Abstract
Like most enzymes, DNA polymerases undergo a large conformational change on the binding of a correct nucleotide. To determine how the conformational change contributes to substrate specificity, we labeled the T7 DNA polymerase with a conformationally sensitive fluorophore at a position that provides a signal coincident with structural changes following nucleotide binding and distinguishes correct base pairs from incorrect ones by the sign of the fluorescence change. Here we describe methods to document that only one site on the polymerase was labeled with the fluorophore based on mass spectral analysis of tryptic peptides. In addition, we show by equilibrium titrations of opposing signals that mismatches and correct bases compete for the same site. This analysis forms an essential basis for characterization of a fluorescently labeled enzyme intended for mechanistic studies. Finally, we show that the labeled enzyme can be used to identify single-nucleotide mutations in a procedure that could be automated.
Keywords: DNA polymerase, Fluorescence labeling, Fidelity, Conformational changes, Induced fit, Single-nucleotide polymorphisms
DNA polymerases successively incorporate nucleotides according to the sequence of the template and can discriminate against mismatched nucleotides with high accuracy [1,2]. Early kinetic studies using T7 DNA polymerase, PolI Klenow fragment, and HIV-RT each suggested a two-step nucleotide binding and recognition sequence with a conformational change step preceding chemistry [3–8]. Structural studies of these enzymes later revealed a large nucleotide-induced conformational change observed in a ternary enzyme–DNA–dNTP complex [9–14]. However, structural data do not define the role that the conformational change plays in nucleotide selectivity, which is a purely kinetic phenomenon dependent on kcat/Km. To understand how polymerases achieve such extraordinary specificity and efficiency of catalysis, we and others have sought a fluorescence signal to monitor the kinetics of the nucleotide-induced conformational change [15–20].
Different methods have been developed using fluorescent signals to resolve the kinetics of the polymerase conformational changes. One method relies on labeling the DNA template with a 2-aminopurine (2-AP)1 at the T + 1 position adjacent (5′) to the templating base. Nucleotide-induced fluorescence changes of the Escherichia coli PolI Klenow fragment, DNA polymerase β, and T4 DNA polymerase were studied using this approach [16–18], leading to the conclusion that the observed change was faster than chemistry. The fluorescence signal is attributed to the unstacking of the 2-AP at the unique T + 1 position where the template undergoes an unusual bend as it enters the polymerase active site. Accordingly, the signal reports changes in DNA structure that could arise during translocation of the DNA or could be correlated with nucleotide-induced changes in protein structure. Another method used for studying the conformational change of the Klenow fragments of Taq polymerase and PolI was based on Förster resonance energy transfer (FRET) between two fluorophores: one on the fingers domain and another on the DNA substrate [19,20]. Again, placing one fluorophore on the DNA introduces ambiguity in the interpretation of results in that the signal could be due to changes in protein structure or to movement in the DNA, a problem that is compounded by the possible partitioning of the DNA between polymerase and exonuclease sites.
Because of ambiguities in the interpretation of signals arising from labels positioned on the DNA, we sought a different method that relied on the placement of a single, environmentally sensitive fluorophore at a position that could sense a change in structure without perturbing the reaction. We previously reported the construction of an E514C mutant of T7 DNA polymerase with eight native cysteines replaced with either alanine or serine (the E514C-8C mutant). This enzyme was then labeled on the nucleotide recognition domain with the fluorophore MDCC (7-diethylamino-3-[([(2-maleimidyl)ethyl]amino)carbonyl]coumarin), and the effects of mutagenesis and labeling on the enzymatic activity were quantified [15]. We showed that the binding of a correct nucleotide (which forms canonical base pairs with the templating base) induced a decrease in fluorescence, whereas the binding of a mismatched nucleotide caused an increase in fluorescence, and we used that signal to monitor the kinetics of the nucleotide-induced conformational change. Here we describe the rationale for choice of fluorophore and selection of the site for labeling, and we report mass spectrometry (MS) analysis of tryptic peptides to show that only a single site was labeled. In addition, we describe a quantitative equilibrium competition assay to demonstrate that both correct and incorrect bases compete for the same site while inducing opposite changes in fluorescence. These methods constitute a minimal set of assays necessary to document the fluorescently labeled protein, and we propose that these assays should be applied as part of the analysis of any fluorescently labeled enzyme. Finally, we explore different DNA sequence contexts and demonstrate that the fluorescence signal can be the basis for a simple assay to detect single-nucleotide polymorphisms (SNPs).
Materials and methods
Construction, purification, and labeling of MDCC–E514C-8C T7 DNA polymerase
The plasmid encoding an exonuclease-deficient (exo−) mutant of T7 DNA polymerase, pG5X, was used for constructing a Cys-light enzyme [21] by removing 8 of the 10 native cysteine residues and adding the mutation E514C to provide a surface-exposed cysteine for site-specific labeling with MDCC as described previously [15]. Thioredoxin was purified and used to reconstitute the native polymerase holoenzyme as described previously [6]. Purified T7 gp5 and E. coli thioredoxin were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and showed a 95% or higher purity by Coomassie blue staining. The enzyme concentration was determined at 280 nm with a molar extinction coefficient of 134,420 M−1 cm−1 calculated using the amino acid sequence [22]. The concentration of MDCC on the protein was measured at 419 nm in methanol with the molar extinction coefficient of 50,000 M−1 cm−1 provided by the manufacturer (Invitrogen). The MDCC labeling efficiency was estimated to be approximately 90 to 95% based on these absorbance measurements.
Tryptic digestion of MDCC–E514C-8C exo−T7 DNA polymerase
The MDCC labeled protein, stored in the final dialysis buffer (40 mM Tris–HCl [pH 7.5], 0.1 mM ethylenediaminetetraacetic acid [EDTA], 50 mM NaCl, 50% glycerol, and 1 mM dithiothreitol [DTT]), was mixed with trypsin (sequencing-grade modified trypsin, Promega) at a ratio of 20:1 (w/w) and then was incubated overnight at 37 °C. The tryptic peptides were separated by reverse phase HPLC.
Reverse phase HPLC
A POROS R2 perfusion column (PerSeptive Biosystems) was used to separate the tryptic peptides. The column was equilibrated with buffer A (0.1% trifluoroacetic acid [TFA], 2% acetonitrile, and doubly distilled H2O [ddH2O]). Then 100 μl of sample was loaded onto the column using an ÄKTA high-performance liquid chromatography (HPLC) instrument (Amersham Pharmacia Biotech). The column was washed with buffer A, and peptides were eluted with an acetonitrile gradient (buffer A to buffer B [0.08% TFA, 80% acetonitrile, and ddH2O] at a 1.2-ml/min flow rate). The eluted peptide peaks were monitored by 220 nm ultraviolet (UV) absorption, and the presence of MDCC was monitored by 425 nm absorption. The fractions corresponding to the absorbance peaks at 425 nm were collected for MS analysis.
MS and tandem MS analysis
The collected samples from HPLC were frozen in liquid nitrogen and dried with a Savant SpeedVac concentrator (Forma Scientific) and then dissolved in 10 μl of a solution containing 50% acetonitrile, 50% H2O, and 1% TFA. The matrix solution was made of α-cyano-4-hydroxycinnamic acid supersaturated in a solution of 70% acetonitrile, 30% H2O, 0.1% TFA, and 5 mM (NH4)2HPO4. The dissolved samples were mixed with the matrix solution at a 1:1 ratio, and 0.5 μl of the mixture was spotted onto a matrix-assisted laser desorption/ionization (MALDI) stainless-steel target. The mass spectra were obtained by an ABI 4700 Proteomics analyzer MALDI tandem time-of-flight (TOF/TOF) instrument (Applied Biosystems). To verify the identities of the ions in the mass spectra, the high-energy collision-induced dissociation (CID) was used to fragment selected ions, producing tandem MS (MS/MS) spectra for the derivation of peptide sequences.
Fluorescence emission profile of MDCC–E514C-8C T7 DNA polymerase at different substrate-bound states
DNA duplexes formed with a 27mer primer (5′-GCC TCG CAG CCG TCC AAC CAA CTC AACdd-3′) and 45mer templates (5′-GGA CGG CAT TGG ATC GAN GTT GAG TTG GTT GGA CGG CTG CGA GGC-3′) with varying bases at position 18 (N) were custom synthesized by IDT and used in the nucleotide binding assays. The enzyme –DNA complex was formed using 200 nM enzyme, 300 nM DNA, 4 μM thioredoxin, and 12.5 mM MgCl2 in the T7 reaction buffer [15]. The fluorescence emission intensity was recorded by exciting the enzyme–DNA complex at 425 nm and monitoring the fluorescence intensities at 460 nm before and after the addition of 1 mM dNTP using a fluorometer from Photon Technology International. No correction for inner filter effects was necessary at these wavelengths.
Equilibrium titration experiments
A solution containing 200 nM MDCC–E514C-8C T7 DNA polymerase in the T7 reaction buffer and 12.5 mM MgCl2 was preincubated in the presence of 300 nM 27ddC/45-18G DNA duplex [15]. Solutions containing nucleotides and equal concentration of MgCl2 were used to titrate the enzyme–DNA complex using a KinTek TMX titration module (http://www.kintek-corp.com). Fluorescence intensities at equilibrium were monitored continuously, while a solution of nucleotide was added at a rate of 4 μl/min, and were corrected for the small dilution. The wavelength of excitation was set at 425 nm, and a 450-nm bandpass filter was used for emission detection. The overall dissociation constant at equilibrium state for nucleotide binding was determined by nonlinear regression to either a quadratic equation or a hyperbolic equation shown below, where A is the amplitude of fluorescence change and [E0] is the enzyme concentration:
Depending on the magnitude of the dissociation constant relative to the enzyme concentration, a Kd less than 1 to 2 μM required a quadratic equation.
Point mutation detection assay
DNA primers and templates were custom synthesized according to the sequences from the human F8 gene (Table 1). The primers were designed to have their 3′ ends at a position located two bases before the positions that contain point mutations. The nucleotide binding assays began with a solution containing 50 nM MDCC–E514C-8C enzyme, 100 nM annealed DNA substrates, and 100 nM ddNTP that correctly base paired with the first template base, for example, ddTTP for 712G and 712A DNA substrates. The solution was incubated at room temperature for at least 1 min, and the emission intensity of the enzyme–DNA complex was recorded at 450 nm by exciting at 425 nm using a fluorometer from Photon Technology International. Next, 1 μM dATP was added to the solution, and the relative fluorescence at 450 nm was recorded. Data are reported as the fluorescence values observed after adding the nucleotide divided by the fluorescence observed prior to adding the nucleotide. The procedure was repeated for each of the other three nucleotides: TTP, dGTP, and dCTP.
Table 1.
DNA primer/template sequences derived from the human F8 gene
| Nucleotide | Synthetic DNA substrate |
|---|---|
| 712G | 5′ ACCTACTCATATCTTTCTCA 3′ TGGATGAGTATAGAAAGAGTACACCTGGACCATTTTCTG 5′ |
| 712A | 5′ ACCTACTCATATCTTTCTCA 3′ TGGATGAGTATAGAAAGAGTATACCTGGACCATTTTCTG 5′ |
| 1807C | 5′ GGGCCAACTAAATCAGATCC 3′ CCCGGTTGATTTAGTCTAGGAGCCACGGACTGGGCGATA 5′ |
| 1807T | 5′ GGGCCAACTAAATCAGATCC 3′ CCCGGTTGATTTAGTCTAGGAACCACGGACTGGGCGATA 5′ |
| 6049C | 5′ ATGGCTCAGGATCAAAGGAT 3′ TACCGAGTCCTAGTTTCCTAAGCTACCATAGACGAGTCG 5′ |
| 6049T | 5′ ATGGCTCAGGATCAAAGGAT 3′ TACCGAGTCCTAGTTTCCTAAACTACCATAGACGAGTCG 5′ |
| 6050G | 5′ ATGGCTCAGGATCAAAGGATT 3′ TACCGAGTCCTAGTTTCCTAAGCTACCATAGACGAGTCG 5′ |
| 6050T | 5′ ATGGCTCAGGATCAAAGGATT 3′ TACCGAGTCCTAGTTTCCTAAGATACCATAGACGAGTCG 5′ |
| 6050A | 5′ ATGGCTCAGGATCAAAGGATT 3′ TACCGAGTCCTAGTTTCCTAAGTTACCATAGACGAGTCG 5′ |
Note. Single-nucleotide substitutions of the template bases are shown with bold characters.
Results
Choice of fluorophore and labeling site
The MDCC label was chosen because in previous studies it showed a sevenfold change in fluorescence when attached to E. coli phosphate binding protein to sense the conformational change on binding phosphate [23]. Moreover, the structure of the labeled phosphate binding protein provided clues as to where the label might be placed on T7 polymerase in that the label sits in a somewhat hydrophobic cleft of the labeled phosphate binding protein [24]. As part of our initial exploration, the MDCC-labeled cysteine from the phosphate binding protein was manually docked on various locations on the surface of the T7 DNA polymerase structure to get a crude estimate of where it might fit on the surface, as illustrated in Fig. 1. Although on initial inspection E514C appeared to be the most promising, we tested several sites for labeling, including the following mutations of surface residues K355C, K502C, E514C, Q539C, and K545C. We first prepared the Cys-light protein by removing eight surface-exposed cysteine residues by the mutations C20S-C88A-C275A-C313A-C451S-C660A-C688A-C703A. Two buried cysteines near the active site (C483 and C622) were retained. The -8C Cys-light enzyme was then characterized by quantifying the rate and amplitude of the pre-steady-state burst of product formation [6] and was shown to have near wild-type activity (data not shown). Each mutation was created in the background of the -8C Cys-light enzyme, purified, and then labeled by reaction with a 20-fold excess of MDCC overnight at 4 °C as described previously [15]. Gel filtration was insufficient to remove excess MDCC, so the labeled protein was purified by single-stranded DNA (ssDNA)–cellulose chromatography. We then characterized each labeled enzyme by looking for a change in fluorescence after nucleotide binding to an E.DNAdd complex formed with dideoxy-terminated DNA. Fluorescence of a solution containing 200 nM enzyme with 300 nM 27/45mer primer/template DNA (Table 2) and 4 μM thioredoxin was monitored before and after the addition of 1 mM dCTP (dG was the templating base). Although each mutant was successfully labeled, only the E514C mutation produced a significant change in fluorescence on nucleotide binding. Experiments to assess the effect of the mutagenesis/labeling strategy on the polymerase activity were reported previously [15]. Here we provide data to show that only one site was labeled, to quantify the fluorescence signal using different templates, and to demonstrate that correct and incorrect bases compete for the same site even though they induce fluorescence changes of the opposite sign.
Fig. 1.

Docking of MDCC on the surface of the recognition domain. The structure of T7 DNA polymerase [10] is shown with MDCC–cysteine (magenta) manually docked on the surface at the position of residue 514C. The recognition domain is shown in blue, the incoming nucleotide is shown in CPK (Corey–Pauling–Koltun) colors with carbon in green, the template strand of the DNA is yellow, and the primer strand is pale green. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.) (Drawn from 1T7P.pdb using Pymol.)
Table 2.
DNA primer/template sequences for nucleotide binding and titration experiments
| Name | Sequence |
|---|---|
| 27ddC | 5′ GCCTCGCAGCCGTCCAACCAACTCAACdd 3′ |
| 45-18A | 5′ GGACGGCATTGGATCGAAGTTGAGTTGGTTGGACGGCTGCGAGGC 3′ |
| 45-18G | 5′ GGACGGCATTGGATCGAGGTTGAGTTGGTTGGACGGCTGCGAGGC 3′ |
| 45-18T | 5′ GGACGGCATTGGATCGATGTTGAGTTGGTTGGACGGCTGCGAGGC 3′ |
| 45-18C | 5′ GGACGGCATTGGATCGACGTTGAGTTGGTTGGACGGCTGCGAGGC 3′ |
Note. The next template bases located after the end of 27ddC primer are indicated with underlines.
Fluorescence properties of MDCC-labeled E514C-8C T7 DNA polymerase
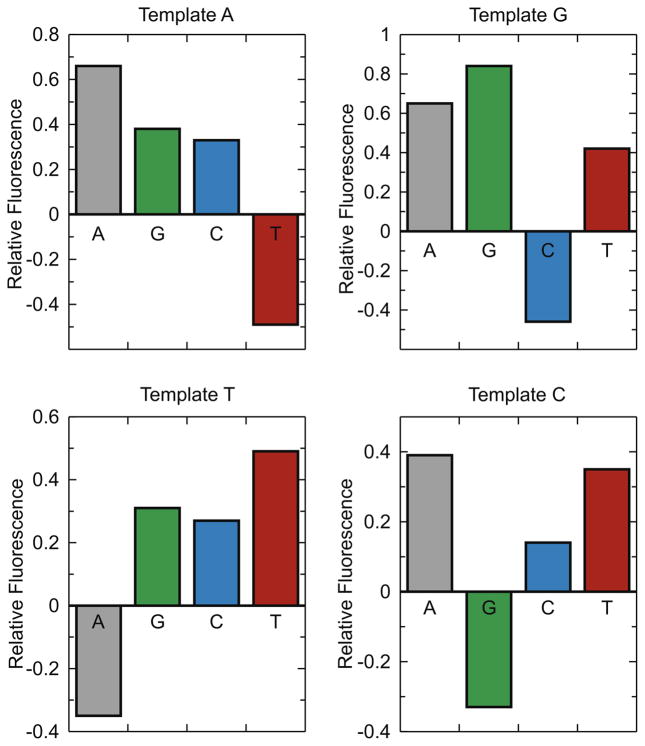
In our previously published studies, the fluorescence intensity of the MDCC–E514C-8C polymerase was shown to decrease 30% when a correct dCTP bound to the enzyme–DNA complex, whereas a mismatch (dGTP) induced a 40% increase in fluorescence [15]. The decrease in fluorescence was correlated with the conformational change of the polymerase as a closed, correct E.DNA.dNTP ternary complex was formed, whereas the increase in signal suggested the presence of a third conformation of the polymerase when binding a mismatched nucleotide. It is necessary to test whether this is a general property of the fluorescently labeled enzyme, strictly dependent on the base-pairing properties between the template bases and the incoming nucleotides and not dependent on our choice of template. We compared the fluorescence changes on the binding of each of the four nucleotides (1 mM concentration) with DNA containing four different template bases to get the results shown in Figs. 2 and 3. Our data indicated that only ternary complexes with correct incoming nucleotides showed a decrease in fluorescence, and in each case the emission maximum showed a red shift from 460 to 466 nm. In contrast, all of the mismatched nucleotide–template pairs induced a higher fluorescence state. Interestingly, the magnitude of the increase in fluorescence after binding a mismatch varied considerably, suggesting possible heterogeneity in the final structural state induced by a mismatch, at least in terms of the structural changes sensed by the fluorophore. Note also that fluorescence quenching was observed only with correct nucleotide–template pairs even with the relatively high nucleotide concentration (1 mM), arguing against nonspecific quenching due to direct interaction between added dNTPs and the MDCC molecule.
Fig. 2.
Emission spectra of MDCC–E514C-8C T7 DNA polymerase. The enzyme–DNA (Table 2, 27ddC/45-18A) complex without dNTP is shown with a dashed line. Each nucleotide was added to a final concentration of 1 mM to form base pairs as designated in the figure. For example, A:A represents a dATP:dA mismatch (with dA in the template), whereas T:A represents a correct TTP:dA base pair where dA is the template base. Note that only the correct base caused a decrease in fluorescence, whereas each mismatch caused an increase but to different extents for different mismatches.
Fig. 3.
Relative fluorescence intensities of the MDCC–E514C-8C T7 DNA polymerase following the binding of 1 mM dNTP. The values listed represent the fractional changes in fluorescence relative to the intensity of the enzyme–DNA complexes (defined as 0). The templating base at position 18 of the 45mer template is dA in 27ddC/45-18A, dG in 27ddC/45-18G, T in 27ddC/45-18T, and dC in 27ddC/45-18C (Table 2). Standard errors in replicate measurements were less than 3% of the observed values (not shown).
Correct and incorrect nucleotide substrates are bound to the same site on the T7 DNA polymerase
We performed equilibrium titration experiments using DNA duplex 27ddC/45-18G. By following the fluorescence changes as dCTP (correct) or dGTP (incorrect) was titrated into a solution containing 200 nM MDCC-labeled enzyme and 300 nM DNA duplex, we obtained the Kd of 0.23 ± 0.004 μM for correct dCTP binding and 198 ± 2.3 μM for incorrect dGTP binding (Fig. 4). If dGTP and dCTP compete for binding to the same site on the polymerase, the apparent Kd for dCTP binding in the presence of competing dGTP can be approximated by the following equation:
Fig. 4.
Fluorescence signal changes on nucleotide binding against a template dG. The preformed enzyme–DNA (Table 2, 27ddC/45-18G) complexes were titrated with up to 5 μM dCTP (blue, Kd,C = 0.23 ± 0.004 μM, obtained by fitting to a quadratic equation) and up to 1 mM dGTP (red, Kd,G = 198 ± 2.3 μM, obtained by fitting to a hyperbola) final concentrations. The titration curve obtained by adding up to 50 μM dCTP into an enzyme–DNA solution containing 1 mM dGTP provided an apparent Kd of 3.56 ± 0.018 μM (green, fit to hyperbola). Note that the x axis is normalized relative to the maximum concentration of the titrating base so that all three titrations can be shown on the same scale. In each trace, the smooth line superimposed on the data represents the fitted curve. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The calculation gives a KdC,app value of 1.39 ± 0.03 μM in the presence of 1 mM dGTP. Our competition experiment showed an apparent Kd of 3.56 ± 0.018 μM for dCTP binding in the presence of 1 mM dGTP. The value obtained from the experiment was in reasonable agreement with the calculation, indicating that both the correct and incorrect nucleotides compete for the same active site. Although detailed structural studies are needed to understand the microscopic environmental changes surrounding the labeling site, the data show that the formation of the closed ternary complex is correlated with the lower fluorescence state. Furthermore, the increase in fluorescence signal associated with incorrect nucleotide binding suggested the presence of one or more unique conformational states when the T7 DNA polymerase bound a mismatched substrate. The role of this structurally undefined state in substrate specificity has been studied using kinetic methods, and its significance was discussed previously [15].
Identification of MDCC-labeled cysteine residues
Although eight of the naturally occurring cysteines were mutated to either alanines or serines, two buried cysteines located near the enzyme active site (C483 and C622) were retained. We reasoned that mutation of these residues may cause changes in enzymatic activity and that they may be sufficiently buried to reduce their reaction with the MDCC. Of course, that can be tested only after establishing the conditions for labeling at the desired E514C site. We used complete tryptic digestion of the labeled enzyme followed by mass spectral analysis to identify the sites of labeling with MDCC. The MDCC-labeled E514C-8C T7 DNA polymerase was digested with trypsin, and the labeled peptide fragments were resolved using reverse phase HPLC. MS analysis of the crude mixture of peptides following tryptic digestion was too complex to resolve the labeled peptides, necessitating the resolution of labeled peptides by HPLC. During chromatography, peptides were monitored by absorbance at 220 nm, whereas the presence of MDCC was monitored at 425 nm (Fig. 5). The number of peaks observed at 220 nm was less than expected, suggesting that some peaks may contain a mixture of several different peptides. Only fractions 12 and 13 showed absorbance at 425 nm, indicating the presence of MDCC. To characterize the peptides in the two fractions, samples were freeze-dried and analyzed by MS. We searched the MS spectrum of fraction 12 (data not shown) and fraction 13 (Fig. 6A) for the possibly modified peptides (Table 3). Only one ion (m/z 1469.8) was identified to match the mass of the MDCC modified peptide, 509NQIAACLPTR518, which contains the C514 residue in either fraction. Because no ions were found to match the masses of peptides containing the other cysteines associated with MDCC, the data suggested that C483 and C622 are not modified with MDCC under our labeling condition (see arrows in Fig. 6A).
Fig. 5.
HPLC profile of peptide fragments from the MDCC–E514C-8C protein after tryptic digestion. Absorbance was measured at 220 nm (black trace) and 425 nm (red trace) during reverse phase HPLC as described in Materials and Methods. Fractions 12 and 13 (indicated by arrows) were selected for MS analysis. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 6.
MS analysis of the sample from fraction 13 shown in Fig. 5. (A) The mass of one ion with m/z 1469.8 matches the calculated mass of the peptide 509NQIAACLPTR518 with one molecule of MDCC (MH+ of 1469.7). (B) MS/MS spectrum of the precursor ion with m/z 1469.8 and assignment of the peaks to b ions and y ions calculated from peptide 509NQIAACLPTR518, which contains MDCC modification at C514. The arrows under the abscissa show locations of ions anticipated to arise from the alternate labeling sites (Table 3) but not seen in the MS. The hypothesized fragmentation site of MDCC is indicated by a dashed line, and the calculated mass of the ion generated from the left portion of MDCC is 244.1 Da.
Table 3.
Tryptic peptide fragments containing cysteines
| Peptide fragment | Cysteine position | Calculated mass (Da) | Mass with MDCC (Da) |
|---|---|---|---|
| 483CLAHFMAR490 | 483 | 948.5 | 1331.6 |
| 509NQIAACLPTR518 | 514 | 1086.6 | 1469.7 |
| 605SPHAALNTLLQSAGALICK623 | 622 | 1980.1 | 2291.2 |
Note. Mass values were calculated using the monoisotopic MH+ masses. The monoisotopic mass of MDCC is 383.15 Da.
To further verify the identity of the ion with m/z 1469.8, this precursor ion was subjected to MS/MS analysis. Because the monoisotopic mass of MDCC is 383.15 Da, the y ions from y5 to y9 and the b ions from b6 to b9 of 509NQIAACLPTR518 should be shifted by a mass of +383.15 Da when the peptide is modified by MDCC at C514. With this correction of the masses, all of the y ions and most of the b ions of the MDCC modified 509NQIAACLPTR518 could be assigned to the peaks in the MS/MS spectrum (Fig. 6B). The b9 ion could not be assigned in the spectrum at first; however, an ion with m/z 1052.6 was proposed to be the adduct of b9 ion and fragmented MDCC that can lose a fragment with a mass of 244.1 Da. The existence of an ion with m/z 244.1 supported this hypothesis; thus, we could finally assign the peak of 1052.6 to the b9 ion with a shifted mass of +140.0 Da. This MS/MS spectral analysis unambiguously confirmed the identity of the MDCC modified 509NQIAACLPTR518, and the modification is at C514. In conclusion, our results suggested that only C514 was labeled with MDCC in our labeling experiment.
Detecting SNPs with MDCC–E514C-8C T7 DNA polymerase
We further examined the fluorescence changes on nucleotide binding using several DNA duplexes with sequences adopted from the human factor VIII (F8) gene to test whether a simple procedure could be devised to detect SNPs. The F8 gene on the X chromosome encodes a protein that is involved in the blood coagulation pathway. Deleterious mutations on the F8 gene cause hemophilia A on approximately 0.02% of males [25]. Mutations on the F8 gene include inversions, point mutations, deletions, and insertions [26]. Nine DNA substrates were selected from four regions of the F8 gene containing at least one polymorphism site associated with mild to severe hemophilia (Table 1). The experiments were designed to allow the incorporation of one dideoxynucleotide before the binding of the dNTP that would base pair with the templating base of interest. As summarized in Fig. 7, results from this experiment displayed that the correct base pairings can be detected by the MDCC fluorescence and that its specificity toward correct base pairing was not lost when the DNA sequences were changed. Unlike the data observed following the addition of 1 mM dNTP (Fig. 3), the addition of only 1 μM dNTP was sufficient to allow for the observation of correct dNTP binding but was insufficient to induce fluorescence changes with mismatched dNTPs. Therefore, the correct base is identified in this screening protocol as the nucleotide that induces the largest decrease in fluorescence.
Fig. 7.
SNP detection experiments using DNA duplexes listed in Table 1. Relative fluorescence changes before and after the addition of 1 μM dNTP are shown in the bar graph. Note that although the magnitude of the fluorescence change varied with the sequence, the dNTP that resulted in the lowest relative fluorescence identified the correct substrate.
Our procedure required the sequential addition of a dideoxynucleotide followed by the test dNTP; however, large-scale screening could employ a DNA test sample that already contained a dideoxynucleotide on the 3′ end of the primer. These data show how the fluorescently labeled DNA polymerase could be used in large-scale, high-throughput screens to search for SNPs that may underlie variations in susceptibility to disease or the effectiveness of certain drugs [27,28]. A simple and fast method for detecting SNPs could be developed using the fluorescence signal from the MDCC–E514C-8C T7 DNA polymerase.
Discussion
In this study, we set forth to characterize the MDCC-labeled T7 DNA polymerase engineered to study the kinetics of nucleotide-induced conformational changes [15]. We used tryptic digestion and MS to verify that only the C514 residue was labeled with the MDCC despite the presence of two extra cysteines in the mutant protein. We took advantage of the enzyme’s unique fluorescence property in distinguishing correct base pairs from incorrect ones and developed a simple method for detecting single-nucleotide variations.
In our previous analysis, the role of conformational changes in specificity was revealed by analysis of the kinetics of the conformational transitions using this fluorescence signal [15]. The data show that the decrease in fluorescence is correlated with the formation of the closed ternary E.DNAdd.dNTP complex, but the trajectory of the conformational change and the point at which the change in fluorescence occurs during the transition from open to closed endpoints have not been established. Moreover, the increase in fluorescence is clearly correlated with a mismatch recognition state that is distinct from either the open state or the closed state, but there is apparent heterogeneity in this state. As seen in Fig. 2, and as revealed by the unusual kinetics of mismatch binding [15], mismatches may induce a variety of conformational states that are distinguished by different fluorescence intensities.
The fluorescence of the modified enzyme was quenched by up to 40% when a correct dNTP was bound to the enzyme–DNA complex. It has been reported that a guanine base could interact nonspecifically with a fluorophore to quench fluorescence [29]. We investigated this possibility by comparing the fluorescence emission spectra using the 16 possible dNTP/template combinations at the same position of the DNA substrate and found that the decrease in signal was strictly dependent on the correct base pairing. Furthermore, binding of any mismatched dNTP resulted in an enhancement of fluorescence signal. We failed to observe any nonspecific interactions between nucleotides and MDCC at 1-mM nucleotide concentrations. Moreover, the addition of a correct dNTP displaces a mismatched nucleotide with an apparent Kd that is consistent with individually measured dissociation constants for the correct and incorrect nucleotides (Fig. 4). These data demonstrate quantitatively that correct and incorrect nucleotides compete for the same site on the T7 DNA polymerase, as measured by their individual effects on the fluorescence signal.
We examined several labeling positions (K355, K502, E514, Q539, and K545) surrounding the fingers domain and found that only MDCC labeling at position 514 provided a significant fluorescence change following nucleotide binding. The position of labeling is located on the surface of the fingers domain, pointing away from the nucleotide binding pocket [10]. Structural studies on polymerases revealed a large conformational change involving this domain following dNTP binding with a dideoxy-terminated primer [9–14]. However, the fingers domain does not move as a rigid body; rather, there are changes in the packing of helices as the domain moves from the open state to the closed state. Presumably, the MDCC label is sensing these changes in structure to detect the three conformational states: open, closed, and mismatch recognition. A red shift of the emission maximum was observed when correct nucleotides bound to enzyme–DNA, suggesting that the formation of closed ternary complex caused the fluorophore to be more exposed to solvent. In contrast, the binding of a mismatch led to a state in which the fluorophore was more buried. Our data show that the decrease in fluorescence signal observed on binding a correct dNTP is correlated with a structural change of the polymerase corresponding to the formation of a closed ternary complex. The increases in fluorescence after binding a mismatch imply the formation of an enzyme conformational state that is distinct from the open enzyme–DNA binary complex and the closed enzyme–DNA–correct dNTP ternary complex that have been characterized structurally. So far, no structural studies of high-fidelity polymerases have succeeded in resolving the three-dimensional structure of the enzyme–DNA–dNTP complex with a mismatched nucleotide. Indeed our kinetic studies imply that the state formed following the binding of a mismatch is a mixed population of at least two conformational states [15], and that may complicate attempts to crystallize the enzyme with a mismatch bound.
The position of C514 is only four residues away from an important active site amino acid, R518, which interacts with the gamma phosphate of the incoming dNTP. We suspected that the base pair sensitive fluorescence changes might be due, in part, to a specific interaction between the incoming dNTP and R518 as well as to additional residues on the O-helix. Observation of nonspecific interactions between the nucleotide and R518 (or its homolog in PolI) in the absence of DNA may be misleading [30] and does not define the structure of the open complex in the presence of DNA. Rather, fidelity studies establish that the incoming dNTP interacts with the DNA template in the open state, and recent analysis of mutants demonstrates that R518 interacts with the incoming dNTP after the conformational change (Z. Jin and K. A. Johnson, unpublished results). If the correct and incorrect incoming nucleotides interact with and position R518 and the O-helix differently, even subtle changes in structure could be translated into different positions of residue 514, resulting in opposite fluorescence changes. The MDCC signal change could be coincident with the large structural change seen crystallographically; alternatively, it is possible that nucleotide binding induces a fast change from the open state to the closed state and then the MDCC fluorophore senses a subsequent more subtle alignment of residues preceding catalysis. In either case, our analysis of the kinetics reveals a new paradigm in which structural changes following nucleotide binding function as a molecular switch to govern selectivity [15]. Ongoing computational and structural studies are intended to resolve how the fluorophore may be sensing multiple conformational states.
Methods for acquiring real-time kinetic data of polymerase conformational changes have relied on fluorescence signals originating from fluorophores covalently attached to the DNA substrate. For example, DNA templates with a 2-AP label were used to study conformational changes of polymerase β and PolI Klenow fragment [16,18]. The interpretation of the 2-AP data relied on the assumption that structural transformation of the enzyme was synchronized with the DNA movement. However, the 2-AP signal is a function of the base-stacking interactions within the DNA molecules, and the direction and magnitude of fluorescence changes are dependent on the position of 2-AP labeling within the DNA template and the starting solution conditions [16]. The conformational change of the KlenTaq polymerase was studied using a FRET signal with donor fluorophore on the DNA substrate and acceptor fluorophore on the fingers domain [19]. This study revealed a fast conformational change step corresponding to the closure of the fingers domain before the chemical reaction, but no signal changes were associated with mismatched nucleotide bindings. One explanation of this observation could be that the KlenTaq does not undergo the conformational change and remains in the open configuration following the incorrect nucleotide binding or that the FRET signal is insensitive to the subtle changes in structure following mismatch binding. Alternatively, one must consider the possibility that the signal arises solely due to repositioning of the DNA on nucleotide binding. Recently, a dual-labeled KlenTaq polymerase provided a FRET signal corresponding to both the closing and opening fingers domain movements in the nucleotide incorporation cycle [31]. However, no fluorescence change was observed when the enzyme encountered a mismatched nucleotide.
We explored the possibility of using the MDCC fluorescence signal to detect single-nucleotide mutations. The method we developed required incorporation of a dideoxynucleotide prior to the binding of the next nucleotide. The bound nucleotide that produces the lowest relative fluorescence among the four dNTPs represents the correct substrate and reveals the identity of the template base. We tested this method with seven synthetic DNA fragments derived from the human F8 gene that is linked to hemophilia. The fluorescence assay successfully identified all of the template variations targeted by designated primers. The method could be simplified by starting the reaction with a DNA primer already terminated with a dideoxynucleotide, but it would still require amplification of genomic DNA samples by PCR followed by annealing with the dideoxy-terminated primer. One could imagine a high-throughput SNP detection system built on the foundation of this simple assay by simultaneously performing the same reaction with thousands of loci-targeting primers immobilized on a DNA chip. The new method may have several advantages over current SNP detection methods [32] that require specially synthesized DNA oligomers with fluorescent labels because our method requires only one fluorescent reagent, the MDCC–E514C-8C T7 DNA polymerase. In addition, the simplicity of the method allows automation and yields high throughput so that the nature of the mutation could be determined directly without sequencing analysis.
Acknowledgments
K.A.J. acknowledges support from the National Institutes of General Medical Sciences (GM071404) and the Welch Foundation (F-1604). K.A.J. is president of KinTek Corporation, which donated the automatic titration module used in these studies.
Footnotes
Abbreviations used: 2-AP, 2-aminopurine; FRET, Förster resonance energy transfer; MDCC, 7-diethylamino-3-[([(2-maleimidyl)ethyl]amino)carbonyl]coumarin; MS, mass spectrometry; SNP, single-nucleotide polymorphism; SDS–PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis; EDTA, ethylenediaminetetraacetic acid; DTT, dithiothreitol; TFA, trifluoroacetic acid; ddH2O, doubly distilled H2O; HPLC, high-performance liquid chromatography; UV, ultraviolet; MALDI, matrix-assisted laser desorption/ionization; TOF/TOF, tandem time-of-flight; CID, collision-induced dissociation; MS/MS, tandem MS; ssDNA, single-stranded DNA.
References
- 1.Johnson KA. Conformational coupling in DNA polymerase fidelity. Annu Rev Biochem. 1993;62:685–713. doi: 10.1146/annurev.bi.62.070193.003345. [DOI] [PubMed] [Google Scholar]
- 2.Joyce CM, Benkovic SJ. DNA polymerase fidelity: Kinetics, structure, and checkpoints. Biochemistry. 2004;43:14317–14324. doi: 10.1021/bi048422z. [DOI] [PubMed] [Google Scholar]
- 3.Kati WM, Johnson KA, Jerva LF, Anderson KS. Mechanism and fidelity of HIV reverse transcriptase. J Biol Chem. 1992;267:25988–25997. [PubMed] [Google Scholar]
- 4.Spence RA, Kati WM, Anderson KS, Johnson KA. Mechanism of inhibition of HIV-1 reverse transcriptase by nonnucleoside inhibitors. Science. 1995;267:988–993. doi: 10.1126/science.7532321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong I, Patel SS, Johnson KA. An induced-fit kinetic mechanism for DNA replication fidelity: direct measurement by single-turnover kinetics. Biochemistry. 1991;30:526–537. doi: 10.1021/bi00216a030. [DOI] [PubMed] [Google Scholar]
- 6.Patel SS, Wong I, Johnson KA. Pre-steady-state kinetic-analysis of processive DNA-replication including complete characterization of an exonuclease-deficient mutant. Biochemistry. 1991;30:511–525. doi: 10.1021/bi00216a029. [DOI] [PubMed] [Google Scholar]
- 7.Wong I, Patel SS, Johnson KA. An induced-fit kinetic mechanism for DNA-replication fidelity: direct measurement by single-turnover kinetics. Biochemistry. 1991;30:526–537. doi: 10.1021/bi00216a030. [DOI] [PubMed] [Google Scholar]
- 8.Mizrahi V, Henrie RN, Marlier JF, Johnson KA, Benkovic SJ. Rate-limiting steps in the DNA polymerase I reaction pathway. Biochemistry. 1985;24:4010–4018. doi: 10.1021/bi00336a031. [DOI] [PubMed] [Google Scholar]
- 9.Doublie S, Sawaya MR, Ellenberger T. An open and closed case for all polymerases. Struct Fold Des. 1999;7:R31–R35. doi: 10.1016/S0969-2126(99)80017-3. [DOI] [PubMed] [Google Scholar]
- 10.Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 11.Huang HF, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-I reverse transcriptase: Implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 12.Pelletier H, Sawaya MR, Wolfle W, Wilson SH, Kraut J. Crystal structures of human DNA polymerase beta complexed with DNA: implications for catalytic mechanism, processivity, and fidelity. Biochemistry. 1996;35:12742–12761. doi: 10.1021/bi952955d. [DOI] [PubMed] [Google Scholar]
- 13.Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sawaya MR, Prasad R, Wilson SH, Kraut J, Pelletier H. Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry. 1997;36:11205–11215. doi: 10.1021/bi9703812. [DOI] [PubMed] [Google Scholar]
- 15.Tsai YC, Johnson KA. A new paradigm for DNA polymerase specificity. Biochemistry. 2006;45:9675–9687. doi: 10.1021/bi060993z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunlap CA, Tsai MD. Use of 2-aminopurine and tryptophan fluorescence as probes in kinetic analyses of DNA polymerase beta. Biochemistry. 2002;41:11226–11235. doi: 10.1021/bi025837g. [DOI] [PubMed] [Google Scholar]
- 17.Hariharan C, Bloom LB, Helquist SA, Kool ET, Reha-Krantz LJ. Dynamics of nucleotide incorporation: snapshots revealed by 2-aminopurine fluorescence studies. Biochemistry. 2006;45:2836–2844. doi: 10.1021/bi051644s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purohit V, Grindley ND, Joyce CM. Use of 2-aminopurine fluorescence to examine conformational changes during nucleotide incorporation by DNA polymerase I. Klenow fragment. Biochemistry. 2003;42:10200–10211. doi: 10.1021/bi0341206. [DOI] [PubMed] [Google Scholar]
- 19.Rothwell PJ, Mitaksov V, Waksman G. Motions of the fingers subdomain of Klentaq1 are fast and not rate limiting: implications for the molecular basis of fidelity in DNA polymerases. Mol Cell. 2005;19:345–355. doi: 10.1016/j.molcel.2005.06.032. [DOI] [PubMed] [Google Scholar]
- 20.Joyce CM, Potapova O, DeLucia AM, Huang XW, Basu VP, Grindley NDF. Fingers-closing and other rapid conformational changes in DNA polymerase I (Klenow fragment) and their role in nucleotide selectivity. Biochemistry. 2008;47:6103–6116. doi: 10.1021/bi7021848. [DOI] [PubMed] [Google Scholar]
- 21.Donlin MJ, Johnson KA. Mutants affecting nucleotide recognition by T7 DNA polymerase. Biochemistry. 1994;33:14908–14917. doi: 10.1021/bi00253a030. [DOI] [PubMed] [Google Scholar]
- 22.Gill SC, Vonhippel PH. Calculation of protein extinction coefficients from amino-acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 23.Brune M, Hunter JL, Corrie JE, Webb MR. Direct, real-time measurement of rapid inorganic phosphate release using a novel fluorescent probe and its application to actomyosin subfragment 1 ATPase. Biochemistry. 1994;33:8262–8271. doi: 10.1021/bi00193a013. [DOI] [PubMed] [Google Scholar]
- 24.Hirshberg M, Henrick K, Haire LL, Vasisht N, Brune M, Corrie JE, Webb MR. Crystal structure of phosphate binding protein labeled with a coumarin fluorophore, a probe for inorganic phosphate. Biochemistry. 1998;37:10381–10385. doi: 10.1021/bi980428z. [DOI] [PubMed] [Google Scholar]
- 25.Hoyer LW. Hemophilia-A. N Engl J Med. 1994;330:38–47. doi: 10.1056/NEJM199401063300108. [DOI] [PubMed] [Google Scholar]
- 26.Tuddenham EGD, Schwaab R, Seehafer J, Millar DS, Gitschier J, Higuchi M, Bidichandani S, Connor JM, Hoyer LW, Yoshioka A, Peake IR, Olek K, Kazazian HH, Lavergne JM, Giannelli F, Antonarakis SE, Cooper DN. Nucleic Acids Res. 2. Vol. 22. 1994. Hemophilia-A: database of nucleotide substitutions, deletions, insertions, and rearrangements of the factor-VIII gene; pp. 3511–3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherry ST, Ward MH, Sirotkin K. DbSNP: database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999;9:677–679. [PubMed] [Google Scholar]
- 28.Twyman RM. SNP discovery and typing technologies for pharmacogenomics. Curr Top Med Chem. 2004;4:1423–1431. doi: 10.2174/1568026043387656. [DOI] [PubMed] [Google Scholar]
- 29.Dohno C, Saito I. Discrimination of single-nucleotide alterations by G-specific fluorescence quenching. Chem Bio Chem. 2005;6:1075–1081. doi: 10.1002/cbic.200400325. [DOI] [PubMed] [Google Scholar]
- 30.Beese LS, Friedman JM, Steitz TA. Crystal structures of the Klenow fragment of DNA polymerase I complexed with deoxynucleoside triphosphate and pyrophosphate. Biochemistry. 1993;32:14095–14101. doi: 10.1021/bi00214a004. [DOI] [PubMed] [Google Scholar]
- 31.Allen WJ, Rothwell PJ, Waksman G. An intramolecular FRET system monitors fingers subdomain opening in Klentaq1. Protein Sci. 2008;17:401–408. doi: 10.1110/ps.073309208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Engle LJ, Simpson CL, Landers JE. Using high-throughput SNP technologies to study cancer. Oncogene. 2006;25:1594–1601. doi: 10.1038/sj.onc.1209368. [DOI] [PubMed] [Google Scholar]