Abstract
The Hsp90/Hsp70-based chaperone machinery plays a well established role in signaling protein function, trafficking and turnover. A number of recent observations also support the notion that Hsp90 and Hsp70 play key roles in the triage of damaged and aberrant proteins for degradation via the ubiquitin-proteasome pathway. In the mid 1990s, it was discovered that Hsp70 is required for ubiquitin-dependent degradation of short lived and abnormal proteins, and it became clear that inhibition of Hsp90 uniformly leads to the proteasomal degradation of Hsp90 client proteins. Subsequently, CHIP and parkin were shown to be Hsp70-binding ubiquitin E3 ligases that direct ubiquitin-charged E2 enzymes to the Hsp70-bound client protein. The stabilizing effect of Hsp90 reflects the interaction of the chaperone with the ligand binding cleft of the client protein. These hydrophobic clefts must be open to allow passage of ligands to binding sites in the protein interior, and they are inherent sites of conformational instability. Hsp90 stabilizes the open state of the cleft and prevents Hsp70-dependent ubiquitination. In the model we present here, progressive oxidative events result in cleft opening as the initial step in protein unfolding, and as long as Hsp90 can interact to stabilize the cleft, it will buffer the effect of oxidative damage. When cleft opening is such that Hsp90 can no longer interact, Hsp70-dependent ubiquitination occurs. We summarize evidence that Hsp90 interacts very dynamically with a variety of proteins that are not classic Hsp90 clients, and we show that this dynamic cycling with Hsp90 protects against CHIP-mediated ubiquitination. Scientific interest to date has focused on stringent regulation of the classic client proteins, which have metastable clefts and are inherently short lived. But, the recognition that Hsp90 cycles dynamically with longer lived proteins with more stable clefts permits extension of the triage model to the quality control of damaged proteins in general.
About 25 years ago, Earl Stadtman noted that oxidative damage to enzymes in some way triggered their degradation (1). Subsequently, it has become clear that the ubiquitin-proteasome pathway is the major route of degradation, but it has not been clear how proteins that have undergone oxidative or other toxic damage are selected for ubiquitination. It has been the prevailing view that E3 ubiquitin ligases perform the role of protein substrate recognition and bring the ubiquitin-charged E2 enzyme to the substrate (reviewed in Refs 2 and 3). However, in the case of proteins that are damaged and unfolding, chaperones appears to be responsible for substrate recognition and chaperone-dependent E3 ligases target the E2 enzyme to the substrate (4).
The major chaperones involved in the protein quality control decision are Hsp90 and Hsp70, which act together in a multichaperone machinery to regulate the function, trafficking and turnover of a wide variety of signaling proteins (5). Over the past decade, both advances in our understanding of how Hsp90 interacts with proteins and the discovery of the role of chaperone-dependent E3 ligases in protein ubiquitination have contributed to a general model of how Hsp90 and Hsp70 work together to select proteins that have undergone oxidative or other toxic damage for degradation. The Hsp90 chaperone machinery also affects the function and trafficking of proteins (5), but this review will focus on the way the machinery functions in protein quality control.
Hsp90 and Hsp70 have essentially opposing roles in the triage of damaged proteins, in that Hsp70 promotes substrate ubiquitination and Hsp90 inhibits ubiquitination. In the model of triage that we develop here, we envision that, as proteins undergo toxic or oxidative damage, their ligand binding clefts open to expose hydrophobic residues as the initial step in unfolding. The Hsp90 chaperone machinery regulates signaling proteins by modulating ligand binding clefts (reviewed in Refs 6 and 7). When cleft opening is such that Hsp90 can no longer interact with the protein to inhibit ubiquitination, E3 ligases interacting with substrate-bound Hsp70 target ubiquitin-charged E2 enzyme to the nascently unfolding substrate. In this way the Hsp90/Hsp70-based chaperone machinery may function as a comprehensive protein management system for quality control of damaged proteins
The Chaperone Machinery
Hsp70 and Hsp90 are conserved, abundant and essential proteins of eukaryotic cells where they are present in the cytoplasm and nucleus, with paralogs being present in mitochondria and endoplasmic reticulum. Both chaperones have ATP binding sites and possess intrinsic ATPase activity that regulates their conformation. In each case, the ATP-bound conformation has a low affinity for binding hydrophobic peptide and ATP hydrolysis is accompanied by a conformational change to a state with high affinity for binding hydrophobic peptide (reviewed in Refs. 8 and 9). The conformational changes that occur during the ATPase cycle of Hsp90 have been reviewed by Wandinger et al. (10). Also, both Hsp70 and Hsp90 possess EEVD motifs at the C terminus that are binding sites for TPR (tetratricopeptide repeat) domains. The TPR cochaperones that bind to Hsp90 play a number of roles in the activation and trafficking of signaling proteins (5,10,11), and they in general will not be considered here. However, one TPR cochaperone, CHIP (C terminus of Hsp70 interacting protein), is a chaperone-binding E3 ubiquitin ligase (4) that is important for quality control of damaged proteins, and it will be a focus of this minireview.
In contrast to the classic model of chaperones interacting with unfolded proteins to facilitate their refolding, the Hsp90 chaperone machinery acts on prefolded proteins in their native conformations to assist the opening and stabilization of ligand binding clefts (7). The Hsp90 chaperone machine acts on proteins in a manner that is not dependent upon protein sequence, size or structure. Rather, Hsp90 interacts with regions where protein folding clefts merge with the charged, hydrophilic surface of the protein. Folding clefts are a general topological feature of proteins in native conformation, and many of these hydrophobic clefts, must open to permit access of ligands, such as steroids, ATP, etc., to binding sites in the protein’s interior.
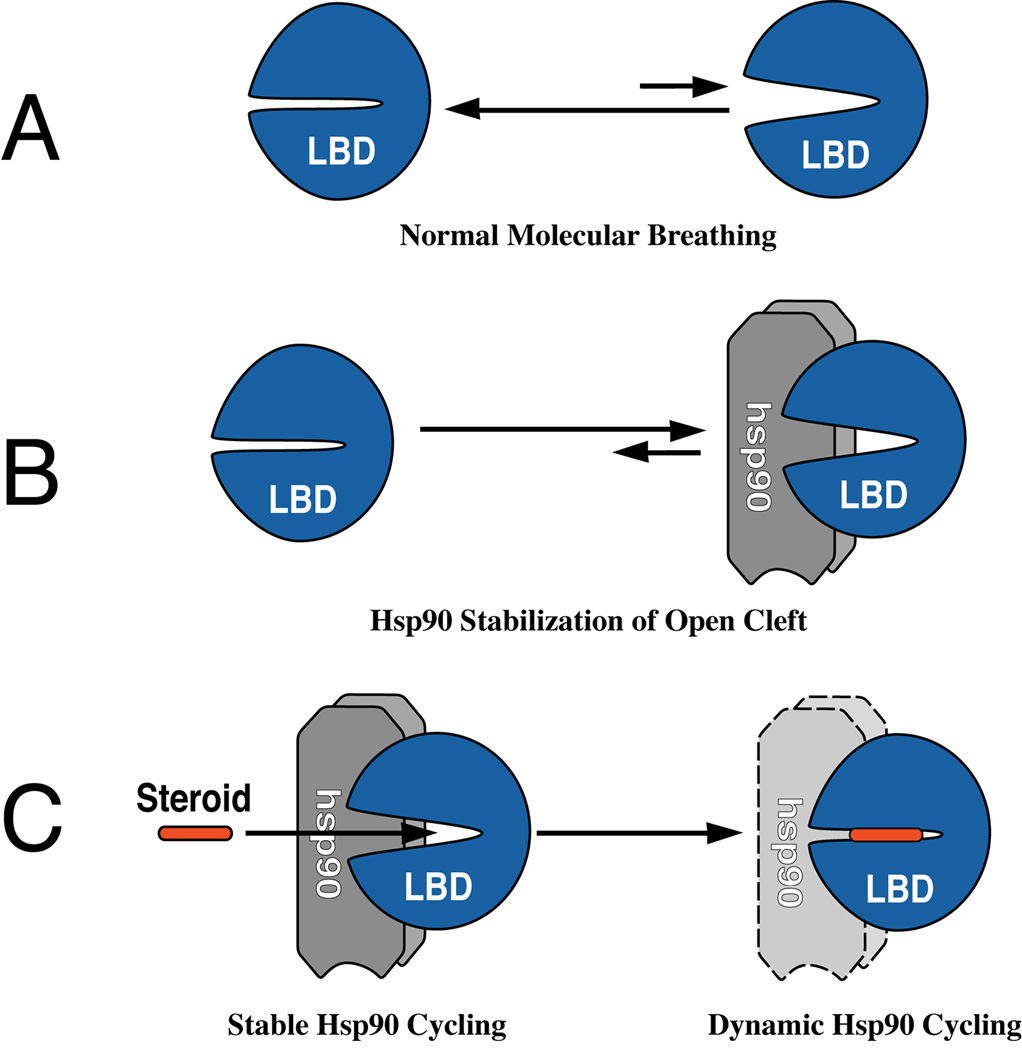
In the absence of the chaperone machinery, these binding clefts are dynamic in that they shift to varying extents between closed and open states (Fig. 1A). When clefts open during this molecular breathing process, hydrophobic residues of the protein interior are exposed to solvent, and continued opening may progress to nascent stages of unfolding. The extent to which the binding cleft is open determines ligand access and protein function, but these clefts are inherent sites of conformational instability. The chaperone machinery assists cleft opening, and Hsp90 binding to the protein stabilizes the open cleft (Fig. 1B), impeding further unfolding and Hsp70-dependent ubiquitination.
Figure 1.
States of the steroid binding cleft in the GR ligand binding domain (LBD) and Hsp90 cycling. (A) In the absence of Hsp90, the cleft in the GR LBD is predominantly closed, opening only very transiently during the course of normal molecular breathing; thus, high concentrations of steroid are required to initiate the hormone effect. (B) When stable complexes are assembled with Hsp90, nearly all the ligand binding clefts are open at any time, and low concentrations of steroid are now sufficient for binding. Under conditions of dynamic cycling, Hsp90 dissociates very rapidly, and the open and closed states of the cleft are more like the non-Hsp90-bound receptor, so higher concentrations of steroid are required for the hormone effect. (C) Steroid binding within the cleft promotes a temperature-dependent collapse of the cleft around the ligand to the closed state which cycles dynamically with Hsp90. Thus, receptors that have bound steroid under physiological conditions in the cell are not recovered in association with Hsp90.
Stable vs. Dynamic Cycling with Hsp90
Hsp90 forms heterocomplexes with proteins in two general ways. The classical Hsp90 ‘client’ proteins, such as many transcription factors and protein kinases, turnover rapidly in the absence of Hsp90 stabilization. These client proteins have metastable clefts that, in the absence of Hsp90 binding, have a high tendency to further unfold, leading to protein degradation. These client proteins are assembled into complexes with Hsp90 that are stable enough to be isolated and analyzed biochemically. Although we call these ‘stable’ Hsp90 heterocomplexes, they are constantly undergoing cycles of assembly and disassembly in the cytoplasm and nucleoplasm (5). These metastable cleft proteins are quite profoundly stabilized when they are complexed with Hsp90; thus, these client proteins are under stringent Hsp90 regulation
Assembly of stable steroid receptor•Hsp90 heterocomplexes proceeds through an ordered series of events in which Hsp70 first binds to the client protein and primes the substrate for interaction with Hsp90 (assembly is presented in detail in Ref. 5). In stable assembly, both Hsp70 and Hsp90 must complete at least one ATPase cycle, and Hsp90 in the final complex is in the ATP-bound state. A third protein required for stable assembly is p23, an Hsp90 cochaperone that binds to the ATP-dependent conformation of Hsp90 and stabilizes the client protein•Hsp90 complex. Other proteins participate in the assembly machinery but only Hsp70, Hsp90 and p23 are required for stable cycling of steroid receptors with Hsp90 (5). Protein kinase•Hsp90 complexes are stabilized by Cdc37, an adaptor protein that binds to both the kinase and the chaperone (12).
A second form of cycling with Hsp90 is much more dynamic in that Hsp90 dissociates very rapidly after heterocomplex assembly and there are very few or no Hsp90 heterocomplexes that can be detected by biochemical techniques. Proteins that engage in ‘dynamic cycling’ with Hsp90 may have ligand binding clefts that are more rigid in their opening and closing dynamics, probably as a result of stabilization by water. In the absence of cycling with Hsp90, these proteins have intrinsically longer half-lives than client proteins that undergo stable cycling. Dynamic cycling proteins, such as the nitric oxide synthase (NOS) enzymes, are further stabilized only a small amount by cycling with Hsp90. Dynamic cycling of proteins with Hsp90 does not appear to require Hsp70 for Hsp90 binding or for Hsp90 to pass through an ATPase cycle. This is inferred from the fact that binding of purified Hsp90 to purified endothelial NOS (eNOS) and neuronal NOS (nNOS) and activation of their enzyme activity has been demonstrated in the absence of ATP and Hsp70 (13–15).
Conversion Between Stable and Dynamic Cycling
Proteins that stably cycle with Hsp90 can be converted to a dynamic cycling mode in a variety of ways. In cells, binding of steroid to the glucocorticoid receptor (GR) converts it to dynamic cycling. The steroid receptors bind their ligands deep within clefts, and steroid binding facilitates a temperature-dependent collapse of the cleft around the ligand to a closed cleft state (16). As illustrated in Figure 1C, this change in the mobility of the ligand binding cleft ‘transforms’ the GR from stable cycling to dynamic cycling with Hsp90 (6,7). Dynamic cycling with Hsp90 is important for the rapid, dynein-dependent movement of the receptor to the nucleus that is triggered by steroids (reviewed in Ref. 11). The GR can also be converted to dynamic cycling by mutation within a short segment in the ligand binding domain that is required for stable Hsp90 heterocomplex assembly (17). This mutation to dynamic cycling with Hsp90 shifts the dose-response curve for dexamethasone-dependent transactivation ∼100-fold to the right (17).
The GR and other stably cycling client proteins can also be converted to dynamic cycling by changing the acetylation state of Hsp90. Hsp90 is normally deacetylated by the cytoplasmic histone deacetylase HDAC6, and in HDAC6 knockdown cells Hsp90 is hyperacetylated (18). The acetylated Hsp90 has decreased ATP binding affinity, and it does not interact properly with p23. Thus, it engages only in dynamic cycling with the GR that, again, is manifest in the cell as a ∼100-fold shift to the right in the dose-response curve for transactivation (19).
The p53 tumor suppressor protein is a transcription factor that is mutated in more than half of all human tumors (20). Some of the p53 mutants are inactive because they form stable heterocomplexes with Hsp90, and are retained in the cytoplasm (21). Wild-type p53 does not form biochemically stable heterocomplexes with Hsp90, but treatment of cells with an Hsp90 inhibitor affects wild-type p53 function (22). Thus, it appears that wild-type p53 undergoes dynamic cycling with Hsp90 and mutation to cytoplasmic localization converts it to stable Hsp90 heterocomplex assembly by the same Hsp90 chaperone machinery that forms stable GR•Hsp90 heterocomplexes (23).
The story developed with ErbB kinases 1 and 2 is an excellent example of how dynamic and stable cycling with Hsp90 confer weak and stringent regulation of protein function and turnover. ErbB-1 is the epidermal growth factor receptor, and ErbB-2 functions as a ligandless coreceptor that heterodimerizes with other members of the ErbB family to amplify signaling. The kinase domain of ErbB-2 is assembled into a stable heterocomplex with Hsp90, whereas little or no Hsp90 is recovered with ErbB-1 (24). When cells are treated with an Hsp90 inhibitor , ErbB-2 is polyubiquitinated and rapidly degraded, whereas ErbB-1 is modestly ubiquitinated and slowly degraded (24,25). Thus, ErbB-2 undergoes stable cycling with Hsp90 and ErbB-1 undergoes dynamic cycling. The difference in behavior is accounted for by a short segment within the highly homologous kinase domains (26,27). This segment lies in close association with the ATP binding cleft, and swapping of an eight-amino acid section between ErbB-1 and ErbB-2 yields an exchange of dynamic versus stable cycling with Hsp90 and the appropriate conversion between weak and stringent Hsp90 regulation of turnover (27).
A comparison of sequences of this segment from multiple stably cycling client kinases and non-clients revealed no motif determining Hsp90 cycling behavior, and it was proposed that recognition by Hsp90 “is most likely based on the surface characteristics of the kinase within this region, as defined by the contribution of adjacent residues in the tertiary structure” (28). We have offered the alternative possibility that the segments determining Hsp90 cycling properties in both steroid receptors and protein kinases are features that determine the mobility of the ligand binding clefts, conferring the metastability required for the chaperone machinery to produce a stable complex with Hsp90 (7). It should be noted that natural, disease-related mutations occurring in the catalytic domains of other protein kinases have altered both their stability and Hsp90 cycling properties (29–31).
The Hsp90 Interactome
Hsp70 (includes Hsp70, Hsc70 and paralogs in organelles) has been found in association with a wide variety of proteins. Indeed, Hsp70 is the most common protein identified in screens designed to fish for other proteins that interact with a candidate protein. It is so commonly found that its presence is often considered to be a sticky artifact unrelated to the candidate protein’s function. It may well be that, at least at some time in the protein life span, the Hsp70 family interacts with all eukaryotic proteins.
Although Hsp90 is equally abundant, it is the prevailing view that Hsp90 interacts with a subset of proteins (9,32,33). Studies in yeast suggest that about 10% of the yeast proteome is regulated by Hsp90 (34) and that many different cellular functions are affected (35,36). Most of the established Hsp90 clients have been discovered in random fashion as various investigators have found that the protein they are studying is stably bound by Hsp90. A comprehensive list of Hsp90 clients is maintained by Didier Picard (http://www.picard.ch/downloads/Hsp90facts.pdf). The list contains proteins for which biochemical evidence for an interaction with cytosolic Hsp90 is available, and proteins where Hsp90 interaction is inferred solely from the effects of Hsp90 inhibitors (e.g. increased degradation, decreased activity) without demonstration of a Hsp90 heterocomplex are not considered.
Both the screens and the biochemical approaches are biased toward detection of stably cycling client proteins but they miss the more subtle effects seen with dynamic cycling. An example is seen with v-src and c-src. v-Src is a mutated protein kinase that causes cellular transformation and it was the first protein shown to interact with Hsp90 (37,38). Hsp90 coprecipitates with v-src but such native complexes are not detected with c-src (39). v-Src is rapidly degraded when cells are treated with an Hsp90 inhibitor but c-src is less susceptible, with a decrease in protein level being seen only after long treatment intervals (39). Thus, c-src appears to cycle dynamically with Hsp90 and v-src engages in stable cycling. Like v-src, p210bcr-abl is a mutated protein that causes cellular transformation and engages in stable cycling with Hsp90, whereas the cellular protein c-abl cycles dynamically (39).
Because we have no idea how many proteins cycle dynamically with Hsp90, we have no idea how large the Hsp90 ‘interactome’ is. Again, as with Hsp70, it could be that the Hsp90 family interacts with all eukaryotic proteins in either a dynamically cycling or stably cycling mode, conferring weak or stringent regulation, respectively. We would propose that dynamic cycling with Hsp90 is a general interaction and that a select subset of metastable cleft proteins that are more conformationally unstable undergo stable cycling and stringent regulation. The metastable cleft proteins inherently turnover more rapidly and their stringent regulation by Hsp90 is of particular importance in pathways of cellular signaling where the activity and amount of enzymes are brought under rapid control.
The Chaperone Machinery and Proteasomal Degradation
It has been widely accepted that the ubiquitin-proteasome system is involved in the degradation of damaged and aberrant proteins, but it has not been clear how the damaged proteins are recognized and targeted for ubiquitination (2). In the mid 1990’s, it was shown that Hsp70 and its cochaperone Hsp40 are required for ubiquitin-dependent degradation of short lived and abnormal proteins (40,41). Overexpression of Hsp70 or Hsp40 decreases the level of abnormal proteins and improves viability in cellular models of certain neurodegenerative diseases characterized by the accumulation of aberrant proteins, such as Parkinson disease, Huntington disease and spinal and bulbar muscular atrophy (42–44). Overexpression of these chaperones also ameliorates the disease phenotype in Drosophila and mouse models of these diseases (45–48, reviewed in Ref. 49).
Hsp90 was connected to ubiquitin-dependent degradation in studies using benzoquinone ansamycins that are quite specific Hsp90 inhibitors. The first of these ansamycins, herbimycin A, was found to reverse v-src transformation of cells to a normal phenotype (50,51), and it was then used as a protein-tyrosine kinase inhibitor (52). In a paper that led to an explosion of work on Hsp90, Whitesell et al. (53) showed that the target of the ansamycins herbimycin A and geldanamycin is Hsp90, not v-src. Treatment with geldanamycin led to a disruption of the v-src•Hsp90 heterocomplex and loss of v-src protein from cells (53). Subsequently, Sepp-Lorenzino et al. (54) showed that herbimycin-induced degradation of several receptor tyrosine kinases occurred via the ubiquitin-proteasome pathway.
Since the mid 1990s, hsp90 inhibition has been shown to promote the proteasomal degradation of several hundred proteins, including many proteins that promote cancer cell growth (55). The ansamycin antibiotics, such as geldanamycin and radicicol, bind in the nucleotide binding pocket near the N-terminus of Hsp90 (56,57). This ATP binding site is structurally unique to the very small GHKL family whose ATP binding domains contain four common motifs that define a “Bergerat fold” for binding ATP (58,59). As most of these proteins are bacterial, geldanamycin and radicicol effects are quite specific for inhibition of Hsp90 family proteins in eukaryotes. A variety of Hsp90 inhibitors that are pharmacologically and toxicologically more appropriate for human use have been synthesized, and one of them, 17-allylamino-17-demethoxygeldanamycin, is in phase II clinical trial for treatment of cancer (60).
Geldanamycin prevents formation of protein aggregates in models of Parkinson disease, Huntington disease and spinal and bulbar muscular atrophy (61–64), and Hsp90 inhibition may prove to be a productive approach to the treatment of several neurodegenerative diseases. Because Hsp90 binding to heat shock factor 1 (HSF1) maintains it in an inactive state (65) and treatment of cells with geldanamycin induces an HSF1-dependent stress response (61–63,65,66), it is often proposed that geldanamycin alleviates the phenotype and accumulation of misfolded proteins in neurodegenerative disease models by inducing a stress response (49,61–63). However, as geldanamycin promotes proteasomal degradation of polyglutamine protein aggregates in Hsf1−/− cells that cannot mount a stress response, this explanation is not valid (67).
The game changing observation that treatment with Hsp90 inhibitors promotes the degradation of Hsp90 client proteins via the ubiquitin-proteasome pathway (53,54) is explained most simply by the notion that Hsp90 binding to a client protein inhibits its degradation, and inhibitors like geldanamycin relieve that inhibition by preventing cycling with Hsp90. The observation of the chaperone effects on client protein turnover are consistent with the two essential components of the chaperone machinery having opposing effects, with Hsp70 promoting ubiquitination and Hsp90 stabilizing the protein against degradation.
Chaperone-Dependent Ubiquitin-Protein Ligases
Ubiquitination occurs via three sequential steps catalyzed by activating (E1), conjugating (E2) and ligase (E3) enzymes, and protein ubiquitination is used to trigger a wide variety of physiological processes (68). A number of E3 ligases play a key role in ubiquitin-mediated protein degradation by serving as the specific recognition factors in the cascade. In some cases, recognition is aided by various chaperones, but it is not known how many E3s function in a chaperone-dependent manner, although several clearly do (69). Two E3s are known to interact with the chaperones involved in the triage of damaged and aberrant proteins. Parkin is an E3 ligase (70) that is targeted to substrate by Hsp70 (71), but it was the discovery of CHIP by Ballinger et al. (72) that germinated a tremendous expansion in our understanding of chaperone-dependent ubiquitination.
Most of the mechanistic information about CHIP comes from the laboratory of Cam Patterson where CHIP was discovered, and Patterson and his colleagues have written several reviews on CHIP function (4,73,74). CHIP is a 35-kDa U-box E3 ligase (sometimes assigned as a member of the RING domain family of E3 ubiquitin ligases), and it binds via an amino-terminal TPR domain to both Hsc/Hsp70 and Hsp90 (72,75). CHIP possesses a carboxy-terminal U-box that interacts with the UBCH5 family of E2 ubiquitin conjugating enzymes (76). The fact that CHIP binds to both Hsp90 and Hsp70 led to the notion that both chaperones could target CHIP to the substrate (4,75).
The notion that Hsp90 is involved in targeting the substrate for ubiquitination cannot account for geldanamycin-induced client protein degradation. Geldanamycin binding to Hsp90 uniformly results in client protein destabilization (77), but geldanamycin prevents client protein cycling with Hsp90 (5). The stable client protein heterocomplexes are assembled in stepwise fashion through an initial ATP-dependent priming interaction with Hsp70 followed by a second ATP-dependent interaction with Hsp90 (5). The interaction with Hsp90 is blocked by geldanamycin, leaving the Hsp70-bound client protein to be ubiquitinated in a CHIP-dependent manner. This explanation is very different from a study of luciferase refolding where it was concluded that geldanamycin binding shifts Hsp90 from refolding to degradation mode (78).
Our conclusion that Hsp90-bound CHIP does not target the client protein for CHIP-dependent ubiquitination does not mean that CHIP interaction with the TPR acceptor site has no effect on Hsp90 function. CHIP binding to the TPR acceptor site on Hsp70 inhibits the chaperone’s ATPase activity, increasing the reactivation of luciferase after thermal inactivation in cells (79). This action requires the TPR domain but not the U-box domain of CHIP. It is possible that CHIP binding to the TPR acceptor site on Hsp90 has a similar effect on Hsp90 function that is independent of its E3 ligase action. Indeed, two other TPR domain proteins that bind to Hsp90, HOP and Cdc37, have been shown to inhibit its ATPase cycle (33). Despite the observations with geldanamycin, several reports (e.g. 80) have concluded that an Hsp90-CHIP complex selectively degrades various Hsp90 client proteins.
Overexpression of CHIP has been shown to increase ubiquitination and degradation of many established Hsp90 client proteins, including the examples cited in this review — GR (75), p53 (81), ErbB-2 (82). Of particular interest to toxicologists is that overexpression of CHIP increases the turnover of two cytochrome P450 enzymes, CYP2E1 (83) and CYP3A4 (84), that metabolize drugs and xenobiotics. CHIP is found in aggregates of aberrant proteins involved in neurodegenerative diseases, such as α-synuclein (Lewy bodies) and polyglutamine proteins (85–89), and overexpression of CHIP suppresses aggregation and protein levels in cellular disease models (85,86,88,89). The importance of CHIP in the neuronal response to aberrant, misfolded proteins is emphasized by the observations that overexpression of CHIP in insect and mouse polyglutamine disease models suppresses toxicity (88,90), and that Huntington disease transgenic mice haploinsufficient for CHIP display an accelerated disease phenotype (86).
Because CHIP has received so much attention, it is often regarded as the most important E3 ligase involved in chaperone-dependent ubiquitination and degradation. CHIP is clearly important, but it should be noted that there is redundancy between CHIP and some other E3 ligases. It was shown, for example, that both the GR and polyglutamine androgen receptor are degraded at the same rate in CHIP−/− and CHIP+/+ mouse embryonic fibroblasts treated with geldanamycin (91). CHIP−/− cytosol also has the same ability as CHIP+/+ cytosol to ubiquitinate a CHIP substrate. Although in this case, the E3 ligases that are acting redundantly to CHIP have not been identified, overexpression of either CHIP or parkin promotes the degradation of nNOS (91) and polyglutamine-expanded ataxin-3 (71,89,91). Consistent with the notion of redundancy, overexpression of E4B, an ubiquitin chain assembly factor that also has E3 ubiquitin ligase activity, promotes the degradation of polyglutamine-expanded ataxin-3 and suppresses neurodegeneration in a Drosophila model of spinocerebellar ataxia (92). In addition to being ubiquitinated by the cytosolic E3 ligase CHIP, CYP3A4 is effectively ubiquitinated by gp78, an endoplasmic reticulum-associated, UBC7-dependent, RING-finger E3 ligase (84). These examples of functional redundancy to CHIP E3 ligase action certainly show that CHIP-dependent degradation of damaged and aberrant proteins is not exclusive, but it does not diminish the widespread enthusiasm for CHIP as a central player in protein triage.
Opposing Actions of Hsp70 and Hsp90 on CHIP-Mediated Ubiquitination
Because CHIP binds via its TPR domain to both Hsp70 and Hsp90, it was logical to propose that both chaperones may have the ability to target substrates for degradation (4,75). However, it was not until the relative effects of the two chaperones on CHIP-mediated nNOS ubiquitination were studied in a purified system that their opposing roles were demonstrated directly (93).
The NOS enzymes, including eNOS, nNOS and iNOS (inducible NOS), are important signaling proteins that function as cytochrome P450-type hemoproteins to catalyze the formation of nitric oxide (NO) and citrulline from L-arginine, O2, and NADPH (94,95). The NOS enzymes are active as homodimers, with each monomer binding tightly 1 eq each of FAD, FMN, tetrahydrobiopterin, and heme. The prosthetic heme is the site of oxygen activation, which is required for the metabolism of L-arginine. NOS activity is Ca2+/calmodulin (CaM)-dependent, and several signaling pathways initiate nNOS and eNOS activity by raising intracellular Ca2+ concentration.
The activity of the NOS enzymes is enhanced by Hsp90, as shown both by studies in intact cells (13, 96–99) and by direct activation assays with purified proteins (13– 15,98–101). Studies with purified proteins show that CaM and Hsp90 increase the binding of each other to both eNOS and nNOS (14,15,99,101). Hsp90 cycles dynamically with NOS, and both direct binding of purified Hsp90 to purified nNOS and eNOS and activation of their activities have been demonstrated in the absence of ATP and Hsp70 (13–15). This stands in contrast to the assembly of stable steroid receptor•Hsp90 heterocomplexes where both Hsp70 and Hsp90 have to pass through a complete ATPase cycle and Hsp70 priming of the receptor is required for Hsp90 binding (5).
Treatment of cells with geldanamycin leads to nNOS degradation via the ubiquitin-proteasome pathway (96,102), and both CHIP and parkin function as E3 ligases for nNOS ubiquitination (91,103). nNOS is a particularly useful model for studying the roles of Hsp90 and Hsp70 in CHIP-mediated degradation and for exploring the relationship between Hsp90 stabilization of the protein and its interaction with the heme/substrate binding cleft. Because the binding of Hsp90 to nNOS does not require Hsp70 priming, the effects of the two chaperones on nNOS ubiquitination can be examined independently. Certain mechanism-based inactivators, such as NG-amino-L-arginine (NAA) and the antihypertensive drug guanabenz, cause accelerated nNOS degradation (102). Many of the inactivators crosslink heme to the enzyme (104,105), a modification that was shown in a myoglobin model to cause opening of the heme binding cleft (106) to yield a more unfolded state of the protein (107). The reaction of the inactivator in the heme/substrate binding cleft triggers nNOS ubiquitination and proteasomal degradation (102,108).
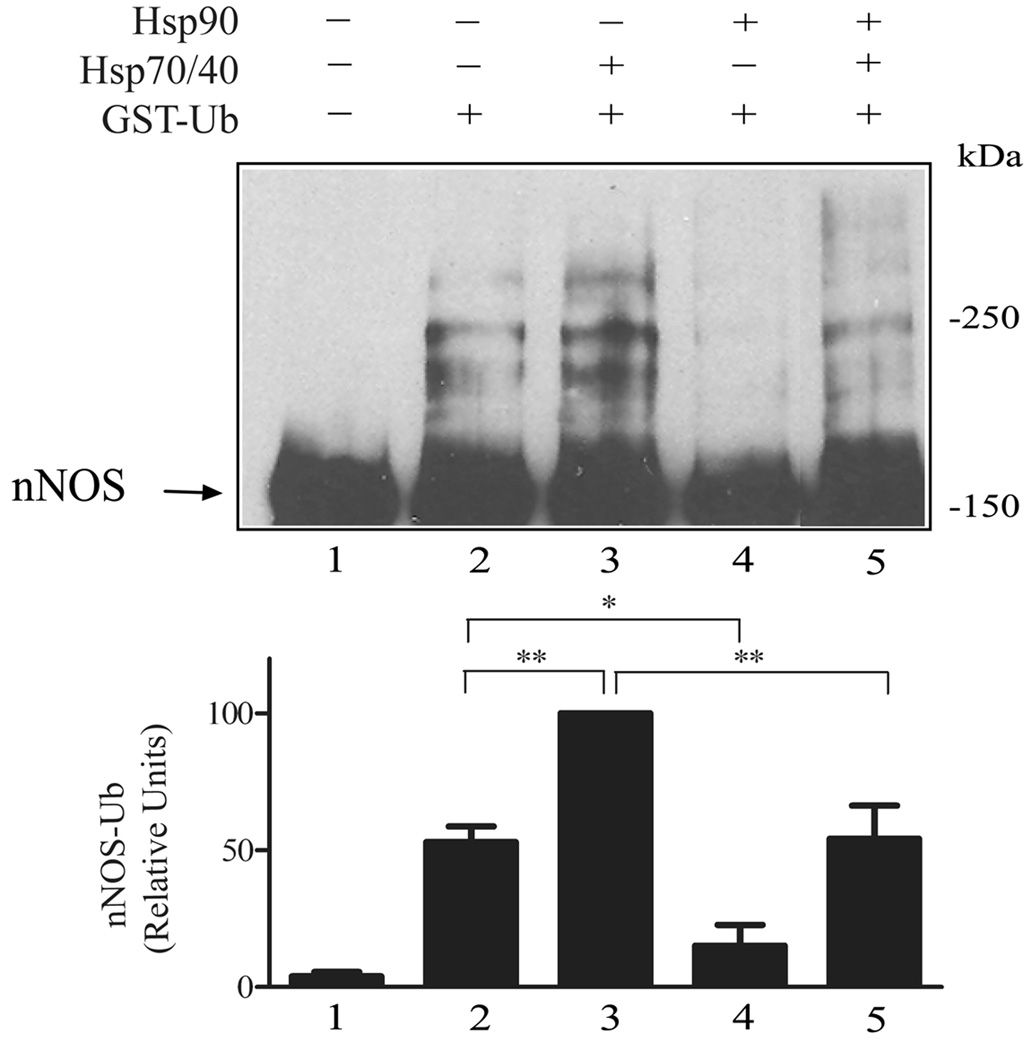
The opposing effects of Hsp90 and Hsp70 on CHIP-mediated nNOS ubiquitination are illustrated in Figure 2. In the absence of chaperones, there is a basal level of polyubiquitination (lane 2) that may reflect the presence of a small amount of insect Hsp70 that copurifies with nNOS (96). Nevertheless, addition of purified rabbit Hsp70 increases nNOS polyubiquitination (lane 3). In contrast, addition of purified rabbit Hsp90 inhibits both basal (lane 4) and Hsp70-stimulated (lane 5) polyubiquitination. Like the established Hsp90 enhancement of NO synthesis from nNOS (14), Hsp90 inhibition of nNOS ubiquitination is promoted by Ca2+/calmodulin, consistent with the two effects ensuing from the same interaction with the enzyme (93).
Figure 2.
Hsp90 inhibits basal and Hsp70-stimulated nNOS ubiquitination. Purified nNOS was preincubated with Hsp70 and its cochaperone Hsp40, or with Hsp90, or with both Hsp70/40 and Hsp90 as indicated and then incubated with a purified ubiquitinating enzyme mixture (E1, E2, CHIP, GST-ubiquitin). The samples were Western blotted by probing with anti-nNOS. The polyubiquitinated bands from three experiments were scanned and relative densities are presented in the bar graph as a % of the Hsp70-stimulated value in lane 3. *p < 0.05, **p < 0.01. The arrow points to the unubiquitinated/monoubiquitinated nNOS in the Western blot. (From Peng et al., 93)
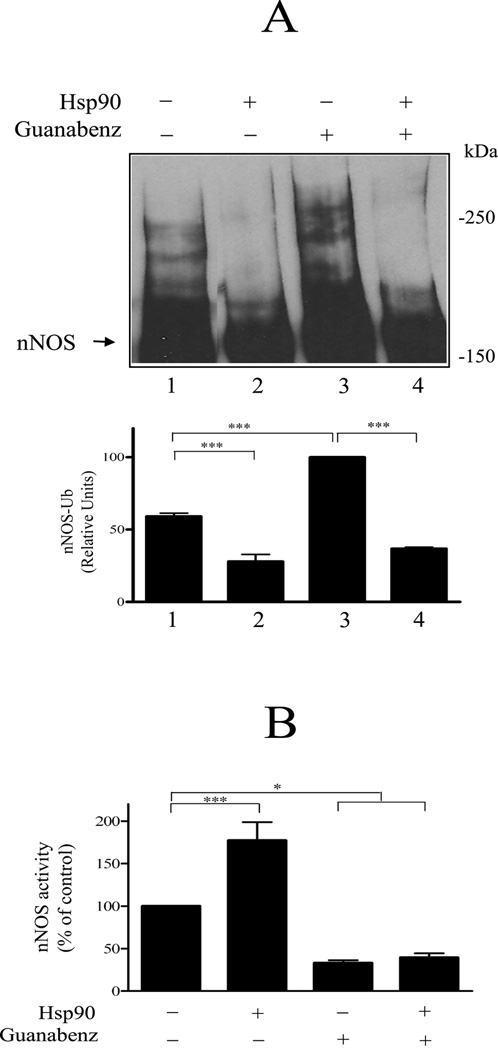
To determine if Hsp90 inhibits ubiquitination triggered by specific attack within the heme/substrate binding cleft, guanabenz-induced ubiquitination was examined (Fig. 3). Guanabenz is an antihypertensive drug that can produce impotence, and it inhibits nNOS activity, with accompanying loss of immunodetectible enzyme (109). Guanabenz enhances the proteasomal degradation of nNOS in cultured cells (102). Guanabenz treatment leads to the oxidation of tetrahydrobiopterin and formation of a pterin-depleted nNOS that is catalytically inactive (110). Loss of tetrahydrobiopterin from its binding site within the heme/substrate binding cleft destabilizes the nNOS dimer and enhances nNOS ubiquitination (111). As shown in Figure 3A, both basal polyubiquitination (lane 1) and the increased ubiquitination that ensues from guanabenz-mediated alteration of the heme/substrate binding cleft (lane 3) are inhibited by Hsp90 (lanes 2 and 4). In contrast, Hsp90 does not inhibit the guanabenz-mediated inhibition of nNOS enzymatic activity. As shown in Figure 3B, in the absence of guanabenz, Hsp90 causes the previously reported (14) enhancement of nNOS activity (lanes 1 and 2). However, preincubation with guanabenz inhibits nNOS activity whether or not Hsp90 is present (lanes 3 and 4). Thus, although CHIP binds equivalently to Hsp70 and Hsp90, Hsp70 promotes substrate ubiquitination by CHIP and Hsp90 inhibits ubiquitination.
Figure 3.
Effect of Hsp90 on guanabenz-mediated nNOS polyubiquitination. A, inhibition of ubiquitination. nNOS was preincubated with Hsp70/40 under conditions required for enzyme activity and in the presence of guanabenz and/or Hsp90 as noted at the top of figure. The samples were then incubated with the ubiquitinating enzyme mixture and Western blotted for nNOS. The relative densities of polyubiquitinated bands are presented as a % of the guanabenz-stimulated value in lane 3. B, guanabenz inhibition of nNOS activity. nNOS was preincubated as in A and nNOS activity was assayed by measuring the NO-mediated conversion of oxyhemoglobin to methemoglobin. Bars represent mean ± S.E. for three experiments. *p < 0.05, ***p < 0.001. (From Peng et al., 93)
The use of mechanism-based inactivators to distort ligand binding clefts and trigger proteasomal degradation is being exploited in the development of drugs that act on specific Hsp90 clients. CI-1033, for example, is a potent, site-specific inhibitor of ErbB-1 and ErbB-2 (112) that induces ErbB-2 ubiquitination and degradation (113). CHIP serves as an E3 ligase for ErbB-2, and both CHIP and Hsp70 are coimmunoadsorbed with ErbB-2 from geldanamycin-treated cells (82), consistent with the notion that site-specific attack of ErbB-2 by CI-1033 triggers the same chaperone-dependent ubiquitination observed with nNOS inactivators. Another example is fulvestrant (ICI 182780), an antiestrogen that causes ubiquitination and proteasomal degradation of the estrogen receptor (ER) (114,115). Fulvestrant was developed for treatment of ER positive breast cancer (116). CHIP is an important E3 ligase for the ER (117), and complexes containing ER, Hsp70 and CHIP have been isolated from cell lysates (118). However, as mentioned above for the GR, degradation of the ER occurs to the same extent in CHIP−/− cells as in CHIP+/+ cells (118), consistent with a redundancy in E3 ligase action.
The Triage Decision
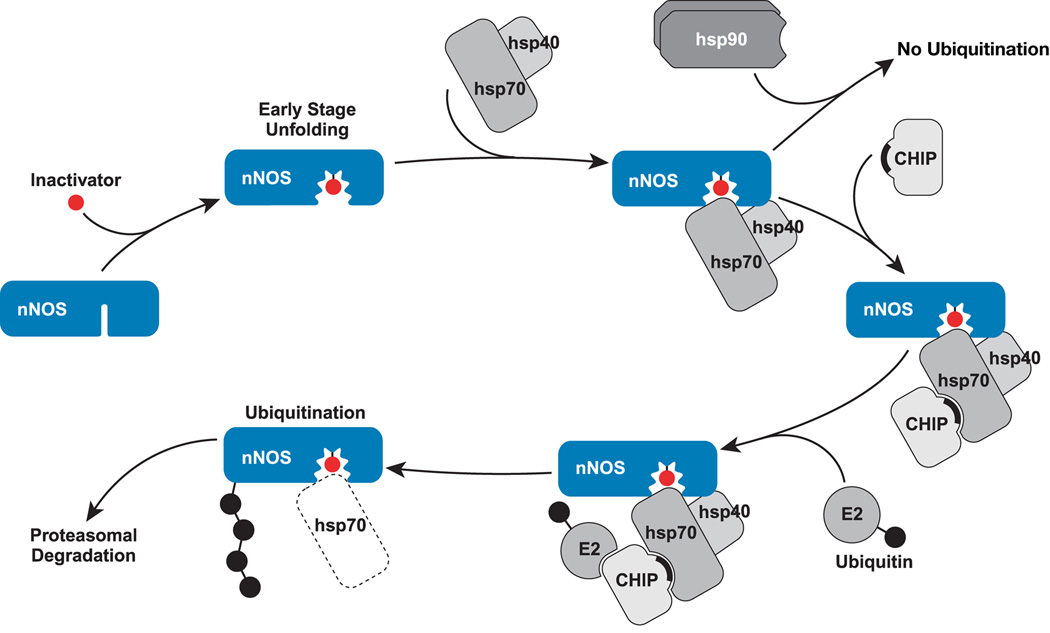
A model of nNOS triage is presented in Figure 4. The effects of guanabenz and NAA (93) serve as examples of toxic damage that is targeted to the ligand binding cleft and triggers ubiquitination of nNOS. As cited above, there is reason to propose that, as nNOS undergoes such toxic damage, the ligand binding cleft opens as the initial step in unfolding of the enzyme. As long as Hsp90 can contact even transiently with the opening cleft, ubiquitination by Hsp70-dependent ubiquitin ligases, like CHIP, is inhibited. But, as suggested by the model in Fig. 4, a point is reached where unfolding of the cleft progresses to a state that cannot cycle with Hsp90, and ubiquitination by Hsp70-dependent E3 ligases is unopposed. We don’t mean to imply in the model that cycling with Hsp90 would prevent binding of CHIP to substrate-bound Hsp70. CHIP coimmunoadsorbs with nNOS•Hsp70 complexes undergoing normal dynamic cycling with Hsp90 (103). Thus, it seems that CHIP TPR interaction with substrate-bound Hsp70 is not affected but that Hsp90 inhibits the subsequent ubiquitination step. Because it is Hsp70-bound CHIP that ubiquitinates the substrate, it is easy to get the impression that Hsp70 makes the triage decision. But, it is really the Hsp90 interaction with the unfolding substrate that determines whether the ubiquitination will proceed at any moment or not. Thus, the key component of the chaperone machinery for making the triage decision is Hsp90.
Figure 4.
Triage of nNOS by the Hsp90/Hsp70-based chaperone machinery after mechanism-based inactivation. Mechanism-based inactivation of nNOS by certain substrates leads to unfolding of the heme/substrate binding cleft to a degree that Hsp90 cannot cycle with the enzyme to inhibit ubiquitination by Hsp70-dependent E3 ligases, such as CHIP. The solid crescent represents the CHIP TPR domain.
Refolding Versus Cleft Stabilization
It was originally thought that Hsp90 and Hsp70 acted to promote refolding of proteins that aggregated in stressed cells, but it has subsequently become clear that Hsp70 promotes the proteasomal degradation of unfolded proteins. In contrast, inhibition of Hsp90 promotes their degradation. Similarly, in assays in vitro where the catalytic activity of an enzyme, such as luciferase, is inactivated by mild heating and recovery of activity is promoted by the chaperones, it has been assumed that Hsp90 and Hsp70 are acting by promoting the refolding of unfolded protein. An entirely different conclusion was reached with the steroid receptors where the focus was on ligand binding activity. When the Hsp90-free progesterone receptor, for example, is submitted to mild heating, it loses its ligand binding activity and the Hsp90/Hsp70-based chaperone machinery and ATP are required to maintain ligand binding activity (119).
The GR immediately loses ligand binding activity when Hsp90 is dissociated at 0°C. The loss of ligand binding activity reflects the collapse of the hydrophobic ligand binding cleft, and the Hsp90 chaperone machinery is required to open and stabilize the cleft in a ligand binding state (7). The analogous event with luciferase would be that mild heating promotes collapse of the ATP binding cleft and that dynamic cycling with the chaperone favors the open state of the cleft (as in Fig. 1B), promoting restoration of luciferase activity. Thus, this traditional assay of refolding may not be assaying refolding, rather it may be assaying the chaperone’s ability to dynamically stabilize a partially unfolded, or open cleft, state of the protein. This would explain why so many proteins that can bind both hydrophobic and charged regions possess some luciferase reactivating (chaperone) activity. If such a simple analysis were applied, it would markedly change the way people in the chaperone field view the action of Hsp90 and perhaps also of Hsp70.
We are proposing here a reinterpretation of early in vitro studies showing that Hsp90 binds to and promotes ATP-independent reactivation of selected enzymes inactivated under mild denaturing conditions (120). It has been suggested that “this appears to be a nonspecific phenomenon largely divorced from Hsp90’s biological interaction with authentic clients in vivo” (33). Instead, we would suggest that these studies reflect the dynamic cycling with Hsp90 that occurs with the ligand binding clefts of many proteins (e.g. nNOS) in the cell. The “authentic clients” are proteins with metastable clefts that are inherently less stable and are stringently regulated by Hsp90. This stringent regulation through stable cycling of Hsp90 requires Hsp70 and is ATP-dependent, but the effect is to stabilize the open state of the metastable cleft. This is the same effect that dynamic cycling of Hsp90 has on ligand binding clefts that are more rigid in their opening and closing dynamics.
As we have discussed above, ligand binding clefts can be mutated to change from one of these Hsp90 cycling modes to the other. Thus, we think the early ATP-independent, in vitro interactions of Hsp90 with stable cleft proteins inactivated by mild heating reflect the same fundamental interaction of Hsp90 with substrate as occurs with metastable cleft proteins. It is reasonable to propose then that the machinery to produce ATP-dependent Hsp90 cycling with “authentic clients” developed with eukaryotes as an essential mechanism from the more primitive and nonessential dynamic Hsp90 effect on ligand binding clefts. This dynamic cycling with ligand binding clefts appears to have arisen in eubacteria where Hsp90 is nonessential and is continued in eukaryotes. Scientific interest, however, has focused almost entirely on the subset of proteins that cycle stably with Hsp90 and are stringently regulated by the chaperone machinery.
Although the fundamental interaction of Hsp90 with metastable clefts may be similar to that of clefts in proteins that cycle dynamically with Hsp90, dynamic cycling of Hsp90 is not sufficient to fully rescue metastable cleft proteins from proteasomal degradation. As mentioned above, acetylated Hsp90 is capable of cycling only dynamically with metastable cleft proteins, such as the GR (19). A number of histone deacetylase inhibitors that have been developed as anticancer drugs induce acetylation of Hsp90, decreasing its affinity for ATP and abrogating stable cycling activity. This is accompanied by the degradation of metastable clients, such as mutant p53, ErbB-2, Raf-1, Bcr-Abl and Akt (121,122).
Hsp90 Cycling and Buffering of Toxic Damage
A major effect of Hsp90 cycling discovered by the Lindquist laboratory is that the chaperone functions as a capacitor of phenotypic variation (123,124, see Ref. 125 for review). Essentially, it was shown that genetic variation can accumulate in genomes and remain phenotypically silent because of Hsp90 cycling. When the buffering capacity of Hsp90 is reduced by mutation of the chaperone or pharmacological inhibition, the phenotypic variation is made evident. The likely mechanistic explanation is that mutations that affect the intrinsic stability of ligand binding clefts are not expressed phenotypically as a change in protein activity or turnover as long as Hsp90 can cycle with the opening cleft. Thus, there is no selection against the mutation, and multiple polymorphisms that affect developmental pathways remain hidden. Thus, as Sangster et al. (125) state: “when Hsp90’s buffer capacity is taxed, the function of normal pathways is perturbed; these hidden variants are revealed, and the phenotype is altered”.
A similar buffering by Hsp90 may determine the level of oxidative damage to proteins required to trigger their degradation. As Stadtman noted (1), these oxidative events have potential consequences in cellular function and ageing. The majority of the active oxygen species that are generated in the cell by P450-type enzymes exit the heme/substrate binding cleft and react with the surface of properly folded enzymes. When the oxidative damage accumulates to the point where the ligand binding cleft in the enzyme cannot move within its normal open and closed states of molecular breathing, more extensive cleft opening occurs as the initial step in unfolding. As long as Hsp90 can interact, even dynamically, to stabilize the cleft, it will buffer the effects of oxidative damage. In this way Hsp90 cycling serves to counteract the effects of oxidative stress on enzyme function as well as to counteract Hsp70-dependent ubiquitination as the first step in proteasomal degradation.
Concluding Remarks
By considering that Hsp90 and Hsp70 act together as a machinery to modulate ligand binding clefts in properly folded proteins, we have taken a very different approach to explaining how the chaperones interact with proteins undergoing toxic and oxidative stress. Using the refolding analysis that is a fundamental assumption of the chaperone field, one is forced into a model in which the chaperones interact with an unfolded state of the stressed protein to promote its proper refolding. In contrast, the ligand binding cleft model predicts that Hsp90 interacts with a stressed protein to stabilize it by preventing inordinate cleft opening that would yield unfolding.
By interacting with clefts, which are a normal topological feature of properly folded proteins, Hsp90 has the theoretical potential to cycle dynamically with virtually any protein. Although Hsp90 is not essential in eubacteria, dynamic cycling with Hsp90 would enhance enzyme activity and stability and serve to buffer bacterial enzymes against oxidative and toxic stress from the environment. When nucleated cells evolved, the genes for Hsp90 and Hsp70 duplicated (126), and Hsp90 became an essential protein (127). In primitive eukaryotes (algae), examples of stable client protein•Hsp90 complexes emerge, suggesting that Hsp90 and Hsp70 may have begun to work together as a machinery at or just after the transition to eukaryotes. Now, in addition to dynamic cycling of Hsp90 with more stable proteins, stable cycling of Hsp90 with inherently instable, metastable cleft proteins served to buffer against oxidative and toxic damage.
Acknowledgments
Preparation of this review and the author’s work reported herein were supported by National Institutes of Health Grants GM77430 and DA022354.
References
- 1.Stadtman ER. Oxidation of proteins by mixed function oxidation systems: implication in protein turnover, ageing and neutrophil function. Trends Biochem Sci. 1986;11:11–12. [Google Scholar]
- 2.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 3.Pickart CM. Back to the future with ubiquitin. Cell. 2004;116:181–190. doi: 10.1016/s0092-8674(03)01074-2. [DOI] [PubMed] [Google Scholar]
- 4.Cyr DM, Hohfeld J, Patterson C. Protein quality control: U-box-containing E3 ubiquitin ligases. Trends Biochem Sci. 2002;27:368–375. doi: 10.1016/s0968-0004(02)02125-4. [DOI] [PubMed] [Google Scholar]
- 5.Pratt WB, Toft DO. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Med. 2003;222:111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- 6.Pratt WB, Morishima Y, Osawa Y. The Hsp90 chaperone machinery acts at protein folding clefts to regulate both signaling protein function and protein quality control. In: Calderwood SK, Sherman MY, Ciocca DR, editors. Heat Shock Proteins in Cancer. Dordrecht, The Netherlands: Springer-Verlag; 2007. pp. 1–30. [Google Scholar]
- 7.Pratt WB, Morishima Y, Osawa Y. The Hsp90 chaperone machinery regulates signaling by modulating ligand binding clefts. J Biol Chem. 2008;283:22885–22889. doi: 10.1074/jbc.R800023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 9.Picard D. Heat shock protein 90, a chaperone for protein folding and regulation. Cell Mol Life Sci. 2002;59:1640–1648. doi: 10.1007/PL00012491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wandinger SK, Richter K, Buchner J. The Hsp90 chaperone machinery. J Biol Chem. 2008;283:18473–18477. doi: 10.1074/jbc.R800007200. [DOI] [PubMed] [Google Scholar]
- 11.Pratt WB, Galigniana MD, Harrell JM, DeFranco DB. Role of hsp90 and the hsp90-binding immunophilins in signaling protein movement. Cell Signal. 2004;16:857–872. doi: 10.1016/j.cellsig.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Pearl LH. Hsp90 and Cdc37 — a chaperone cancer conspiracy. Curr Opin Genet Dev. 2005;15:55–61. doi: 10.1016/j.gde.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Cardena G, Fan R, Shah V, Sorrentino R, Cirino G, Papapetropoulos A, Sessa WC. Dynamic activation of endothelial nitric oxide synthase by Hsp90. Nature. 1998;392:821–824. doi: 10.1038/33934. [DOI] [PubMed] [Google Scholar]
- 14.Song Y, Zweier JL, Xia Y. Heat-shock protein 90 augments neuronal nitric oxide activity by enhancing Ca2+/calmodulin binding. Biochem J. 2001;355:357–360. doi: 10.1042/0264-6021:3550357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song Y, Zweier JL, Xia Y. Determination of the enhancing action of HSP90 on neuronal nitric oxide synthase by EPR spectroscopy. Am J Physiol Cell Physiol. 2001;281:C1819–C1824. doi: 10.1152/ajpcell.2001.281.6.C1819. [DOI] [PubMed] [Google Scholar]
- 16.Gee AC, Katzenellenbogen JA. Probing conformational changes in the estrogen receptor: Evidence for a partially unfolded intermediate facilitating ligand binding and release. Mol Endocrinol. 2001;15:421–428. doi: 10.1210/mend.15.3.0602. [DOI] [PubMed] [Google Scholar]
- 17.Kaul S, Murphy PJM, Chen J, Brown L, Pratt WB, Simons SS. Mutations at positions 547–553 of rat glucocorticoid receptors reveal that hsp90 binding requires the presence, but not defined composition, of a seven-amino acid sequence of the amino terminus of the ligand binding domain. J Biol Chem. 2002;277:36223–36232. doi: 10.1074/jbc.M206748200. [DOI] [PubMed] [Google Scholar]
- 18.Kovacs JJ, Murphy PJM, Gaillard S, Zhao X, Wu JT, Nicchitta CV, Yoshida M, Toft DO, Pratt WB, Yao TP. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell. 2005;18:601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 19.Murphy PJM, Morishima Y, Kovacs JJ, Yao TP, Pratt WB. Regulation of the dynamics of hsp90 action on the glucocorticoid receptor by acetylation/deacetylation of the chaperone. J Biol Chem. 2005;280:33792–33799. doi: 10.1074/jbc.M506997200. [DOI] [PubMed] [Google Scholar]
- 20.Hollstein M, Hergenhahn M, Yang Q, Bartsch H, Wang ZQ, Hainaut P. New approaches to understanding p53 gene tumor mutation spectra. Mutat Res. 1999;431:199–209. doi: 10.1016/s0027-5107(99)00162-1. [DOI] [PubMed] [Google Scholar]
- 21.Blagosklonny MV, Toretsky J, Bohen S, Neckers L. Mutant conformation of p53 translated in vitro or in vivo requires functional HSP90. Proc Natl Acad Sci USA. 1996;93:8379–8383. doi: 10.1073/pnas.93.16.8379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walerych D, Kudla G, Gutkowska M, Wawrzynow B, Muller L, King FW, Helwak A, Boros J, Zylicz A, Zylicz M. Hsp90 chaperones wild-type p53 tumor suppressor protein. J Biol Chem. 2004;279:48836–48845. doi: 10.1074/jbc.M407601200. [DOI] [PubMed] [Google Scholar]
- 23.Galigniana MD, Harrell JM, O’Hagen HM, Ljungman M, Pratt WB. Hsp90-binding immunophilins link p53 to dynein during p53 transport to the nucleus. J Biol Chem. 2004;279:22483–22489. doi: 10.1074/jbc.M402223200. [DOI] [PubMed] [Google Scholar]
- 24.Xu W, Mimnaugh E, Roser MFN, Nicchitta C, Marcu M, Yarden Y, Neckers L. Sensitivity of mature ErbB-2 to geldanamycin is conferred by its kinase domain and is mediated by the chaperone protein Hsp90. J. Biol. Chem. 2001;276:3702–3708. doi: 10.1074/jbc.M006864200. [DOI] [PubMed] [Google Scholar]
- 25.Citri A, Alroy I, Lavi S, Rubin C, Xu W, Grammatikakis N, Patterson C, Neckers L, Fry DW, Yarden Y. Drug induced ubiquitination and degradation of ErbB receptor tyrosine kinases: implications for cancer therapy. EMBO J. 2002;21:2407–2417. doi: 10.1093/emboj/21.10.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tikhomirov O, Carpenter G. Identification of ErbB-2 kinase domain motifs required for geldanamycin-induced degradation. Cancer Res. 2003;63:39–43. [PubMed] [Google Scholar]
- 27.Citri A, Gan J, Mosesson Y, Vereb G, Szollosi J, Yarden Y. Hsp90 restrains ErbB2/HER2 signalling by limiting heterodimer formation. EMBO Rep. 2004;5:1165–1170. doi: 10.1038/sj.embor.7400300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Citri A, Harari D, Shohat G, Ramakrishnan P, Gan J, Lavi S, Eisenstein M, Kimchi A, Wallach D, Pietrokovski S, Yarden Y. Hsp90 recognizes a common surface on client kinases. J Biol Chem. 2006;281:14361–14369. doi: 10.1074/jbc.M512613200. [DOI] [PubMed] [Google Scholar]
- 29.Imamura T, Haruta T, Takata Y, Usui I, Iwata M, Ishihara H, Ishiki M, Ishibashi O, Ueno E, Sasaoka T, Kobayashi M. Involvement of heat shock protein 90 in the degradation of mutant insulin receptors by the proteasome. J Biol Chem. 1998;273:11183–11188. doi: 10.1074/jbc.273.18.11183. [DOI] [PubMed] [Google Scholar]
- 30.Matsuda S, Suzuki-Fujimoto T, Minowa A, Ueno H, Katamura K, Koyasu S. Temperature-sensitive ZAP70 mutants degrading through a proteasome-independent pathway. Restoration of a kinase domain mutant by Cdc37. J Biol Chem. 1999;274:34515–34518. doi: 10.1074/jbc.274.49.34515. [DOI] [PubMed] [Google Scholar]
- 31.Simizu S, Osada H. Mutations in the Plk gene lead to instability of Plk protein in human tumour cell lines. Nat Cell Biol. 2000;2:852–854. doi: 10.1038/35041102. [DOI] [PubMed] [Google Scholar]
- 32.Whitesell L, Lindquist SL. Hsp90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 33.Pearl LH, Podromou C. Structure and mechanism of the Hsp90 chaperone machinery. Annu Rev Biochem. 2006;75:271–294. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- 34.Zhao R, Davey M, Hsu YC, Kaplanek P, Tong A, Parsons AB, Krogan N, Cagney G, Mai D, Greenblatt J, Boone C, Emili A, Houry WA. Negotiating the chaperone network: An integrative map of physical and genetic interactions mediated by the Hsp90 chaperone. Cell. 2005;120:715–727. doi: 10.1016/j.cell.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 35.Millson SH, Truman AW, King V, Prodromou C, Pearl LH, Piper PW. A two-hybrid screen of the yeast proteome for Hsp90 interactions uncovers a novel Hsp90 chaperone requirement in the activity of a stress-activated mitogen-activated protein kinase, Slt2p (Mpk1p) Eukaryotic Cell. 2005;4:849–860. doi: 10.1128/EC.4.5.849-860.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McClellan AJ, Xia Y, Deutschbauer AM, Davis RW, Gerstein M, Frydman J. Diverse cellular functions of the Hsp90 molecular chaperone uncovered using systems approaches. Cell. 2007;131:121–135. doi: 10.1016/j.cell.2007.07.036. [DOI] [PubMed] [Google Scholar]
- 37.Brugge JS, Erikson E, Erikson RL. The specific interaction of the Rous sarcoma virus transformed protein pp60v-src with two cellular proteins. Cell. 1981;25:363–372. doi: 10.1016/0092-8674(81)90055-6. [DOI] [PubMed] [Google Scholar]
- 38.Oppermann H, Levinson W, Bishop JM. A cellular protein that associates with the transforming protein of Rous sarcoma virus is also a heat shock protein. Proc Natl Acad Sci USA. 1981;78:1067–1071. doi: 10.1073/pnas.78.2.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.An WG, Schulte TW, Neckers LM. The heat shock protein 90 antagonist geldanamycin alters chaperone association with p210bcr-abl and v-src proteins before their degradation by the proteasome. Cell Growth Diff. 2000;11:355–360. [PubMed] [Google Scholar]
- 40.Lee DH, Sherman MY, Goldberg AL. Involvement of the molecular chaperone Ydj1 in the ubiquitin-dependent degradation of short-lived and abnormal proteins in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4773–4781. doi: 10.1128/mcb.16.9.4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bercovich B, Stancoviski I, Mayer A, Blumenfeld N, Laszlo A, Schwartz AL, Ciechanover A. Ubiquitin-dependent degradation of certain protein substrates in vitro requires the molecular chaperone Hsp70. J Biol Chem. 1997;272:9002–9010. doi: 10.1074/jbc.272.14.9002. [DOI] [PubMed] [Google Scholar]
- 42.Klucken J, Shin Y, Masliah E, Hyman BT, McLean PJ. Hsp70 reduces α-synuclein aggregation and toxicity. J Biol Chem. 2004;279:25497–25502. doi: 10.1074/jbc.M400255200. [DOI] [PubMed] [Google Scholar]
- 43.Jana NR, Tanaka M, Wang G, Nukina N. Polyglutamine length-dependent interaction of Hsp40 and Hsp70 family chaperones with truncated N-terminal huntingtin: their role in suppression of aggregation and cellular toxicity. Hum Mol Genet. 2000;9:2009–2018. doi: 10.1093/hmg/9.13.2009. [DOI] [PubMed] [Google Scholar]
- 44.Baily CK, Andriola IFM, Kampinga HH, Merry DE. Molecular chaperones enhance the degradation of expanded polyglutamine repeat androgen receptor in a cellular model of spinal and bulbar muscular atrophy. Hum Mol Genet. 2002;11:515–523. doi: 10.1093/hmg/11.5.515. [DOI] [PubMed] [Google Scholar]
- 45.Auluck PK, Chan HYE, Trojanowski JQ, Lee VMY, Bonini NM. Chaperone suppression of α-synuclein toxicity in a Drosophila model for Parkinson’s disease. Science. 2002;295:865–868. doi: 10.1126/science.1067389. [DOI] [PubMed] [Google Scholar]
- 46.Warrick JM, Chan HYE, Gray-Board GL, Chai Y, Paulson HL, Bonini NM. Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat Genet. 1999;23:425–428. doi: 10.1038/70532. [DOI] [PubMed] [Google Scholar]
- 47.Chan HYE, Warrick JM, Gray-Board GL, Paulson HL, Bonini NM. Mechanisms of chaperone suppression of polyglutamine disease: selectivity, synergy and modulation of protein solubility in Drosophila. Hum Mol Genet. 2000;9:2811–2820. doi: 10.1093/hmg/9.19.2811. [DOI] [PubMed] [Google Scholar]
- 48.Adachi H, Katsuno M, Minamiyama M, Sang C, Pagoulatos G, Angelidis C, Kusakabe M, Yoshiki A, Kobayashi Y, Doyu M, Sobue G. Heat shock protein 70 chaperone overexpression ameliorates phenotypes of the spinal and bulbar muscular atrophy transgenic mouse model by reducing nuclear-localized mutant androgen receptor protein. J Neurosci. 2003;23:2203–2211. doi: 10.1523/JNEUROSCI.23-06-02203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muchowski PJ, Wacker JL. Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci. 2005;6:11–22. doi: 10.1038/nrn1587. [DOI] [PubMed] [Google Scholar]
- 50.Uehara Y, Hori M, Takeuchi T, Umezawa H. Screening of agents which convert “transformed morphology” of Rous sarcoma virus-infected rat kidney cells to “normal morphology”: identification of an active agent as herbimycin and its inhibition of intracellular src kinase. Jpn J Cancer Res. 1985;76:672–675. [PubMed] [Google Scholar]
- 51.Uehara Y, Hori M, Takeuchi T, Umezawa H. Phenotypic change from transformed to normal induced by benzoquinoid ansamycins accompanies inactivation of p60src in rat kidney cells infected with Rous sarcoma virus. Mol Cell Biol. 1986;6:2198–2206. doi: 10.1128/mcb.6.6.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uehara Y, Fukazawa H. Use and selectivity of herbimycin A, an inhibitor of protein-tyrosine kinases. Methods Enzymol. 1991;201:370–379. doi: 10.1016/0076-6879(91)01033-x. [DOI] [PubMed] [Google Scholar]
- 53.Whitesell L, Mimnaugh EG, DeCosta B, Myers CE, Neckers LM. Inhibition of heat shock protein HSP90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: Essential role for stress proteins in oncogenic transformation. Proc Natl Acad Sci USA. 1994;91:8324–8328. doi: 10.1073/pnas.91.18.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sepp-Lorenzino L, Ma Z, Lebwohl DE, Vinitsky A, Rosen N. Herbimycin A induces the 20 S proteasome- and ubiquitin-dependent degradation of receptor tyrosine kinases. J Biol Chem. 1995;270:16580–16587. doi: 10.1074/jbc.270.28.16580. [DOI] [PubMed] [Google Scholar]
- 55.Neckers L. Heat shock protein 90: the cancer chaperone. In: Calderwood SK, Sherman MY, Ciocca DR, editors. Heat Shock Proteins in Cancer. Dordrecht, The Netherlands: Springer-Verlag; 2007. pp. 231–252. [Google Scholar]
- 56.Stebbins CE, Russo AA, Schneider C, Rosen N, Hartl FU, Pavleich NP. Crystal structure of an hsp90-geldanamycin complex: Targeting of a protein chaperone by an antitumor agent. Cell. 1997;89:239–250. doi: 10.1016/s0092-8674(00)80203-2. [DOI] [PubMed] [Google Scholar]
- 57.Prodromou C, Roe SM, O’Brien R, Ladbury JE, Piper PW, Pearl LH. Identification and structural characterization of the ATP/ADP-binding site in the hsp90 molecular chaperone. Cell. 1997;90:65–75. doi: 10.1016/s0092-8674(00)80314-1. [DOI] [PubMed] [Google Scholar]
- 58.Dutta R, Inouye M. GHKL, an emergent ATPase/kinase superfamily. TIBS. 2000;25:24–28. doi: 10.1016/s0968-0004(99)01503-0. [DOI] [PubMed] [Google Scholar]
- 59.Bergerat A, deMassy B, Gadelle D, Varoutas PC, Nicolas A, Forterre P. An atypical topoisomerase II from archaea with implications for meiotic recombination. Nature. 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- 60.Holmes JL, Sharp SY, Workman P. Drugging the Hsp90 chaperone machine for cancer treatment. In: Calderwood SK, Sherman MY, Ciocca DR, editors. Heat Shock Proteins in Cancer. Dordrecht, The Netherlands: Springer-Verlag; 2007. pp. 295–329. [Google Scholar]
- 61.Auluck PK, Bonini NM. Pharmacological prevention of Parkinson disease in Drosophila. Nat Med. 2002;8:1185–1186. doi: 10.1038/nm1102-1185. [DOI] [PubMed] [Google Scholar]
- 62.Hay DG, Sathasivam K, Tobaben S, Stahl B, Marber M, Mestril R, Mahal A, Smith DL, Woodman B, Bates GP. Progressive decrease in chaperone protein levels in a mouse model of Huntington’s disease and induction of stress protein as a therapeutic approach. Hum Mol Genet. 2004;13:1389–1405. doi: 10.1093/hmg/ddh144. [DOI] [PubMed] [Google Scholar]
- 63.Sittler A, Lurz R, Lueder G, Priller J, Lehrach H, Hayer-Hartl MK, Hartl FU, Wanker EE. Geldanamycin activates a heat shock response and inhibits huntingtin aggregation in a cell culture model of Huntington’s disease. Hum Mol Genet. 2001;10:1307–1315. doi: 10.1093/hmg/10.12.1307. [DOI] [PubMed] [Google Scholar]
- 64.Waza M, Adachi H, Katsuno M, Minamiyama M, Sang C, Tanaka F, Inukai A, Doyu M, Sobue G. 17-AAG, an Hsp90 inhibitor, ameliorates polyglutamine-mediated motor neuron degeneration. Nat Med. 2005;11:1088–1095. doi: 10.1038/nm1298. [DOI] [PubMed] [Google Scholar]
- 65.Zou J, Guo Y, Guettouche T, Smith DF, Voellmy R. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell. 1998;94:471–480. doi: 10.1016/s0092-8674(00)81588-3. [DOI] [PubMed] [Google Scholar]
- 66.Bagatell R, Paine-Murrieta GD, Taylor CW, Pulcini EJ, Akinaga S, Benjamin IJ, Whitesell L. Induction of a heat shock factor 1-dependent stress response alters the cytotoxicity activity of hsp90-binding agents. Clin Cancer Res. 2000;6:3312–3318. [PubMed] [Google Scholar]
- 67.Thomas M, Harrell JM, Morishima Y, Peng HM, Pratt WB, Lieberman AP. Pharmacologic and genetic inhibition of hsp90-dependent trafficking reduces aggregation and promotes degradation of the expanded glutamine androgen receptor without stress protein induction. Hum Mol Genet. 2006;15:1876–1883. doi: 10.1093/hmg/ddl110. [DOI] [PubMed] [Google Scholar]
- 68.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 69.Hatakeyama S, Matsumoto M, Yada M, Nakayama KI. Interaction of U-box-type ubiquitin-protein ligases (E3s) with molecular chaperones. Genes Cells. 2004;9:533–548. doi: 10.1111/j.1356-9597.2004.00742.x. [DOI] [PubMed] [Google Scholar]
- 70.Zhang Y, Gao J, Chung JJJ, Huang H, Dawson VL, Dawson TM. Parkin functions as an E2-dependent ubiquitin-protein ligase and promotes the degradation of the synaptic vesicle-associated protein CDCrel-1. Proc Natl Acad Sci USA. 2000;97:13354–13359. doi: 10.1073/pnas.240347797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tsai YC, Fishman PS, Thakor NV, Oyler GA. Parkin facilitates the elimination of expanded polyglutamine proteins and leads to preservation of proteasome function. J Biol Chem. 2003;278:22044–22055. doi: 10.1074/jbc.M212235200. [DOI] [PubMed] [Google Scholar]
- 72.Ballinger CA, Connell P, Wu Y, Hu Z, Thompson LJ, Yin LY, Patterson C. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol Cell Biol. 1999;19:4535–4545. doi: 10.1128/mcb.19.6.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hohfeld J, Cyr DM, Patterson C. From cradle to the grave: molecular chaperones that may choose between folding and degradation. EMBO Reports. 2001;2:885–890. doi: 10.1093/embo-reports/kve206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McDonough H, Patterson C. CHIP: a link between the chaperone and proteasome systems. Cell Stress Chap. 2003;8:303–308. doi: 10.1379/1466-1268(2003)008<0303:calbtc>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, Hohfeld J, Patterson C. The co-chaperone CHIP regulates protein triage decisions mediated by heat shock proteins. Nat Cell Biol. 2001;3:93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 76.Jiang J, Ballinger CA, Wu Y, Dai Q, Cyr DM, Hohfeld J, Patterson C. CHIP is a U-box dependent E3 ubiquitin ligase: identification of hsc70 as a target for ubiquitylation. J Biol Chem. 2001;276:42938–42944. doi: 10.1074/jbc.M101968200. [DOI] [PubMed] [Google Scholar]
- 77.Isaacs JS, Xu W, Neckers L. Heat shock protein 90 as a molecular target for cancer therapeutics. Cancer Cell. 2003;3:213–217. doi: 10.1016/s1535-6108(03)00029-1. [DOI] [PubMed] [Google Scholar]
- 78.Schneider C, Sepp-Lorenzino L, Nimmesgern E, Ouerfelli O, Danishefsky S, Rosen N, Hartl FU. Pharmacological shifting of a balance between protein refolding and degradation mediated by Hsp90. Proc Natl Acad Sci USA. 1996;93:14536–14541. doi: 10.1073/pnas.93.25.14536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kampinga HH, Kanon B, Salomons FA, Kabakov AE, Patterson C. Overexpression of the cochaperone CHIP enhances Hsp70-dependent folding activity in mammalian cells. Mol Cell Biol. 2003;23:4948–4958. doi: 10.1128/MCB.23.14.4948-4958.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dickey CA, Kamal A, Lundgren K, Klosak N, Bailey RM, Dunmore J, Ash P, Shoraka S, Zlatkovic J, Eckman CB, Patterson C, Dickson DW, Nahman NS, Hutton M, Burrows F, Petrucelli L. The high affinity HSP90-CHIP complex recognizes and selectively degrades phosphorylated tau client proteins. J Clin Invest. 2007;117:648–658. doi: 10.1172/JCI29715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Esser C, Scheffner M, Hohfeld J. The chaperone-associated ubiquitin ligase CHIP is able to target p53 for proteasomal degradation. J Biol Chem. 2005;280:27443–27448. doi: 10.1074/jbc.M501574200. [DOI] [PubMed] [Google Scholar]
- 82.Zhou P, Fernandes N, Dodge IL, Reddi AL, Rao N, Safran H, DiPetrillo TA, Wazer DE, Band V, Band H. ErbB2 degradation mediated by the co-chaperone protein CHIP. J Biol Chem. 2003;278:13829–13837. doi: 10.1074/jbc.M209640200. [DOI] [PubMed] [Google Scholar]
- 83.Morishima Y, Peng HM, Lin H, Hollenberg PF, Sunahara RK, Osawa Y, Pratt WB. Regulation of cytochrome P450 2E1 by heat shock protein 90-dependent stabilization and CHIP-dependent proteasomal degradation. Biochemistry. 2005;44:16333–16340. doi: 10.1021/bi0515570. [DOI] [PubMed] [Google Scholar]
- 84.Pabarcus MK, Hoe N, Sadeghi S, Patterson C, Wiertz E, Correia MA. CYP3A4 ubiquitination by gp78 (the tumor autocrine motility factor receptor, AMFR) and CHIP E3 ligases. Arch Biochem Biophys. 2009;483:66–74. doi: 10.1016/j.abb.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shin Y, Klucken J, Patterson C, Hyman BT, McLean PJ. The co-chaperone carboxyl terminus of hsp70-interacting protein (CHIP) mediates α-synuclein degradation decisions between proteasomal and lysosomal pathways. J Biol Chem. 2005;280:23727–23734. doi: 10.1074/jbc.M503326200. [DOI] [PubMed] [Google Scholar]
- 86.Miller VM, Nelson RF, Gouvion CM, Williams A, Rodriguez-Lebron E, Harper SQ, Davidson BL, Rebagliati MR, Paulson HL. CHIP suppresses polyglutamine aggregation and toxicity in vitro and in vivo. J Neurosci. 2005;25:9152–9161. doi: 10.1523/JNEUROSCI.3001-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thomas M, Dadgar N, Aphale A, Harrell JM, Kunkel R, Pratt WB, Lieberman AP. Androgen receptor acetylation site mutations cause trafficking defects, misfolding, and aggregation similar to expanded glutamine tracts. J Biol Chem. 2004;279:8389–8395. doi: 10.1074/jbc.M311761200. [DOI] [PubMed] [Google Scholar]
- 88.Al-Ramahi I, Lam YC, Chen HK, de Gouyon B, Zhang M, Perez AM, Branco J, de Haro M, Patterson C, Zoghbi HY, Botas J. CHIP protects from the neurotoxicity of expanded and wild-type ataxin-1 and promotes their ubiquitination and degradation. J Biol Chem. 2006;281:26714–26724. doi: 10.1074/jbc.M601603200. [DOI] [PubMed] [Google Scholar]
- 89.Jana NR, Dikshit P, Goswami A, Kotliarova S, Murata S, Tanaka K, Nukina N. Co-chaperone CHIP associates with expanded polyglutamine protein and promotes their degradation by proteasomes. J Biol Chem. 2005;280:11635–11640. doi: 10.1074/jbc.M412042200. [DOI] [PubMed] [Google Scholar]
- 90.Adachi H, Waza M, Tokui K, Katsuno M, Minamiyama M, Tanaka F, Doyu M, Sobue G. CHIP overexpression reduces mutant androgen receptor protein and ameliorates phenotypes of the spinal and bulbar muscular atrophy transgenic mouse model. J Neurosci. 2007;27:5115–5126. doi: 10.1523/JNEUROSCI.1242-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Morishima Y, Wang AM, Yu Z, Pratt WB, Osawa Y, Lieberman AP. CHIP deletion reveals functional redundancy of E3 ligases in promoting degradation of both signaling proteins and expanded glutamine proteins. Hum Mol Genet. 2008;17:3942–3952. doi: 10.1093/hmg/ddn296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Matsumoto M, Yada M, Hatakeyama S, Ishimoto H, Tanimura T, Tsuji S, Kakizuka A, Kitagawa M, Nakayama KI. Molecular clearance of ataxin-3 is regulated by a mammalian E4. EMBO J. 2004;23:659–669. doi: 10.1038/sj.emboj.7600081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Peng HM, Morishima Y, Clapp KM, Lau M, Pratt WB, Osawa Y. Dynamic cycling with Hsp90 stabilizes neuronal nitric oxide synthase through calmodulin-dependent inhibition of ubiquitination. Biochemistry. 2009 doi: 10.1021/bi901058g. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Marletta MA. Nitric oxide synthase structure and mechanism. J Biol Chem. 1993;268:12231–12234. [PubMed] [Google Scholar]
- 95.Stuehr DJ. Mammalian nitric oxide synthases. Biochim Biophys Acta. 1999;1411:217–230. doi: 10.1016/s0005-2728(99)00016-x. [DOI] [PubMed] [Google Scholar]
- 96.Bender AT, Silverstein AM, Demady DR, Kanelakis KC, Noguchi S, Pratt WB, Osawa Y. Neuronal nitric oxide synthase is regulated by the hsp90-based chaperone system in vivo. J Biol Chem. 1999;274:1472–1478. doi: 10.1074/jbc.274.3.1472. [DOI] [PubMed] [Google Scholar]
- 97.Brouet A, Sonveaux P, Dessy C, Ballingand JL, Feron O. Hsp90 ensures the transition from the early Ca2+-dependent to the late phosphorylation-dependent activation of the endothelial nitric-oxide synthase in vascular endothelial growth factor-exposed endothelial cells. J Biol Chem. 2001;276:32663–32669. doi: 10.1074/jbc.M101371200. [DOI] [PubMed] [Google Scholar]
- 98.Yoshida M, Xia Y. Heat shock protein 90 as an endogenous protein enhancer of inducible nitric oxide synthase. J Biol Chem. 2003;278:36953–36958. doi: 10.1074/jbc.M305214200. [DOI] [PubMed] [Google Scholar]
- 99.Gratton JP, Fontana J, O’Connor DS, Garcia-Cardena G, McCabe TJ, Sessa WC. Reconstitution of an endothelial nitric-oxide synthase (eNOS), hsp90, and caveolin-1 complex in vitro: evidence that hsp90 facilitates calmodulin stimulated displacement of eNOS from caveolin-1. J Biol Chem. 2000;275:22268–22272. doi: 10.1074/jbc.M001644200. [DOI] [PubMed] [Google Scholar]
- 100.Takahashi S, Mendelsohn ME. Synergistic activation of endothelial nitric-oxide synthase (eNOS) by HSP90 and Akt. J Biol Chem. 2003;278:30821–30827. doi: 10.1074/jbc.M304471200. [DOI] [PubMed] [Google Scholar]
- 101.Takahashi S, Mendelsohn ME. Calmodulin-dependent and -independent activation of endothelial nitric-oxide synthase by heat shock protein 90. J Biol Chem. 2000;278:9339–9344. doi: 10.1074/jbc.M212651200. [DOI] [PubMed] [Google Scholar]
- 102.Noguchi S, Jianmongkol S, Bender AT, Kamada Y, Demady DR, Osawa Y. Guanabenz-mediated inactivation and enhanced proteolytic degradation of neuronal nitric oxide synthase. J Biol Chem. 2000;275:2376–2380. doi: 10.1074/jbc.275.4.2376. [DOI] [PubMed] [Google Scholar]
- 103.Peng HM, Morishima Y, Jenkins GJ, Dunbar AY, Lau M, Patterson C, Pratt WB, Osawa Y. Ubiquitination of neuronal nitric-oxide synthase by CHIP, a chaperone-dependent E3 ligase. J Biol Chem. 2004;279:52970–52977. doi: 10.1074/jbc.M406926200. [DOI] [PubMed] [Google Scholar]
- 104.Vuletich JL, Lowe ER, Jianmongkol S, Kamada Y, Kent UM, Bender AT, Demady DR, Hollenberg PF, Osawa Y. Alteration of the heme prosthetic group of neuronal nitric-oxide synthase during inactivation by NG-amino-L-arginine in vitro and in vivo. Mol Pharmacol. 2002;62:110–118. doi: 10.1124/mol.62.1.110. [DOI] [PubMed] [Google Scholar]
- 105.Osawa Y, Lowe ER, Everett AC, Dunbar AY, Billecke SS. Proteolytic degradation of nitric oxide synthase: effect of inhibitors and role of hsp90-based chaperones. J Pharmacol Exptl Ther. 2003;304:493–497. doi: 10.1124/jpet.102.035055. [DOI] [PubMed] [Google Scholar]
- 106.Osawa Y, Darbyshire JF, Steinbach PJ, Brooks BR. Metabolism-based transformation of myoglobin to an oxidase by BrCCl3 and molecular modeling of the oxidase form. J Biol Chem. 1993;268:2953–2959. [PubMed] [Google Scholar]
- 107.Osawa Y, Pohl LR. Covalent bonding of the prosthetic heme to protein: a potential mechanism for the suicide inactivation or activation of hemoproteins. Chem Res Toxicol. 1989;2:131–141. doi: 10.1021/tx00009a001. [DOI] [PubMed] [Google Scholar]
- 108.Bender AT, Demady DR, Osawa Y. Ubiquitination of neuronal nitric oxide synthase in vitro and in vivo. J Biol Chem. 2000;275:17407–17411. doi: 10.1074/jbc.M000155200. [DOI] [PubMed] [Google Scholar]
- 109.Nakatsuka M, Nakatsuka K, Osawa Y. Metabolism based inactivation of penile nitric oxide synthase activity by guanabenz. Drug Metab Disp. 1998;26:497–501. [PubMed] [Google Scholar]
- 110.Dunbar AY, Jenkins GJ, Jianmongkol S, Nakatsuka M, Lowe ER, Lau M, Osawa Y. Tetrahydrobiopterin protects against guanabenz-mediated inhibition of neuronal NO synthase in vitro and in vivo. Drug Metab Disp. 2006;34:1448–1456. doi: 10.1124/dmd.106.009951. [DOI] [PubMed] [Google Scholar]
- 111.Kamada Y, Jenkins GJ, Lau M, Dunbar AY, Lowe ER, Osawa Y. Tetrahydrobiopterin depletion and ubiquitylation of neuronal NO-synthase. Mol Brain Res. 2005;142:19–27. doi: 10.1016/j.molbrainres.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 112.Fry D. Site-directed irreversible inhibitors of the erbB family of receptor tyrosine kinases as novel chemotherapeutic agents for cancer. Anticancer Drug Des. 2000;15:3–16. [PubMed] [Google Scholar]
- 113.Citri A, Alroy I, Lavi S, Rubin C, Xu W, Grammatikakis N, Patterson C, Neckers L, Fry DW, Yarden Y. Drug-induced ubiquitylation and degradation of ErbB receptor tyrosine kinases: implications for cancer therapy. EMBO J. 2002;21:2407–2417. doi: 10.1093/emboj/21.10.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dauvois S, Danielian PS, White R, Parker MG. Antiestrogen ICI 164,384 reduces cellular estrogen receptor content by increasing its turnover. Proc Natl Acad Sci USA. 1992;89:4037–4041. doi: 10.1073/pnas.89.9.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wijayaratne AL, McDonnell DP. The human estrogen receptor-α is a ubiquitinated protein whose stability is affected differentially by agonists, antagonists, and selective estrogen receptor modulators. J Biol Chem. 2001;276:35684–35692. doi: 10.1074/jbc.M101097200. [DOI] [PubMed] [Google Scholar]
- 116.Osborne CK, Wakeling A, Nicholson RI. Fulvestrant: an oestrogen receptor antagonist with a novel mechanism of action. Br J Cancer. 2004;90:S2–S6. doi: 10.1038/sj.bjc.6601629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fan M, Park A, Nephew KP. CHIP (carboxy terminus of Hsc70-interacting protein) promotes basal and geldanamycin-induced degradation of estrogen receptor-alpha. Mol Endocrinol. 2005;19:2901–2914. doi: 10.1210/me.2005-0111. [DOI] [PubMed] [Google Scholar]
- 118.Tateishi Y, Kawabe Y, Chiba T, Murata S, Ichikawa K, Murayama A, Tanaka K, Baba T, Kato S, Yanagisawa J. Ligand-dependent switching of ubiquitin-proteasome pathways for estrogen receptor. EMBO J. 2004;23:4813–4823. doi: 10.1038/sj.emboj.7600472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Smith DF. Dynamics of heat shock protein 90-progesterone receptor binding and the disactivation loop model for steroid receptor complexes. Mol Endocrinol. 1993;7:1418–1429. doi: 10.1210/mend.7.11.7906860. [DOI] [PubMed] [Google Scholar]
- 120.Wiech H, Buchner J, Zimmerman R, Jakob U. Hsp90 chaperones protein folding in vitro. Nature. 1992;350:169–170. doi: 10.1038/358169a0. [DOI] [PubMed] [Google Scholar]
- 121.Yu X, Guo S, Marcu MG, Neckers L, Nguyen DM, Chen GA, Schrump DS. Modulation of p53, ErbB1, ErbB2, and Raf-1 expression in lung cancer cells by depsipeptide FR901228. J Natl Cancer Inst. 2002;94:504–513. doi: 10.1093/jnci/94.7.504. [DOI] [PubMed] [Google Scholar]
- 122.Bali P, Pranpat M, Bradner J, Balasis M, Fiskus W, Guo F, Rocha K, Kurnaraswamy S, Boyapalle S, Atadja P, Seto E, Bhalla K. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: A novel basis for antilukemic activity of histone deacetylase inhibitors. J Biol Chem. 2005;280:26729–26734. doi: 10.1074/jbc.C500186200. [DOI] [PubMed] [Google Scholar]
- 123.Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- 124.Queitsch C, Sangster TA, Lindquist S. Hsp90 as a capacitor of phenotypic variation. Nature. 2002;417:618–624. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- 125.Sangster TA, Lindquist S, Queitsch C. Under cover: causes, effects and implications of Hsp90-mediated genetic capacitance. BioEssays. 2004;26:348–362. doi: 10.1002/bies.20020. [DOI] [PubMed] [Google Scholar]
- 126.Gupta RS, Golding B. The origin of the eukaryotic cell. Trends Biochem Sci. 1996;21:166–171. [PubMed] [Google Scholar]
- 127.Parsell DA, Lindquist S. The function of heat shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu Rev Genet. 1993;27:437–496. doi: 10.1146/annurev.ge.27.120193.002253. [DOI] [PubMed] [Google Scholar]