Abstract
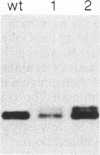
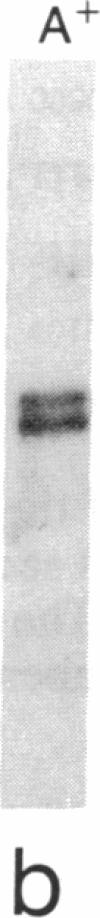
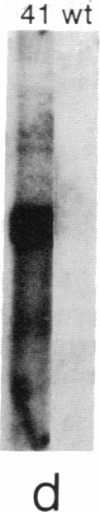
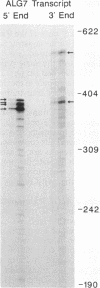
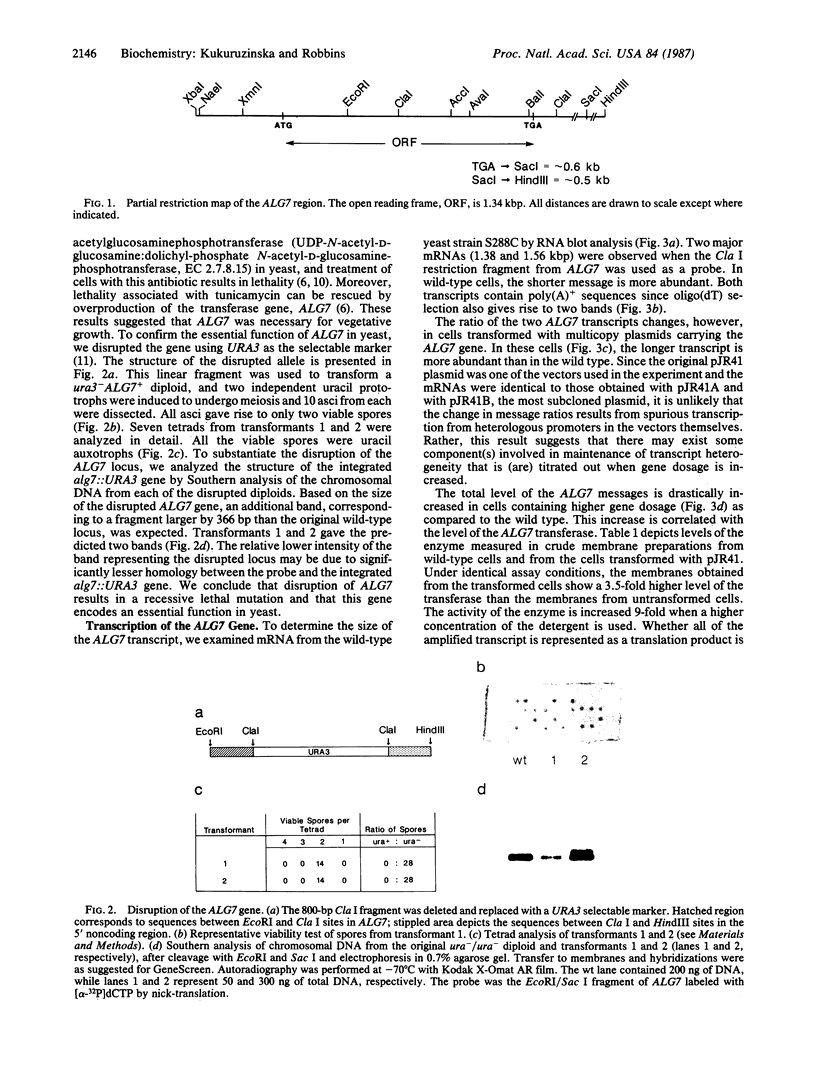
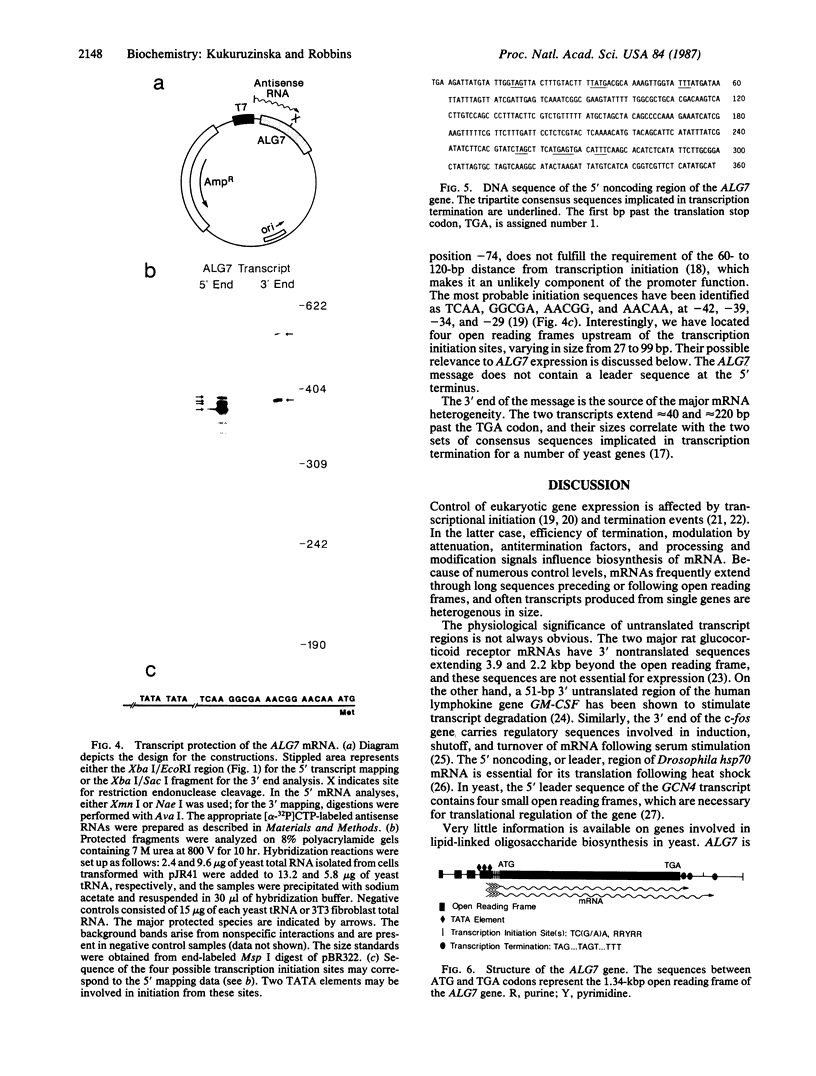
The first enzyme in the lipid-linked oligosaccharide biosynthetic pathway, UDP-N-acetylglucosamine-dolichyl-phosphate N-acetylglucosaminephosphotransferase (UDP-N-acetyl-D-glucosamine:dolichyl-phosphate-N-acetyl- D-glucosaminephosphotransferase, EC 2.7.8.15), is encoded by the ALG7 gene. We show that this gene is essential for cell growth, since a null mutation constructed with standard gene disruption techniques results in cell lethality. The ALG7 gene is transcribed into two major messages, approximately 1.38 and 1.56 kilobase pairs (kbp) in size, and this heterogeneity has been mapped to the 3' untranslated region. Two sets of tripartite sequences implicated in transcription termination begin 15 bp and 256 bp past the translation stop codon, TGA. The ratios of the two major transcripts change with gene dosage, with the longer mRNA becoming more abundant in cells containing higher levels of the ALG7 gene. Changes in transcript ratios are also observed in mutants defective in lipid-linked sugar-donor biosynthesis. In addition, there is 5' heterogeneity in the ALG7 mRNAs. The transcripts start at four initiation sites located within a 20-bp region. Two potentially functional TATA elements have been identified at positions -157 and -139, which may be involved in initiation from multiple sites. These features point to numerous factors that may be involved in the regulation of the expression of the ALG7 gene.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andreadis A., Hsu Y. P., Kohlhaw G. B., Schimmel P. Nucleotide sequence of yeast LEU2 shows 5'-noncoding region has sequences cognate to leucine. Cell. 1982 Dec;31(2 Pt 1):319–325. doi: 10.1016/0092-8674(82)90125-8. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Mason T. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell. 1983 Apr;32(4):1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- Hahn S., Hoar E. T., Guarente L. Each of three "TATA elements" specifies a subset of the transcription initiation sites at the CYC-1 promoter of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8562–8566. doi: 10.1073/pnas.82.24.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Kelly J. D., Cohen E. H. Transcription terminates in yeast distal to a control sequence. Cell. 1983 Jun;33(2):607–614. doi: 10.1016/0092-8674(83)90441-5. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G., Fink G. R. Repeated DNA sequences upstream from HIS1 also occur at several other co-regulated genes in Saccharomyces cerevisiae. J Biol Chem. 1983 Apr 25;258(8):5238–5247. [PubMed] [Google Scholar]
- Hubbard S. C., Ivatt R. J. Synthesis and processing of asparagine-linked oligosaccharides. Annu Rev Biochem. 1981;50:555–583. doi: 10.1146/annurev.bi.50.070181.003011. [DOI] [PubMed] [Google Scholar]
- Huffaker T. C., Robbins P. W. Temperature-sensitive yeast mutants deficient in asparagine-linked glycosylation. J Biol Chem. 1982 Mar 25;257(6):3203–3210. [PubMed] [Google Scholar]
- Huffaker T. C., Robbins P. W. Yeast mutants deficient in protein glycosylation. Proc Natl Acad Sci U S A. 1983 Dec;80(24):7466–7470. doi: 10.1073/pnas.80.24.7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo S. C., Lampen J. O. Tunicamycin--an inhibitor of yeast glycoprotein synthesis. Biochem Biophys Res Commun. 1974 May 7;58(1):287–295. doi: 10.1016/0006-291x(74)90925-5. [DOI] [PubMed] [Google Scholar]
- Leff S. E., Rosenfeld M. G., Evans R. M. Complex transcriptional units: diversity in gene expression by alternative RNA processing. Annu Rev Biochem. 1986;55:1091–1117. doi: 10.1146/annurev.bi.55.070186.005303. [DOI] [PubMed] [Google Scholar]
- Lehle L., Tanner W. The specific site of tunicamycin inhibition in the formation of dolichol-bound N-acetylglucosamine derivatives. FEBS Lett. 1976 Nov 15;72(1):167–170. doi: 10.1016/0014-5793(76)80922-2. [DOI] [PubMed] [Google Scholar]
- McGarry T. J., Lindquist S. The preferential translation of Drosophila hsp70 mRNA requires sequences in the untranslated leader. Cell. 1985 Oct;42(3):903–911. doi: 10.1016/0092-8674(85)90286-7. [DOI] [PubMed] [Google Scholar]
- Miesfeld R., Rusconi S., Godowski P. J., Maler B. A., Okret S., Wikström A. C., Gustafsson J. A., Yamamoto K. R. Genetic complementation of a glucocorticoid receptor deficiency by expression of cloned receptor cDNA. Cell. 1986 Aug 1;46(3):389–399. doi: 10.1016/0092-8674(86)90659-8. [DOI] [PubMed] [Google Scholar]
- Mueller P. P., Hinnebusch A. G. Multiple upstream AUG codons mediate translational control of GCN4. Cell. 1986 Apr 25;45(2):201–207. doi: 10.1016/0092-8674(86)90384-3. [DOI] [PubMed] [Google Scholar]
- Nagawa F., Fink G. R. The relationship between the "TATA" sequence and transcription initiation sites at the HIS4 gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8557–8561. doi: 10.1073/pnas.82.24.8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- Ravoet A. M., Amar-Costesec A., Godelaine D., Beaufay H. Quantitative assay and subcellular distribution of enzymes acting on dolichyl phosphate in rat liver. J Cell Biol. 1981 Dec;91(3 Pt 1):679–688. doi: 10.1083/jcb.91.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rine J., Hansen W., Hardeman E., Davis R. W. Targeted selection of recombinant clones through gene dosage effects. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6750–6754. doi: 10.1073/pnas.80.22.6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman J. H., Hunter C. P., Valls L. A., Stevens T. H. Overproduction-induced mislocalization of a yeast vacuolar protein allows isolation of its structural gene. Proc Natl Acad Sci U S A. 1986 May;83(10):3248–3252. doi: 10.1073/pnas.83.10.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Runge K. W., Huffaker T. C., Robbins P. W. Two yeast mutations in glucosylation steps of the asparagine glycosylation pathway. J Biol Chem. 1984 Jan 10;259(1):412–417. [PubMed] [Google Scholar]
- Shaw G., Kamen R. A conserved AU sequence from the 3' untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986 Aug 29;46(5):659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Stevens T. H., Rothman J. H., Payne G. S., Schekman R. Gene dosage-dependent secretion of yeast vacuolar carboxypeptidase Y. J Cell Biol. 1986 May;102(5):1551–1557. doi: 10.1083/jcb.102.5.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Genetic properties and chromatin structure of the yeast gal regulatory element: an enhancer-like sequence. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7865–7869. doi: 10.1073/pnas.81.24.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R. Transient accumulation of c-fos RNA following serum stimulation requires a conserved 5' element and c-fos 3' sequences. Cell. 1985 Oct;42(3):889–902. doi: 10.1016/0092-8674(85)90285-5. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]