal thrombin inhibitor from Triatoma brasiliensis (Hemiptera: Reduviidae) with an important role in blood intake. Int J Parasitol. 2007;37:1351–8. doi: 10.1016/j.ijpara.2007.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Yin Y, Zhang B, Wang Z, Peng G, Cao Y, Xia Y. Cloning of a novel protease required for the molting of Locusta migratoria manilensis. Dev Growth Differ. 2007;49:611–21. doi: 10.1111/j.1440-169X.2007.00957.x. [DOI] [PubMed] [Google Scholar]
- Chen XG, Mathur G, James AA. Gene expression studies in mosquitoes. Adv Genet. 2008;64:19–50. doi: 10.1016/S0065-2660(08)00802-X. full_text. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isoe J, Rascón AA Jr, Kunz S, Miesfeld RL. Molecular genetic analysis of midgut serine proteases in Aedes aegypti mosquitoes. Insect Biochem Mol Biol. 2009;39:903–12. doi: 10.1016/j.ibmb.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan P, Gireesh-Babu P, Rajendran KV, Chaudhari A. RNA interference-based therapeutics for shrimp viral diseases. Dis Aquat Organ. 2009;86:263–72. doi: 10.3354/dao02126. [DOI] [PubMed] [Google Scholar]
- Bellés X. Beyond Drosophila: RNAi in vivo and functional genomics in insects. Annu Rev Entomol. 2010;55:111–28. doi: 10.1146/annurev-ento-112408-085301. [DOI] [PubMed] [Google Scholar]
- Huvenne H, Smagghe G. Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: a review. J Insect Physiol. 2010;56:227–35. doi: 10.1016/j.jinsphys.2009.10.004. [DOI] [PubMed] [Google Scholar]
- Lingel A, Sattler M. Novel modes of protein-RNA recognition in the RNAi pathway. Curr Opin Struct Biol. 2005;15:107–15. doi: 10.1016/j.sbi.2005.01.010. [DOI] [PubMed] [Google Scholar]
- de la Fuente J, Maritz-Olivier C, Naranjo V, Ayoubi P, Nijhof AM, Almazán C, Canales M, Pérez de la Lastra JM, Galindo RC, Blouin EF, Gortazar C, Jongejan F, Kocan KM. Evidence of the role of tick subolesin in gene expression. BMC Genomics. 2008;9:372. doi: 10.1186/1471-2164-9-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brackney DE, Foy BD, Olson KE. The effects of midgut serine proteases on dengue virus type 2 infectivity of Aedes aegypti. Am J Trop Med Hyg. 2008;79:267–74. [PMC free article] [PubMed] [Google Scholar]
- Shah PK, Tripathi LP, Jensen LJ, Gahnim M, Mason C, Furlong EE, Rodrigues V, White KP, Bork P, Sowdhamini R. Enhanced function annotations for Drosophila serine proteases: a case study for systematic annotation of multi-member gene families. Gene. 2008;407:199–215. doi: 10.1016/j.gene.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Marshall JL, Huestis DL, Hiromasa Y, Wheeler S, Oppert C, Marshall SA, Tomich JM, Oppert B. Identification, RNAi knockdown, and functional analysis of an ejaculate protein that mediates a postmating, prezygotic phenotype in a cricket. PLoS One. 2009;4:e7537. doi: 10.1371/journal.pone.0007537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amparyup P, Wiriyaukaradecha K, Charoensapsri W, Tassanakajon A. A clip domain serine proteinase plays a role in antibacterial defense but is not required for prophenoloxidase activation in shrimp. Dev Comp Immunol. 2010;34:168–76. doi: 10.1016/j.dci.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Azzolini SS, Santos JM, Souza AF, Torquato RJ, Hirata IY, Andreotti R, Tanaka AS. Purification, characterization, and cloning of a serine proteinase inhibitor from the ectoparasite Haematobia irritans irritans (Diptera: Muscidae) Exp Parasitol. 2004;106:103–109. doi: 10.1016/j.exppara.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Azzolini SS, Sasaki SD, Campos IT, S Torquato RJ, Juliano MA, Tanaka AS. The role of HiTI, a serine protease inhibitor from Haematobia irritans irritans (Diptera: Muscidae) in the control of fly and bacterial proteases. Exp Parasitol. 2005;111:30–36. doi: 10.1016/j.exppara.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Michel K, Suwanchaichinda C, Morlais I, Lambrechts L, Cohuet A, Awono-Ambene PH, Simard F, Fontenille D, Kanost MR, Kafatos FC. Increased melanizing activity in Anopheles gambiae does not affect development of Plasmodium falciparum. Proc Natl Acad Sci USA. 2006;103:16858–63. doi: 10.1073/pnas.0608033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne BM, Gruber M, Ab G. The evolution of egg yolk proteins. Prog Biophys Mol Biol. 1989;53:33–69. doi: 10.1016/0079-6107(89)90005-9. [DOI] [PubMed] [Google Scholar]
- Mitchell RD, Ross E, Osgood C, Sonenshine DE, Donohue KV, Khalil SM, Thompson DM, Michael Roe R. Molecular characterization, tissue-specific expression and RNAi knockdown of the first vitellogenin receptor from a tick. Insect Biochem Mol Biol. 2007;37:375–88. doi: 10.1016/j.ibmb.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Boldbaatar D, Battsetseg B, Matsuo T, Hatta T, Umemiya-Shirafuji R, Xuan X, Fujisaki K. Tick vitellogenin receptor reveals critical role in oocyte development and transovarial transmission of Babesia parasite. Biochem Cell Biol. 2008;86:331–44. doi: 10.1139/O08-071. [DOI] [PubMed] [Google Scholar]
- Lu HL, Vinson SB, Pietrantonio PV. Oocyte membrane localization of vitellogenin receptor coincides with queen flying age, and receptor silencing by RNAi disrupts egg formation in fire ant virgin queens. FEBS J. 2009;276:3110–23. doi: 10.1111/j.1742-4658.2009.07029.x. [DOI] [PubMed] [Google Scholar]
- Amdam GV, Norberg K, Page RE Jr, Erber J, Scheiner R. Downregulation of vitellogenin gene activity increases the gustatory responsiveness of honey bee workers (Apis mellifera) Behav Brain Res. 2006;169:201–5. doi: 10.1016/j.bbr.2006.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco Antonio DS, Guidugli-Lazzarini KR, do Nascimento AM, Simões ZL, Hartfelder K. RNAi-mediated silencing of vitellogenin gene function turns honeybee (Apis mellifera) workers into extremely precocious foragers. Naturwissenschaften. 2008;95:953–61. doi: 10.1007/s00114-008-0413-9. [DOI] [PubMed] [Google Scholar]
- Sorokin AV, Kim ER, Ovchinnikov LP. Proteasome system of protein degradation and processing. Biochem (Mosc) 2009;74:1411–42. doi: 10.1134/S000629790913001X. [DOI] [PubMed] [Google Scholar]
- Steller H. Regulation of apoptosis in Drosophila. Cell Death Differ. 2008;15:1132–1138. doi: 10.1038/cdd.2008.50. [DOI] [PubMed] [Google Scholar]
- de la Fuente J, Blouin EF, Manzano-Roman R, Naranjo V, Almazán C, Perez de la Lastra JM, Zivkovic Z, Jongejan F, Kocan KM. Functional genomic studies of tick cells in response to infection with the cattle pathogen, Anaplasma marginale. Genomics. 2007;90:712–722. doi: 10.1016/j.ygeno.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Hajdusek O, Sojka D, Kopacek P, Buresova V, Franta Z, Sauman I, Winzerling J, Grubhoffer L. Knockdown of proteins involved in iron metabolism limits tick reproduction and development. Proc Natl Acad Sci USA. 2009;106:1033–8. doi: 10.1073/pnas.0807961106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyenbach KW, Wieczorek H. The v-type H+ ATPase: molecular structure and function, physiological roles and regulation. J Exp Biol. 2006;209:577–589. doi: 10.1242/jeb.02014. [DOI] [PubMed] [Google Scholar]
- de Vito P. The sodium/hydrogen exchanger: A possible mediator of immunity. Cell Immunol. 2006;240:69–85. doi: 10.1016/j.cellimm.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Whyard S, Singh AD, Wong S. Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochem Mol Biol. 2009;39:824–32. doi: 10.1016/j.ibmb.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Hao L, Sakurai A, Watanabe T, Sorensen E, Nidom CA, Newton MA, Ahlquist P, Kawaoka Y. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature. 2008;454:890–893. doi: 10.1038/nature07151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocan KM, Zivkovic Z, Blouin EF, Naranjo V, Almazán C, Mitra R, de la Fuente J. Silencing of genes involved in Anaplasma marginale-tick interactions affects the pathogen developmentalcycle in Dermacentor variabilis. BMC Developmenl Biol. 2009;9:42. doi: 10.1186/1471-213X-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren J, Masson P, Realini CA, Young P. Use of RNA interference and complementation to study the function of the Drosophila and human 26S proteasome subunit S13. Mol Cell Biol. 2003;23:5320–30. doi: 10.1128/MCB.23.15.5320-5330.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wójcik C, DeMartino GN. Analysis of Drosophila 26S proteasome using RNA interference. J Biol Chem. 2002;277:6188–97. doi: 10.1074/jbc.M109996200. [DOI] [PubMed] [Google Scholar]
- Hornok S, Földvári G, Elek V, Naranjo V, Farkas R, de la Fuente J. Molecular identification of Anaplasma marginale and rickettsial endosymbionts in blood-sucking flies (Diptera: Tabanidae, Muscidae) and hard ticks (Acari: Ixodidae) Vet Parasitol. 2008;154:354–9. doi: 10.1016/j.vetpar.2008.03.019. [DOI] [PubMed] [Google Scholar]
- Zhang B, McGraw E, Floate KD, James P, Jorgensen W, Rothwell J. Wolbachia infection in Australasian and North American populations of Haematobia irritans (Diptera: Muscidae) Vet Parasitol. 2009;162:350–3. doi: 10.1016/j.vetpar.2009.03.012. [DOI] [PubMed] [Google Scholar]
- Habayeb MS, Ekengren SK, Hultmark D. Nora virus, a persistent virus in Drosophila, defines a new picorna-like virus family. J Gen Virol. 2006;87:3045–3051. doi: 10.1099/vir.0.81997-0. [DOI] [PubMed] [Google Scholar]
- Eleftherianos I, Millichap PJ, ffrench-Constant RH, Reynolds SE. RNAi suppression of recognition protein mediated immune responses in the tobacco hornworm Manduca sexta causes increased susceptibility to the insect pathogen Photorhabdus. Dev Comp Immunol. 2006;30:1099–107. doi: 10.1016/j.dci.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Ayres JS, Freitag N, Schneider DS. Identification of Drosophila mutants altering defense of and endurance to Listeria monocytogenes infection. Genetics. 2008;178:1807–15. doi: 10.1534/genetics.107.083782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han-Ching Wang K, Tseng CW, Lin HY, Chen IT, Chen YH, Chen YM, Chen TY, Yang HL. RNAi knock-down of the Litopenaeus vannamei Toll gene (LvToll) significantly increases mortality and reduces bacterial clearance after challenge with Vibrio harveyi. Dev Comp Immunol. 2010;34:49–58. doi: 10.1016/j.dci.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Schetinger MR, Morsch VM, Bonan CD, Wyse AT. NTPDase and 5'-nucleotidase activities in physiological and disease conditions: new perspectives for human health. Biofactors. 2007;31:77–98. doi: 10.1002/biof.5520310205. [DOI] [PubMed] [Google Scholar]
- Liyou N, Hamilton S, Mckenna R, Elvin C, Willadsen P. Localisation and functional studies on the 5'-nucleotidase of the cattle tick Boophilus microplus. Exp Appl Acarol. 2000;24:235–46. doi: 10.1023/A:1006452325210. [DOI] [PubMed] [Google Scholar]
- Faudry E, Lozzi SP, Santana JM, D'Souza-Ault M, Kieffer S, Felix CR, Ricart CA, Sousa MV, Vernet T, Teixeira AR. Triatoma infestans apyrases belong to the 5'-nucleotidase family. J Biol Chem. 2004;279:19607–13. doi: 10.1074/jbc.M401681200. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Garfield M, Rowton ED, Pham VM. Identification of the most abundant secreted proteins from the salivary glands of the sand fly Lutzomyia longipalpis, vector of Leishmania chagasi. J Exp Biol. 2004;207:3717–29. doi: 10.1242/jeb.01185. [DOI] [PubMed] [Google Scholar]
- Van Den Abbeele J, Caljon G, Dierick JF, Moens L, De Ridder K, Coosemans M. The Glossina morsitans tsetse fly saliva: general characteristics and identification of novel salivary proteins. Insect Biochem Mol Biol. 2007;37:1075–85. doi: 10.1016/j.ibmb.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Francischetti IM, Valenzuela JG, Pham VM, Garfield MK, Ribeiro JM. Toward a catalog for the transcripts and proteins (sialome) from the salivary gland of the malaria vector Anopheles gambiae. J Exp Biol. 2002;205:2429–51. doi: 10.1242/jeb.205.16.2429. [DOI] [PubMed] [Google Scholar]
- Francischetti IM, Meng Z, Mans BJ, Gudderra N, Hall M, Veenstra TD, Pham VM, Kotsyfakis M, Ribeiro JM. An insight into the salivary transcriptome and proteome of the soft tick and vector of epizootic bovine abortion, Ornithodoros coriaceus. J Proteomics. 2008;71:493–512. doi: 10.1016/j.jprot.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt CD, Harris RL, Hoffman RA. Mass rearing of the horn fly, Haematobia irritans (Díptera: Muscidae), in the laboratory. Ann Entomol Soc Am. 1967;60:508–510. [Google Scholar]
- Ririe KM, Rasmussen RP, Wittwer CT. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal Biochem. 1997;245:154–160. doi: 10.1006/abio.1996.9916. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta CT) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Assembling and analysis of H. irritans high quality ESTs.